Abstract
Microbes can defend their host against virulent infections, but direct evidence for the adaptive origin of microbe-mediated protection is lacking. Using experimental evolution of a novel, tripartite interaction, we demonstrate that mildly pathogenic bacteria (Enterococcus faecalis) living in worms (Caenorhabditis elegans) rapidly evolved to defend their animal hosts against infection by a more virulent pathogen (Staphylococcus aureus), crossing the parasitism–mutualism continuum. Host protection evolved in all six, independently selected populations in response to within-host bacterial interactions and without direct selection for host health. Microbe-mediated protection was also effective against a broad spectrum of pathogenic S. aureus isolates. Genomic analysis implied that the mechanistic basis for E. faecalis-mediated protection was through increased production of antimicrobial superoxide, which was confirmed by biochemical assays. Our results indicate that microbes living within a host may make the evolutionary transition to mutualism in response to pathogen attack, and that microbiome evolution warrants consideration as a driver of infection outcome.
Introduction
Microbes can have effects on host biology far beyond their core impacts on digestion (Dillon et al., 2000; Cerf-Bensussan and Gaboriau-Routhiau, 2010; Brucker and Bordenstein, 2013; Lize et al., 2014). Microbes can cause infectious disease, but they can also act to protect hosts from pathogens, a phenomenon observed in a range of animals (Dillon et al., 2005; Dong et al., 2009; Jaenike et al., 2010; Koch and Schmid-Hempel, 2011), including humans (Kamada et al., 2013), and in plants at the root–soil interface (Mendes et al., 2011; May and Nelson, 2014). These protective microbes provide an important complement to the host's defence systems (Abt and Artis, 2013; Hooper et al., 2012; McFall-Ngai et al., 2013). As pathogens invade the host, they can not only be targeted by the host immune system, but also interact with pathogenic and commensal microbial species already present within the host (McFall-Ngai et al., 2013). Resident microbes can therefore provide strong protection against virulent pathogens, and corresponding microbial traits might be evolutionarily advantageous. Evolution of this nature would represent microbes evolving along the parasitism–mutualism continuum (Chamberlain et al., 2014).
The large population sizes and short generation times of microbes also create the potential for the rapid evolution of such defences. Can microbes evolve to protect their host in response to virulent pathogen challenge, and, in doing so make an evolutionary transition to mutualism? It is well established that infecting pathogens can undergo rapid adaptation (Brockhurst and Koskella, 2013) in response to transmission opportunity and mode (Messenger et al., 1999), prior immune exposure (Mackinnon and Read, 2004) and multi-strain coinfection (Garbutt et al., 2011) with host defences known to reciprocally evolve to pathogen adaptation (Schulte et al., 2010; Morran et al., 2011). Despite this, evolutionary responses by resident microbes against pathogen infection have not before been considered.
Here, we use experimental evolution to test whether a mildly pathogenic, resident microbe (Enterococcus faecalis) can evolve to defend its host (Caenorhabditis elegans) against infection by a more virulent pathogen (Staphylococcus aureus), and thus cross the parasitism–mutualism continuum. E. faecalis and S. aureus frequently occur in animal and human microbiomes (Holden et al., 2004; Martin-Vivaldi et al., 2010; Lawley et al., 2012; Cruz et al., 2013; Kommineni et al., 2015) wherein they can be pathogenic or commensal. Both bacteria can colonise the gut of C. elegans (Garsin et al., 2001), a model animal system for investigating natural and lab-based host–microbiota associations (Cabreiro and Gems, 2013; Clark and Hodgkin, 2013; Petersen et al., 2015) and their evolutionary consequences (reviewed in Gray and Cutter, 2014). Within the lifetime of an individual nematode, both S. aureus and E. faecalis can be harmful. S. aureus is highly virulent, causing 100% host mortality after approximately 2 days of exposure (Sifri et al., 2003) by lysing the cells lining the gut wall of nematode hosts (Gravato-Nobre and Hodgkin, 2005). By contrast, E. faecalis is lethal to C. elegans only over the longer term, requiring more than 7 days of exposure (and no food) to cause total host population reduction (Sifri et al., 2002) and is only mildly pathogenic in shorter-term infections. In our experimental set-up, involving 2-day colonisations (described in Figure 1), S. aureus is a highly virulent pathogen in single infection, whereas E. faecalis is a mildly pathogenic resident of the nematode gut, causing c. <1% mortality of the host. E. faecalis is under selection in this state.
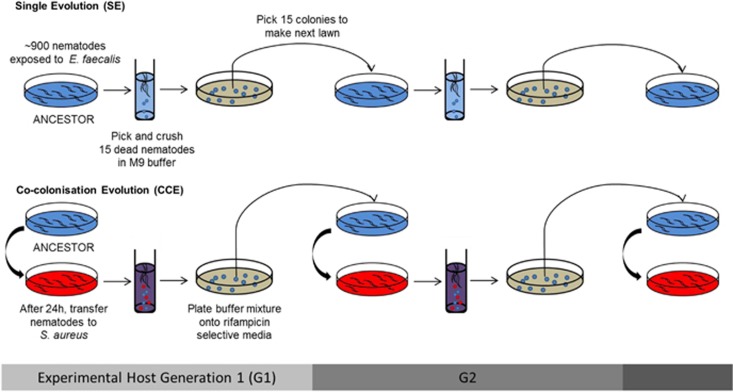
Figure 1.
Experimental procedure for experimental evolution of E. faecalis within C. elegans populations. Treatments are shown for a single replicate population. Six populations of E. faecalis were independently passaged from a single clone ancestor for 15 experimental host generations through nematode hosts under one of two different selection regimes: (i) SE repeated passage of E. faecalis alone in C. elegans and (ii) CCE repeated passage of E. faecalis in C. elegans with a fixed, non-evolving S. aureus isolate. In treatment (i), nematodes were only exposed to E. faecalis, while in (ii), nematodes were exposed to E. faecalis first, so that the microbe could establish residency, and then to S. aureus. We enforced within-host interactions between the bacterial species by propagating E. faecalis cells harvested from bacteria-killed nematodes, a method that also avoided direct selection against virulence and for host health. All replicate populations were passaged at the same time during the experiment.
We tested whether E. faecalis resident within C. elegans evolved to protect against S. aureus infection where its host was challenged with the pathogen over 15 experimental host generations. Our experiments examined the following interaction: resident E. faecalis was allowed to evolve inside hosts in the presence/absence of a genetically fixed pathogen (supplied from ancestral culture each host generation; experimental procedure in Figure 1), and the properties of E. faecalis were compared between the two treatments. Both treatments consisted of six replicate populations started from a single clone of E. faecalis that were then independently passaged, and thus any adaptive evolution that occurred was due to de novo mutation and selection. We passaged E. faecalis from dead hosts to observe evolutionary processes arising from species interactions within hosts, rather than imposing direct selection for host health. We found that host protection against S. aureus by resident E. faecalis evolved rapidly within nematode hosts in all replicate populations. Genomic and subsequent biochemical analyses pointed to increased production of antimicrobial superoxide as the mechanism. Our results indicate that resident microbes, even mildly pathogenic ones, can rapidly evolve to defend their hosts in response to virulent pathogenic infection.
Materials and methods
Nematode host and bacteria
C. elegans is a nematode that constantly interacts with microbes in its natural habitat (Felix and Braendle, 2010), and it can act as a predator or host for numerous species (Cabreiro and Gems, 2013; Clark and Hodgkin, 2013; Petersen et al., 2015). These animals are thus an established model for microbial colonisation and pathogenesis (Gravato-Nobre and Hodgkin, 2005) and their gut can be co-colonised by multiple pathogens and commensals (Peleg et al., 2008; Portal-Celhay and Blaser, 2012; Montalvo-Katz et al., 2013; Hodgkin et al., 2013).
The N2 wild-type nematode strain used herein was obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN, USA). We used the E. faecalis lab strain OG1RF (Garsin et al., 2001), an isolate from the human digestive tract, and S. aureus strain MSSA476 (Holden et al., 2004), a disease-causing pathogen.
Experimental evolution
A single, randomly selected clone of E. faecalis was the ancestor for all evolving populations, and stock of a single clone of S. aureus was used. Thus, only E. faecalis was permitted to evolve in response to species interactions whereby they inhabited the C. elegans gut alone (single evolution, SE) or with S. aureus (co-colonisation evolution, CCE; Figure 1). Nematodes also remained evolutionarily static throughout the experiment. A stock population of N2 wild-type nematodes was derived by isolating a single hermaphroditic female every generation from the population for five generations to ensure genetic homogeneity. Stock populations of the ‘isofemale' line were routinely maintained on nematode growth medium plates seeded with 50 ul of Escherichia coli OP50 in Luria-Bertani broth and kept at 20 °C. The nematodes digest E. coli after this bacterium is consumed, and it does not accumulate in the gut.
Exposure, transfer and selection
Bacteria were cultured in Todd-Hewitt (TH) broth at 28 °C overnight. Lawns of S. aureus liquid culture (60 μl) were plated onto 9 cm petri plates with Tryptone Soy Broth (TSB) agar, and lawns of E. faecalis culture (60 μl) were also plated on TSB with 100 μg ml−1 rifampicin (in both experimental evolution treatments). This antibiotic is used to select for E. faecalis OG1RF from mixed cultures. Bacterial lawns were placed at 28 °C overnight and then cooled at room temperature for several hours prior to use.
For a given replicate, approximately 900 L4 (larval) individuals, previously feeding on E. coli, were transferred in M9 buffer to an exposure plate with E. faecalis. In the CCE treatment, after 24 h, all nematodes were washed off the plate with M9 buffer and centrifuged at 3000 r.p.m. for 3 min. The supernatant was discarded, and then 5 ml M9 buffer was added to the test tube. This washing procedure was repeated five times to clean excess bacterial cells off the nematode cuticle. Nematodes were in the M9 buffer for <10 min at any given point in time. Nematodes were then transferred to the second exposure plate with S. aureus from a frozen culture stock. During exposures, nematodes were placed at 25 °C. E. faecalis populations evolved in the absence of S. aureus during the SE treatment were simply maintained in C. elegans on their plate without transfer during that period.
Twenty-four hours later, 15 bacteria-killed nematode carcasses were picked from a single replicate population and placed in a 1.5 ml centrifuge tube with 1 ml M9 buffer. The tube was centrifuged at 3000 r.p.m. for 3 min, the supernatant was discarded, and 1 ml M9 buffer was added. The wash process was repeated five more times. After the final rinse, the nematode pellet was crushed with a pestle to release the pathogens from inside the carcass. The suspension was streaked onto selective media—TSB agar with 100 μg ml−1 rifampicin to isolate E. faecalis—and individual colonies were grown up at 28 °C overnight. Subsequently, 15 colonies of E. faecalis were picked from a given replicate population and mixed together in 5 ml THB overnight at 28 °C overnight. This liquid culture was then used to make a lawn for the next generation of exposures for that replicate. This procedure was identical for both experimental evolution treatments to control against possible impacts of rifampicin.
The liquid cultures of an ancestral colony (prior to selection) and evolved E. faecalis populations were frozen at −80 °C in 20% glycerol every five generations.
Host mortality and bacterial fitness assays
Host mortality was assayed simultaneously for each population in each treatment at the end of the evolution experiment. We exposed approximately 200 L4 nematodes from the C. elegans stock to the ancestral and each of the 12 evolved populations of E. faecalis (from the G5, G10 and G15 experimental host generations). If populations were then tested with S. aureus, nematodes were washed off the E. faecalis exposure plate with M9 buffer into a 15 ml test tube, washed and transferred to the S. aureus exposure plate as described above. After 24 h of exposure, we counted the total number of dead nematodes. Nematodes were considered dead if they did not respond to touch with a platinum wire, as is standard in assays of C. elegans death. Simultaneously, approximately 200 nematodes were placed on each of six control plates with E. coli OP50, but no mortality was observed after 24 h. We also tested for within-population variation in the protective effect exhibited by CCE E. faecalis. Four colonies from each of the six replicate populations at G15 were randomly picked, separately grown in THB media and plated. Host mortality in the presence of S. aureus was tested as above.
We tested the generality of this protective effect against six pathogenic, genetically divergent S. aureus isolates (COL-MRSA, MSSA SH-1000, Newman, N13-MSSA, Mu50 MRSA, MRSA 252), in addition to MSSA476. All isolates were cultured the same way as described above. Similar to the methods above, approximately 50 L4 nematodes from the C. elegans stock were exposed to only S. aureus, or initially to populations of E. faecalis (ancestral or CCE G15) and then to S. aureus. After 24 h of exposure at 25 °C, we counted the total number of dead nematodes.
To examine the fitness differences of E. faecalis (ancestor vs SE at G15 vs CCE at G15) in co-colonised nematode hosts with S. aureus, we measured the number of colony-forming units (cfus) of E. faecalis and S. aureus. Five dead nematodes were picked from a plate, placed into 1 ml M9 buffer and washed repeatedly. After the final rinse, the nematode pellet was crushed with a pestle to release the bacterial cells from inside each carcass. The mixture was spread onto selective media to separate E. faecalis and S. aureus colonies (TSB with 100ug ml−1 rifampicin and Mannitol Salt Agar, respectively), and colonies were counted.
Mechanism of pathogen suppression
Genomic analysis
To investigate the genetic basis of host protection conferred by E. faecalis, whole-genome resequencing was used for a randomly selected E. faecalis clone from each replicate at G15. The phenotype of that clone was confirmed as being consistent with population-level effects on nematodes as assessed above. DNA was isolated using either a DNeasy blood and tissue kit using standard methods for Gram-positive bacteria or using a modified CTAB extraction (von der Schulenburg et al., 2001), and importantly, the addition of 10 mg ml−1 of Lysozyme (for E. faecalis) or Lyostaphin (for S. aureus) to the digestion step in both protocols was required. Illumina (San Diego, CA, USA) TruSeq Nano libraries were prepared from 200 ng of DNA according to the manufacturer's protocol and 250 bp paired-end reads generated on an Illumina MiSeq using v2 chemistry. Reads were trimmed for the presence of Illumina adaptor sequences using Cutadapt v1.2.1 and for a minimum quality score of Q20 using Sickle v1.200. The resulting reads (between 395 Mb and 715 Mb per sample) were then mapped to either E. faecalis OGIRF (NC_017316) or S. aureus MSSA4776 (NC_002953.3 and NC_005951, for main chromosome and plasmid, respectively) using BWA-MEM, duplicate reads were removed using Picard, local realignment and single nucleotide polymorphism calling was performed in GATK and structural variants detected using Breakdancer. Variants found in experimental but not ancestral clones were identified, and SnpEFF was used to predict their functional effects.
The genes with revealed single nucleotide polymorphisms were identified in the SEED database by ‘blasting' the corresponding sequences against a collection of E. faecalis genomes. The gene annotations were confirmed or suggested by analysis of the sequences and the 20000 bp window neighbourhoods of the corresponding genes. The composition of gene loci of the top 10 homologues in other bacteria was also analysed. STRING database and software was used to reconstruct gene connectivity networks for the detected genes. This application automatically assembles the data on gene positional associations in genomes, genetic, regulatory and physical protein interactions for the input genes that satisfy a set of confidence thresholds.
In vitro biochemical assays
We assessed for a difference in the ability of ancestral and evolved E. faecalis to produce superoxides as the mechanism of host protection. Ancestral E. faecalis, SE E. faecalis (at G15) and CCE E. faecalis (at G5 and G15) were grown overnight to stationary phase in TSB. Wells in an opaque, black 96-well plate with a transparent bottom were then inoculated with 5 μl from each overnight culture. Three technical replicates of each replicate population were made. Replicate populations that failed to grow properly in liquid culture were excluded from the analysis. The wells were prepared with 95 μl TSB and 100 μl of a reaction mixture from a superoxide ion assay kit (Sigma Aldrich, St Louis, MO, USA) containing luminol: a reagent that becomes luminescent following oxidation by superoxide, allowing the quantitative and relativistic measure of superoxide production. The inoculated reaction mixtures were monitored over 10 h (for which the kit was optimised by Sigma-Aldrich) and measured every 2.5 min for both OD600 and luminescence in a Synergy 2 plate reader (BioTek, Winooski, VT, USA). The actual luminescence produced by a sample is sensitive to starting conditions as it is proportional to bacterial biomass concentration. Bacterial growth is sensitive to several factors (that is, media concentration, population size) and is exponential, translating small differences in growth rate to large differences in luminescence. We thus simultaneously measured bacterial biomass concentration (OD600) and controlled for it in our luminescence data.
To examine the impact of superoxide production by evolved E. faecalis on S. aureus (and whether this was the source of suppression), we tested the degree to which the evolved enhanced suppression could be removed by the action of catalase (CAT) and superoxide dismutase (SOD). Superoxide dismutase converts superoxide into hydrogen peroxide, and CAT converts hydrogen peroxide into water and oxygen. Alone, SOD would remove superoxides by simply replacing it with harmful hydrogen peroxide. Likewise, CAT on its own would only remove the problems caused by hydrogen peroxide without affecting superoxides. Together, these enzymes create a pathway converting harmful superoxide into harmless products. If exogenous superoxides were responsible for S. aureus growth inhibition, we would therefore expect this inhibition to be lifted only when both enzymes are administered. Overnight cultures of all ancestral and CCE populations of E. faecalis, as well as S. aureus, were grown separately in TSB (standardised to an OD600 of 1.00±0.05). A solution of TSB was prepared with 0.25 M potassium phosphate buffer containing Superoxide Dismutase E.C. 1.15.1.1 (SOD) from bovine erythrocytes (Sigma-Aldrich) and Catalase E.C. 1.11.1.6 (CAT) from bovine liver (Sigma-Aldrich) each at 0.25 mg ml−1. An enzyme-free solution of TSB was also prepared as a control, containing the 0.25 M potassium phosphate buffer alone. The ancestor and CCE E. faecalis (two technical replicates of each replicate population) were mixed in equal ratios with S. aureus. From the liquid culture, 6 μl was used to inoculate wells in a 96-well plate with 196 μl of the TSB solution alongside wells of an S. aureus control (S. aureus only). The experiment was run in duplicate on the enzyme-free and enzyme-containing media. Cultures were shaken for 24 h at 30 °C, after which cfu counts were performed.
Statistical analyses
All statistical analyses were conducted in SPSS 20.0.
Host mortality and bacterial fitness assays
Mortality data met assumptions of normality and equal variances. We performed an analysis of variance (ANOVA) on untransformed data to test for the difference in nematode mortality caused by E. faecalis and S. aureus independent colonisation, as well as their co-colonisation.
For the evolution experiment, we examined changes in nematode mortality every G5 in both the SE and CCE selection regimes. We performed a generalised linear model with binomial distribution (and maximum likelihood estimates) on host mortality data in the evolution treatments (with and without the presence of S. aureus) over time. Treatment and time (experimental generations) were fixed effects. A separate ANOVA was conducted to test for variation among isolates in within-population protective effects.
To examine the spectrum of host protection, we quantified nematode mortality upon co-colonisation by E. faecalis and a diverse range of S. aureus isolates including both laboratory and human disease isolates (Figure 4). An ANOVA was performed on host mortality data collected from single infections of S. aureus across the seven isolates to examine the variability in nematode mortality they produced. We then performed a generalised linear model with binomial distribution (and maximum likelihood estimates) on host mortality data with treatment (‘alone', ‘with ancestral E. faecalis' and ‘with CCE faecalis') and S. aureus isolate as fixed effects.
The number of E. faecalis and S. aureus viable cfus was square-root transformed to meet parametric assumptions. Separate ANOVAs were performed on transformed cfu values for each of E. faecalis and S. aureus within a dead, co-colonised nematode to test the effects of treatment on bacterial fitness. Least square mean contrasts were performed to test for differences between treatments.
Mechanism of superoxide production and pathogen suppression
Mean superoxide production was compared between ancestral and evolved E. faecalis populations from the in vivo experiment during the exponential growth phase (6–10 h) of the bacteria using t-tests as the data met assumptions of normality. Luminescence measurements were controlled for OD600.
S. aureus growth in liquid culture was compared in the presence and absence of E. faecalis. We performed a generalised linear model with Poisson loglinear model (and maximum likelihood estimates) on S. aureus cfu values with treatment (‘alone', ‘with ancestral E. faecalis' and ‘with CCE E. faecalis'), and enzymes (presence and absence) as fixed effects. Their interaction was also evaluated.
Results
Changes in host mortality due to within-host microbial evolution
Ancestral E. faecalis is mildly pathogenic within the 2-day exposure window of this experiment, with <1% of nematodes dying after colonisation. In contrast, 52% of worms exposed to S. aureus died after exposure. Colonisation of worms with E. faecalis before exposure to S. aureus results in intermediate rates of nematode mortality, indicating that resident E. faecalis has the potential to suppress S. aureus virulence (Figure 2a; ANOVA: F2,18=51.908, P<0.001).
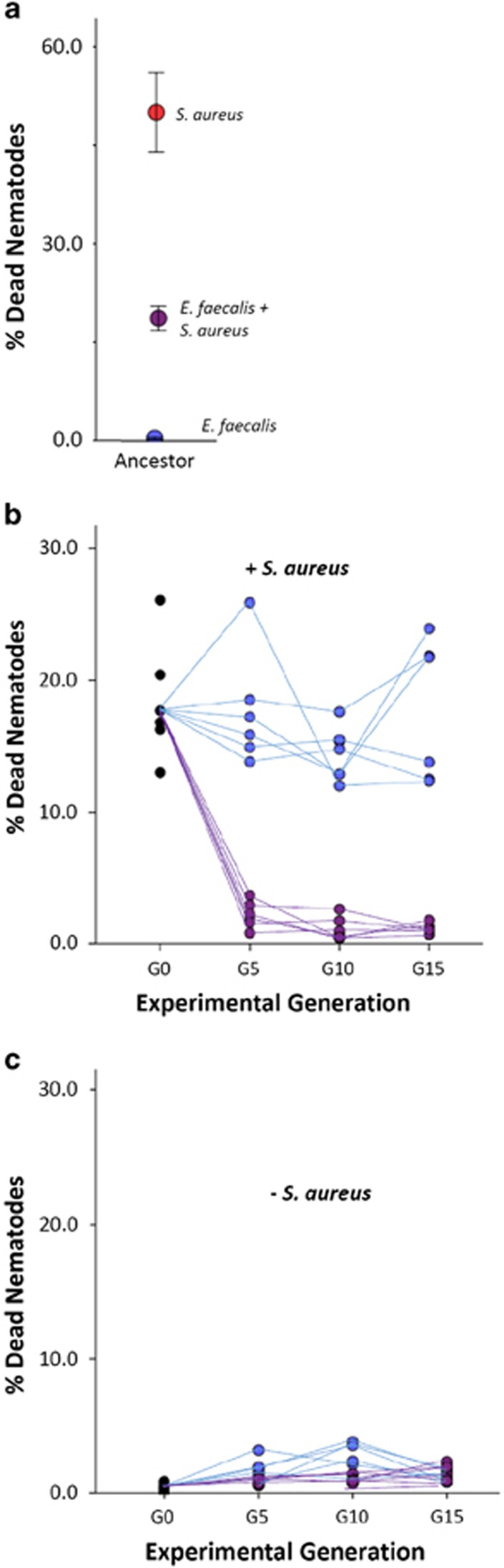
Figure 2.
Effects of resident microbes on hosts over evolutionary time. (a) Host mortality with ancestral E. faecalis (blue circle) and S. aureus (red circle) separate and co-colonising (purple circle) in the nematode. The intermediate level of virulence from co-colonising bacteria species suggested the potential for E. faecalis to suppress pathogenic S. aureus. Error bars, 1 s.e. (b, c) Populations of E. faecalis were evolved under two different selection regimes: SE and CCE for 15 experimental host generations. To assess the ability of E. faecalis to protect hosts from S. aureus, host mortality in the (b) presence and (c) absence of S. aureus was quantified every G5 for SE (blue circles) and CCE (purple circles) E. faecalis. Lines connect each of the six replicate populations per treatment across time.
At the end of the evolution experiment, we assayed the protective ability of E. faecalis evolved in nematodes, either alone or with S. aureus co-colonisation (Figure 1). All of the six replicate CCE E. faecalis populations evolved to further suppress its virulence. Whereas 18% of worms died within 24 h of S. aureus exposure in the presence of the ancestral E. faecalis resident, this declined to 1% mortality, on average, in the presence of resident CCE E. faecalis from G5 onwards (Figure 2b). Although there is some among-population variation in the mortality rates caused by SE E. faecalis upon challenge with S. aureus, none of the replicate populations evolved significantly enhanced protective ability relative to the ancestor (Figure 2b; Generalised Linear Model, Treatment: Wald χ2=280.723, P<0.001; Time: Wald χ2=97.230, P<0.001). When four colonies within each replicate population of the CCE treatment were tested at G15, an equivalently enhanced protective effect was observed (ANOVA: F5,24=0.318, P=0.895).
Lower host mortality on S. aureus exposure was not associated with a reduction in E. faecalis virulence. Rather, whilst mortality remained generally low (<2%) in all replicate populations, E. faecalis evolved in both treatments to increase nematode mortality over time when tested alone (Figure 2c; Generalised Linear Model: Treatment: Wald χ2=9.126, P=0.003; Time: Wald χ2=22.510, P<0.001). Thus, although highly beneficial to hosts when tested in the presence of S. aureus, on its own, CCE E. faecalis remained mildly pathogenic and costly for the nematode host to possess. This result clearly demonstrates the context dependence of the fitness effects of this protective microbe upon hosts (Chamberlain et al., 2014).
Spectrum of host protection
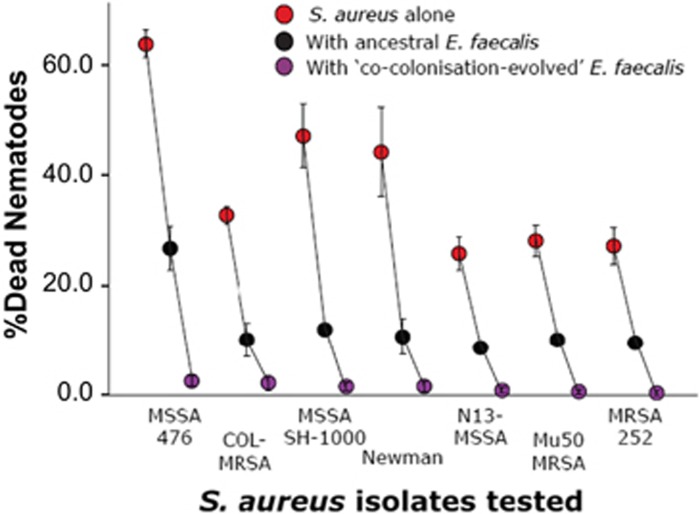
All CCE E. faecalis populations at G15 were effective at protecting nematode hosts against a broad spectrum of S. aureus isolates (Figure 3). In single infections, these S. aureus isolates exhibited variation in their virulence towards C. elegans (Figure 3; 26–65% mean nematode mortality; ANOVA F6,42=10.505, P<0.001). In co-colonised hosts, all S. aureus isolates with ancestral E. faecalis produced intermediate rates of host mortality, whereas with all replicate populations of CCE E. faecalis, nematode mortality was dramatically reduced to 0–1%. Both treatment and isolate affected the virulence of pathogens on hosts (Generalised Linear Model, Treatment: Wald χ2=370.961, P<0.001; Isolate: Wald χ2=303.650, P<0.001).
Figure 3.
Generality of host protection by evolved E. faecalis against seven S. aureus isolates. Host mortality was evaluated after 24 h of exposure to S. aureus. Nematodes were exposed to S. aureus alone (red circles) or were previously colonised by ancestral E. faecalis (black circles) or CCE E. faecalis at G15 (purple circles). MSSA476 was used in the evolution experiment. Error bars, 1 s.e.
Host protection and microbial growth within hosts
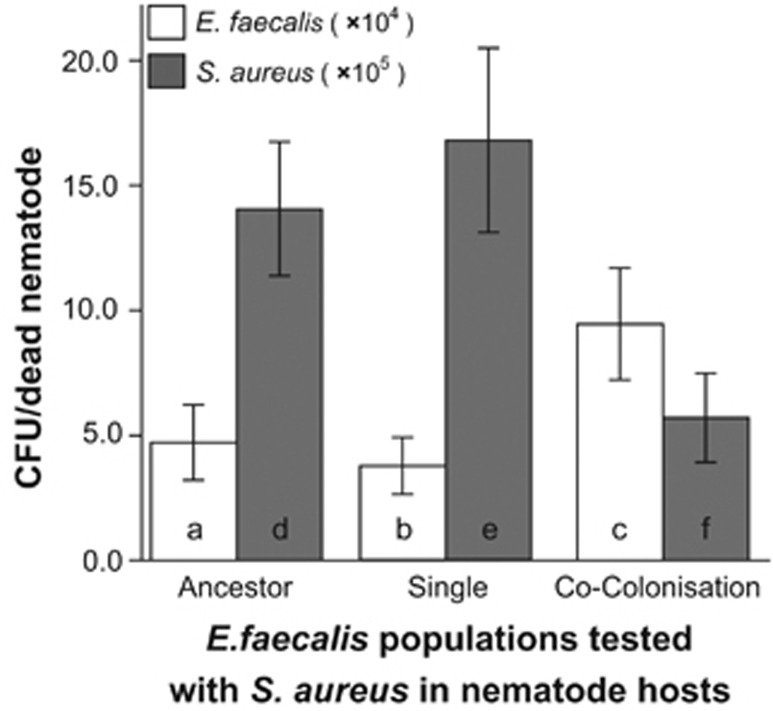
We examined whether the evolved E. faecalis suppression of S. aureus virulence was associated with increased E. faecalis proliferation and reduced S. aureus growth (Figure 4). Compared with ancestral E. faecalis, CCE E. faecalis (assayed at G15) suppressed S. aureus viable bacterial counts more (Figure 4; d vs f) and accumulated marginally more within nematodes (Figure 4; a vs c). By contrast, SE E. faecalis populations did not grow to higher density on average than the ancestor when interacting with S. aureus (Figure 4; a vs b). These SE populations were also associated with higher within-host growth of S. aureus compared with CCE E. faecalis (Figure 4; Analysis for S. aureus cfu: ANOVA across the three treatments: F2,18=4.072, P=0.039; Least Square difference d>f, P=0.038; Least Square difference e>f, P=0.019; Analysis for E. faecalis cfu: ANOVA across all three treatments: F2,18=3.603, P=0.053; Least Square difference a<c, P=0.057; b<c, P=0.023 a=b, P=0.649).
Figure 4.
Fitness (cfus/nematode) of resident E. faecalis populations and infecting S. aureus. S. aureus is co-colonising with ancestral, SE, or CCE E. faecalis populations. Error bars, 1 s.e.
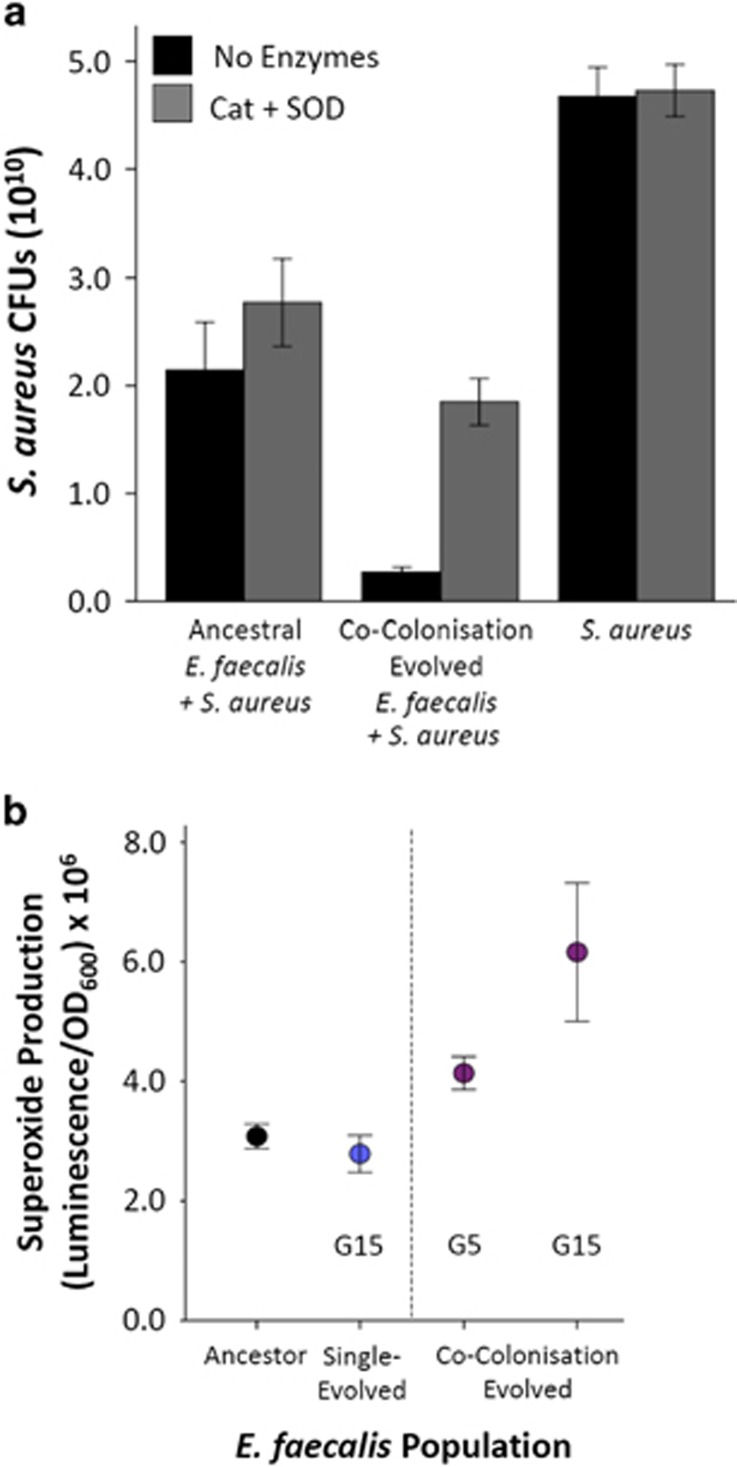
Suppression of S. aureus may occur either directly from the presence of E. faecalis, be mediated by E. faecalis-induced alterations to host biology or be a product of both direct and host-mediated effects. We assessed the importance of direct suppression using in vitro experiments. In vitro experiments recapitulated in vivo assays showing that CCE E. faecalis populations were better able to suppress S. aureus growth in liquid culture than ancestral E. faecalis (Figure 5a; Generalised Linear Model, Treatment: Wald χ2=3.18 × 1011, P<0.001).
Figure 5.
Evolved mechanism of suppression of S. aureus by E. faecalis. (a) Suppression and enzyme-mediated lifting of suppression of S. aureus outside the host. S. aureus cfus were counted when the pathogen was grown alone and co-cultured with ancestral or CCE E. faecalis. Counts were also made upon the addition of CAT and SOD, enzymes that remove the presence of reactive oxygen species. Error bars, 1 s.e. (b) Mean superoxide production (measure of luminescence controlling for OD600) across exponential growth phase of ancestral, SE and CCE E. faecalis (the latter at G5 and G15).
Genomic and biochemical analysis of the mechanism underpinning protection
To investigate the genetic basis of E. faecalis-mediated protection, we whole-genome resequenced a randomly picked clone of ancestral E. faecalis and evolved E. faecalis from each of the 12 replicate populations at G15. Each evolved E. faecalis clone exhibited a unique set of between one and three mutations (Supplementary Table 1). Consistent with the distinct phenotypes that evolved under the two contrasting treatments, the SE and CCE regimes selected for substitutions in different, functionally distinct gene sets. Six of 12 mutations in the CCE E. faecalis clones—1 per clone per replicated population—were putatively associated with superoxide production, but no mutations associated with this pathway were observed in clones from the SE treatment. E. faecalis is known to produce extracellular superoxide (Huycke et al., 2011), mediated by dehydrogenase and fumarate reductase. Mutations were found in two NADH dehydrogenases and four genes associated with the respiratory complex function or purine biosynthesis. Purine biosynthesis represents the major pathway for fumarate production which, if blocked, leads to superoxide production (Supplementary Table 1; Supplementary Figure 1).
We therefore hypothesised that enhanced production of antimicrobial reactive oxygen species was the mechanism behind E. faecalis-mediated defence. In accordance with this hypothesis, all CCE E. faecalis populations produced more superoxide per cell than the ancestor, in both the G5 and G15 generations (Figure 5b; t-test: Ancestor vs G5, t=−3.056, df=31.385, P=0.005; Ancestor vs G15, t=−2.619, df=14.888, P=0.019). Moreover, there was no difference in superoxide production between SE and ancestral E. faecalis (t=0.788, df=20.329, P=0.440) suggesting that this trait only evolved during S. aureus challenge. The addition of CAT and SOD enzymes to growth media ablated the suppression of S. aureus growth by E. faecalis during in vitro interactions (Figure 5a; Generalised Linear Model, Enzymes: Wald χ2=8.49 × 1010, P<0.001), and had a greater effect at reducing suppression during interactions with CCE E. faecalis compared with the ancestor (Generalised Linear Model, Treatment × Enzymes: Wald χ2=7.24 × 1010, P<0.001). Together these data strongly point to increased superoxide production by evolved CCE E. faecalis as the mechanism of suppression of S. aureus.
Discussion
The role of microbes in protecting their host against virulent pathogens has traditionally focused on ecological sources of protection, namely niche occupancy and competition for resources (for example, in insects, Gerardo and Parker, 2014). We hypothesised that owing to the high evolutionary potential of microbes—associated with their short generation times and large within-host population size—rapid de novo microbial evolution could have a role in shaping host resistance against infection. We observed the evolution of host-protective effects during microbial experimental evolution within nematode hosts in all independently passaged CCE populations, thus confirming the potential for this process to occur. Thus, E. faecalis, a microbe that has been observed in natural microbiomes to possess protective traits (Martin-Vivaldi et al., 2010; Kommineni et al., 2015), evolved across the parasite-mutualist continuum to become a host-protective mutualist upon pathogen attack. Notably, these host-protective effects evolved without any direct selection against host mortality. Instead, a beneficial relationship between the host and the resident bacterium emerged out of interactions with a virulent pathogen and selective processes acting upon the resident microbial populations. Although CCE E. faecalis populations evolved the ability to attenuate the high mortality caused by S. aureus, they also retained mild pathology against C. elegans when colonising alone, demonstrating the context dependence of their fitness effects on the host (Chamberlain et al., 2014). In an environment where such virulent infection is common, E. faecalis would therefore now represent a net mutualist with respect to its impact on host fitness. This result reflects observations of other protective microbes found naturally, which defend their host whilst retaining pathogenicity (Martinez et al., 2014; Polin et al., 2014).
The mechanisms of microbe-mediated protection observed in nature are remarkably diverse (Gerardo and Parker, 2014). Although niche occupation (Dillon et al., 2005), resource competition and immune system mediation (Abt and Artis, 2013; Hooper et al., 2012; McFall-Ngai et al., 2013) may still have a role in our system, the genomic evidence indicates that selection acted predominantly through direct E. faecalis–S. aureus interactions during host colonisation. Further experiments, however, are required to determine whether the microbial interactions observed to evolve here are adaptive to the host environment or whether similar evolutionary outcomes would arise in vitro. Regardless, we observed parallel evolution of the superoxide production pathway in CCE E. faecalis across all replicate populations, and we were able to ablate the evolved suppression through enzymatic treatment to remove superoxide radicals. These data strongly suggest that antimicrobial superoxides, which may act to directly suppress S. aureus or act indirectly via oxidation of the S. aureus auto-inducing pheromone (Rothfork et al., 2004), are a key mechanism in the evolved protective phenotype. The lack of genotype specificity we observed is also consistent with a superoxide-mediated suppression system, which represents a broad-spectrum form of microbial suppression. While C. elegans itself produces reactive oxygen species in response to pathogen infection (Chávez et al., 2007), reactive oxygen species produced by resident bacteria may also be a common means of broad-spectrum protection against infection, and one that is thus likely to be evolutionary labile in its activity. For instance, lactic acid bacteria in the guts of honeybees are able to suppress a range of pathogens, including S. aureus and Pseudomonas aeruginosa via reactive oxygen species production (Olofsson et al., 2014). That our experimental treatment drove the evolution of a broad-spectrum defence mechanism, as opposed to more specific mechanisms of suppression (for example, bacteriocin secretion), is also consistent with observations from natural disease systems showing that microbes can protect against a diversity of pathogen isolates (Koch and Schmid-Hempel, 2011) and species (Martinez et al., 2014).
The extent of the protective phenotype that evolved here, and the rate at which it evolved, were striking. Despite being regularly attacked by pathogens, if nematode hosts were colonised by evolving E. faecalis, they were almost universally protected against pathogens that would otherwise quickly kill most of the population. Moreover, although the evolution in our experiment occurred during passage through a number of individual worms, the time frame for the evolution of protection by E. faecalis was just 5 days of co-colonisation. This short timescale presents the possibility of the evolution of microbe-mediated protection within the lifetime of a longer-lived host, such as a mammal or tree, in which evolution is likely potentiated by larger population sizes.
Future research will need to establish how within-host evolution of microbial species would alter disease progression. A key simplification in our experiment is that the virulent pathogen is genetically fixed, thus mimicking a spillover zoonotic infection whereby the pathogen normally circulates in a different host species. An example is Salmonella. Some isolates of this bacterium commonly reside within the microbiomes of livestock animals, but can cause serious infections if transmitted to humans. Within a host individual, however, it is possible that pathogen evolution would also occur on a similar timescale, obviating any evolved protective abilities in resident microbial species and setting the stage for coevolutionary interactions. Our experiment also considers only a binary microbial interaction, whereas natural microbial communities are often highly diverse. The impacts of the evolution of the microbiome on pathogen attack (Mueller and Sachs, 2015) and on interactions within the microbiome also warrant consideration. Notwithstanding this, the potential for evolution of interactions between resident microbes and pathogens is clear, and future research on microbiome–pathogen relationships should go beyond ecological responses to examine evolved ones.
Acknowledgments
We are grateful to MA Félix and L Morran for advice on C. elegans. Thanks to M Phillipo for assistance in the lab. Sequence data are available from the European Nucleotide Archive under accession PRJEB7382. Funding was provided by a Royal Society Newton International fellowship to KCK. The doi for our data i; doi:10.5061/dryad.nd848.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abt MC, Artis D. (2013). The dynamic influence of commensal bacteria on the immune response to pathogens. Curr Opin Microbiol 16: 4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst MA, Koskella B. (2013). Experimental coevolution of species interactions. Trends Ecol Evol 28: 367–375. [DOI] [PubMed] [Google Scholar]
- Brucker RM, Bordenstein SR. (2013). The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science 341: 667–669. [DOI] [PubMed] [Google Scholar]
- Cabreiro F, Gems D. (2013). Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans. EMBO Mol Med 5: 1300–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerf-Bensussan N, Gaboriau-Routhiau V. (2010). The immune system and the gut microbiota: friends or foes? Nat Rev Immunol 10: 735–744. [DOI] [PubMed] [Google Scholar]
- Chamberlain SA, Bronstein JL, Rudgers JA. (2014). How context dependent are species interactions? Ecol Lett 17: 881–890. [DOI] [PubMed] [Google Scholar]
- Chávez V, Mohri-Shiomi A, Maadani A, Vega LA, Garsin DA. (2007). Oxidative stress enzymes are required for daf-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics 176: 1567–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LC, Hodgkin J. (2013). Commensals, probiotics, and pathogens in the Caenorhabditis elegans model. Cell Microbiol 16: 27–38. [DOI] [PubMed] [Google Scholar]
- Cruz MR, Graham CE, Gagliano BC, Lorenz MC, Garsin DA. (2013). Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect Immun 81: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RJ, Vennard CT, Buckling A, Charnley AK. (2005). Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett 8: 1291–1298. [Google Scholar]
- Dillon RJ, Vennard CT, Charnley AK. (2000). Pheromones: exploitation of gut bacteria in the locust. Nature 403: 851–853. [DOI] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. (2009). Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathogens 5: e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix M, Braendle C. (2010). The natural history of Caenorhabditis elegans. Curr Biol 20: R965–R969. [DOI] [PubMed] [Google Scholar]
- Garbutt J, Bonsall MB, Wright DJ, Raymond B. (2011). Antagonistic competition moderates virulence in Bacillus thuringiensis. Ecol Lett 14: 765–772. [DOI] [PubMed] [Google Scholar]
- Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE et al. (2001). A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci USA 98: 10892–10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerardo NM, Parker BJ. (2014). Mechanisms of symbiont-conferred protection against natural enemies: an ecological and evolutionary framework. Curr Opin Insect Sci 4: 8–14. [DOI] [PubMed] [Google Scholar]
- Gravato-Nobre MJ, Hodgkin J. (2005). Caenorhabditis elegans as a model for innate immunity to pathogens. Cell Microbiol 7: 741–751. [DOI] [PubMed] [Google Scholar]
- Gray JC, Cutter AD. (2014). Mainstreaming Caenorhabditis elegans in experimental evoution. Proc R Soc B 281: 20133055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Felix M-A, Clark LC, Stroud D, Gravato-Nobre MJ. (2013). Two Leucobacter strains exert complementary virulence on Caenorhabditis including death by worm-star formation. Curr Biol 23: 2157–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC et al. (2004). Complete genomes of two clinical Staphylococcus aureus strains: evidence for rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA 101: 9786–9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. (2012). Interactions between the microbiota and the immune system. Science 336: 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huycke MM, Moore D, Joyce W, Wise P, Shepard L, Kotake Y et al. (2011). Extracellular superoxide production by Enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol Microbiol 42: 729–740. [DOI] [PubMed] [Google Scholar]
- Jaenike J, Unkless R, Cockburn SN, Boelio LM, Perlman SJ. (2010). Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329: 212–215. [DOI] [PubMed] [Google Scholar]
- Kamada N, Seo S-U, Chen GY, Nunez G. (2013). Role of gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 13: 321–335. [DOI] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P. (2011). Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci USA 108: 19288–19292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P et al. (2015). Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526: 719–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C et al. (2012). Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PloS Pathogens 8: e1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lize A, McKay R, Lewis Z. (2014). Kin recognition in Drosophila: the importance of ecology and gut microbiota. ISME J 8: 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon MJ, Read AF. (2004). Immunity promotes virulence evolution in a malaria model. PLoS Biol 2: E230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Vivaldi M, Pena A, Peralta-Sánchez JM, Sánchez L, Ananou S, Ruiz-Rodriguez M et al. (2010). Antimicrobial chemicals in hoopoe preen secretions are produced by symbiotic bacteria. Proc R Soc B 277: 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Longdon B, Bauer S, Chan Y, Miller W, Bourtzis K et al. (2014). Symbionts commonly provide broad spectrum resistance to viruses in insects: a comparative analysis of Wolbachia strains. PloS Pathogens 10: e1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May G, Nelson P. (2014). Defensive mutualisms: do microbial interactions within hosts drive the evolution of defensive traits? Funct Ecol 28: 356–363. [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA 110: 3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332: 1097–1100. [DOI] [PubMed] [Google Scholar]
- Messenger SL, Molineux IJ, Bull JJ. (1999). Virulence evolution in a virus obeys a trade-off. Proc R Soc B 266: 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvo-Katz S, Huang H, Appel MD, Berg M, Shapira M. (2013). Association with soil bacteria enhances p38-dependent infection resistance in Caenorhabditis elegans. Infect Immun 81: 514–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran LT, Schmidt OG, Gelarden IA, Parrish RCI, Lively CM. (2011). Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science 333: 216–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller UG, Sachs JL. (2015). Engineering microbiomes to improve plant and animal health. Trends Microbiol 23: 606–617. [DOI] [PubMed] [Google Scholar]
- Olofsson TC, Butler E, Markowicz P, Lindholm C, Larsson P, Vasquez A. (2014). Lactic acid bacterial symbionts in honeybees - an unknown key to honey's antimicrobial and therapeutic activities. Int Wound J e-pub ahead of print 8 September 2014.. [DOI] [PMC free article] [PubMed]
- Peleg AY, Tampakakis E, Fuchs BB, Eliopoulos GM, Moellering RC, Mylonakis E. (2008). Prokaryote-eukaryote interactions identified by using C. elegans. Proc Natl Acad Sci USA 105: 14585–14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C, Dirksen P, Schulenburg H. (2015). Why we need more ecology for genetic models such as C. elegans. Trends Genet 31: 120–127. [DOI] [PubMed] [Google Scholar]
- Polin S, Simon J-C, Outreman Y. (2014). An ecological cost associated with protective symbionts of aphids. Ecol Evol 4: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal-Celhay C, Blaser MJ. (2012). Competition and resilience between founder and introduced bacteria in the Caenorhabditis elegans gut. Infect Immun 80: 1288–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfork JM, Timmins GS, Harris MN, Chen X, Lusis AJ, Otto M et al. (2004). Inactivation of a bacterial virulence pheromone by phagocyte-derived oxidants: new role for the NADPH oxidase in host defense. Proc Natl Acad Sci USA 101: 13867–13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. (2010). Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc Natl Acad Sci USA 107: 7359–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifri CD, Begun J, Ausubel FM, Calderwood SB. (2003). Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect Immun 71: 2208–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifri CD, Mylonakis E, Singh KV, Qin X, Garsin DA, Murray BE et al. (2002). Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect Immun 70: 5647–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Schulenburg JH, Hancock JM, Pagnamenta A, Sloggett JJ, Majerus ME, Hurst GDD. (2001). Extreme length and length variation in the first ribsosomal internal transcribed spacer of ladybird beetles (Coleoptera: Coccinellidae). Mol Biol Evol 18: 648–660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.