Abstract
Although little is known regarding microbial life within our planet's rock-hosted deep subseafloor biosphere, boreholes drilled through deep ocean sediment and into the underlying basaltic crust provide invaluable windows of access that have been used previously to document the presence of microorganisms within fluids percolating through the deep ocean crust. In this study, the analysis of 1.7 million small subunit ribosomal RNA genes amplified and sequenced from marine sediment, bottom seawater and basalt-hosted deep subseafloor fluids that span multiple years and locations on the Juan de Fuca Ridge flank was used to quantitatively delineate a subseafloor microbiome comprised of distinct bacteria and archaea. Hot, anoxic crustal fluids tapped by newly installed seafloor sampling observatories at boreholes U1362A and U1362B contained abundant bacterial lineages of phylogenetically unique Nitrospirae, Aminicenantes, Calescamantes and Chloroflexi. Although less abundant, the domain Archaea was dominated by unique, uncultivated lineages of marine benthic group E, the Terrestrial Hot Spring Crenarchaeotic Group, the Bathyarchaeota and relatives of cultivated, sulfate-reducing Archaeoglobi. Consistent with recent geochemical measurements and bioenergetic predictions, the potential importance of methane cycling and sulfate reduction were imprinted within the basalt-hosted deep subseafloor crustal fluid microbial community. This unique window of access to the deep ocean subsurface basement reveals a microbial landscape that exhibits previously undetected spatial heterogeneity.
Introduction
The volume of oceanic lithosphere habitable by microbial life is thought to be a substantial portion of the Earth's crust—one that potentially extends thousands of meters below the seafloor (Edwards et al., 2012; Biddle et al., 2014). Although the actual limit for life is likely quite deep, extrusive basalts buried within aging igneous oceanic crust are thought to be the most conducive subsurface environments for microbial life owing to their high porosity and extensive hydrothermal circulation (Baross et al., 2004). Here the uppermost 500 m of subseafloor rock-hosted aquifer is estimated to contain a volume of fluid equivalent to ~2% of the current world ocean and circulates a volume equivalent to the entire ocean every ~100 000 years (Johnson and Pruis, 2003).
Mid-ocean ridge environments are the locus of extensive hydrothermal circulation deep into the oceanic crust that ultimately fuels a variety of novel subseafloor microbial habitats (Summit and Baross, 2001; Edwards et al., 2005). Although the mid-ocean ridges themselves make up only a small portion of the seafloor, hydrothermally heated fluid also circulates through the sediment-covered deep ocean crust along the ridge flanks and basins. Fluid transport that is generally driven by differences in pressure between recharging and discharging fluids at exposed rocky outcrops protruding through sedimented regions can occur laterally over long distances in ridge flank and basin systems (>50 km; Fisher et al., 2003) and at timescales ranging from decades to several thousands of years (Elderfield et al., 1999; Fisher et al., 2003; Fisher and Wheat, 2010). In contrast to fluid circulation within mid-ocean ridges, the hydrothermal circulation through the sediment-covered deep ocean crust within ridge flanks and basins occurs through a more vast interconnected network of fluid flow paths that can be highly complicated in directional nature (Wilcock and Fisher, 2004). This flow can be isolated from deep seawater for extended time periods owing to thick, impermeable layers of overlying sediment.
Although many studies have investigated the sedimentary microbiology of the global ocean (for example, Parkes et al., 1994; Kallmeyer et al., 2012; D'Hondt et al., 2015), few have investigated aspects of microbial life in the igneous subsurface crust (Cowen et al., 2003; Huber et al., 2006; Orcutt et al., 2011; Smith et al., 2011; Lever et al., 2013; Jungbluth et al., 2013a, 2014). This is not altogether surprising given the difficulties in adequately sampling this unique environment: the characterization of life inhabiting the basaltic deep subseafloor biosphere has consistently faced formidable challenges in the form of access and obtaining samples of high integrity that lack chemical and/or microbial contamination (Cowen, 2004; Biddle et al., 2014). Until recently, a lack of access necessitated the use of discharging hydrothermal fluids and exposed basalt as indirect ‘windows' into the deep subsurface (for example, Deming and Baross, 1993; Butterfield et al., 1997). However, progressive improvements in the design and performance of subseafloor borehole ‘observatories', and the instrumentation used to sample them, have increasingly made fluid sampling from the basalt-hosted deep subseafloor via boreholes a more consistent, efficient and pristine process (Wheat et al., 2011; Cowen et al., 2012).
Along the eastern flank of the Juan de Fuca Ridge (JdFR), an array of boreholes intermittently drilled through the sediment layer and into subseafloor basalt currently encompasses several different types of subseafloor observatories, also known as ‘CORKs' (Circulation Obviation Retrofit Kits) (Becker and Davis, 2005; Wheat et al., 2011). These include early generation instrument and sampling platforms at boreholes 1024C, 1025C and 1027C that deliver warm crustal fluids directly through a potentially reactive steel casing that is open at depth in the formation, allowing fluids to be accessed through a spigot on the CORK at the seafloor, and second-generation CORK observatories at boreholes 1026B and U1301A that include stainless steel fluid delivery lines running exterior to the CORK casing, where they can then be accessed through a spigot at the seafloor wellhead (Fisher et al., 2005). Borehole 1026B did initially have an early generation CORK in place that was subsequently replaced with a second-generation version in 2004 (Fisher et al., 2005). In 2010, a new generation of subseafloor borehole observatories were drilled and installed at boreholes U1362A and U1362B that substitute potentially corrosive components with alternative construction materials such as fiberglass, titanium and Teflon and incorporate other design features such as wide-diameter Tefzel fluid delivery lines for microbiological sampling that aim to reduce the impact of borehole observatories on in-formation biological processes and facilitate the retrieval of pristine crustal fluid samples (Wheat et al., 2011; Cowen et al., 2012). In addition to variation in CORK construction, the CORK observatories along the JdFR flank also reside in spatially and topographically distinct locations that access crustal fluids at different states of alteration (for example, from oxic to anoxic), at different temperatures (~39 °C at borehole 1025C and ~65 °C at boreholes 1026B, U1301A and U1362A and U1362B), and tap different depth horizons within the basaltic crust.
Although CORK-mediated deep subseafloor microbiology extends back nearly 15 years (Cowen et al., 2003), the number of independent samples that have been examined remains sparse. Two of the earliest studies took advantage of the first-generation CORK initially installed at overpressured borehole 1026B to collect single fluid samples emanating from the spigot of the wellhead (Cowen et al., 2003; Huber et al., 2006). More recently, the microbiology of basalt-hosted crustal fluids has been investigated via single samples obtained from the first-generation CORK at borehole 1025C (Jungbluth et al., 2014), the stainless steel fluid delivery line of the second-generation CORK subsequently installed at borehole 1026B (Jungbluth et al., 2014), and single samples collected annually from 2008 to 2010 via the stainless steel fluid delivery line of the CORK at borehole U1301A (Jungbluth et al., 2013a). The phylogenetic structure of microbial communities within these seven samples was characterized by relatively low throughput small subunit ribosomal RNA (SSU rRNA) gene cloning and sequencing which, when coupled with few samples overall, prohibited a rigorous evaluation or statistical analysis of community structure between samples, boreholes or potential source or seed communities inhabiting nearby seawater and sediments.
In 2011, a new iteration of instrumentation for clean sampling was used to collect fresh fluids from the CORK and stainless steel fluid delivery line at borehole U1301A and the first set of fluid samples intended for microbiological analysis from the Tefzel fluid delivery lines of CORKs at boreholes U1362A and U1362B along the JdFR flank (Cowen et al., 2012; Lin et al., 2012). The goals of this study were to use high throughput SSU rRNA gene sequencing and epifluorescence microscopy to compare the taxonomic diversity and cellular abundance of microorganisms within this new set of samples to those previously obtained from the first- and second-generation CORKs in this area and to identify common and abundant microbial taxa residing in anoxic, deep subseafloor basement fluids. In total, a suite of 64 basalt-hosted deep subsurface crustal fluid, bottom seawater and sediment samples was compared in an effort to quantify relationships between the microbial communities inhabiting closely arrayed but disparate points of access to the basalt-hosted aquifer below the JdFR flank, as well as between basalt-hosted deep subsurface communities and those occupying their most closely adjoining habitats—sediments and bottom seawater (Table 1). Crustal fluids collected simultaneously from three boreholes drilled within a ~1-km radius allowed us to compare microbial communities from independent, yet closely arranged, sampling points in the deep ocean crust in what is currently the only location where this comparison is feasible.
Table 1. Environmental DNA samples analyzed in this study.
| Borehole | Crustal age (Ma) | Depth | Fluid line | Sample |
Number of samples by year |
Total | |||
|---|---|---|---|---|---|---|---|---|---|
| 2008 | 2009a | 2010 | 2011 | ||||||
| 1025C | 1.2 | 0–46 msb | None | Basalt-hosted | — | — | 1 | — | 1 |
| U1301A | 3.5 | 8–107 msb | Stainless | Basalt-hosted | 4 | 4 | 9 | 3 | 20 |
| U1362A | 3.5 | 193–292 msb | Tefzel | Basalt-hosted | — | — | — | 3 | 3 |
| U1362B | 3.5 | 29–117 msb | Tefzel | Basalt-hosted | — | — | — | 4 | 4 |
| U1363 suite | NA | ~1–225 mbsf | NA | Sediment | — | — | 11 | — | 11 |
| NA | NA | ~2500–2650 mbsl | NA | Seawater | 2 | 3 | 10 | 5 | 20 |
Abbreviations: Ma, million years old; mbsf: meters below seafloor; mbsl: meters below sea-level; msb, meters sub-basement; NA, not applicable.
Samples from CORKs 1026B and U1301B were excluded because no high-quality samples were collected or analyzed.
Materials and methods
Additional details regarding the Materials and Methods are provided as Supplementary Information.
Sample collection and preparation
During R/V Atlantis cruises AT15-35 (28 July–13 August 2008), AT15-51 (20 August–6 September 2009), AT15-66 (13 June–1 July 2010) and AT18-07 (28 June–14 July 2011), fluid samples were collected from CORK observatories located east of the JdFR (Table 1, Supplementary Table S1 and Supplementary Figures S1A–C) via HOV Alvin and ROV Jason-II submersibles equipped with borehole-attachment devices and sampling equipment to conduct fluid sampling via dedicated fluid delivery lines of the CORK observatories (Cowen et al., 2012). One borehole observatory (1025C) is located in cooler (~39 °C) 1.2 million year-old ocean crust while all other observatories sampled for this study (1026B, U1301A, U1301B, U1362A, U1362B) are located within ~1 km radius in warmer (~65 °C) 3.5 million year-old ocean crust (Table 1 and Supplementary Figure S1B). The borehole observatories penetrate up to 260 m of sediments and up to another 292 m of basement rock (Supplementary Figure S1C). Fluids were sampled from the CORK observatory at borehole U1301A (47°45.209′N, 127°45.833′W) over four consecutive years (2008–2011) from the stainless steel microbiological sampling line (Fisher et al., 2005; Wheat et al., 2011); 2008–2010 samples have been described previously (Jungbluth et al., 2013a). The stainless steel line at U1301A taps an interval between a packer set at 8 m sub-basement (msb; that is, below the sediment–basement interface) and the top of the fill in the bottom of the borehole at 107 msb (Supplementary Table S1). Fluids from the CORK observatories at boreholes 1025C (47°53.247′N, 128°38.919′W) and 1026B (47°45.7571′N, 127°45.5482′W) were sampled in 2010 as previously described (Jungbluth et al., 2014). The CORK at borehole 1025C does not have a fluid delivery line, but the hole is cased to a depth of 5 msb. Thus samples derived from 1025C originate from an interval spanning 5–34 msb (top of the fill in the bottom of the hole) (Supplementary Table S1). The stainless steel line at 1026B samples an interval between 9 msb (end of hole casing) and 32 msb (top of the fill in the bottom of the hole) (Supplementary Table S1). Fluids from the CORK observatories at boreholes U1301B (47°45.2286′N, 127°45.8262′W) and 1026B were sampled in 2009 but ultimately yielded only samples contaminated with seawater. During the course of this study, two new boreholes, U1362A (47°45.6628′N, 127°45.6720′W) and U1362B (47°45.4997′N, 127°45.7312′W), were drilled and fitted with lateral CORKs (L-CORKs) equipped with both stainless steel and Tefzel-lined fluid delivery lines (Expedition 327 Scientists, 2011). Following 1 year of equilibration within the new boreholes, U1362A and U1362B were sampled in July 2011 from Tefzel fluid delivery lines that tap basement intervals spanning 193–280 msb (U1362A) and 30–110 msb (U1362B). These intervals are bounded by inflatable packers above and top of the fill in the bottom of the respective boreholes at depth (Supplementary Figure S1C; Supplementary Table S1). The fluid flow path of our sampling instrumentation usually contained a custom flowcell with an in situ oxygen optode (Aanderaa Data Systems, Bergen, Norway), temperature sensor (Sea-Bird Electronics Inc., Bellingham, WA, USA) and a custom fluid flow sensor to allow for real-time assessment of the integrity of fluids and the plumbing flow path (for example, Cowen et al., 2012). During most sampling events, temperature and oxygen were monitored in situ in order to rapidly assess the likely quality of samples; elevated fluid temperature and draw down of oxygen relative to bottom seawater concentrations were indicative of high-integrity crustal fluids. The majority of fluid and sediment pore water samples investigated for microbiology here were also subsampled and characterized by an extensive suite of biogeochemical measurements (Supplementary Table S2; Lin et al., 2012, 2014).
In some instances, the ROV Jason-II was also equipped with a 5-liter Niskin bottle (General Oceanics, Miami, FL, USA) to collect seawater. Hydrocasts were used to collect additional seawater and were conducted using a conductivity–temperature–depth rosette water sampler equipped with 24 10-liter sampling bottles (General Oceanics). Surficial and deep sediments were collected from the periphery of an exposed rocky seamount during IODP Expedition 327 (Jungbluth et al., 2013b).
SSU rRNA gene sequencing and analysis
A barcoding approach using primers targeting the V4 region was used during the amplification of SSU rRNA genes for sequencing on an Illumina MiSeq DNA sequencer (Illumina, San Diego, CA, USA) (Caporaso et al., 2011). A total of 1 734 012 reads (150-bp average length) were processed using a combination of programs and scripts, including QIIME (Caporaso et al., 2010) and mothur (Schloss et al., 2009) (Supplementary Figure S2). SSU rRNA gene cloning and sequencing was performed on a small subset of samples. The isolation of single cells by flow cytometry, amplification of their genomes, and identification via SSU rRNA gene sequencing was performed from a sample collected from borehole U1362A (SSF19 in Supplementary Table S1) following previously described methods (Swan et al., 2011).
Community analyses
Sequence clustering was performed using several methods, including the use of unpaired and paired sequence reads, in order to evaluate and select an appropriate method for subsequent analyses (Supplementary Methods; Supplementary Tables S3 and S4; Supplementary Figure S2). The lowest sequence diversity (for example, Chao1, observed species, Shannon diversity) was found using distribution-based clustering with the default cutoff value (d=0.1); other parameter values selected for this clustering method had diversity levels comparable to those found using traditional clustering methods that do not incorporate information about read distribution across samples (for example, UPARSE, UCLUST, mothur; Supplementary Figure S3). As expected, analyses performed with unique reads resulted in the highest diversity with respect to Chao1, observed species and Shannon indices (Supplementary Figure S3). Procrustes shape analysis was performed using the Bray–Curtis dissimilarity index and the abundance weighted Jaccard distance to assess differences in read-pairing and clustering techniques; no major observable differences in the clustering patterns were detected when comparing forward and reverse reads, forward reads versus paired reads or forward reads clustered using the different techniques (Supplementary Figure S4). Thus, subsequent community analyses were ultimately performed with the unique read analysis method and either unpaired forward reads or paired-end reads generated using the merge-illumina-pairs script (Eren et al., 2013) owing to our interest in potential ecological differences between very closely related sequences. The longer length paired reads were used for analyses involving taxonomically assigned sequences, including principal coordinate analysis (PCoA), phylogenetic and unique read (heatmap) analysis and indicator species analysis. Unpaired forward reads were used for all other analyses owing to the higher number of sequences per library, including unweighted paired group mean average (UPGMA) of Bray–Curtis distances, distance-based Redundancy Analysis (db-RDA), jackknifed β-diversity, mantel tests, adonis, analysis of similarity (ANOSIM), multi-response permutation procedures (MRPP), permutational analysis of variance (PERMANOVA), heterogeneity of dispersion (PERMDISP), venn diagram and α-diversity. Alpha-diversity analyses (Chao1, observed species, Good's coverage, Shannon, Simpson evenness) were performed in QIIME using 100 randomly generated sets of rarefied reads sampled to an even depth (n=6108) and for each clustering method described above. PCoA and Procrustes analyses were performed in QIIME following removal of singleton operational taxonomic units (OTUs; OTUs with one member) and rarefying samples to the depth of the lowest sample in each comparison. Jackknifed beta diversity analysis was performed in QIIME using 100 sequences randomly selected from each sample, and the results were visualized using EMPeror (Vázquez-Baeza et al., 2013). UPGMA analysis was generated in QIIME using read clusters rarefied to an even sampling depth (n=6108) across all samples. Venn diagrams were generated in mothur (Schloss et al., 2009) and plotted in R (R Core Team, 2014) using venneuler (Wilkinson, 2011). Read clusters taxonomically assigned as described in Supplementary Material were imported into R for generation of OTU heatmaps and Dufrêne-Legendre indicator species analysis. Heatmaps and indicator taxa plots were generated using R packages phyloseq (McMurdie and Holmes, 2013), vegan (Oksanen et al., 2013) and labdsv (Roberts, 2013). Following identification of reads to be included in the heatmap and indicator taxa plots, the taxonomic identity of those reads was manually curated by adding filtered sequences into the SILVA SSU Ref 99 version 115 database and base tree using the parsimony insertion tools within ARB (Ludwig et al., 2004).
Results
Borehole fluid sampling and integrity
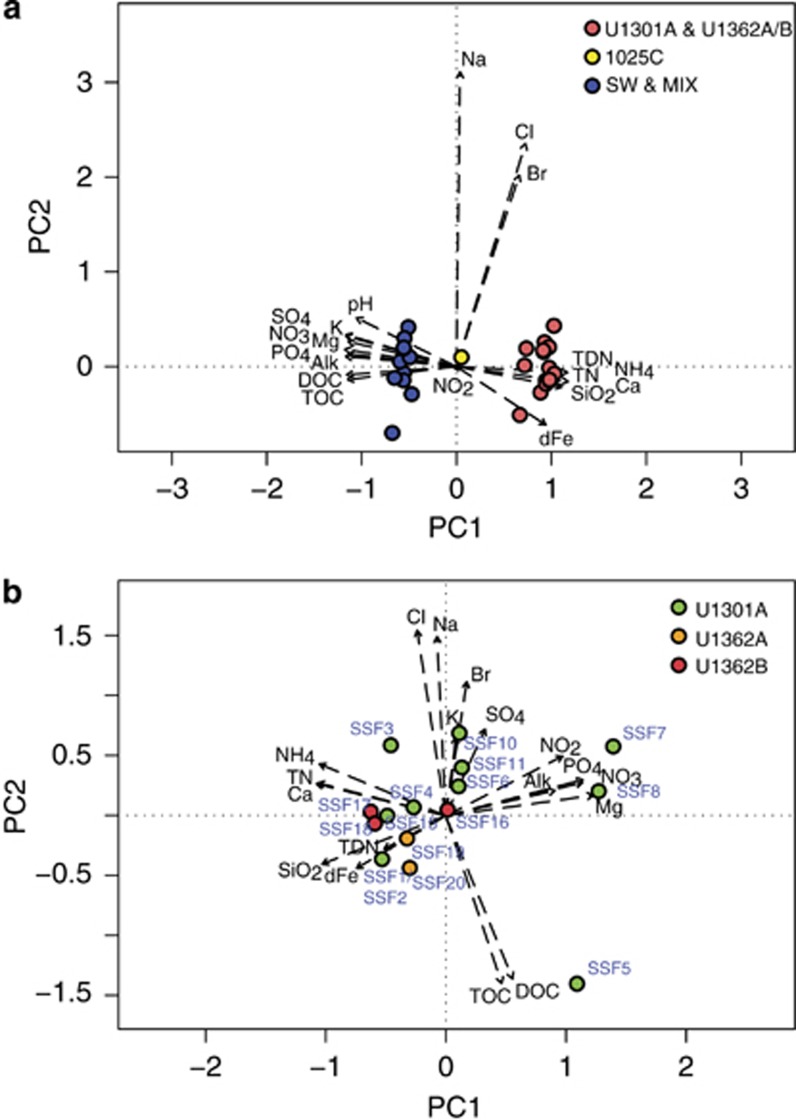
Subseafloor crustal fluids were sampled from CORKs installed within five different boreholes along the JdFR flank (Supplementary Figures S1A and B) during the summers of 2008–2011 using pumping equipment and samplers capable of collecting whole water and filtered particulates in situ (Supplementary Table S1). The chemical characteristics of the majority of borehole fluid samples are typical of highly altered crustal fluids, including near-neutral pH, elevated calcium, silicate, and ammonium and depleted in dissolved organic carbon, total organic carbon, carbon dioxide, magnesium, potassium, nitrate, sulfate, and phosphate, relative to surrounding bottom seawater (Figures 1a and b; Supplementary Table S2; Lin et al., 2012, 2014). Based on nitrate concentrations, seawater contamination within the ‘high-integrity' group of deep subseafloor samples from boreholes U1301A, U1362A and U1362B ranged from undetectable (0%) up to 8% seawater. The geochemical characteristics of fluids retrieved from Hole 1025C are intermediate between those of older, highly altered fluids and deep seawater, which is consistent with the younger age of the ocean crust at this site (~1.2 million years; Figure 1a). Within the subset of fluid samples from ~3.5 million year old (Ma) ocean crust possessing chemical characteristics that were consistent with highly altered crustal fluids, some minor chemical variability was observed (Figure 1b), indicating either variability in crustal fluids in situ or minor seawater intrusion into otherwise high-integrity crustal fluid samples. A subset of fluid samples retrieved from the boreholes was ultimately determined to be highly contaminated with ⩾90% of seawater (low-quality subsurface fluids labeled as ‘MIX' in Figure 1a and Supplementary Table S1). Although no chemical data are available for the handful of samples filtered in situ (SSF12, SSF21–SSF24) or samples SSF13 and SSF14 owing to the small volume of recovered fluids, depleted oxygen and elevated temperature measured in these fluids at the seafloor during sampling provided a strong indication that they were high-quality crustal fluid samples.
Figure 1.
Pristine deep subseafloor basement fluids are readily distinguishable from seawater and mixed samples based on geochemistry. Principal components analysis biplot of (a) all samples and (b) borehole U1301A, U1362A, and U1362B high-quality deep subsurface fluid samples based on eigenvalues derived from their corresponding chemical data. Dashed arrows indicate the direction of increase for measured chemical parameters while arrow length indicates the degree of correlation with the ordination axis. Only the first two principal component axes are shown. Details of individual samples are described in Supplementary Tables S1 and S2. Alk, alkalinity; DOC, dissolved organic carbon; TOC, total organic carbon; TDN, total dissolved nitrogen; TN, total nitrogen; dFe, dissolved iron.
Microbial cell abundance
The microscopic examination of borehole fluids with high chemical integrity revealed microbial cell abundances ranging from 2.6 × 103 to 5.1 × 104 cells ml−1, which is up to ~30-fold lower than is characteristic of nearby bottom seawater (7.7 × 104 cells ml−1 average; Supplementary Table S1). In general, microbial cell abundances in samples with no chemical evidence for seawater contamination were ⩽1.6 × 104 cells ml−1. Overall, microbial cell abundances were strongly correlated with magnesium concentrations (Supplementary Figure S5). Magnesium is depleted in pristine crustal fluids relative to bottom seawater in this system and may be used as a proxy for seawater intrusion (for example, Mottl and Wheat, 1994; Lin et al., 2012). High-integrity fluid samples collected from recently installed CORKs at boreholes U1362A and U1362B containing Tefzel-lined fluid delivery lines revealed cellular abundances that ranged from 2.6 × 103 cells ml−1 in a sample with relatively low total alkalinity to 2.6 × 104 cells ml−1 in a sample elevated in dissolved iron. At borehole U1301A, four crustal fluid samples that yielded relatively high cellular abundance values contained sodium and chloride concentrations on the extreme low (SSF5) and high (SSF10) ends of the range, sample SSF5 contained an anomalously high concentration of dissolved organic carbon, and two samples were highly depleted in silicate (SSF7 and SSF8) (Supplementary Table S2). These samples have no clear chemical indication of seawater contamination, however, suggesting that variability in microbial biomass is potentially associated with natural variability in basement fluid biogeochemistry (Figure 1b).
Microbial community structure
A total of ~1.7 million SSU rRNA genes were sequenced in the forward and reverse directions, resulting in 150 base pair (bp) long reads and an expected overlap of ~24 nucleotides. The average unpaired forward reads per sample was 25 029, and around 54% were successfully paired (13 488 reads).
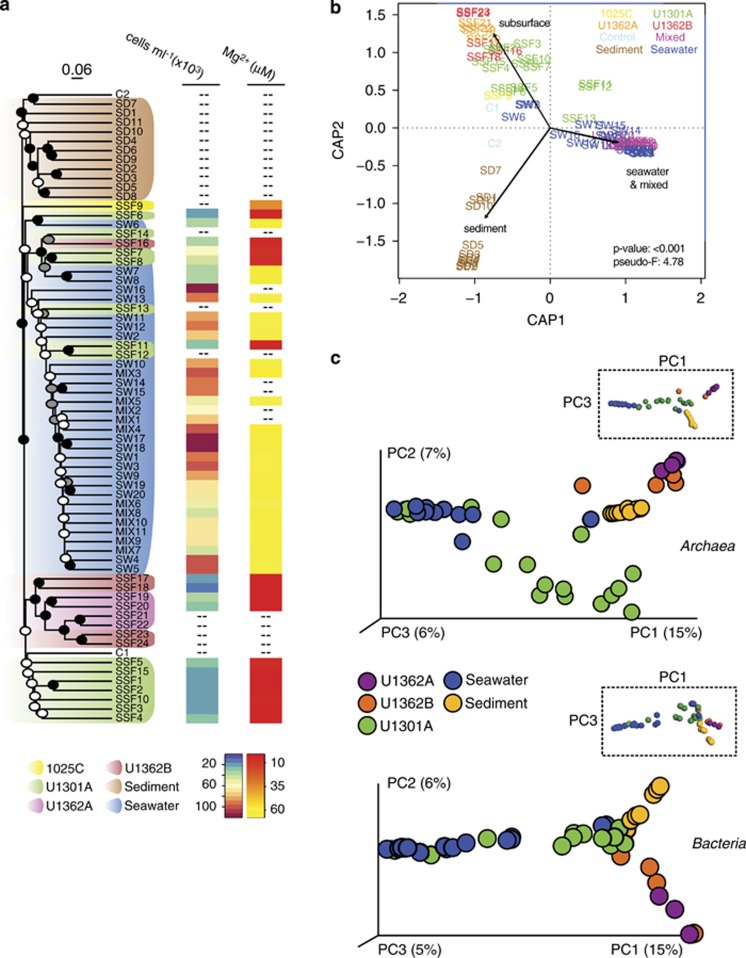
The unique nature of high-integrity crustal fluids with respect to geochemistry and microbial cell abundances was also reflected in the characteristics of their microbial communities: deep subseafloor crustal fluids contained microbial communities that were different from nearby sediments, surrounding bottom seawater and seawater-contaminated samples (Figures 2a–c, Supplementary Figures S6A and B). Average linkage analysis revealed differences in the microbial communities that generally corresponded with differences in the chemical characteristics of samples: all fluid samples that were pumped from boreholes but identified as chemically indistinguishable from bottom seawater clustered with other bottom seawater samples, as did a subset of borehole fluid samples that exhibited lesser degrees of apparent seawater contamination (for example, elevated magnesium and cell concentrations: samples SSF7, SSF8, SSF11, SSF12, SSF13, and SSF16 in Figures 2a and b; Supplementary Table S2). Grouping samples by type (subsurface, sediment, seawater and mixed, control) was statistically significant using db-RDA (P-value⩽0.001; α=0.05; pseudo-F=4.78) (Figure 2b) and was highly consistent with the UPGMA analysis of Bray–Curtis distances (Figure 2a). The db-RDA analysis also revealed sediments from deeper horizons (that is, 25–225 m below the seafloor; SD1, SD7, SD10, SD11) to be most similar to subsurface fluid samples (Figure 2b). In the different analyses, it was also consistently shown that microbial communities within crustal fluids retrieved from borehole U1301A were distinct from those sampled from boreholes U1362A and U1362B (Figures 2a–c).
Figure 2.
Mid-ocean ridge flank basement fluids harbor a distinct microbial assemblage. (a) Dendogram depicting relationships between subsurface fluid samples and nearby sediments, seawater and controls. The tree shown was the first replicate tree constructed with the UPGMA algorithm from Bray–Curtis dissimilarity indices calculated using unique unpaired forward Illumina tag reads and rarefying to an even sequence depth across samples (6108 reads). Support for nodes on the dendogram was determined by analysis of 100 UPGMA replicate dendograms constructed from randomly generated rarefied libraries (6108 reads) and is indicated by black (100%), gray (>80%) and white (>50%) circles. The scale bar corresponds to a Bray–Curtis dissimilarity value of 0.06. Where available, microbial cell abundances and magnesium concentrations are indicated for each sample. (b) Distance-based redundancy analysis showing grouping of samples based on the categories ‘subsurface', ‘sediment', ‘seawater' and ‘mixed'. (c) PCoA of Bray–Curtis dissimiliarity indices using rarefied unique paired Illumina tag reads representing the domains Archaea and Bacteria. Rarefaction of archaeal sequences was performed to the lowest number of sequences detected in the subsurface fluid samples (190 reads) and excluded samples C1, C2, SW6 and SW8. Rarefaction of bacterial sequences was performed to the lowest number of sequences (2803 reads) represented in a single sample. Low-quality subsurface samples are not shown. Plot axes are scaled according to the percentage variation explained via the top three principal components. Rotated plots highlighting P1 versus P3 are shown inset to reveal additional features.
Microbial community structure was also explored in higher-dimensional space via PCoA analysis of the entire SSU rRNA gene sequence data set, as well as by separately analyzing sequences from bacteria and archaea (Figure 2c). The results were highly consistent with that from the average linkage analysis and support the distinction of three separate microbial community end members: sediments, seawater, and subsurface oceanic crustal fluids (Figure 2c). The stability of clusters generated with PCoA analysis of the entire sequence data set was assessed using jackknifed UPGMA clustering, which supported a similar, three end-member result (Supplementary Figure S6A). Sequences found in more than one end-member environment were a minor contribution to the total, providing further evidence that a distinct microbial community exists in deep subsurface ocean crustal fluids (Supplementary Figure S6B).
Relationships between environmental parameters and microbial community structure were identified using Mantel correlation analysis. Microbial community structure of all samples was significantly (P=0.001) associated with chemical characteristics: pH, alkalinity, and the concentrations of calcium, magnesium, potassium, silicate, ammonium, phosphate, nitrate, sulfate, dissolved iron, dissolved organic carbon and total dissolved nitrogen (Supplementary Table S5). The Mantel analysis was repeated with only subsurface fluid samples, which revealed that the concentrations of calcium, magnesium, ammonium, nitrate, dissolved iron and, to a lesser degree, potassium were correlated with differences in the observed microbial community structure (Supplementary Table S5). Grouping samples by type into subsurface, sediment and seawater categories is statistically significant using the rank-based ANOSIM test (P-value=0.001; α=0.05); an intermediate–high value for the R statistic (R=0.6859) indicates dissimilarity between the categorical groupings. Other statistical tests (adonis, MRPP, PERMANOVA) performed using the same categorical groups were also significant using α=0.05. Analysis of group dispersion patterns using PERMDISP revealed marginally significant (that is, P-value<0.1) differences between the environments sampled (Supplementary Table S6).
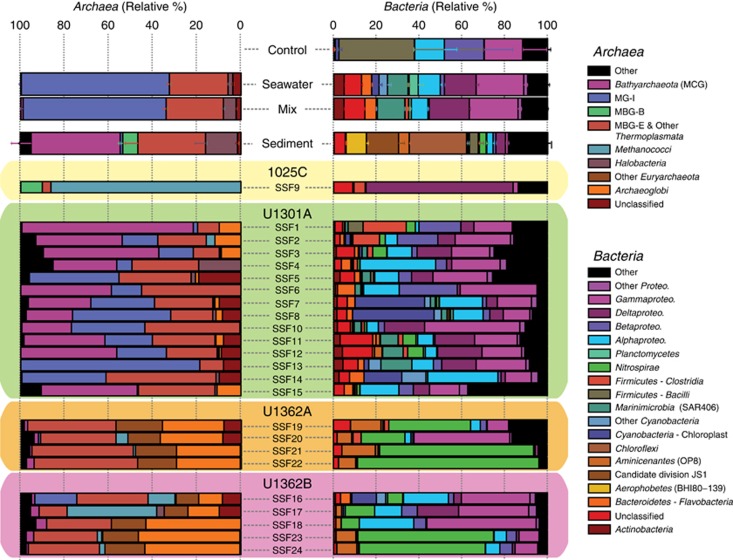
From a taxonomic perspective, the microbial communities inhabiting pristine crustal fluids are also distinct from overlying sediments and bottom seawater and are dominated by a variety of uncultivated archaeal and bacterial lineages that include marine benthic group E (MBG-E), Archaeoglobi, Bathyarchaeota, Nitrospirae, Calescamantes, Aminicenantes and Thermotoga lineage EM3 (Figures 3 and 4a and b). Many of the abundant groups found in fluids from boreholes U1362A and U1362B have previously been detected in other crustal fluid samples characterized from older borehole observatories on the JdFR flank such as 1026B and U1301A, though rarely in high abundance (Cowen et al., 2003; Huber et al., 2006; Nakagawa et al., 2006; Orcutt et al., 2011; Jungbluth et al., 2013a). In general, microbial communities recovered from borehole U1301A fluids were not as distinct from seawater microbial communities as fluids from boreholes U1362A and U1362B (Figures 2a–c). The taxonomic composition of microbial communities recovered from borehole 1025C fluids was highly similar to communities characterized from the same samples previously via rRNA gene cloning and Sanger sequencing (Jungbluth et al., 2014), although the deeper sequencing employed here uncovered additional taxonomic diversity.
Figure 3.
Class-level taxonomic distributions of archaea and bacteria are distinct between deep subsurface fluid microbial communities and seawater, mixed (seawater and deep subsurface fluid), sediment and control samples. Deep subsurface fluid samples are plotted individually; the taxonomic distribution of other samples is averaged and the s.d. of the mean is indicated. Archaeal sequences in controls were either present in very low (<0.01%) abundance (sample C1) or absent (sample C2) and so are not included.
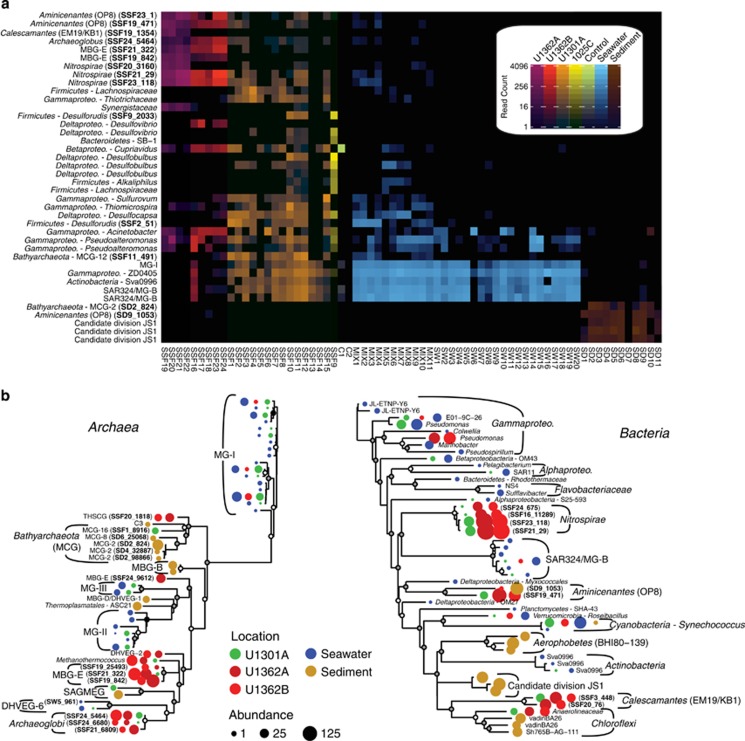
Figure 4.
The abundant microbial lineages found within deep subseafloor basement fluids are rare or absent within sediments or seawater. (a) Heatmap of unique Illumina tag sequences rarefied to an even sampling depth (3250 reads) representing the 10 most abundant bacterial and archaeal lineages within borehole 1025C, U1301A, U1362A and U1362B deep subsurface fluids, and the 5 most abundant lineages within control, sediment and seawater samples. The scale bar indicates the total number of reads detected in the rarified samples. (b) Dufrêne–Legendre indicator species analysis of archaea and bacteria based on unique reads rarefied as in Figure 2 and using 1000 randomized iterations to calculate probabilities. The abundance of unique reads corresponding to each unique taxonomic lineage/sample source combination is indicated by circle size.
An indicator taxa analysis revealed that lineages within both the archaea (Terrestrial Hot Spring Crenarchaeotic Group (THSCG), MBG-E, Deep-sea Hydrothermal Vent Euryarchaeotic Group 2 (DHVEG-2), Methanothermococcus, Archaeaoglobi, Bathyarchaeota) and bacteria (Gammaproteobacteria, Nitrospirae, Aminicenantes, Calescamantes, Chloroflexi) are characteristic of subsurface crustal fluids (Figure 4b). Indicator taxa from sediment, seawater and subsurface fluids overlapped very little (Figure 4b). In particular, overlap between subsurface fluids and seawater was limited to lineages commonly found in seawater (for example, MG-I, MG-II, Pseudomonas, OM43, Cyanobacteria) and are most likely explained by trace amounts of seawater contamination in the subsurface fluid samples (Figure 4b). The indicator taxa found to be characteristic of subsurface fluids originate from multiple boreholes (Figure 4b). Overall, microbial communities inhabiting nearby bottom seawater and sediments or found in mixed samples and controls are taxonomically distinct from subsurface fluids (Figures 3 and 4a and b).
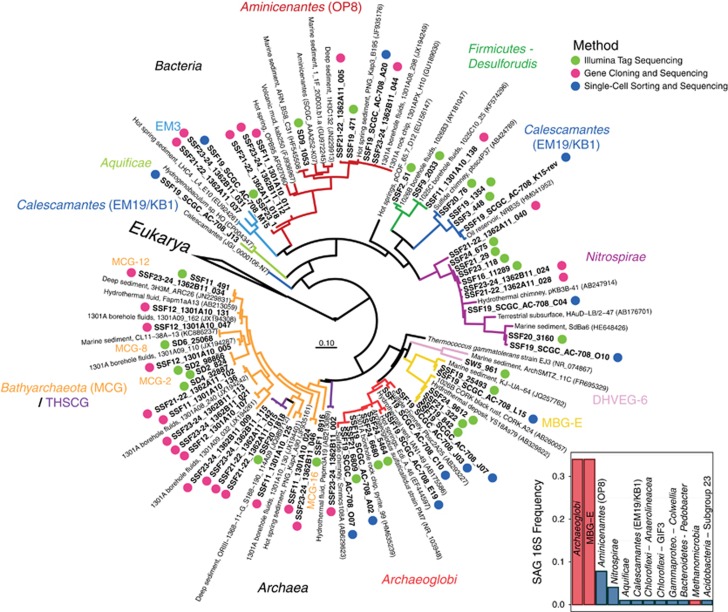
rRNA genes sequenced from whole-genome-amplified single cells isolated from U1362A crustal fluids was used as a complementary means to assess the taxonomic diversity of deep subsurface microorganisms, providing additional evidence for the prevalence of Archaeoglobi, MBG-E, Nitrospirae, Calescamantes and Aminicenantes (Figure 5). Archaea, specifically Archaeoglobi and MBG-E, dominated the taxonomic diversity of the flow-sorted sample (Figure 5). In addition, gene cloning and sequencing employing distinct SSU rRNA gene primers was used to generate near full-length SSU rRNA gene sequences from a handful of high-quality subsurface samples (Figure 5 and Supplementary Figure S7); this method revealed Bathyarchaeota and Aminicenantes in relatively high abundance (Figure 5 and Supplementary Figure S7). Overlap between the three SSU rRNA gene data sets (Illumina tag, single cell, gene cloning) was found for lineages of Nitrospirae and Aminicenantes, while Illumina-based sequencing detected all of the lineages recovered via single-cell flow sorting except for a single SAG from the phylum Aquificae (Figure 5). Although diverse members of the Bathyarchaeota were detected within the modest environmental gene clone libraries, this approach failed to detect Archaeoglobi, MBG-E and Calescamantes identified via the other two methods (Figure 5). Interestingly, Thermotoga lineage EM3 was only detected within crustal fluids from boreholes U1362A and U1362B via gene cloning and sequencing (Figure 5).
Figure 5.
The abundant microorganisms inhabiting deep subseafloor basement fluids are phylogenetically diverse and span multiple unclassified bacterial and archaeal lineages. Phylogenetic comparison between Illumina tag-sequenced SSU rRNA gene amplicons from deep subsurface fluid samples, SSU rRNA genes PCR-amplified and sequenced from the whole-genome amplicons of single cells sorted from the deep Tefzel bioline of the CORK observatory in borehole U1362A and SSU rRNA genes PCR-amplified, cloned and sequenced individually from several deep subsurface fluid samples (Supplementary Table S1). SSU rRNA genes recovered in this study are in bold font, while SSU rRNA genes from cultivated Eukarya were used as outgroups. The scale bar corresponds to 0.1 substitutions per nucleotide position. Borehole U1362A fluid taxonomic diversity and group abundance identified via flow sorting and SSU rRNA gene cloning analysis is shown in the inset histogram. Archaea are highlighted in red (n=57) and bacteria in blue (n=17).
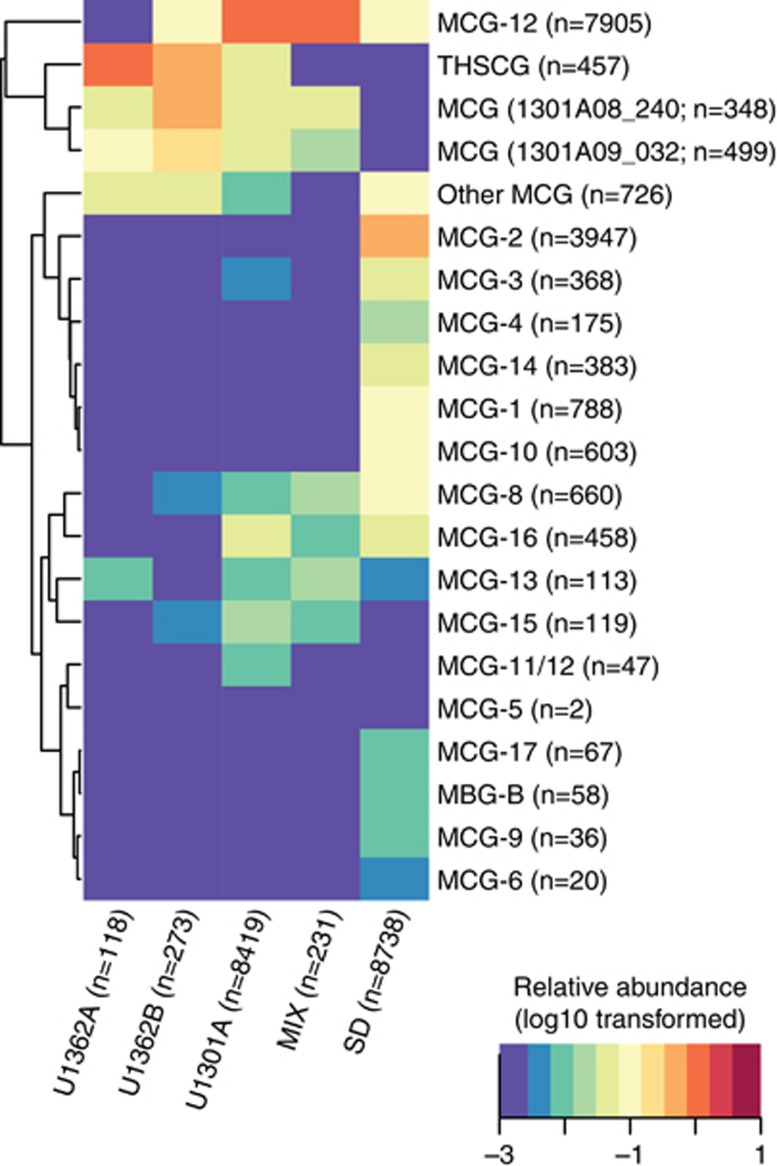
Phylogenetic analyses of Illumina tag sequences from sediments and deep subsurface fluids related to the Aminicenantes and Bathyarchaeota partitioned into sub-lineages based on environment (Figure 5). Most Aminicenantes-related tag sequences from sediments were closely related to sequences previously recovered from sediments, while those from deep subsurface fluids were often closely related to either (1) lineages previously recovered from deep subsurface crustal fluids (for example, 1301A08_298 in Figure 5; Jungbluth et al., 2013a) or (2) new reference SSU rRNA gene sequences from boreholes U1362A and U1362B recovered in this study via traditional gene cloning or single-cell isolation coupled with whole-genome amplification (Figure 5). The diverse Illumina tag sequences related to archaeal group Bathyarchaeaota recovered in this study were analyzed via fine-scale phylogenetic classification using PhyloAssigner and a custom database consistent with Bathyarchaeota/MCG naming schemes described previously (Kubo et al., 2012), supplemented with novel groups from deep subsurface basement fluids characterized previously (Cowen et al., 2003; Orcutt et al., 2011; Jungbluth et al., 2013a). The Bathyarchaeota are a highly diverse group present in both sediment and deep subsurface fluids, though significant lineage-specific partitioning was observed based on habitat type and, in the case of U1301A, borehole (Figure 6). Although the Bathyarchaeota was not a particularly abundant lineage within borehole U1362A and U1362B fluids, those sequences that were detected were predominantly from two novel lineages detected previously from crustal fluids (1301A08_240 and 1301A09_032; 15), as well as other novel and as-yet unnamed Bathyarchaeota lineages (‘Other MCG' in Figure 6). A more detailed phylogenetic analysis using a hand-curated database of Bathyarchaeota SSU rRNA gene sequences (Kubo et al., 2012) revealed that a portion of the crustal fluid Illumina tag sequences classified by the SINA tool as Bathyarchaeota were actually associated with the MBG-B and THSCG archaeal lineages. The gene cloning and sequencing approach recovered a much wider diversity of Bathyarchaeaota from deep subsurface fluids than was recovered via the Illumina amplicon sequencing approach, and Bathyarchaeota as a whole were conspicuously absent from the single-cell sorting and whole-genome amplification analysis (Figure 5).
Figure 6.
Taxonomic analysis of unique reads identified as Bathyarchaeaota using the SILVA database and PhyloAssigner. Naming convention is adopted from Kubo et al. (2012) but modified to include two divergent lineages (clones 1301A08_240 and 1301A09_032) detected previously in subsurface crustal fluids (Jungbluth et al., 2013a). The log10-transformed relative abundance of reads by location is indicated in the heatmap and used to construct the dendogram. The total number of reads detected from each location and ‘MCG' lineage is indicated.
Discussion
Of the few routes available to study microbial life within fluids contained in the Earth's vast sediment-buried subseafloor basement, hydrologically sealed boreholes that penetrate through the sediment and into the basaltic crust currently provide the only direct means to do so. Globally, only a handful of boreholes drilled into the basaltic subseafloor have considered microbiological sampling in their design, the most prominent of which are the CORK-fitted boreholes along the JdFR flank of the Northeast Pacific Ocean and their more recently installed counterparts at North Pond on the Mid-Atlantic Ridge. In addition to ocean basin-scale geographic partitioning, these two sets of subseafloor observatories provide a valuable contrast in proximal environmental setting in that the Juan de Fuca CORK-instrumented borehole array lies on a rapidly sedimented ridge flank with relatively sluggish fluid flow horizontally and no appreciable vertical fluid advection, while the North Pond array is covered by a relatively thin sediment layer that allows vertical advection and limits the extent of geothermal heating of the aquifer. One prominent result is that basalt-hosted subseafloor fluids sampled via the JdFR CORK platforms exhibit chemical characteristics that are expected of fluids that have reacted extensively with basaltic rock and have been transformed by microbial activity, including exhaustion of oxygen and nitrate as electron acceptors. By contrast, similarly sampled fluids from North Pond contain both oxygen and nitrate at concentrations near those of bottom seawater (Orcutt et al., 2013).
Until recently, restricted physical access and semi-continuous successional changes in the development of CORK platforms and the instrumentation required to interface with them have limited successful microbiological sampling at deep subseafloor borehole observatories. Thus a robust and statistically significant characterization of basalt-hosted subseafloor aquifer microbial assemblages has been elusive. To date, three of the four studies describing microbial communities within basement fluids collected via JdFR CORK observatories have included single samples that are difficult to interpret within the context of seawater, sediment and other potentially contaminating sources (Cowen et al., 2003; Huber et al., 2006; Jungbluth et al., 2014). The fourth study investigated one sample per year for three years, all at borehole U1301A (Jungbluth et al., 2013a). The current study is the first to investigate microbial community structure within basalt-hosted deep subseafloor aquifers via multiple independent, high-integrity basement fluid samples collected over the course of several years and from multiple CORK-instrumented borehole locations along the JdFR flank, including fluids from the most recent generation of CORK observatories that are equipped with titanium sampling ports and Tefzel-lined fluid delivery lines that deliver water from depth horizons of nearly 300 m below the sediment–rock interface. Thus these samples span unprecedented spatial, temporal and CORK infrastructure gradients that allowed us to quantify relationships between the microbial communities sampled via several different points of access into the basalt-hosted aquifer below the JdFR flank, as well as between basalt-hosted communities and microorganisms occupying their most closely adjoining habitats—sediments and bottom seawater. The chemical characteristics of fluid samples interrogated here are consistent with previous studies (for example, Mottl et al., 1998; Wheat et al., 2000; Lin et al., 2012, 2014) though provide valuable refinement of their natural range. In addition, the large number of high-integrity subsurface crustal fluid samples provides a robust assessment of the biomass supported by highly reacted fluids within the deep ocean crustal biosphere.
Using a combination of methodologies, we have shown that the microbial communities inhabiting deep subseafloor crustal fluids from the warm, anoxic basement aquifer along the JdFR flank are conspicuously distinct from their counterparts inhabiting surrounding bottom seawater and sediments. Over 85% of the total unique Illumina SSU rRNA gene sequencing reads from deep subseafloor fluids were unique to this environment compared with seawater, sediments and experimental controls, with an even higher percentage recovered from pristine samples collected from the new CORK observatories at boreholes U1362A and U1362B. Many of the microbial lineages characteristic of the subsurface fluids collected here, such as the Nitrospirae, Aminicenantes and MBG-E, are rare in cold seafloor sediments but instead have relatives that are most often detected in studies of hydrothermally influenced sediments or deposits (for example, Takai and Horikoshi, 1999; Suzuki et al., 2004; Nercessian et al., 2005). The Archaeoglobi are also not common to sediment but are instead affiliated primarily with high-temperature hydrothermal systems. At the location of the JdFR flank boreholes, the temperatures typical for growth of Archaeoglobus are encountered only in the basement environment, where fluids in the upper basement are ~65 °C (Davis and Becker, 2002). Microbial lineages related to Thermotoga and Aquifex were also found within crustal fluids, providing additional evidence that the fluid samples characterized here originate from deep within the Earth's geothermally heated crust. Overall, the novel microbial diversity and unique community structure reveals a distinct microbial assemblage in this anoxic basaltic aquifer and harbors members with temperature optima inferred from the closest neighbors that are consistent with a system that is thermogenically driven.
Most of the ubiquitous and abundant lineages within basalt-hosted subseafloor fluids currently have poorly characterized metabolisms owing to a lack of relevant cultivated isolates or genomic information to use for inference. Exceptions are lineages of Methanococci and a subset of the Archaeoglobi detected here; their close relationship with cultivated and characterized strains reveals that they likely maintain methanogenic and sulfate-reducing metabolic lifestyles, respectively. Evidence for sulfate reduction has also been corroborated by the successful identification of dissimilatory sulfite reductase (dsr) genes related to Archaeoglobus (Robador et al., 2015) and other taxa (Lever et al., 2013), which is consistent with the SSU rRNA gene-based findings described here, and is also consistent with the depletion of oxygen and nitrate as terminal electron acceptors. Although detected in basaltic rocks from U1301B by Lever et al. (2013), Methanosarcinales were not detected in U1301A fluids despite the close proximity (~35 m) of these two locations; however, this lineage was identified in low abundances (<0.2%) in all samples of U1362A crustal fluids. In contrast to the reports from Lever et al. (2013), anaerobic methanotrophic archaea (ANME) groups were undetected in borehole crustal fluids, with the exception of one unique sequence related to ANME-2c found in U1362B fluid sample SSF24. Although phylogenetically unique and deeply branching lineages of Nitrospirae were also major indicator taxa for subseafloor crustal fluid samples, their significant evolutionary divergence from the closest relatives limits inferences regarding their potential primary metabolic characteristics. However, it is noted that sulfate reduction is characteristic of some members of the Nitrospirae (Sekiguchi et al., 2008), and putative deeply branching Nitrospirae-related dsr genes were recently identified within some of the same samples characterized in our study (Robador et al., 2015). Of the remaining ubiquitous and highly abundant microbial lineages inhabiting deep subseafloor fluids of the JdFR flank, little is known regarding their potential physiological characteristics. Recently, single-cell genomics revealed some insight regarding the potential for amino-acid degradation within the candidate bacterial phylum Aminicenantes (OP8) (Rinke et al., 2013), although the Aminicenantes-related lineages we describe here diverge significantly from those with genomic information. Despite single-cell genome sequencing efforts (Rinke et al., 2013), the potential metabolic characteristics of the candidate phylum Calescamantes remains unknown. This also remains the case for members of the uncultivated archaeal lineage MBG-E, where no isolated strains or metabolic inferences from genomic information are available.
The question of whether the variability we have documented in the structure of microbial communities inhabiting crustal fluids collected from the JdFR is a reflection of variability in the natural state or artificially induced over the course of sampling and analysis remains unresolved. For borehole U1301A, the consistent detection of microbial lineages indicative of seawater contamination, such as SAR11, SAR324, and planktonic marine Thaumarchaeota and Euryarchaeota, reveal that seawater invasion regularly occurs during sampling at this borehole owing to either insufficient sealing between borehole and sampling instrumentation or seawater intrusion into the borehole prior to connection to the fluid delivery line. Although we found no evidence for connectivity issues, borehole U1301A was underpressured and drew in surrounding bottom ocean seawater for several years after drilling (Wheat et al., 2010), making the later a plausible explanation. In contrast, boreholes U1362A and U1362B were positively pressured and producing fluids following drilling in <1 year and had little-to-no evidence of seawater intrusion. Thus we can conclude that differences between U1301A and U1362A/U1362B are due, at least in part, to artificial factors outside of natural variability of microbial communities in situ. Because we sampled two non-overlapping basement horizons at boreholes U1362A and U1362B, we were able to investigate vertical stratification of microbial communities within the basaltic subseafloor crust. Although each horizon at U1362A and U1362B has not been characterized with replicate, identically processed samples in sufficient numbers to quantitatively evaluate the natural variability within each borehole and horizon, the samples described in this study reveal differences in community structure that may derive from vertical stratification or horizontal segregation of fluid flow in this region of the ocean basement.
Overall, this study provides strong evidence for the dominance of a limited number of bacteria and archaea within deep subseafloor crustal fluids accessed through multiple boreholes and multiple horizons of the anoxic, basalt-hosted aquifer within the eastern flank of the JdFR. The vast majority of abundant lineages, which includes some within-candidate bacterial (Aminicenantes, Calescamantes) and archaeal (marine benthic group E, Terrestrial Hot Spring Crenarchaeotic Group) phyla, as well as divergent lineages only distantly related to the characterized taxa (Nitrospirae Chloroflexi, Archaeoglobi), are of unknown physiology. However, severe depletion in dissolved organic carbon, oxygen, nitrate and cellular abundances coupled with hydrogen and methane in sufficient quantities to fuel microbial metabolism create an environmental setting where it is likely that chemolithotrophy and methane cycling coupled with sulfate reduction are traits of the unusual microbial lineages prevalent in this system (Lin et al., 2014). Although the pervasiveness of oxygen within the global network of geothermally heated deep subseafloor basalt-hosted aquifers is not yet known in other regions where oxygen is extinguished in a similar manner to the eastern flank of the JdFR, we postulate the existence of microbial communities containing lineages closely related to those found here.
Acknowledgments
This study is dedicated to the memory of our friend, colleague, mentor and co-author, James P Cowen, whose determination and enthusiasm were driving forces in the adaptation of seafloor borehole observatories for microbiology. We thank the captain and crew, A Fisher, K Becker, CG Wheat and other members of the science teams on board R/V Atlantis cruises AT15-35, AT15-51, AT15-66 and AT18-07. We also thank the pilots and crew of human-occupied vehicle Alvin and remote-operated vehicle Jason II and Brian Glazer, Ryan Matsumoto, Michael Matzinger, Michelle Jungbluth, Alberto Robador, Jennifer Murphy, Chih-Chiang Hseih, Natalie Hamada, Karen Meech and Joshua Bninski for sampling, technical and other assistance. This research was supported by funding from National Science Foundation Microbial Observatories grant MCB06-04014 (to JC and MSR), a Schlanger Ocean Drilling Fellowship (to SPJ), which is part of the NSF-sponsored US Science Support Program for IODP that is administered by the Consortium for Ocean Leadership, the UH NASA Astrobiology Institute and the Center for Dark Energy Biosphere Investigations (C-DEBI) (OCE-0939564), a National Science Foundation-funded Science and Technology Centers of Excellence. This study used samples and data provided by the Integrated Ocean Drilling Program. This is SOEST contribution 9539, HIMB contribution 1636 and C-DEBI contribution 289.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Baross JA, Wilcock WSD, Kelley DS, DeLong EF, Cary SC. (2004). The subsurface biosphere at mid-ocean ridges: issues and challenges. In: Wilcock WSD, DeLong EF, Kelley DS, Baross JA, Cary SC (eds), The Subseafloor Biosphere at Mid-Ocean Ridges. American Geophysical Union: Washington, DC, USA, pp 1–11. [Google Scholar]
- Becker K, Davis EE. (2005). A review of CORK designs and operations during the Ocean Drilling Program. In: Fisher AT, Urabe T, Klaus A, Expedition 301 Scientists (eds), Proceedings of the Integrated Ocean Drilling Program. Integrated Ocean Drilling Program Management International, Inc.: Tokyo, Japan, pp 1–28. [Google Scholar]
- Biddle JF, Jungbluth SP, Lever MA, Rappé MS. (2014). Life in the oceanic crust. In: Kallmeyer J, Wagner D (eds), Microbial Life of the Deep Biosphere. DeGruyter: Berlin, Germany, pp 29–62. [Google Scholar]
- Butterfield DA, Jonasson IR, Massoth GJ, Feely RA, Roe KK, Embley RE et al. (1997). Seafloor eruptions and evolution of hydrothermal fluid chemistry. Philos Trans R Soc A 355: 369–386. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108: 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen JP. (2004). The microbial biosphere of sediment-buried oceanic basement. Res Microbiol 155: 497–506. [DOI] [PubMed] [Google Scholar]
- Cowen JP, Copson DA, Jolly J, Hsieh C-C, Lin H-T, Glazer BT et al. (2012). Advanced instrument system for real-time and time-series microbial geochemical sampling of the deep (basaltic) crustal biosphere. Deep-Sea Res Pt I 61: 43–56. [Google Scholar]
- Cowen JP, Giovannoni SJ, Kenig F, Johnson HP, Butterfield D, Rappé MS et al. (2003). Fluids from aging ocean crust that support microbial life. Science 299: 120–123. [DOI] [PubMed] [Google Scholar]
- Davis EE, Becker K. (2002). Observations of natural-state fluid pressures and temperatures in young oceanic crust and inferences regarding hydrothermal circulation. Earth Planet Sci Lett 204: 231–248. [Google Scholar]
- D'Hondt S, Inagaki F, Zarikian CA, Abrams LJ, Dubois N, Engelhardt T et al. (2015). Presence of oxygen and aerobic communities from sea floor to basement in deep-sea sediments. Nat Geosci 8: 299–304. [Google Scholar]
- Deming JW, Baross JA. (1993). Deep-sea smokers: windows to a subsurface biosphere? Geochim Cosmochim Acta 57: 3219–3230. [DOI] [PubMed] [Google Scholar]
- Edwards KJ, Bach W, McCollom TM. (2005). Geomicrobiology in oceanography: microbe–mineral interactions at and below the seafloor. Trends Microbiol 13: 449–456. [DOI] [PubMed] [Google Scholar]
- Edwards KJ, Fisher AT, Wheat CG. (2012). The deep subsurface biosphere in igneous ocean crust: frontier habitats for microbiological exploration. Front Microbiol 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elderfield H, Wheat CG, Mottl MJ, Monnin C, Spiro B. (1999). Fluid and geochmical transport through oceanic crust: a transect across the eastern flank of the Juan de Fuca Ridge. Earth Planet Sci Lett 172: 151–165. [Google Scholar]
- Eren AM, Vineis JH, Morrison HG, Sogin ML. (2013). A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS One 8: e66643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expedition 327 Scientists. (2011). Expedition 327 summary. In: Fisher AT, Tsuji T, Petronotis K, Expedition 327 Scientists (eds), Proceedings of the Integrated Ocean Drilling Program. Integrated Ocean Drilling Program Management International, Inc.: College Station, TX, USA, pp 1–32. [Google Scholar]
- Fisher AT, Davis EE, Hutnak M, Spiess V, Zühlsdorff L, Cherkaoui A et al. (2003). Hydrothermal recharge and discharge across 50 km guided by seamounts on a young ridge flank. Nature 421: 618–621. [DOI] [PubMed] [Google Scholar]
- Fisher AT, Wheat CG, Becker K, Davis EE, Jannasch H, Schroeder D et al. (2005) Scientific and technical design and deployment of long-term subseafloor observatories for hydrogeologic and related experiments, IODP Expedition 301, eastern flank of Juan de Fuca Ridge. In: Fisher AT, Urabe T, Klaus A, Expedition 301 Scientists (eds), Proceedings of the Integrated Ocean Drilling Program. Integrated Ocean Drilling Program Management International, Inc.: Tokyo, Japan, pp 1–39. [Google Scholar]
- Fisher AT, Wheat CG. (2010). Seamounts as conduits for massive fluid, heat and solute fluxes on ridge flanks. Oceanography 23: 74–87. [Google Scholar]
- Huber JA, Johnson HP, Butterfield DA, Baross JA. (2006). Microbial life in ridge flank crustal fluids. Environ Microbiol 8: 88–99. [DOI] [PubMed] [Google Scholar]
- Johnson HP, Pruis MJ. (2003). Fluxes of fluid and heat from the oceanic crustal reservoir. Earth Planet Sci Lett 216: 565–574. [Google Scholar]
- Jungbluth SP, Grote J, Lin H-T, Cowen JP, Rappé MS. (2013. a). Microbial diversity within basement fluids of the sediment-buried Juan de Fuca Ridge flank. ISME J 7: 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungbluth SP, Johnson LGH, Cowen JP, Rappé MS. (2013. b). Data report: microbial diversity in sediment near Grizzly Bare Seamount from Holes U1363B and U1363G. In: Fisher AT, Tsuji T, Petronotis K, Expedition 327 Scientists (eds), Proceedings of the Integrated Ocean Drilling Program. Integrated Ocean Drilling Program Management International, Inc.: Tokyo, Japan. [Google Scholar]
- Jungbluth SP, Lin H-T, Cowen JP, Glazer BT, Rappé MS. (2014). Phylogenetic diversity of microorganisms in subseafloor crustal fluids from boreholes 1025C and 1026B along the Juan de Fuca Ridge flank. Front Microbiol 5: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D'Hondt S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA 109: 16213–16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K, Lloyd KG, F Biddle J, Amann R, Teske A, Knittel K. (2012). Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J 6: 1949–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever MA, Rouxel O, Alt JC, Shimizu N, Ono S, Coggon RM et al. (2013). Evidence for microbial carbon and sulfur cycling in deeply buried ridge flank basalt. Science 339: 1305–1308. [DOI] [PubMed] [Google Scholar]
- Lin H-T, Cowen JP, Olson EJ, Amend JP, Lilley MD. (2012). Inorganic chemistry, gas compositions and dissolved organic carbon in fluids from sedimented young basaltic crust on the Juan de Fuca Ridge flanks. Geochim Cosmochim Acta 85: 213–227. [Google Scholar]
- Lin H-T, Cowen JP, Olson EJ, Lilley MD, Jungbluth SP, Wilson S et al. (2014). Dissolved hydrogen and methane in the oceanic basaltic biosphere. Earth Planet Sci Lett 405: 62–73. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottl MJ, Wheat CG. (1994). Hydrothermal circulation through mid-ocean ridge flanks: fluxes of heat and magnesium. Geochim Cosmochim Acta 58: 2225–2237. [Google Scholar]
- Mottl MJ, Wheat G, Baker E, Becker N, Davis E, Feely R et al. (1998). Warm springs discovered on 3.5 Ma oceanic crust, eastern flank of the Juan de Fuca Ridge. Geology 26: 51–54. [Google Scholar]
- Nakagawa S, Inagaki F, Suzuki Y, Steinsbu BO, Lever MA, Takai K et al. (2006). Microbial community in black rust exposed to hot ridge flank crustal fluids. Appl Environ Microbiol 72: 6789–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nercessian O, Fouquet Y, Pierre C, Prieur D, Jeanthon C. (2005). Diversity of Bacteria and Archaea associated with a carbonate-rich metalliferous sediment sample from the Rainbow vent field on the Mid-Atlantic Ridge. Environ Microbiol 7: 698–714. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2013). vegan: Community Ecology Package. Available from http://cran.r-project.org/web/packages/vegan/index.html.
- Orcutt BN, Bach W, Becker K, Fisher AT, Hentscher M, Toner BM et al. (2011). Colonization of subsurface microbial observatories deployed in young ocean crust. ISME J 5: 692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orcutt BN, Wheat CG, Rouxel O, Hulme S, Edwards KJ, Bach W et al. (2013). Oxygen consumption rates in subseafloor basaltic crust derived from a reaction transport model. Nat Comm 4: 2539. [DOI] [PubMed] [Google Scholar]
- Parkes RJ, Cragg BA, Bale SJ, Getliff JM, Goodman K, Rochelle PA et al. (1994). Deep bacterial biosphere in Pacific Ocean sediments. Nature 371: 410–413. [Google Scholar]
- R Core Team. (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, Available from http://www.r-project.org. [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Robador A, Jungbluth SP, LaRowe DE, Bowers RM, Rappé MS, Amend JP et al. (2015). Activity and phylogenetic diversity of sulfate-reducing microorganisms in low-temperature subsurface fluids within the upper oceanic crust. Front Microbiol 5: 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DW. (2013), labdsv: Ordination and Multivariate Analysis for Ecology. Available from http://cran.r-project.org/web/packages/labdsv/.
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi Y, Muramatsu M, Imachi H, Narihiro T, Ohashi A, Harada H et al. (2008). Thermodesulfovibrio aggregans sp. nov. and Thermodesulfovibrio thiophilus sp. nov., anaerobic, thermophilic, sulfate-reducing bacteria isolated from thermophilic methanogenic sludge, and emended description of the genus Thermodesulfovibrio. Int J Syst Evol Microbiol 58: 2541–2548. [DOI] [PubMed] [Google Scholar]
- Smith A, Popa R, Fisk M, Nielsen M, Wheat CG, Jannasch HW et al. (2011). In situ enrichment of ocean crust microbes on igneous minerals and glasses using an osmotic flow-through device. Geochem Geophys Geosys 12: Q06007. [Google Scholar]
- Summit M, Baross JA. (2001). A novel microbial habitat in the mid-ocean ridge subseafloor. Proc Natl Acad Sci USA 98: 2158–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Inagaki F, Takai K, Nealson KH, Horikoshi K. (2004). Microbial diversity in inactive chimney structures from deep-sea hydrothermal systems. Microb Ecol 47: 186–196. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautrotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Takai K, Horikoshi K. (1999). Genetic diversity of Archaea in deep-sea hydrothermal vent environments. Genetics 152: 1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. (2013). EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat CG, Elderfield H, Mottl MJ, Monnin C. (2000). Chemical composition of basement fluids within an oceanic ridge flank: implications for along-strike and across-strike hydrothermal circulation. J Geophys Res 105: 13437–13447. [Google Scholar]
- Wheat CG, Jannasch HW, Fisher AT, Becker K, Sharkey J, Hulme S. (2010). Subseafloor seawater-basalt-microbe reactions: continuous sampling of borehole fluids in a ridge flank environment. Geochem Geophys Geosys 11: Q07011. [Google Scholar]
- Wheat CG, Jannasch HW, Kastner M, Hulme S, Cowen J, Edwards KJ et al. (2011). Fluid sampling from oceanic borehole observatories: design and methods for CORK activities (1990-2010). In: Fisher AT, Tsuji T, Petronotis K, Expedition 327 Scientists (eds), Proceedings of the Integrated Ocean Drilling Program. Integrated Ocean Drilling Program Management International, Inc.: Tokyo, Japan, pp 1–36. [Google Scholar]
- Wilcock WSD, Fisher AT. (2004). Geophysical constraints on the sub-seafloor environment near mid-ocean ridges. In: Wilcock WSD, DeLong EF, Kelley DS, Baross JA, Cary SC (eds). The Subseafloor Biosphere at Mid-Ocean Ridges. American Geophysical Union: Washington, DC, USA, pp 51–74. [Google Scholar]
- Wilkinson L. (2011). venneuler: Venn and Euler Diagrams. Available from http://cran.r-project.org/web/packages/venneuler/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.