Abstract
Prochlorococcus, the smallest known photosynthetic bacterium, is abundant in the ocean's surface layer despite large variation in environmental conditions. There are several genetically divergent lineages within Prochlorococcus and superimposed on this phylogenetic diversity is extensive gene gain and loss. The environmental role in shaping the global ocean distribution of genome diversity in Prochlorococcus is largely unknown, particularly in a framework that considers the vertical and lateral mechanisms of evolution. Here we show that Prochlorococcus field populations from a global circumnavigation harbor extensive genome diversity across the surface ocean, but this diversity is not randomly distributed. We observed a significant correspondence between phylogenetic and gene content diversity, including regional differences in both phylogenetic composition and gene content that were related to environmental factors. Several gene families were strongly associated with specific regions and environmental factors, including the identification of a set of genes related to lower nutrient and temperature regions. Metagenomic assemblies of natural Prochlorococcus genomes reinforced this association by providing linkage of genes across genomic backbones. Overall, our results show that the phylogeography in Prochlorococcus taxonomy is echoed in its genome content. Thus environmental variation shapes the functional capabilities and associated ecosystem role of the globally abundant Prochlorococcus.
Introduction
Prochlorococcus is the most abundant marine phytoplankton and an important contributor to global primary production (Bouman et al., 2006; Flombaum et al., 2013). Within the group, there are multiple phylogenetically distinct lineages and the distribution of this diversity has been linked to environmental factors, such as light, temperature and iron availability (Moore et al., 1998; West and Scanlan, 1999; Johnson et al., 2006; Zwirglmaier et al., 2008; Rusch et al., 2010). It has been suggested that pan-genome variability allows Prochlorococcus as a genus to proliferate across many environments (Partensky et al., 1999; Kettler et al., 2007), but it is unclear how the mechanisms of descent contribute to their evolution and the diversity of local populations. Vertical descent, specifically the transfer of genes through cellular division, and horizontal gene transfer, encompassing the several mechanisms of moving genes between organisms without division, are two ways that genes can arrive in an organism. Based on a ‘genomic backbones' concept (Kashtan et al., 2014), where a stable niche partitioning of sub-populations of Prochlorococcus is proposed, it should follow that phylogeny will largely correspond to the biogeography of the overall gene content. However, the genomic presence of many genes is highly variable as gene gain or loss may be a driving force in the evolution of Prochlorococcus (Kettler et al., 2007). Variables affecting simple traits such as nutrient acquisition may better explain the flexible gene content than variables associated with complex traits such as light acquisition and DNA repair that require more exchanged genes (Martiny et al., 2013; Brown et al., 2014). For example, some of these flexible genes are found in association with phosphate acquisition (Martiny et al., 2006), where the genomes of Prochlorococcus cells in environments with lower phosphate concentrations are enriched in phosphate-acquisition genes (Martiny et al., 2009a). Although a few sets of flexible genes have well-characterized distributions, we know little about the global biogeography of Prochlorococcus genome content diversity for the whole of the pan genome. Understanding the distribution of phylogenetic and genomic diversity across environments will provide key insight into the selective pressures in the context of vertical inheritance structuring Prochlorococcus genome content and community composition. This will ultimately lead to clarification of the global biogeography of key biogeochemical functions of this marine phytoplankton lineage.
Previous investigations of Prochlorococcus biogeography have characterized phylogenetic diversity using specific genetic markers that reflect genome content to an unknown extent (Bouman et al., 2006; Johnson et al., 2006; Zwirglmaier et al., 2008). Metagenomics has also been employed to examine Prochlorococcus genomic diversity at smaller regional scales (Hewson et al., 2009; Rusch et al., 2010; Thompson et al., 2013) but not yet on a global scale. Here we examine the biogeography of Prochlorococcus genome diversity, both in terms of phylogenetic and genome content, globally. We first quantify the biogeography of the phylogenetic diversity based on a set of single-copy core genes. We expect that phylogenetic diversity will largely be driven by factors delineating the major ecotype clades, specifically light followed by temperature (Moore et al., 1998; Johnson et al., 2006; Coleman and Chisholm, 2007; Martiny et al., 2009c). Next we analyze the distribution of the flexible genome content by asking whether there are geographic patterns in gene distributions, whether these patterns are driven by environmental factors and whether there are specific genes that are linked to ocean regions. Finally, we determine the extent to which variation in phylogenetic diversity corresponds with differences in the flexible gene content. Although Prochlorococcus has been well characterized phylogenetically, identifying the factors controlling its genome content diversity across ocean environments is central to understanding the ecological role of phylotypes within a key lineage in marine ecosystems.
Materials and methods
We searched for sample sequences highly similar to known Prochlorococcus genes (Biller et al., 2014; see Prochlorococcus sequence calling below) and used geographic and environmental data to define the spatial distribution of the phylogenetic lineages and flexible orthologous genes. Subsequent analyses used 87 metagenomes for the phylogenetic, core gene distribution and 56 metagenomes for the non-core or flexible gene content analyses.
Metagenomic samples
Metagenomic samples from 226 Global Ocean Sampling expedition (GOS) sites were analyzed in this study (Genbank bioproject PRJNA13694 and European Bioinformatics Institute accession numbers ERX913362–ERX913706). Environmental variables were either measured while sampling (temperature, sample depth, ocean depth, latitude and longitude) or determined from composite monthly objectively analyzed means (nitrate and phosphate) for the relevant locations using the World Ocean Atlas 2009 (Garcia et al., 2010) at a 1-degree and 5-m depth resolution. If the sample's coordinates rounded to a land location, we averaged the values from the adjacent oceanic grid points. Temperature ranged from 12.1 to 37.6 °C, sample depth ranged from the surface to 62 m, ocean depth ranged from 0.33 m to 5800 m, nitrate ranged from 0 to 7.54 μmol l−1 and phosphate ranged from 0.002 to 0.953 μmol l−1. We defined regions first by separating between the oceans and then by using the 0.3-μmol l−1 phosphate contours to delineate equatorial regions (Supplementary Table S1).
Reference database
Our reference database was built from 2 Prochlorococcus metagenomic assemblies (high-light III (HLIII) and HLIV—nearly complete except for genomic islands), 41 Prochlorococcus and 15 Synechococcus fully sequenced genomes (Biller et al., 2014). We used ProPortal v.4 (Kelly et al., 2012) clusters annotated using the RAST server (Aziz et al., 2008) to define our orthologous groups. The non-core genome consists of 8027 genes found in at least one but not all of the Prochlorococcus whole genomes. The core genome consists of 504 single-copy genes, which are present in each of the Prochlorococcus and Synechococcus genomes and metagenomic assemblies. Prochlorococcus alone had a core genome of 975 genes (Supplementary Figure S1). Among the full Prochlorococcus genomes, 91.5% are single copy.
Prochlorococcus sequence calling
All pyrosequencing and Sanger metagenomic sequences were co-assembled using the CELERA assembler (Miller et al., 2008) at 92% nucleotide identity. This threshold allowed for consensus assemblies at the species and strain level (Swan et al., 2013) with reasonable computation times. The resulting scaffolds encompass 3 Gbp of contiguous DNA sequence, while 85% of the sequence reads could be mapped back to the assembly. Open reading frames on scaffolds were called using MetaGene (Noguchi et al., 2006). To determine the putative phylogenetic origin of the scaffolds, each predicted peptide was phylogenetically annotated using Automated Phylogenetic Inference System (APIS) (Dupont et al., 2014), which annotates according to the position of the peptide within a phylogenetic tree. Thus a peptide 99% similar to a Prochlorococcus protein will be annotated as Prochlorococcus (with the associated taxonomic tree), while a peptide that branches basally within the phylogenetic tree next to Cyanobacteria would only be annotated as Cyanobacteria. The scaffolds were taxonomically annotated at the lowest level for which >50% of the open reading frames had agreement in the APIS calls. All reads mapped to scaffolds defined as Cyanobacteria were then matched against protein-coding regions from our reference database using BLASTn (Camacho et al., 2009), e-value<10−5. To increase confidence in called sequences, we included Prochlorococcus' nearest evolutionary neighbor, Synechococcus, as an outgroup in the sequence calling, but all analyses only examine Prochlorococcus-related sequences.
Phylogenetic analysis
We built a reference phylogeny using the 504 core orthologous groups aligned separately in protein space using TranslatorX (Abascal et al., 2010) with ClustalW (Larkin et al., 2007) and then concatenated the gene alignments (544 614 bp) to build the maximum likelihood tree using Phylip's DNAML (default parameters, constant rate variation model with WH5701 as outgroup and re-rooted with Synechococcus to have a clearer distinction between genera). We analyzed 100 bootstrap resamplings using SEQBOOT (Felsenstein, 2005). After mapping to protein-coding regions using BLASTn, we used hmmalign from HMMER 3.1b1 (Eddy, 2011) to align sequences to the individual gene reference alignments of the corresponding orthologous group PPlacer (Matsen et al., 2010) coupled with RAxML (GTRGAMMA model) (Stamatakis, 2014) to generate the appropriate phylogenetic model statistics and map the sequences onto the reference phylogeny one core gene at a time, and finally we collapsed single gene branch abundance matrices yielding a phylogenetic distribution across samples (Supplementary Figure S2). Clades were defined by previous annotations (Berube et al., 2015) except HLII, which was split into clades with members <10% different (DNADIST, F84 model) and bootstrap values >75 (Felsenstein, 2005); deep branches were grouped as in Supplementary Figure S2. We excluded samples with <500 sequences and rarefied to account for sampling depth biases. After determining dissimilarity (Bray–Curtis), we clustered the samples hierarchically using the ‘average' clustering algorithm (R Development Core Team, 2013). We bootstrapped the data to determine the robustness of the cluster signal. We measured the association between sampling regions and phylogenetic clades or deep groups, using a point biserial correlation coefficient (r.g.) corrected for differences in sampling sizes (alpha=0.05; function ‘multipatt' De Cáceres and Legendre, 2009; Supplementary Table S2). We used the permutation multivariate analysis of variance test (function adonis; Oksanen et al., 2013) to test if regions explain clade abundance variation. We used a test for homogeneity of multivariate dispersion (betadisper, TukeyHSD; Oksanen et al., 2013; R Development Core Team, 2013). There was a significant difference in spread between a few of the pairwise combinations in the phylogenetic analysis (Supplementary Figure S4). This suggested possible differences in both dispersion and location.
Gene content analysis
Sequences of high similarity to non-core clusters (best hit, BLASTn, e-value<10−5; Camacho et al., 2009) were used in the gene content analysis. We placed a threshold of 1500 sequences per sample and rarefied to account for sampling depth biases. We used the permutation multivariate analysis of variance test (function adonis; Oksanen et al., 2013) to test if regions explain abundance variation. Using sequence similarity to non-core genes (BLASTp), we identified the categories of Clusters of Orthologous Groups of proteins (COGs) from cyanobacterial eggNOG database (Powell et al., 2014). We measured the association between sampling regions and either COG categories using a point biserial correlation coefficient (r.g.) (Supplementary Table S3) or orthologous genes using an indicator value index (indval.g) (Supplementary Table S4), both corrected for differences in sampling sizes (function ‘multipatt', alpha=0.05; De Cáceres and Legendre, 2009) but not corrected for multiple testing.
Environmental analysis
We used Canonical Correspondence Analysis to determine the most significant variables with forward selection for both the phylogenetic and the genome content signal (Canoco, v4.5; ter Braak, 1986). Ordination plots were visualized with the ‘vegan' package in R (Oksanen et al., 2013).
Temperature-related genes
We identified 85 potentially low-temperature adaptive genes by first examining orthologous groups with a negative correlation to temperature (r2<−0.3, P<0.027; Supplementary Table S5). From this set, three of the COGs with the strongest negative correlation temperature and three of the COGs unique or nearly unique to the HLI clade were used in a further analysis. To strengthen the validity of each putatively low-temperature gene, we identified scaffolds assembled from the entire GOS data set with the gene present and identified the nearest two genes closest on each metagenomic assembly (Supplementary Figure S5). We compared the distribution of all reads mapped to assemblies with the lower-temperature gene against all reads mapped to assemblies with a neighbor present but missing the lower-temperature gene of interest. We normalized read counts to total Prochlorococcus reads in each respective region allowing us to compare across regions, and we normalized to the number of reads in each set of assemblies to compare across COGs. We also identified whether a read mapped to a member of the HLI clade or not based on our previous BLASTn calls (Camacho et al., 2009).
Phylogenetic versus gene content comparison
To compare the phylogenetic variance with the gene content variance we used the Mantel test in ‘vegan' package (function mantel) (Oksanen et al., 2013) to test for a correlation between the phylogenetic and gene content sample dissimilarity matrices.
Results
We analyzed a complete global circumnavigation of 226 metagenomic samples (Figure 1 and Supplementary Table S1). With the recent addition of 29 strains to the set of fully sequenced Prochlorococcus genomes (Biller et al., 2014), we also created a maximum likelihood phylogeny (Figure 2a). We first quantified the biogeography of phylogenetic diversity by mapping sequence reads onto this detailed phylogeny and, despite the presence of every clade in every region, we observed unequal lineage representation both in terms of relative abundance and regional distribution (Supplementary Figure S1). As predicted, Prochlorococcus phylogenetic community composition varied significantly between regions (permutational multivariate analysis of variance (ANOVA), R2=0.47, P=0.0001; Figures 2b and c).
Figure 1.
Sample map. Samples color coded by region and outlined in black if included in the gene content analysis. See Supplementary Table S1 for metadata of each sample site.
Figure 2.
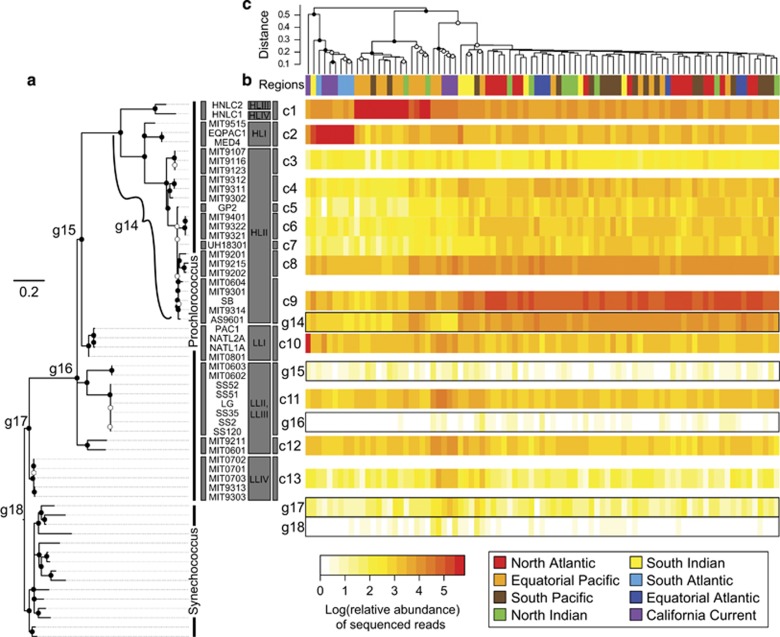
Phylogenetic diversity across ocean samples. (a) Maximum likelihood phylogeny using core genes from 41 Prochlorococcus strains, 2 Prochlorococcus metagenomic assemblies and 15 Synechococcus strains. Bootstrap values out of 100 resamplings are indicated by circles, filled in have 100% support and empty have at least 50% support. (b) Heatmap of samples versus the clades denoted in the phylogeny, c1–c13, and deep phylogenetic group(s), g14–g18. Relative abundances of sequences were log transformed and clustered hierarchically. (c) Prochlorococcus phylogenetic variation across metagenomic sites clustered using Bray–Curtis dissimilarity of PPlacer placed sequences for samples passing a 500-sequence threshold and rarefied. Node values are bootstrap support for the clade out of 100 resamplings, with filled in circles representing >75% support and empty circles representing >25% support for the group. Sample sites are color coded by region (Figure 1).
Prochlorococcus lineages varied in relative frequency among ocean basins. The strongest association was between clade c2, (HLI) and the South Atlantic Ocean and California Current regions (indicator analysis, r.g.=0.80, P=0.001). We also found that sequences mapping to clade c1 (HLIII and HLIV) significantly dominated the Equatorial Pacific Ocean samples (indicator analysis, r.g.=0.53, P=0.008) and was consistent with a past analysis of these samples from the region (Rusch et al., 2010). Among HLII clades, c9 (including strain MIT9301) was the most frequent subclade and together with c5, c6 and c7 common across all regions except the California Current, South Atlantic Ocean and Equatorial Pacific Ocean (Supplementary Table S2). Although clade c4 was within the HLII group, it corresponded to the same set of regions but was found at lower frequency in the North Atlantic Ocean (indicator analysis, r.g.=0.64, P=0.001). Another HLII group, clade c8, was positively correlated with all regions except the South Atlantic Ocean (indicator analysis, r.g.=0.55, P=0.001). Most LL clades (c10, c12, c13) correlated significantly with the California Current (Supplementary Table S2), a data set with five samples collected from the subsurface chlorophyll a maximum (Supplementary Table S1; Dupont et al., 2015). All other clades did not differ in their relative abundance among regions.
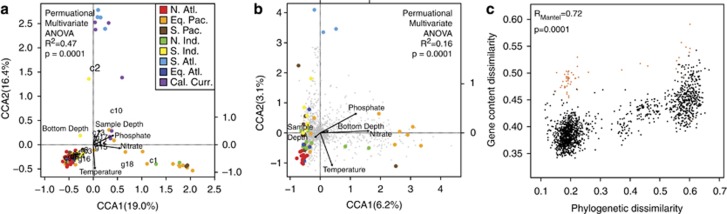
In a canonical correspondence analysis, nitrate and temperature accounted for most of the variation in phylogenetic diversity among regions. In contrast, phosphate, sample depth (proxy for light availability) and bottom depth (proxy for coastal influence) had smaller significant effects (Table 1). The majority of the Equatorial Pacific Ocean samples grouped with a few from the North Indian and South Pacific Ocean samples along the first canonical axis (CCA1). This group of samples predominantly corresponded to environments with higher nitrate (and presumably lower iron) and an elevated frequency of clade c1 (Figure 3a). The California Current and the South Atlantic Ocean samples negatively corresponded with temperature and positively with sample depth along the second canonical axis (CCA2) and clade c2 (the lower-temperature-adapted HLI clade; Johnson et al., 2006).
Table 1. Partial canonical correspondence analysis of five environmental variables.
| Variables |
Phylogenetic analysis |
Gene content analysis |
||
|---|---|---|---|---|
| Conditional effects | P-value | Conditional effects | P-value | |
| Nitrate | 16.7% | 0.0001 | 6.2% | 0.0001 |
| Temperature | 13.3% | 0.0001 | 2.7% | 0.0002 |
| Sample depth | 4.4% | 0.0038 | 2.4% | 0.0052 |
| Bottom depth | 3.3% | 0.0033 | 1.9% | 0.0697 |
| Phosphate | 3.3% | 0.0035 | 2.0% | 0.1092 |
| Total | 41.0% | 15.2% | ||
Figure 3.
Influence of environmental factors on the distribution of Prochlorococcus (a) phylogenetic and (b) gene content diversity. The gene content diversity does not contain any California Current samples owing to the increased sample size threshold. The canonical correspondence analysis triplot showing sample sites (colored circles) and vectors of environmental parameters: temperature (in situ), nitrate (World Ocean Atlas (WOA)), phosphate (WOA), depth and bottom depth. The numbers c1–c13 and g14–g18 refers to clades defined in Figure 2a and gray pluses refer to orthologous genes. (c) Pairwise comparisons between sample sites using phylogenetic and gene content dissimilarity. Correlation estimated based on Mantel test with a Pearson correlation. If GS068 is removed, Rmantel increases to 0.82 (P=0.001); comparisons involving GS068 are highlighted in orange and contribute to the bimodality of the plot.
We then determined the biogeography of the Prochlorococcus gene content using the relative frequency of 1663 recovered orthologous groups of non-core genes between sites having at least 1500 reads. Gene content displayed clear biogeographic patterns across regions (permutational multivariate ANOVA, R2=0.16, P=0.0001). Similar to phylogenetic diversity, gene frequencies were most significantly related to nitrate concentrations and temperature (CCA analysis: P=0.0001 and P=0.0002, respectively; Table 1). The additional variables (bottom depth, phosphate and sample depth) accounted for smaller proportions of variation. Nevertheless, the North Atlantic Ocean samples appeared to cluster together in a negative association with phosphate concentrations (Figure 3b), although phosphate was not a significant environmental factor in our forward selection analysis (CCA analysis: P=0.1092; Table 1). The phylogenetic and gene content diversity of Prochlorococcus populations was significantly correlated (RMantel=0.72, P=0.001; Figure 3c), and thus sample sites more similar in their phylogeny were more similar in their gene content. Of the 1663 flexible genes, 360 were significantly correlated with the phylogenetic diversity (RMantel>0, P<0.05) and 49 strongly correlated (RMantel>0.5, P<0.05). Another 584 genes were positively correlated but not significant, likely because of a lack of information due to rarefaction. The rest had an insignificant negative correlation (RMantel>−0.17, P⩾0.05).
To understand regional differences in Prochlorococcus functional potential, we grouped non-core genes into functional categories as defined by the COGs (Tatusov et al., 1997). The COG categories differentiated significantly with respect to region (permutational multivariate ANOVA, R2=0.17, P=0.0069). Populations from the North and South Atlantic Oceans were enriched for nucleotide transport and metabolism (COG group F, r.g.=0.564, 0.006) and the South Atlantic, North Indian and South Pacific Oceans for amino-acid transport and metabolism (COG group E; r.g.=0.048, P=0.035) (Supplementary Table S3).
Of the 202 non-core gene clusters significantly differentiating among ocean regions (indicator analysis, P<0.05), 81 (40%) had hypothetical functions (Supplementary Table S4). Irrespective of any knowledge about gene function, it is notable that significantly differentiated genes were more often found in genomic islands (34.2%) than all recruited flexible gene clusters (17.2%). It is known that genomic islands have a role in Prochlorococcus gene gain and loss (Coleman et al., 2006). Moreover, genes in the island regions of Prochlorococcus genomes are among the most dynamic in terms of abundance in the GOS data set (Rusch et al., 2007).
Next we wanted to define possible environmental drivers associated with non-core genes. Some of the most significantly differentiating genes (6 out of the 29, indicator analysis, P=0.001) were related to phosphate acquisition (Martiny et al., 2006; Scanlan et al., 2009). Of all significant known phosphate-acquisition-related genes, most were positively associated with the North Atlantic Ocean alone (ptrA-a transcriptional regulator related to stress response to phosphorus starvation) or the combined regions of the North Atlantic Ocean and the Equatorial Atlantic Ocean (chrA—a response regulator, phoR—a phosphate regulon sensor histidine kinase, phoA—an alkaline phosphatase, mfs—a major facilitator superfamily transporter, arsA—an arsenate reductase and a gene expressed in MED4 during phosphate starvation—PMM0720; Indval.g, P<0.05; Supplementary Table S4). In a biogeochemical context, these regions had lower average annual phosphate concentrations than other regions according to the World Ocean Atlas (Garcia et al., 2010).
Nitrogen assimilation genes significant in the indicator analysis were almost always associated with the Equatorial Pacific Ocean (Supplementary Table S4; Martiny et al., 2009b). Most were negatively associated specifically with the Equatorial Pacific Ocean and South Atlantic Ocean (napA—a nitrate/nitrite transporter and moaA—a molybdenum cofactor biosynthesis protein necessary for nitrate reductase) or with these two regions and a third (North Atlantic Ocean: narB—an assimilatory nitrate reductase; South Pacific Ocean: moeA—molydobpterin biosynthesis protein, a cofactor necessary for nitrate reductase; North Indian Ocean: an agmatinase; Supplementary Table S4). However, a few nitrogen-related genes were positively associated with the Equatorial Pacific Ocean (an alkyl hydroperoxide reductase subunit C-like protein, a glutamate N-acetyltransferase involved in the urea cycle) and the Equatorial Pacific and South Atlantic Oceans (a leucine dehydrogenase—which frees ammonium ions from branched-chain amino acids and an ammonium transporter; Supplementary Table S4).
A set of iron-requiring proteins were frequently under-represented in the Equatorial Pacific Ocean—perhaps as an adaptation to lower iron availability (Rusch et al., 2010). In our analysis, we recovered a small subset of these orthologous genes (5 out of the 17 non-core genes) (Rusch et al., 2010) as significantly differentiating between regions. Of these, all were negatively associated with only the Equatorial Pacific Ocean (an Fe–S oxidoreductase and a transglutaminase) or the Equatorial Pacific and other regions (a transglutaminase, a cytochrome oxidase C subunit and a metal-binding protein homologous to CopG—a protein involved in copper homeostasis).
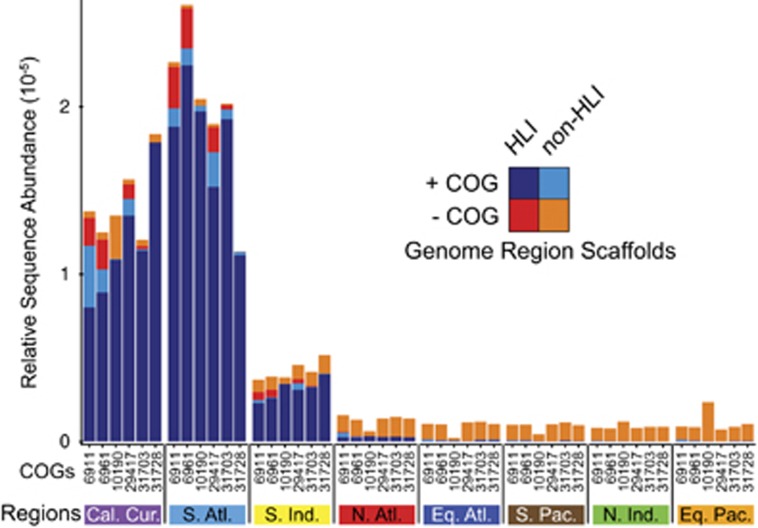
Nitrate and temperature were the largest drivers in Prochlorococcus community composition, and while the former has tangible physiological and genomic underpinnings, the role of temperature on Prochlorococcus genome content is largely unknown. Using a metagenomic assembly approach to further understand genes underpinning adaptation to lower temperature, we identified a set of orthologous groups negatively correlated to low temperature (Supplementary Table S5, see Materials and methods). A subset of the most negatively correlated (COGs 6961, 10190 and 6911) and most unique to the cold-adapted c2 (HLI) clade (COGs 29417, 31703 and 31728) were found on assembled scaffolds composed of more sequences from lower-temperature sites in the California Current and the South Atlantic Ocean than other warmer regions (Figure 4). Further, these sequences mostly mapped to the lower-temperature-adapted c2 (HLI) lineage using BLASTn (Camacho et al., 2009). In contrast, the syntenic assemblies lacking the lower-temperature genes generally recruited sequences evenly across all geographic regions (Figure 4, −COG). Of the four genes most negatively correlated with temperature, three were annotated: a possible trypsin 2OG-Fe(II) oxygenase, a dihydroorotase, and a hydrolase—a pseudouridine-5' phosphatase which dephosphorylates a potential intermediate in rRNA degradation. One of the COGs unique to the HLI clade, COG 29417, annotated as a possible glycoprotein. In several of these cases, the genomic neighborhood is fairly conserved across assemblies, while in other cases, the genomic context in the assemblies varies more relative to the cultured strains. (Supplementary Figure S5). Although the detailed functions are unclear, the geographical distribution of the individual genes and the structure of the genomic neighborhood suggested that specific genes contribute to the adaption of Prochlorococcus to growth at lower ocean temperatures.
Figure 4.
Distribution of sequences making up metagenomic assemblies with and without lower-temperature genes. The corresponding sequenced reads for each cold temperature gene were mapped to metagenomic assemblies comprised of pooled sequences from the entire data set (+COG). The two closest neighboring genes were then identified on each metagenomic assembly and used to find assemblies without the lower-temperature gene. Sequences on each of these sets of assemblies, with (+COG) and without (−COG) the cold temperature gene, were distributed across regions. Sequences were additionally subdivided based on sequence similarity to HLI genomes (+HLI) or not (−HLI).
Discussion
Questions regarding the relationship between phylogeny and functional gene diversity are critical to understanding the ecological and evolutionary mechanisms structuring microbial populations (Martiny et al., 2009c; Burke et al., 2011; Raes et al., 2011; Wiedenbeck and Cohan, 2011; Brown et al., 2014; Dupont et al., 2014). Increasing amounts of evidence suggest that using the whole genome rather than simply a single-gene classification is often needed to define microbial lineages and, more importantly, determine its role in the environment (Brown et al., 2014). Here we observe a positive correlation between phylogenetic and gene content diversity across Prochlorococcus populations. This correlation can be driven by at least three mechanisms that may not be exclusive: (i) genome content is phylogenetically structured, that is, the set of genes an individual has depends on its taxonomy, (ii) the same environmental selective forces shape the distribution of phylotypes and specific genes, or (iii) horizontal gene transfer events could be more prevalent between closely related lineages than observed. Considered together, the geographic patterns of the flexible genes follow those of core genes (Figure 3c), but there are many examples of flexible genes that are more variable and have distinct patterns. In support of a primarily vertically driven evolutionary history, the gene content of 12 Prochlorococcus strains is largely congruent with a molecular phylogeny based on the core genes, suggesting a strong role of phylogenetic descent (Kettler et al., 2007). In addition, core gene alleles share evolutionary history with distinct sets of flexible genes in specific field populations of Prochlorococcus (Kashtan et al., 2014). Populations with these core gene alleles can change in abundance depending on environmental conditions owing to differential fitness of the allele or the associated flexible genes. In our analysis, phylogeny and gene content is not perfectly correlated. This may be because of sampling error but can also be due to fine-scale diversity existing within the defined clades or due to particular sets of genes that vary independent of taxonomy (Kashtan et al., 2014; Pittera et al., 2014; see below). Divergent genes can be associated with variables that are not measured, such as trace metals, vitamins or ocean mixing. Traditionally, horizontal gene transfer events are identified by finding topologically incongruent gene trees; however, knowing the organismal origin is necessary, but adequate coverage with metagenomic reads can be difficult to obtain especially in highly complex communities (Guo et al., 2015). Network analyses provide an alternative method to gene phylogenies (Dagan et al., 2008) but still require enough genomic context to identify the taxonomic origin to adequately capture the evolutionary dynamics. Other analyses have been undertaken to identify metagenomic community recombination rates by utilizing metagenomic assemblies (Konstantinidis and DeLong, 2008; Johnson and Slatkin, 2009). Although we do not explicitly identify lateral transfer events, the correlation between gene content and phylogenetic signal is indicative of predominantly vertical inheritance with some level of gene gain and loss accounting for the incongruence between sample phylogeny and genome content diversity.
It is evident here that Prochlorococcus has a clear phylogeography, as demonstrated by changes in the lineage distribution of core genes across oceanic regions; consistent with previous observations (Bouman et al., 2006; Johnson et al., 2006; Zinser et al., 2007; Zwirglmaier et al., 2008; Martiny et al., 2009c). In addition, <10% of sequences associated with deeper HL nodes (g14), indicating that the current genomes capture a majority of the core gene variation in the surface ocean. Moreover, sequences associated with deeper branches, for example, g14, are distributed similarly to sequences associated with their corresponding tips of the tree (Supplementary Figure S2). This indicates a reasonable coverage of diversity within the most abundant Prochlorococcus clade.
Although nitrate and temperature explained the most phylogenetic variation, we did not expect to see the same environmental parameters explaining the genome content variability, as temperature preference separates ecotypes deeper in their evolutionary history at roughly 95% sequence similarity of internal transcribed spacer sequences (Martiny et al., 2009c), for example, HLI is adapted to cooler temperatures than HLII (Johnson et al., 2006). Nevertheless, temperature remains a strong factor influencing the distribution of the Prochlorococcus flexible gene content. The importance of temperature in this data set for both the phylogenetic and gene content analyses of Prochlorococcus echoes that of the SAR11 group (Brown et al., 2012), where temperature has a significant effect both on the abundance of phylotypes and the distribution of temperature-related genes. Several Prochlorococcus genes significantly associated with colder regions in our read-based analysis are also disproportionately found on assemblies made up of sequences predominantly from colder climates. It is puzzling that some of these temperature-related genes are found in several genomes rather than only the cold-adapted HLI strains, although the main signal from the distribution of sequences on assemblies with the gene clearly come from sequences mapping to HLI genomes. This suggests multiple levels of adaptation to differences in temperature, including ecotype differentiation as well as genome variability within each ecotype. Additionally, the greater variability in the genomic context of environmental assemblies versus sequenced strains suggests that the assemblies are capturing new genomic backbones to some extent. Thus strains in culture may not fully represent Prochlorococcus diversity as it relates to temperature adaptation—as was recently observed for nitrogen assimilation (Martiny et al., 2009b; Berube et al., 2015). In Synechococcus, variation has been observed in the thermal niches of strains consistent with the thermal range of their isolation location. Rather than following a strict phylogenetic split between high and low temperature, thermal niche variation occurs within clades across its phylogeny (Pittera et al., 2014).
Phosphate-acquisition genes follow past descriptions based on fewer samples in marine bacteria other than Prochlorococcus, whereby the relative abundance is negatively correlated to nutrient concentrations (Martiny et al., 2011). Field studies of Prochlorococcus are also suggestive of a similar pattern in phosphorus (Martiny et al., 2009a; Thompson et al., 2013). Here we observed a positive association of P-uptake genes with regions low in phosphate. That the COG groups (Tatusov et al., 1997) significantly differentiate on categories related to transport and metabolism indicates that broad regional differences among Prochlorococcus are likely related to nutrient supply stoichiometry (Wu et al., 2000).
Our analysis also identified novel patterns at the global level, suggesting undiscovered links between environmental conditions and the genetic diversity of Prochlorococcus. For example, many genes differentially distributed along the nitrogen gradient. This includes previously identified genes responsible for nitrate uptake present in lower nitrogen waters (Martiny et al., 2009b) as well as a higher frequency of genes related to reduced nitrogen uptake in Equatorial Pacific and South Atlantic Oceans. In an environmental context, average annual nitrate concentrations of these two regions were higher than other regions (Garcia et al., 2010), and our data suggest that there appears to be a shift in nitrogen assimilation between oxidized and reduced forms of nitrogen, in contrast to phosphate, which simply drives the acquisition or loss of specialized transport. Specifically, significant nitrate/nitrite genes were positively associated with low nitrate regions, whereas significant ammonium/ammonia genes positively associated with regions high in nitrate. We hypothesize that having nitrate uptake genes in regions with nitrogen stress enables those organisms to more efficiently access energetically costly nitrogen compounds. In this system of low nitrogen, organisms are competing heavily for ammonia and urea; the capability to utilize nitrate when it is sporadically available makes them competitive against others. In high nitrogen regions, a decent flow of reduced nitrogen from a variety of sources is available (grazers, leakage of urea from diatoms, degradation), but eukaryotes will be competing heavily for nitrate. Ammonium-related genes here allow individuals to solely rely on energetically cheap reduced nitrogen sources.
A large proportion of the genes differing between regions are still annotated as hypothetical. These genes define Prochlorococcus' biogeography, but we lack the ability to interpret their physiological and ecological role. Of particular interest is developing our understanding of the South Atlantic Ocean, a region that has been limited in its exploration. Eighty-one non-core genes associate with this region alone, and most of these are conserved hypothetical protein (66.6%), with only seven genes found uniquely in a single Prochlorococcus genome.
It is clear that Prochlorococcus harbors extensive genome diversity across the global surface ocean, and our results demonstrate that this diversity is not randomly distributed. Instead, Prochlorococcus genome diversity displays clear regional biogeographic patterns with a strong relationship between its taxonomy and gene content. This diversity, including adaptations to local environmental conditions implied by our results, will likely influence future ocean and atmospheric conditions through changes in phytoplankton community structures (Flombaum et al., 2013). In particular, understanding the relative strengths of different evolutionary mechanisms will further inform how these dynamic processes will occur. Considering the high abundance of Prochlorococcus, such interactions can have a large impact on future ocean ecosystems and global biogeochemical cycles.
Acknowledgments
We thank J Martiny and S Hatosy for many helpful comments. AGK was supported by the National Science Foundation-Graduate Research Fellowship Program (DGE-1321846) and the National Institute of Biomedical Imaging and Bioengineering, National Research Service Award EB009418 from the University of California, Irvine, Center for Complex Biological Systems. The Global Ocean Sampling project was supported by US Department of Energy, Office of Science, Office of Biological and Environmental Research (DE-FC02-02ER63453), the Gordon and Betty Moore Foundation, the Beyster Family fund of the San Diego Foundation and the Life Technologies Foundation. ACM was supported by the National Science Foundation (OCE-0928544 and OCE-1046297).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abascal F, Zardoya R, Telford MJ. (2010). TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38: W7–W13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube PM, Biller SJ, Kent AG, Berta-Thompson JW, Roggensack SE, Roache-Johnson KH et al. (2015). Physiology and evolution of nitrate acquisition in Prochlorococcus. ISME J 9: 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Berube PM, Berta-Thompson JW, Kelly L, Roggensack SE, Awad L et al. (2014). Genomes of diverse isolates of the marine cyanobacterium Prochlorococcus. Sci Data 1: 140034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouman HA, Ulloa O, Scanlan DJ, Zwirglmaier K, Li WKW, Platt T et al. (2006). Oceanographic basis of the global surface distribution of Prochlorococcus ecotypes. Science 312: 918–921. [DOI] [PubMed] [Google Scholar]
- ter Braak CJF. (1986). Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67: 1167–1179. [Google Scholar]
- Brown MV, Lauro FM, DeMaere MZ, Muir L, Wilkins D, Thomas T et al. (2012). Global biogeography of SAR11 marine bacteria. Mol Syst Biol 8: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MV., Ostrowski M, Grzymski JJ, Lauro FM. (2014). A trait based perspective on the biogeography of common and abundant marine bacterioplankton clades. Mar Genomics 15: 17–28. [DOI] [PubMed] [Google Scholar]
- Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. (2011). Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA 108: 14288–14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, Chisholm SW. (2007). Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol 15: 398–407. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF et al. (2006). Genomic islands and the ecology and evolution of Prochlorococcus. Science 311: 1768–1770. [DOI] [PubMed] [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W. (2008). Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci USA 105: 10039–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cáceres M, Legendre P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90: 3566–3574. [DOI] [PubMed] [Google Scholar]
- Dupont CL, Larsson J, Yooseph S, Ininbergs K, Goll J, Asplund-Samuelsson J et al. (2014). Functional tradeoffs underpin salinity-driven divergence in microbial community composition. PLoS One 9: e89549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont CL, McCrow JP, Valas R, Moustafa A, Walworth N, Goodenough U et al. (2015). Genomes and gene expression across light and productivity gradients in eastern subtropical Pacific microbial communities. ISME J 9: 1076–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (2011). Accelerated profile HMM searches. PLoS Comput Biol 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (2005). PHYLIP (Phylogeny Inference Package) version 3.69.
- Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N et al. (2013). Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA 110: 9824–9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia HE, Locarnini RA, Boyer TP, Antonov JI, Zweng MM, Baranova OK et al. (2010). World Ocean Atlas 2009, Volume 4: Nutrients (Phosphate, Nitrate, and Silicate). In: Levitus S (ed) NOAA Atlas NESDIS 71. US Government Printing Office: Washington, DC, USA, pp 398. [Google Scholar]
- Guo J, Wang Q, Wang X, Wang F, Yao J, Zhu H. (2015). Horizontal gene transfer in an acid mine drainage microbial community. BMC Genomics 16: 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson I, Paerl RW, Tripp HJ, Zehr JP, Karl DM. (2009). Metagenomic potential of microbial assemblages in the surface waters of the central Pacific Ocean tracks variability in oceanic habitat. Limnol Oceanogr 54: 1981–1994. [Google Scholar]
- Johnson PL, Slatkin M. (2009). Inference of microbial recombination rates from metagenomic data. PLoS Genet 5: e1000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311: 1737–1740. [DOI] [PubMed] [Google Scholar]
- Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A et al. (2014). Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344: 416–420. [DOI] [PubMed] [Google Scholar]
- Kelly L, Huang KH, Ding H, Chisholm SW. (2012). ProPortal: a resource for integrated systems biology of Prochlorococcus and its phage. Nucleic Acids Res 40: D632–D640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S et al. (2007). Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3: 2515–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, DeLong EF. (2008). Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J 2: 1052–1065. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Coleman ML, Chisholm SW. (2006). Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc Natl Acad Sci USA 103: 12552–12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Huang Y, Li W. (2011). Adaptation to nutrient availability in marine microorganisms by gene gain and loss. In: de Bruijn FJ (ed) Handbook of Molecular Microbial Ecology II: Metagenomics in Different Habitats vol. II John Wiley & Sons, Inc.: Hoboken, NJ, USA, pp 269–276. [Google Scholar]
- Martiny AC, Huang Y, Li W. (2009. a). Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol 11: 1340–1347. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Kathuria S, Berube PM. (2009. b). Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. Proc Natl Acad Sci USA 106: 10787–10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Tai APK, Veneziano D, Primeau F, Chisholm SW. (2009. a). Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ Microbiol 11: 823–832. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Treseder K, Pusch G. (2013). Phylogenetic conservatism of functional traits in microorganisms. ISME J 7: 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsen FA, Kodner RB, Armbrust EV. (2010). pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11: 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Delcher AL, Koren S, Venter E, Walenz BP, Brownley A et al. (2008). Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24: 2818–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L, Rocap G, Chisholm S. (1998). Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 576: 220–223. [DOI] [PubMed] [Google Scholar]
- Noguchi H, Park J, Takagi T. (2006). MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 34: 5623–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet F, Kindt R, Legendre P, Minchin P, O'Hara R et al. (2013). vegan: Community Ecology Package. R package version 2.0-10. R Packag version 1 (http://cran.r-project.org).
- Partensky F, Hess WR, Vaulot D. (1999). Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev 63: 106–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittera J, Humily F, Thorel M, Grulois D, Garczarek L, Six C. (2014). Connecting thermal physiology and latitudinal niche partitioning in marine Synechococcus. ISME J 8: 1221–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J et al. (2014). EggNOG v4.0: Nested orthology inference across 3686 organisms. Nucleic Acids Res 42: 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. (http://www.R-project.org/).
- Raes J, Letunic I, Yamada T, Jensen LJ, Bork P. (2011). Toward molecular trait-based ecology through integration of biogeochemical, geographical and metagenomic data. Mol Syst Biol 7: 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: 398–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. (2010). Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc Natl Acad Sci USA 107: 16184–16189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR et al. (2009). Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, González JM et al. (2013). Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA 110: 11463–11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R, Koonin EV, Lipman DJ. (1997). A genomic perspective on protein families. Science 278: 631–637. [DOI] [PubMed] [Google Scholar]
- Thompson LR, Field C, Romanuk T, Kamanda Ngugi D, Siam R, El Dorry H et al. (2013). Patterns of ecological specialization among microbial populations in the Red Sea and diverse oligotrophic marine environments. Ecol Evol 3: 1780–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West NJ, Scanlan DJ. (1999). Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl Environ Microbiol 65: 2585–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenbeck J, Cohan FM. (2011). Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev 35: 957–976. [DOI] [PubMed] [Google Scholar]
- Wu J, Sunda W, Boyle EA, Karl DM. (2000). Phosphate depletion in the western North Atlantic Ocean. Science 289: 759–762. [DOI] [PubMed] [Google Scholar]
- Zinser ER, Johnson ZI, Coe A, Karaca E, Veneziano D, Chisholm SW. (2007). Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol Oceanogr 52: 2205–2220. [Google Scholar]
- Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D et al. (2008). Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol 10: 147–161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.