Abstract
The chemolithoautotrophic microbial community of the rocky subseafloor potentially provides a large amount of organic carbon to the deep ocean, yet our understanding of the activity and metabolic complexity of subseafloor organisms remains poorly described. A combination of metagenomic, metatranscriptomic, and RNA stable isotope probing (RNA-SIP) analyses were used to identify the metabolic potential, expression patterns, and active autotrophic bacteria and archaea and their pathways present in low-temperature hydrothermal fluids from Axial Seamount, an active submarine volcano. Metagenomic and metatranscriptomic results showed the presence of genes and transcripts for sulfur, hydrogen, and ammonium oxidation, oxygen respiration, denitrification, and methanogenesis, as well as multiple carbon fixation pathways. In RNA-SIP experiments across a range of temperatures under reducing conditions, the enriched 13C fractions showed differences in taxonomic and functional diversity. At 30 °C and 55 °C, Epsilonproteobacteria were dominant, oxidizing hydrogen and primarily reducing nitrate. Methanogenic archaea were also present at 55 °C, and were the only autotrophs present at 80 °C. Correspondingly, the predominant CO2 fixation pathways changed from the reductive tricarboxylic acid (rTCA) cycle to the reductive acetyl-CoA pathway with increasing temperature. By coupling RNA-SIP with meta-omics, this study demonstrates the presence and activity of distinct chemolithoautotrophic communities across a thermal gradient of a deep-sea hydrothermal vent.
Introduction
Venting fluids at deep-sea hydrothermal systems export members of the extensive microbial communities of the rocky subseafloor. Within the fluids circulating through the seafloor, chemolithoautotrophs convert CO2 to biomass using the geochemical energy created from chemical disequilibria when hydrothermal fluid mixes with seawater (Amend et al., 2011). Previous studies found high rates of carbon fixation and concentrations of organic carbon and ATP in venting fluids and plume waters (Karl et al., 1980; Jannasch and Wirsen, 1981; Jannasch et al., 1989; Wirsen et al., 1993). Energetic modeling suggests that subseafloor mixing zones are likely to be the most productive of vent habitats with respect to microbial growth, potentially providing an important source of carbon to the deep ocean (McCollom, 2005).
16S ribosomal RNA (rRNA) gene surveys and culture-dependent approaches have revealed the extensive taxonomic diversity and metabolic plasticity of chemolithoautotrophs at deep-sea hydrothermal vents (Reysenbach and Shock, 2002; Sievert and Vetriani, 2012). Further studies of diffuse hydrothermal fluids have shown that the population structure of the subseafloor community can vary with time (Huber et al., 2002, 2003; Perner et al., 2009), space (Opatkiewicz et al., 2009; Huber et al., 2010), and geochemical gradients (Huber et al., 2007; Perner et al., 2010; Akerman et al., 2013). In addition, the genomes of key autotrophs from hydrothermal vents have revealed the broad array of CO2 fixation pathways and energy acquisition strategies used by these organisms (Takai et al., 2005; Campbell et al., 2009; Yamamoto and Takai, 2011; Meyer and Huber, 2014).
Recent metagenomic and metatranscriptomic studies from hydrothermal plumes and diffuse hydrothermal fluids have provided additional insights into the metabolic potential and gene expression patterns of the microbial communities in hydrothermal systems (for example, Lesniewski et al., 2012; Anantharaman et al., 2013; Baker et al., 2013; Anderson et al., 2014; Urich et al., 2014). Studies of hydrothermal vent plumes, which are a mixture of deep-water and hydrothermal vent microbes (Dick et al., 2013), demonstrated the metabolic versatility of plume microbial communities, with genes for ammonium, sulfur, and methane oxidation highly expressed (Lesniewski et al., 2012; Anantharaman et al., 2013) and presence of both chemolithoautotrophic populations such as SUP05 and heterotrophic archaea (Dick et al., 2013; Li et al., 2015). Targeting the subseafloor chemolithoautotrophic community is especially important for determining their role in the primary production of biomass by dark fixation. However, as these subsurface communities are difficult to sample, studies of their microbiology have relied on sampling vent fluids, which represent a mixture of subseafloor hydrothermal fluids and deep seawater consisting of 75–99% seawater and 1–25% hot hydrothermal end-member. This results in an extremely diverse microbial community (Huber et al., 2007) where the subseafloor microbial signal can be dampened by the overwhelming signature of seawater mixing.
Stable isotope probing (SIP) is one powerful tool for focusing on specific active communities through incorporation of isotopically labeled substrates under near natural conditions (Dumont and Murrell, 2005). During a SIP experiment, the active community will incorporate the labeled substrate and increase in biomass relative to the non-active fraction of the community. The active community can then be identified using various molecular techniques (Dumont and Murrell, 2005; Neufeld et al., 2007). Recently, SIP studies have expanded from purely taxonomic identification (for example, Lueders et al., 2004; Bernard et al., 2007; Glaubitz et al., 2010) to using DNA-SIP to construct metagenomic libraries of the labeled populations (Dumont et al., 2006; Neufeld et al., 2008). SIP studies have also coupled RNA-SIP with metatranscriptomics to link actively transcribed genes to specific microbial populations (Huang et al., 2009; Dumont et al., 2013). The first study to attempt mRNA-SIP used reverse transcriptase-PCR and primers specific to naphthalene degrading genes to understand the specific bacterial species that were degrading this compound in groundwater (Huang et al., 2009). A more recent study was able to construct a metatranscriptome from a SIP experiment of lake sediments looking at aerobic methanotrophs and showed the presence of transcripts for methane oxidation, carbon assimilation and nitrogen metabolism (Dumont et al., 2013).
Although SIP presents an excellent way to link community structure and function, there are limitations that must be considered. The active microbial community must be able to take up the labeled isotope and incorporate it into nucleic acids within a given incubation time under particular experimental conditions. Too short of an incubation may bias against microbes with slow growth rates and lead to incomplete labeling of the community, while too long of a time may lead to nonspecific labeling of the community (that is, cross-feeding) (Radajewski et al., 2003). It is also important to differentiate between assimilation and metabolism of a labeled isotope. For example, the general uptake of 13C-labeled bicarbonate into anaplerotic pathways of the TCA cycle may result in nonspecific labeling of the community (Yakimov et al., 2014). Thus, it is important to take these limitations into account when analyzing SIP results. In this study, traditional metagenomics and metatranscriptomic analyses of diffuse vent fluids were coupled with RNA-SIP metatranscriptomics to determine which autotrophs were active under relevant subseafloor conditions, what metabolisms and carbon fixation pathways were used, and how these organisms function across different temperatures representing subseafloor thermal gradients.
Experiments were carried out at Axial Seamount, a submarine volcano located on the Juan de Fuca Ridge and a site of long term geological, chemical, and biological study (Chadwick et al., 2010). 16S rRNA and functional gene surveys have shown that diverse Epsilonproteobacteria dominate across diffuse vents at Axial and their community structure can often be linked to the chemistry of the vent sampled (Huber et al., 2007; Opatkiewicz et al., 2009; Akerman et al., 2013; Meyer et al., 2013). Epsilonproteobacteria were first isolated from hydrothermal vents in 2001 (Campbell et al., 2001) and all isolates to date are chemolithoautotrophic and either moderately thermophilic hydrogen oxidizers or mesophilic hydrogen and sulfur oxidizers (Campbell et al., 2006). Recent studies of hydrothermal vent Epsilonproteobacteria also show that many are capable of nitrate reduction and denitrification (Campbell et al., 2006; Vetriani et al., 2014), as well as nitrogen fixation (Meyer and Huber, 2014). In addition to Epsilonproteobacteria, Gammaproteobacteria, specifically SUP05 populations, were also shown to be abundant and active within both diffuse fluids and plumes at Axial (Bourbonnais et al., 2012; Akerman et al., 2013; Anderson et al., 2013). Although SUP05 remains uncultivated from the deep ocean, genomic evidence indicates these bacteria possess genes to carry out both sulfide and hydrogen oxidation, similar to many Epsilonproteobacteria (Takai et al., 2005; Campbell et al., 2006; Anantharaman et al., 2013; Meyer and Huber, 2014). Hydrogen oxidation is a more energetically favorable reaction compared with sulfur oxidation and thus may be an alternate energy pathway for these chemolithoautotrophs in venting fluids (Amend et al. 2011).
In addition to the dominance of sulfur and hydrogen-oxidizing microbes, Axial Seamount has also been a study site of subseafloor methanogenic archaea, with the detection, quantification, and isolation of many mesophilic, thermophilic, and hyperthermophilic strains from diffuse fluids (Huber et al., 2002; Ver Eecke et al., 2012; Meyer et al., 2013; Ver Eecke et al., 2013), including isolation of the first nitrogen-fixing hyperthermophilic methanogen (Mehta and Baross, 2006). Growth experiments with methanogenic archaea isolated from diffuse fluids at Axial indicate they require concentrations of hydrogen 10 times lower than previous estimates and have the potential for syntrophic relationships with hydrogen producing heterotrophs (Ver Eecke et al., 2012, 2013).
Building upon previous studies, we used metagenomics and metatranscriptomics to first establish the metabolic potential and gene expression patterns of the microbial community from a singular vent at Axial Seamount, Marker 113. We then used RNA-SIP combined with metatranscriptomics to separate out the chemolithoautotrophic signature in the mixed fluids in order to determine the key taxa and metabolisms representative of the warm, reducing subseafloor habitat. Results showed the metabolic complexity of the vent microbial community and highlight the diverse metabolic pathways of the potential members of the subseafloor chemolithoautotrophic community across different temperatures.
Materials and methods
Fluid collection and vent chemistry
Diffuse hydrothermal vent fluids (24 °C) were collected from Marker 113 vent (Ver Eecke et al., 2012; 45.9228, −129.9881) at Axial Seamount on 27 September 2013 on board the R/V Falkor using the deep-sea research submarine ROV ROPOS. Sampling depth at Marker 113 vent was 1521 m. Fluids were collected 2 cm above the seafloor using the hydrothermal fluid and particle sampler (HFPS) (Butterfield et al., 2004) and the large volume water sampler (LVWS) (Wommack et al., 2004). Both instruments had integrated temperature sensors at the point of intake to allow for constant temperature monitoring while drawing fluids. For RNA-SIP experiments, 4–5 l of fluid were pumped into either an acid-washed 4-l Tedlar bag using the HFPS or into a 5-l acid-washed plastic carboy using the LVWS. For DNA and RNA collection, the HFPS was used to pump 3 l of diffuse fluid at a rate of 100–150 ml min−1 through a 0.22 μm, 47 mm GWSP filter (Millipore, Billerica, MA, USA) that was then preserved in situ with RNALater (Ambion, Grand Island, NY, USA) as previously described in Akerman et al. (2013). Fluid samples were also collected for chemistry and analyzed for alkalinity, hydrogen sulfide, ammonia, methane, and hydrogen concentrations following methods described in Butterfield et al. (2004). The oxygen concentration was measured using a Seabird (Bellevue, WA, USA) 63 Optical oxygen sensor, and the pH was measured using an AMT (Rostock, Germany) deep-sea glass pH electrode, both plumbed to the HFPS. For total cell counts, fluids collected via the HFPS were preserved in 3–4% paraformaldehyde, stained with acridine orange, and counted via epifluorescent microscopy. For virus-like particle counts, fluids collected via the LVWS were preserved in 1% formalin, stained with 2.5x SYBR Gold (Invitrogen, Grand Island, NY, USA), and counted via epifluorescent microscopy (Table 1).
Table 1. Comparison of Marker 113 and seawater chemistry characteristics.
| Fluid Characteristic | Marker 113 | Seawater |
|---|---|---|
| Temperature, °C | 24.1 | 2.4 |
| Percent seawater | 96 | 100 |
| Magnesium, mmol kg−1 | 49.1 | 51.5 |
| Total CO2, mmol kg−1 | 4.6 | 2.3 |
| Hydrogen sulfide, mmol l−1 | 0.6 | 0.0 |
| Ammonia, μmol l−1 | 3.5 | <0.6 |
| Silica, μmol l−1 | 636 | 155 |
| pH | 6.2 | 7.8 |
| Hydrogen, μmol kg−1 | <1.0 | 0.0 |
| Methane, μmol kg−1 | 13.2 | 0.0 |
| Oxygen, μmol l−1 | 7.0 | 28.3 |
| Microbial cells per ml | 4.4 × 105 | 1.8 × 104 |
| Virus-like particles per ml | 1.6 × 108 | ND |
Abbreviation: ND, no data.
Percent seawater was calculated via the conservative tracer magnesium (Mg): [Mg]vent/[Mg]seawater × 100. Most seawater data are reported from Akerman et al. (2013). Seawater magnesium and total CO2 were measured from a 2013 sample, whereas oxygen data were reported from a 2015 CTD cast at 1520 m.
RNA-SIP experimental set-up
For HFPS samples, fluid was pumped using a peristaltic pump from the Tedlar bag to six evacuated 500 ml Pyrex bottles, each filled nearly to capacity (530 ml). For LVWS samples, fluid was pumped from the carboy into each of four evacuated 1 liter Pyrex bottles to nearly full capacity (1060 ml). Before filling, 13C-labeled sodium bicarbonate (Cambridge Isotope Laboratories, Tewksbury, MA, USA) or 12C sodium bicarbonate (Sigma, St Louis, MO, USA) was added to each bottle for a final concentration of 10 mM additional bicarbonate. During filling, steps were taken to minimize introduction of oxygen and outgassing of fluid. After filling, 1–2.5 ml of 10% HCl was added to ensure a pH similar to vent conditions (pH <6.5). Pure H2 was added by syringe to each bottle: 20 ml (900 μmol) to each HFPS bottle and 60 ml to each LVWS bottle. 13C- and 12C-bicarbonate amended HFPS bottles were each incubated at 30 °C, 55 °C, and 80 °C for 36 h. A pair of 13C- and 12C-bicarbonate amended LVWS bottles were each incubated at 80 °C, one set for 18 h; the other for 36 h. After incubation, the fluid in each bottle was filtered through 0.22 μm Sterivex filters (Millipore), preserved in RNALater (Ambion), and frozen at −80 °C.
Isopycnic centrifugation, fractionation and RNA-SIP library preparation
RNA from SIP experiments was extracted using the mirVana miRNA isolation kit (Ambion) with an added bead-beating step using RNA PowerSoil beads (MoBio, Carlsbad, CA, USA). A total volume of 100 μl was extracted and was then DNase treated using the Turbo-DNase kit (Ambion). Gradient preparation, isopycnic centrifugation, and gradient fractionation were performed as described in Lueders (2010). For each gradient sample, 5.1 ml of CsTFA (~2 g ml−1, GE Healthcare Life Sciences, Piscataway, NJ, USA), 185 μl formamide, and a mixture of 750 ng RNA and gradient buffer solution (0.1 m Tris-HCl, 0.1 m KCl, 0.1 mM EDTA) up to 1 ml were first mixed in a 15 ml tube. Once mixed, the refractive index was measured for each sample to ensure a median density of ~1.80 g ml−1. Samples were then loaded into 4.9 ml OptiSeal tubes (Beckman Coulter, Brea, CA, USA), placed into a VTi 65.2 vertical rotor (Beckman Coulter) and spun at 37 000 r.p.m. at 20 °C for 64 h using an Optima L-80 XP ultracentrifuge (Beckman Coulter). Each gradient was fractionated into 12 tubes of approximately 410 μl each and the refractory index of each fraction was measured to determine density. RNA was precipitated with isopropanol and the pellet was washed with 70% ethanol as described in Lueders (2010). RNA concentration of each fraction was determined using the RiboGreen quantification kit (Invitrogen) and a Gemini XPS plate reader (Molecular Devices, Sunnyvale, CA, USA).
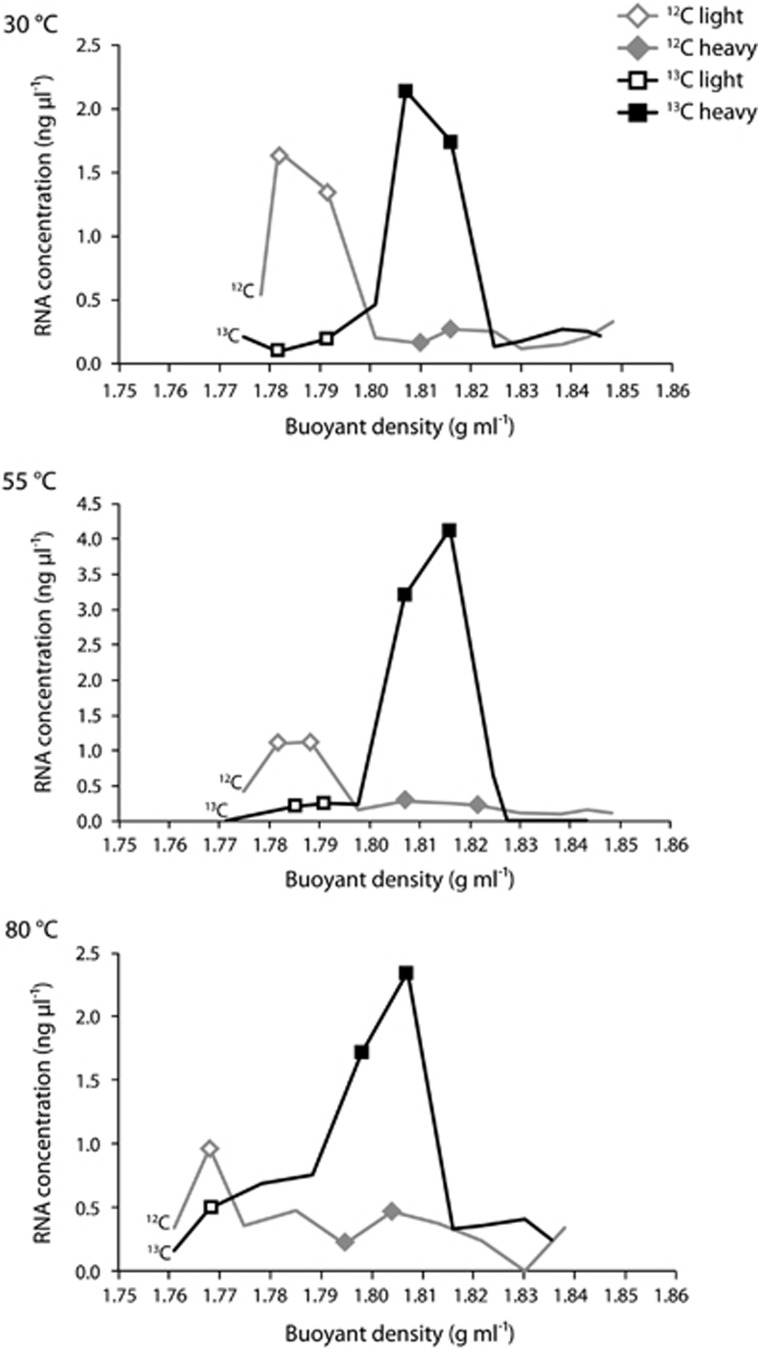
Figure 1 depicts the density fractions chosen for sequencing. At each temperature, four metatranscriptomic libraries were constructed, two from the 12C-control, named 12C-light and 12C-heavy, and two from the 13C-experiment, named 13C-light and 13C-heavy. The two density fractions with the highest RNA concentration in the 12C-control incubation were combined to construct the 12C-light library, which represents the natural community. Similarly, two density fractions with the highest RNA concentration in the 13C-experiment incubation were combined to construct the 13C-heavy library, which represents the labeled autotrophic community. The other two libraries from each temperature, 12C-heavy and 13C-light, were constructed to act as additional controls. To construct the 12C-heavy library, density fractions were chosen that corresponded to the same density fractions used to construct the 13C-heavy library. The 12C-heavy library represents the heavier density unlabeled community and acts as a key comparison with the 13C-heavy labeled community to confirm that the populations in the 13C-heavy library have taken up the labeled isotope and are not naturally found at heavier nucleic acid densities. To construct the 13C-light library, density fractions were chosen that corresponded to the same density fractions used to construct the 12C-light library. The 13C-light library represents the lighter density community in the 13C-experiment, which includes both labeled and unlabeled populations that are found at lighter densities. For all libraries, double-stranded complementary DNA was constructed and metatranscriptomic library preparation and analysis was carried out as described in the Supplementary methods. Raw read data are available through the European Nucleotide Archive for all 20 RNA-SIP metatranscriptomes, under sample accession numbers ERS743595 through ERS743614.
Figure 1.
Fractionation plots of RNA-SIP experiments for each of the three temperatures. Plots depict the RNA concentration (ng μl−1) vs buoyant density (g ml−1) for each fraction. RNA from fractions denoted by a symbol was used for metatranscriptomic library preparation and sequencing.
Metagenomic and metatranscriptomic library preparation and analysis
The 47 mm flat filters were first cut in half with a sterile razor, with half used for DNA and half used for RNA extraction. RNA and DNA were extracted and prepared for sequencing as described in the Supplementary methods. For all metagenomic and metatranscriptomic libraries, paired-end partially overlapping reads were merged and quality filtered using custom Illumina utility scripts (https://github.com/meren/illumina-utils). Merged reads were assembled using CLC Genomics Workbench (v 7.0) and default settings. Assembled contigs from each library were submitted to the DOE Joint Genome Institute's Integrated Microbial Genome Expert Review (IMG/ER). A detailed description of open reading frame mapping, rRNA identification, and statistical analyses can be found in the Supplementary methods. Raw metagenomic and metatranscriptomic read data are available through the European Nucleotide Archive under sample accession numbers ERS614323 and ERS614324 for the Marker 113 metagenome and metatranscriptome, respectively.
Results
At Marker 113, the average temperature of the diffuse fluids collected at 2 cm above the seafloor was 24 °C. Gas and chemical analysis of the diffuse fluid showed H2 concentrations were below the limit of detection (<1 μmol kg−1), whereas the CH4 concentration (13.2 μmol kg−1) equaled or exceeded CH4 concentrations measured elsewhere at Axial (Butterfield et al., 2004; Ver Eecke et al., 2012). The concentration of O2 in the fluid was 7 μM (Table 1). The natural DIC concentration measured at Marker 113 was 4.6 mmol kg−1. We extrapolated the percent seawater of the diffuse fluid using the conservative tracer magnesium (Mg), where percent seawater was calculated as [Mg]vent/[Mg]seawater × 100 (Table 1). The percent seawater of the diffuse fluid at Marker 113 was ~96% seawater, meaning it contained ~4% hydrothermal fluid. From these measurements, we estimated the amount of DIC in the end-member fluid (100% hydrothermal fluid) to ~60 mmol kg−1. The concentrations of prokaryotic cells and virus-like particles were 4.4 × 105 ml−1 and 1.6 × 108 ml−1, respectively (Table 1).
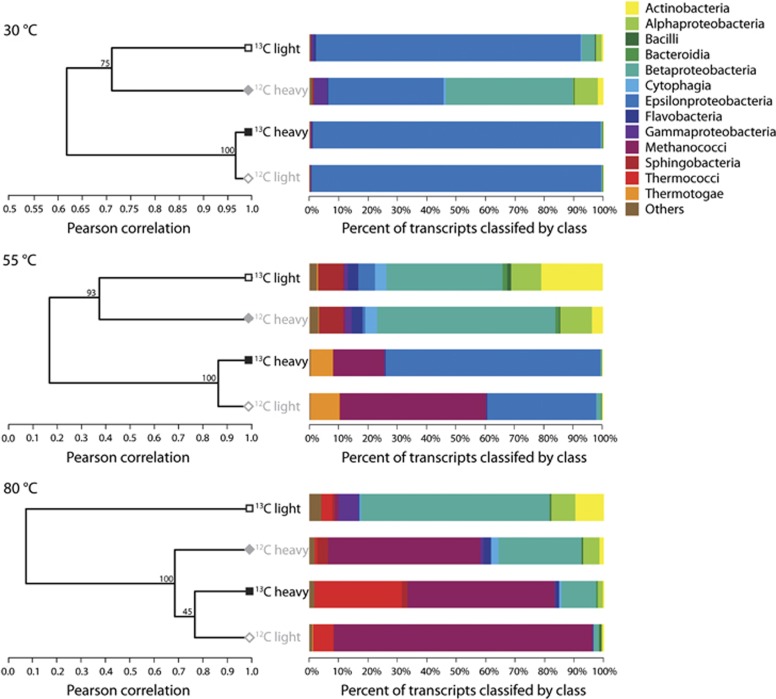
For the RNA-SIP experiments, there appeared to be uptake of the 13C-labeled bicarbonate at all three incubation temperatures, resulting in a higher peak density of the community RNA in the 13C-experiment compared with the 12C-control (Figure 1). On average, the peak RNA density for the controls was 1.78 (±0.009), whereas the average peak RNA density for the 13C experiments was 1.81 (±0.007). Of the three 80 °C SIP experiments, only the 18-h LVWS experiment was used for analysis. This is the 80 °C experiment shown in Figure 1 and throughout the article. The number of sequencing reads and percent total rRNA for each experiment is shown in Supplementary Table S1. Hierarchical clustering of the four libraries for each temperature based on the relative abundance of transcripts annotated to the KEGG Ontology (KO) database showed that 12C-light and 13C-heavy libraries clustered together, separating from the 12C-heavy and 13C-light libraries at all three temperatures. This indicates the functional similarities between the two communities (Figure 2). In addition, 12C-heavy libraries clustered separately from the 13C-heavy libraries at all temperatures, demonstrating the 13C-heavy libraries were functionally different from the communities at the same densities in the 12C-heavy controls (Figure 2). In the 55 °C experiment, the 12C-light and 13C-heavy communities consisted of Epsilonproteobacteria and Methanococci, whereas the 12C-heavy and 13C-light libraries consisted mainly of Actinobacteria, Alphaproteobacteria, and Betaproteobacteria. At 30 °C, Epsilonproteobacteria were present in all four libraries, but in highest abundance in the 12C-light and 13C-heavy libraries at 99% and 98.6% of the community, respectively. At 80 °C, the 13C-light library was taxonomically very different from the other three fractions, with a high percentage of Betaproteobacteria, as well as the occurrence of Actinobacteria, and Alpha- and Gammaproteobacteria. In contrast, Methanococci were dominant in the other three 80 °C fractions. Although Methanococci occurred in both the 12C-light and 13C-heavy libraries, this group was also in the 12C-heavy library. However, based on annotated transcripts there was still separation between the 12C-heavy and 13C-heavy communities, which indicated that the 13C-heavy library was representative of the labeled autotrophic community. In addition, the presence of transcripts attributed to the class Thermococci, a known class of heterotrophic species, in both the 12C-light and 13C-heavy libraries indicated the possibility of nonspecific labeling in the experiment. Finally, Betaproteobacteria were present in annotated transcripts for SIP metatranscriptomic libraries across all three temperatures (Figure 2). These transcripts were taxonomically classified and represented by various heterotrophic genera including Burkholderia, Ralstonia, and Curvibacter. In all SIP metatranscriptomes, Betaproteobacteria comprised a small percentage, between 0.0003% and 0.2%, of 16S rRNA sequences yet they accounted for a high proportion of the annotated transcripts in a few RNA-SIP metatranscriptomes.
Figure 2.
Hierarchical clustering of the four SIP library fractions for each temperature, based on the relative abundance of KO annotated transcripts. The bar graphs display the taxonomy of annotated transcripts at the class level.
In the RNA-SIP experiments, the majority of RNA was found in only a few fractions across the density gradient, with some fractions having no RNA (Figure 1). This suggests that the majority of the community took up the 13C-labeled bicarbonate, which is not entirely surprising given vent systems are dominated by autotrophs and hydrogen amendment pushed these experiments toward autotrophy (Orcutt et al., 2011). Owing to the presence of RNA in only a few fractions, taxonomic analysis of all density fractions was not possible and thus we were not able to completely verify separation of 12C and 13C-labeled RNA in our experiments. Consequently, the results of the RNA-SIP experiments more resemble enrichment experiments when compared with SIP studies of soil, sediment, and marine surface waters (for example, Lueders et al., 2004; Neufeld et al., 2008; Dumont et al., 2013) and should be treated as such.
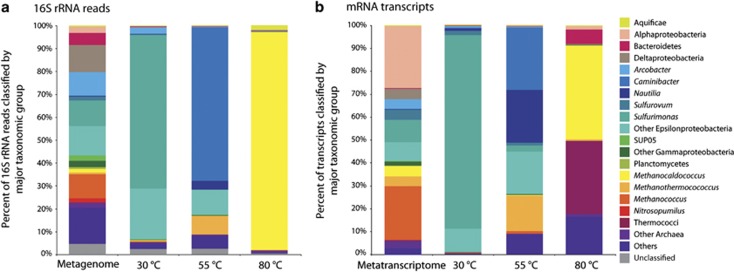
Comparison of the 16S rRNA community composition between the Marker 113 metagenome and the 13C-heavy metatranscriptomes showed that while the un-manipulated diffuse fluid community was highly diverse, a different taxonomic group became dominant under hydrogen amended conditions at each incubation temperature (Figure 3a). In the Marker 113 metagenome, there was a mix of both known vent and seawater communities (Figure 3a). The most dominant bacterial groups included Delta-, Epsilon-, and Gammaproteobacteria, which made up 12%, 35%, and 6.5% of the community, respectively. Archaeal 16S rRNA gene sequences comprised 17% of all 16S rRNA gene sequences in the metagenome, with 9% of all 16S rRNA gene sequences classifying to the genus Methanococcus (Figure 3a). The 16S rRNA of the SIP metatranscriptomes displayed much less diversity. At 30 °C, the autotrophic community consisted mostly of Sulfurimonas, which are known mesophilic Epsilonproteobacteria at vent systems, and made up 66% of the 16S rRNA sequences (Figure 3a). At 55 °C, Epsilonproteobacteria were again dominant, with the thermophilic hydrogen-oxidizing genera Caminibacter being the most prevalent. In addition, 9% of 55 °C 16S rRNA sequences were classified to the thermophilic methanogen Methanothermococcus. At 80 °C, methanogens, specifically Methanocaldococcus species, comprised 95% of the 16S rRNA sequences (Figure 3a).
Figure 3.
16S rRNA gene (a) and annotated transcript taxonomy (b), classified by major taxonomic group for the 13C-heavy metatranscriptome at each of the three incubation temperatures. Included for comparison is the Marker 113 metagenome and metatranscriptome.
The taxonomic classification of the transcripts from the Marker 113 metatranscriptome and the three 13C-heavy metatranscriptomes was similar to the 16S rRNA community data, but also highlighted distinct differences. Transcripts associated with Alphaproteobacteria, Epsilonproteobacteria, and the mesophilic methanogenic genus Methanococcus comprised the majority of the Marker 113 metatranscriptome (Figure 3b). At 30 °C, the taxonomic composition of the transcripts was similar to the 16S rRNA, with the dominance of Sulfurimonas (Figure 3b). Some differences between 16S rRNA and transcript community composition were seen at both 55 °C and 80 °C. In addition to Caminibacter, Nautilia classified transcripts were prevalent at 55 °C, as were transcripts classified as Methanothermococcus, a thermophilic methanogen (15% of all annotated transcripts, Figure 3b). At 80 °C, Methanocaldococcus transcripts accounted for 50% of annotated transcripts, with a majority of the rest classified as other archaeal groups including Thermococci (Figure 3b).
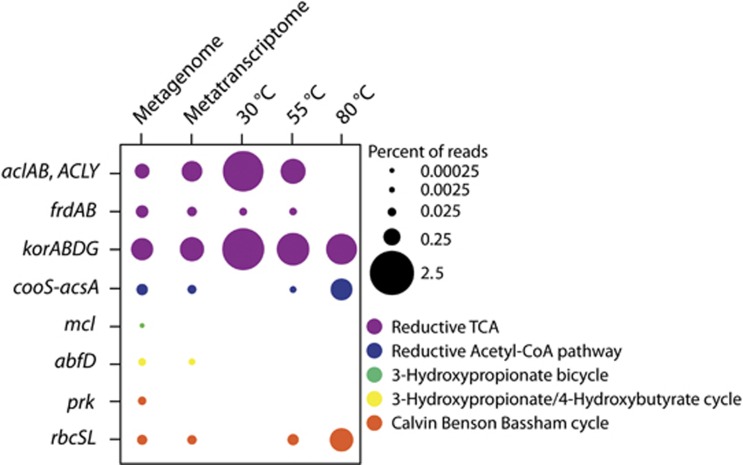
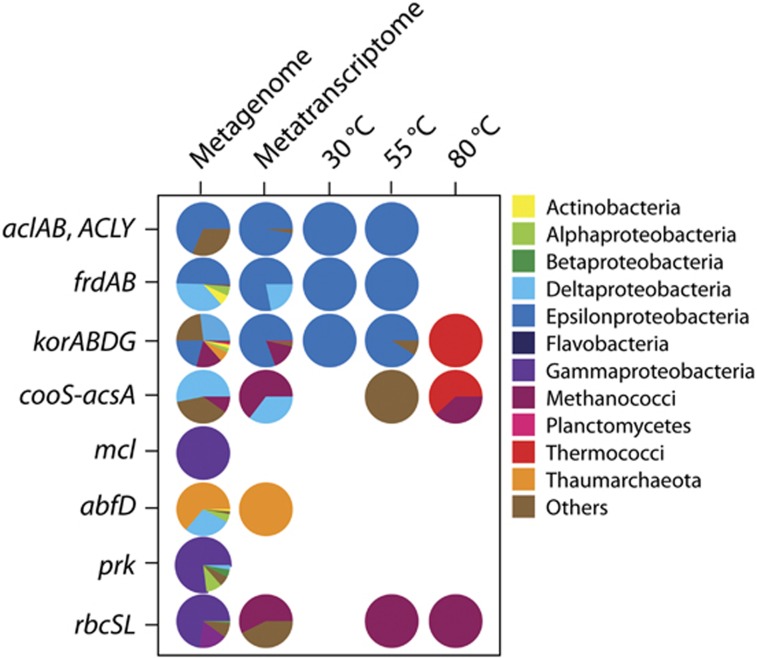
Genes were present for a number of carbon fixation pathways in the Marker 113 metagenome, metatranscriptome, and SIP metatranscriptomes. In the metagenome, genes were present for five carbon fixation pathways: the reductive TCA (rTCA) cycle, reductive acetyl-CoA pathway, 3-hydroxypropionate bicycle, 3-hydroxypropionate/4-hydroxybutyrate cycle, and the Calvin Benson Bassham (CBB) cycle. Transcripts were present for all pathways except the 3-hydroxypropionate bicycle (Figure 4). The rTCA cycle was the most abundant carbon fixation pathway in the Marker 113 metagenome, metatranscriptome, and the 30 °C and 55 °C SIP metatranscriptomes (Figure 4). Transcripts for rTCA cycle genes comprised over 4% of all annotated transcripts at 30 °C and were all taxonomically identified as Epsilonproteobacteria, specifically Sulfurimonas, Sulfurovum, and Sulfuricurvum species (Figures 4 and 5). Reductive TCA cycle transcripts comprised about 2% of total annotated transcripts at 55 °C and were mostly assigned to Caminibacter and Nautilia species (Figures 4 and 5). Transcripts for the reductive acetyl-CoA pathway, the pathway through which methanogens fix carbon, were observed at 80 °C, making up 1% of total annotated transcripts. In addition, transcripts for the rTCA cycle were present and comprised about 0.5% of all annotated transcripts in the 80 °C experiment (Figure 4). Transcripts from this experiment were attributed to methanogens and Thermococci (Figure 5). Rubisco gene transcripts (CBB cycle) were also present at 55 °C and 80 °C and assigned to methanogen genera, specifically Methanothermococcus and Methanocaldococcus (Figures 4 and 5).
Figure 4.
Percentage of reads annotated to genes for carbon fixation pathways for the Marker 113 metagenome, metatranscriptome and the 13C-heavy metatranscriptomes from each of the three incubation temperatures. acl, ATP-citrate lyase; frd, fumarate reductase; kor, 2-oxoglutarate ferredoxin oxidoreductase; cooS-acsA, carbon-monoxide dehydrogenase; mcl, malyl-CoA lyase; abfD, 4-hydroxybutyryl-CoA dehydratase; prk, phosphoribulokinase; rbc, ribulose-bisphosphate carboxylase.
Figure 5.
Taxonomy of reads annotated to genes for carbon fixation pathways, classified by class, for the Marker 113 metagenome, metatranscriptome, and the three 13C-heavy metatranscriptomes. Gene names are identical to those listed in Figure 4.
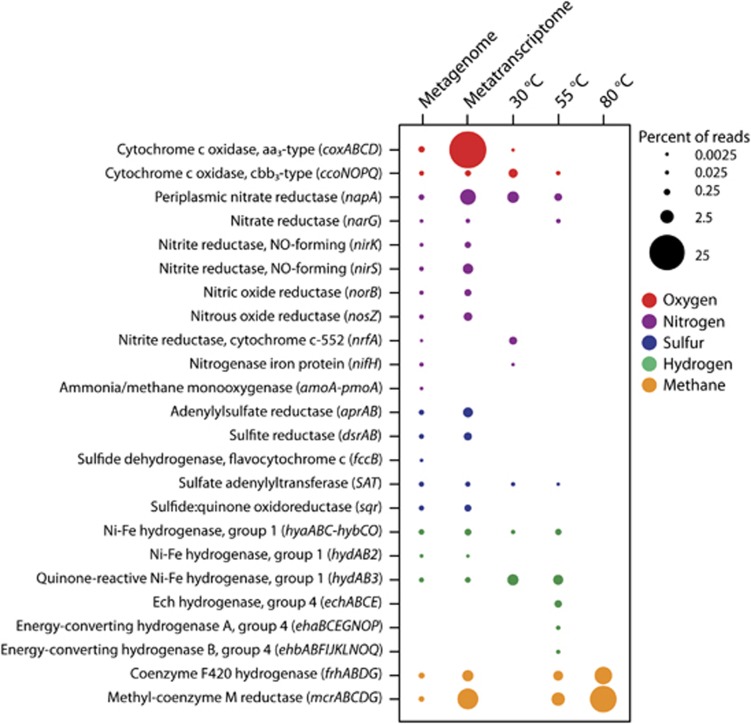
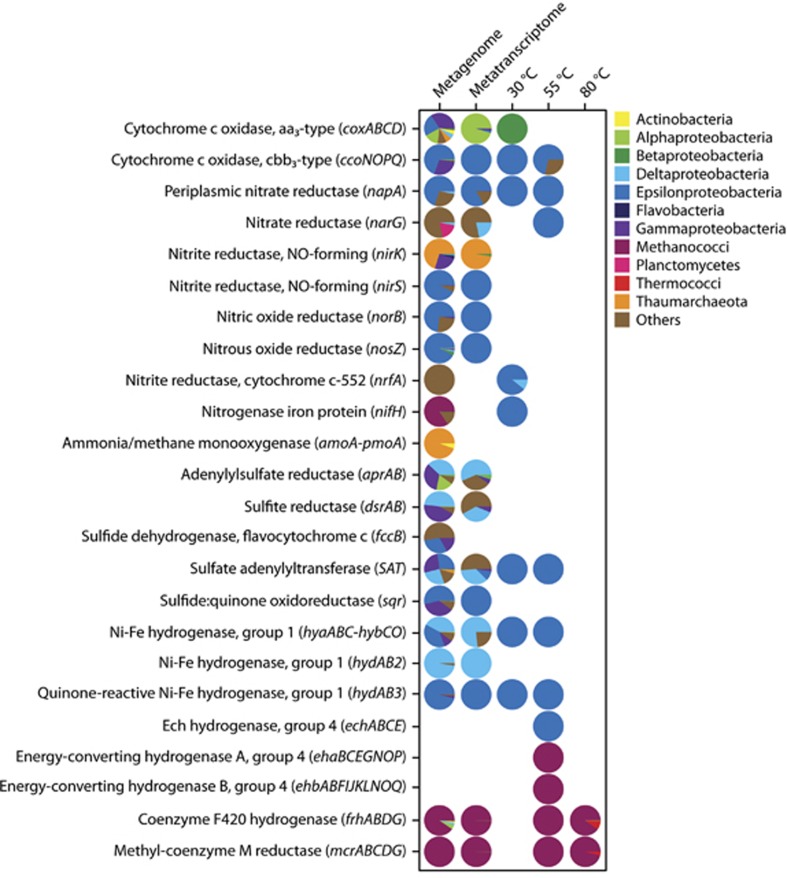
Genes were also present and expressed for many different metabolic pathways. The genes included in Figure 6 were chosen based on the inferred metabolisms of the groups seen in the 16S rRNA and metatranscriptomic sequences and focused on oxygen, nitrogen, sulfur, hydrogen, and methane metabolisms. In the metagenome, genes attributed to oxygen respiration, nitrate reduction, denitrification, ammonium oxidation, nitrogen fixation, sulfur oxidation and reduction, hydrogen oxidation, and methanogenesis were all present (Figure 6). Expression was highest for the cytochrome c oxidase aa3-type gene complex (cox), which comprised over 25% of all annotated transcripts in the metatranscriptome (Figure 6). Transcripts for the coxABCD genes were classified mostly as Alpha-, Gamma-, and Epsilonproteobacteria (Figure 7). Other genes that were highly expressed in the metatranscriptome were napA, a periplasmic nitrate reductase, and genes involved in methanogenesis (Figure 6). Transcripts for methanogenesis genes made up almost 10% of all annotated transcripts, with the majority (58%) attributed to the mesophilic Methanococcus genus (Figure 7). Along with napA, other denitrification genes were also expressed, including genes for nitrite, nitric oxide, and nitrous oxide reductases, with 85% of denitrification gene transcripts classified as Epsilonproteobacteria (Figures 6 and 7). For sulfur metabolism, transcripts for sulfate reduction/oxidation genes dsrAB, aprAB, and SAT were classified as Gamma- and Deltaproteobacteria, as well as the archaeal genera Archaeoglobus and Caldivirga (Figure 7). The sulfur metabolism transcripts classified as Epsilonproteobacteria were for the genes SAT and sqr (Figure 7).
Figure 6.
Percentage of reads annotated to genes for key oxygen, nitrogen, sulfur, hydrogen, and methane metabolism genes for the Marker 113 metagenome, metatranscriptome, and the 13C-heavy metatranscriptomes from each of the three incubation temperatures.
Figure 7.
Taxonomy of reads annotated to genes for oxygen, nitrogen, sulfur, hydrogen, and methane metabolism genes, classified by class, for the Marker 113 metagenome, metatranscriptome, and the 13C-heavy metatranscriptomes from each of the three incubation temperatures.
At 30 °C, the SIP metatranscriptome revealed a transition in the community from the use of oxygen to the use of nitrate as a terminal electron acceptor. Transcripts were present for both cytochrome c oxidase genes as well as for the nitrate reductase gene napA (Figure 6). Of the two cytochrome c oxidase gene complexes present, the high-affinity cbb3 type, ccoNOPQ, was more highly expressed. Hydrogen oxidation was the main autotrophic metabolism, as hydrogenase genes hya, hyb, and hyd were all expressed (Figure 6). Transcripts for hydrogen oxidation genes, napA, as well as for the cbb3-type cytochrome c oxidase gene complex ccoNOPQ were all classified as Epsilonproteobacteria, specifically as Sulfurimonas, Sulfurovum, Sulfuricurvum, and Arcobactor species (Figure 7). At 55 °C, there was still some evidence of oxygen respiration, with limited expression of the cbb3-type cytochrome c oxidase gene, but nitrate reduction transcripts for napA and narG genes were more abundant (Figure 6). Hydrogen oxidation and methanogenesis were the main autotrophic metabolisms. Hydrogenase genes, hya, hyb, hyd, and ech were expressed and mostly classified as known thermophilic hydrogen oxidizers Caminibacter and Nautilia (Figures 6 and 7). Methanogenesis genes were also expressed at 55 °C and classified as the thermophilic methanogen Methanothermococcus (Figures 6 and 7). Methanogenesis was the dominant metabolism at 80 °C, with 18.5% of all annotated transcripts attributed to the mcr and frh gene complexes (Figure 6). Nearly 95% of transcripts annotated to genes for methanogenesis were classified as the hyperthermophile Methanocaldococcus (Figure 7).
Discussion
The microbial communities in low-temperature diffuse hydrothermal fluids are metabolically and taxonomically complex because of the mixing of deep seawater and subseafloor vent communities. This mixing can make it difficult to tease apart the vent, seafloor, and subseafloor signatures from those of deep-water microorganisms growing in background seawater near the seafloor (Akerman et al., 2013). In this work, we set out to characterize the hydrogenotrophic chemolithoautotrophic community present in vent fluids using both meta-omics and RNA-SIP. Through metagenomics and metatranscriptomics, we first determined the taxonomic diversity and range of metabolic processes in un-manipulated diffuse fluids. We then used RNA-SIP metatranscriptomics in hydrogen-enriched incubations to specifically target the subseafloor chemolithoautotrophic community and to identify the key taxa and metabolisms occurring across different temperatures representative of the warm, reducing subseafloor habitat. Analysis of un-manipulated fluids showed a phylogenetically and metabolically diverse group of bacteria and archaea from both background seawater and vent environments including members of the subseafloor community. However, under experimental conditions at elevated temperatures, hydrogen amendment, and reducing conditions, diversity was greatly reduced. Still, distinct genera and metabolisms dominated the subseafloor autotrophic community when comparing different temperature regimes.
In the shipboard experimental treatment of vent fluids, there were a number factors that needed to be addressed, including the introduction of oxygen, outgassing of methane and hydrogen, pH changes, limited access to samples, and timing of incubations. Hydrogen concentrations at Marker 113 are low, yet previous work has shown that subseafloor hyperthermophilic methanogenic archaea are abundant there and require a minimum hydrogen concentration of ~17–23 μM for growth (Ver Eecke et al. 2012). Given these known constraints, hydrogen was added to the SIP experiments at this level in order to mimic likely energy-rich, reducing, and warm subseafloor conditions. When hydrogen was not added, there was no label uptake at any temperature (30 °C, 55 °C and 80 °C) and little RNA was recovered (data not shown).
The length of incubations is also a challenge to SIP experiments, as it is important to minimize cross-feeding, the uptake of the labeled isotope by a non-target community. The incubation time of the experiments was estimated from previous work with cultured representatives from mesophilic, thermophilic, and hyperthermophilic subseafloor communities (Mehta and Baross, 2006; Ver Eecke et al., 2012; Meyer and Huber, 2014). In our 80 °C experiments, the 36-h incubations resulted in almost all transcripts assigned to heterotrophic bacteria, indicating the consumption of labeled organic matter and an incubation time that was too long to capture the autotrophic community (Supplementary Figure S1). Cross-feeding also appeared to occur, but to a lesser extent, in the 80 °C 18-h incubation, with about 30% of annotated transcripts assigned to the heterotrophic phylum Thermococci, and specifically to the genera Thermococcus, Palaeococcus, and Pyrococcus. A possible syntrophic relationship between hyperthermophilic heterotrophs like Thermococcus and methanogens has been previously observed (Ver Eecke et al., 2012) and may explain the labeling of this heterotrophic community even in our shorter 18-h incubation. The length of our experiments plus amendment with hydrogen resulted in RNA being concentrated in only a few density fractions (Figure 1) and thus we were unable to taxonomically analyze all fractions to verify separation of 12C and 13C-labeled RNA. Our RNA-SIP experiments instead resemble experimental enrichments with a labeled substrate, in this case 13C-labeled bicarbonate. In density fractions with low RNA concentrations, mainly in the 12C-heavy and 13C-light controls, we found a high abundance of transcripts for a common laboratory contaminant. Betaproteobacteria comprised a large percentage of the annotated transcripts from many of the 12C-heavy and 13C-light metranscriptomes, regardless of temperature. This group has recently been shown to be present as a common lab contaminant in extraction kit reagents and is abundant in sequence data of samples with low microbial biomass (Salter et al., 2014), consistent with our findings.
Given the challenges of carrying out RNA-SIP under simulated subseafloor conditions, it was important to compare our experimental treatments to the Marker 113 metagenome and metatranscriptome of un-manipulated fluids, which represent the potential and active metabolic processes occurring within mixed seawater-vent fluids as they exit the seafloor. The Marker 113 metagenome was comprised of genes for a diverse array of aerobic and anaerobic metabolic processes including sulfur oxidation and reduction, hydrogen oxidation, denitrification, and methanogenesis (Figure 6). Transcripts for all of these processes were also observed in the Marker 113 metatranscriptome. In addition, genes for many different carbon fixation pathways were observed and expressed, again showing the metabolic flexibility of vent communities (Figure 4).
Epsilonproteobacteria comprised over one-quarter of the 16S rRNA gene sequences in the metagenome and 8% of annotated transcripts in the metatranscriptome. Epsilonproteobacteria are an abundant and metabolically versatile group in many habitats at deep-sea hydrothermal vents, including diffuse vent fluids (Huber et al., 2003; Takai et al., 2003; Campbell et al., 2006). The metabolic flexibility of the Epsilonproteobacteria was observed in both the Marker 113 metagenome and metatranscriptome, with genes and transcripts for both hydrogen and sulfur oxidation present. The measured oxygen at Marker 113 was just below 7 μM, indicating a low oxygen environment at the point of venting and the potential for use of oxygen or nitrate as terminal electron acceptors. Epsilonproteobacteria appeared to take advantage of these conditions, as both cytochrome c oxidase and denitrification genes that classified as Epsilonproteobacteria were present and expressed. In fact, in the Marker 113 metatranscriptome, all transcripts for the cbb3-type cytochrome c oxidase, along with 85% of all denitrification gene transcripts, were classified as Epsilonproteobacteria, demonstrating the genomic plasticity of this key phylum and the ability of these bacteria to flourish across redox gradients at hydrothermal vents and within the underlying seafloor.
Methanogenic archaea are also important autotrophs at Marker 113. Long-term chemistry measurements at Marker 113 have shown anomalously high methane concentrations in the diffuse vent fluids, which suggest a biologically derived source and prominence of methanogenesis at this site (Ver Eecke et al., 2012). Previous work using quantitative PCR and sequencing of the mcrA gene for methanogenesis have shown the presence of different methanogen genera at a wide range of temperatures at Marker 113 (Ver Eecke et al., 2012). In this study, we observed the expression of genes associated with mesophilic, thermophilic, and hyperthermophilic methanogens including Methanococcus, Methanothermococcus, and Methanocaldococcus genera, respectively. In the Marker 113 metatranscriptome, 66% of archaeal transcripts and nearly half the transcripts for the mcr gene complex (mcrABG) for methanogenesis were classified as the mesophilic group Methanococcus. Methanogens are thus present and active across a wide temperature range and are important contributors to deep-sea primary production at this site.
In response to the hydrogen enrichment of diffuse vent fluids, the main autotrophic metabolism in the 30 °C and 55 °C SIP experiments was hydrogen oxidation coupled with use of oxygen or nitrate as a terminal electron acceptor. Molecular hydrogen is a key energy source for hydrothermal plume, diffuse fluid, subseafloor, and vent symbiont communities (Petersen et al., 2011; Wankel et al., 2011; Anantharaman et al., 2013). Within the 55 °C experiment, hydrogenase gene transcripts were classified as Caminibacter and Nautilia, both known thermophilic, hydrogen oxidizing, nitrate (or sulfur) reducing Epsilonproteobacteria genera (Meyer and Huber, 2014). At 30 °C, hydrogenase gene transcripts were classified mostly as Sulfurimonas and Sulfuricurvum, specifically as Sulfurimonas sp. GD1, Sulfurimonas denitrificans and Sulfuricurvum kujiense, all of which possess the capability to oxidize hydrogen (or sulfide) using either nitrate or oxygen. Although there are many mesophilic Epsilonproteobacteria that use both oxygen and nitrate, there is currently only one vent-associated cultured representative from the Sulfurimonas genus that has been shown to both oxidize hydrogen and reduce nitrate, Sulfurimonas paralvinellae, isolated from a vent at the Mid-Okinawa Trough (Takai et al., 2006). Sulfurimonas paralvinellae is a facultative anaerobe with optimal growth observed with nitrate as the sole electron acceptor (Takai et al., 2006). The results seen in the 30 °C SIP metatranscriptome suggest a similar metabolism for subseafloor Sulfurimonas species at Axial Seamount.
Likely, the little oxygen present in the fluids was consumed during the 36-h enrichment with hydrogen in all incubations. Accordingly, the transition between aerobic and anaerobic metabolisms was seen in the 30 °C, and to a lesser extent in the 55 °C SIP experiments. In both the 30 °C and 55 °C, the cbb3-type cytochrome c oxidase gene was the cytochrome oxidase gene most highly expressed. The cbb3-type cytochrome c oxidase is present in bacteria only and has been shown to be a high-affinity cytochrome c oxidase, with the highest activity under low oxygen conditions (Ekici et al., 2012). The depletion of oxygen probably induced the expression of genes for nitrate reduction at 30 °C and 55 °C by Epsilonproteobacteria. Transcripts for nitrate reduction via the napA gene comprised about 2% of the total annotated transcripts in the 30 °C experiment and 0.5% in the 55 °C experiment. Nitrate reduction via the nap operon has been shown to be highly conserved and widespread throughout the Epsilonproteobacteria (Vetriani et al., 2014). NapA has been shown to be a high-affinity nitrate reductase and only expressed during low nitrate conditions (Potter et al., 1999), which may explain the prevalence of this gene within the Epsilonproteobacteria, as hydrothermal fluid is depleted in nitrate compared with deep seawater (Vetriani et al., 2014).
At 30 °C, other mesophilic autotrophic groups, specifically SUP05, were not detected. However, we did not add sulfide to the enrichments, which may have been limiting further growth of sulfide-oxidizing autotrophs. SUP05 is a key sulfur and hydrogen oxidizer in the deep sea, oxygen minimum zones, and hydrothermal plumes (Walsh et al., 2009; Stewart et al., 2012; Anantharaman et al., 2013) and is present and active at Axial Seamount and specifically at Marker 113 (Akerman et al., 2013; Anderson et al., 2013). Sequences identified to the SUP05 group were present in low concentrations in both the metagenome and metatranscriptome but were absent in the 30 °C SIP metatranscriptome. Given their ubiquity in the deep ocean and hydrothermal plumes where temperatures rarely exceed 5 °C, SUP05 populations likely have a lower growth optima temperature than 30 °C and thus are not expected to be present in our SIP experiments. In addition, another explanation could be the possible lack of nitrate reduction genes in SUP05 populations. A recent genomic study of two SUP05 isolates from a hydrothermal plume revealed they did not possess genes for nitrate reduction (Anantharaman et al., 2013). In our SIP experiment, the main metabolic process occurring at 30 °C was hydrogen oxidation using oxygen or nitrate. This lack of nitrate reduction genes could also explain why SUP05 was not present in our mesophilic SIP experiment.
In the Marker 113 metatranscriptome, most transcripts identified as methanogenic archaea were classified as the putatively mesophilic Methanococcus genus, but in our SIP experiments, methanogenesis at higher temperatures was observed. Methanogenesis was the only autotrophic metabolism observed at 80 °C, with nearly 20% of transcripts annotated to methanogenesis genes and all were classified as the hyperthermophilic genus, Methanocaldococcus. At 55 °C, along with the thermophilic Epsilonproteobacteria, Methanothermococcus, a thermophilic methanogen, was present, making up 16% of the community. Although carbon fixation gene transcripts for the reductive acetyl-CoA pathway were observed at 55 °C and 80 °C, transcripts for Rubisco genes were also present and were all classified as Methanothermococcus and Methanocaldococcus. These methanogens are not using Rubisco for carbon fixation as they lack other important enzymes in the CBB cycle, namely phosphoribulokinase, but they have been shown to actively synthesize the enzyme (Finn and Tabita, 2004). The function of Rubsico in methanogens is not fully understood, but it could have a role in purine recycling (Finn and Tabita, 2004). These experiments show that at Marker 113, thermophilic and hyperthemophilic subseafloor methanogens are active in subseafloor carbon cycling.
In conclusion, metagenomic and metatranscriptomic analyses revealed the high taxonomic diversity and extensive metabolic potential of microbial communities found in venting fluids at hydrothermal vents. RNA-SIP experiments under simulated subseafloor conditions revealed that different phylogenetic groups and metabolisms dominated at different temperature regimes. Future experiments at geochemically diverse sites will help to further determine the extent of taxonomic and metabolic diversity across hydrothermal vents and the key autotrophic metabolisms that occur under different redox and geothermal conditions.
Acknowledgments
This work was funded by the Gordon and Betty Moore Foundation Grant GBMF3297 and NSF Center for Dark Energy Biosphere Investigations (C-DEBI) (OCE-0939564). The data collected in this study is based upon work supported by the Schmidt Ocean Institute during cruise FK010-2013 aboard R/V Falkor. We thank the captain and crew of the R/V Falkor, as well as the entire ROV ROPOS group. Chris Algar, Dave Butterfield, Ben Larson, Jim Holden, Giora Proskurowski, Lisa Zeigler Allen, Kevin Roe, Lucy Stewart, and Begum Topcuoglu provided critical support in sample collection and experimental design and execution. We would also like to thank the ISME J editor and reviewers for the detailed and helpful comments on the manuscript. This is C-DEBI contribution number 267.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Akerman NH, Butterfield DA, Huber JA. (2013). Phylogenetic diversity and functional gene patterns of sulfur-oxidizing subseafloor Epsilonproteobacteria in diffuse hydrothermal vent fluids. Front Microbiol 4: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amend JP, McCollom TM, Hentscher M, Bach W. (2011). Catabolic and anabolic energy for chemolithoautotrophs in deep-sea hydrothermal systems hosted in different rock types. Geochim Cosmochim Acta 75: 5736–5748. [Google Scholar]
- Anantharaman K, Breier JA, Sheik CS, Dick GJ. (2013). Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc Natl Acad Sci USA 110: 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RE, Beltran MT, Hallam SJ, Baross JA. (2013). Microbial community structure across fluid gradients in the Juan de Fuca Ridge hydrothermal system. FEMS Microbiol Ecol 83: 324–339. [DOI] [PubMed] [Google Scholar]
- Anderson RE, Sogin ML, Baross JA. (2014). Evolutionary strategies of viruses, bacteria and archaea in hydrothermal vent ecosystems revealed through metagenomics. PLoS One 9: e109696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Sheik CS, Taylor CA, Jain S, Bhasi A, Cavalcoli JD et al. (2013). Community transcriptomic assembly reveals microbes that contribute to deep-sea carbon and nitrogen cycling. ISME J 7: 1962–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard L, Mougel C, Maron P-A, Nowak V, Leveque J, Henault C et al. (2007). Dynamics and identification of soil microbial populations actively assimilating carbon from C-13-labelled wheat residue as estimated by DNA- and RNA-SIP techniques. Environ Microbiol 9: 752–764. [DOI] [PubMed] [Google Scholar]
- Bourbonnais A, Juniper SK, Butterfield DA, Devol AH, Kuypers MMM, Lavik G et al. (2012). Activity and abundance of denitrifying bacteria in the subsurface biosphere of diffuse hydrothermal vents of the Juan de Fuca Ridge. Biogeosciences 9: 4661–4678. [Google Scholar]
- Butterfield DA, Roe KK, Lilley MD, Huber JA, Baross JA, Embley RW et al. (2004) Mixing, reaction and microbial activity in the sub-seafloor revealed by temporal and spatial variation in diffuse flow vents at Axial volcano. The Subseafloor Biosphere at Mid-Ocean Ridges. American Geophysical Union: Washington, DC, USA, pp 269–289. [Google Scholar]
- Campbell BJ, Jeanthon C, Kostka JE, Luther GW, Cary SC. (2001). Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl Environ Microbiol 67: 4566–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Engel AS, Porter ML, Takai K. (2006). The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol 4: 458–468. [DOI] [PubMed] [Google Scholar]
- Campbell BJ, Smith JL, Hanson TE, Klotz MG, Stein LY, Lee CK et al. (2009). Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet 5: e1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick WW, Butterfield DA, Embley RW, Tunnicliffe V, Huber JA, Nooner SL et al. (2010). Axial Seamount. Oceanography 23: 38–39. [Google Scholar]
- Dick GJ, Anantharaman K, Baker BJ, Li M, Reed DC, Sheik CS. (2013). The microbiology of deep-sea hydrothermal vent plumes: ecological and biogeographic linkages to seafloor and water column habitats. Front Microbiol 4: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont MG, Murrell JC. (2005). Stable isotope probing - linking microbial identity to function. Nat Rev Microbiol 3: 499–504. [DOI] [PubMed] [Google Scholar]
- Dumont MG, Radajewski SM, Miguez CB, McDonald IR, Murrell JC. (2006). Identification of a complete methane monooxygenase operon from soil by combining stable isotope probing and metagenomic analysis. Environ Microbiol 8: 1240–1250. [DOI] [PubMed] [Google Scholar]
- Dumont MG, Pommerenke B, Casper P. (2013). Using stable isotope probing to obtain a targeted metatranscriptome of aerobic methanotrophs in lake sediment. Environ Microbiol Rep 5: 757–764. [DOI] [PubMed] [Google Scholar]
- Ekici S, Pawlik G, Lohmeyer E, Koch HG, Daldal F. (2012). Biogenesis of cbb(3)-type cytochrome c oxidase in Rhodobacter capsulatus. Biochimica Et Biophysica Acta-Bioenerget 1817: 898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn MW, Tabita FR. (2004). Modified pathway to synthesize ribulose 1,5-bisphosphate in methanogenic archaea. J Bacteriol 186: 6360–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz S, Labrenz M, Jost G, Jurgens K. (2010). Diversity of active chemolithoautotrophic prokaryotes in the sulfidic zone of a Black Sea pelagic redoxcline as determined by rRNA-based stable isotope probing. FEMS Microbiol Ecol 74: 32–41. [DOI] [PubMed] [Google Scholar]
- Huang WE, Ferguson A, Singer AC, Lawson K, Thompson IP, Kalin RM et al. (2009). Resolving genetic functions within microbial populations: in situ analyses using rRNA and mRNA stable isotope probing coupled with single-cell Raman-fluorescence in situ hybridization. Appl Environ Microbiol 75: 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JA, Butterfield DA, Baross JA. (2002). Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl Environ Microbiol 68: 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JA, Butterfield DA, Baross JA. (2003). Bacterial diversity in a subseafloor habitat following a deep-sea volcanic eruption. FEMS Microbiol Ecol 43: 393–409. [DOI] [PubMed] [Google Scholar]
- Huber JA, Mark Welch D, Morrison HG, Huse SM, Neal PR, Butterfield DA et al. (2007). Microbial population structures in the deep marine biosphere. Science 318: 97–100. [DOI] [PubMed] [Google Scholar]
- Huber JA, Cantin HV, Huse SM, Welch DBM, Sogin ML, Butterfield DA. (2010). Isolated communities of Epsilonproteobacteria in hydrothermal vent fluids of the Mariana Arc seamounts. FEMS Microbiol Ecol 73: 538–549. [DOI] [PubMed] [Google Scholar]
- Jannasch HW, Wirsen CO. (1981). Morphological survey of microbial mats near deep-sea thermal vents. Appl Environ Microbiol 41: 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannasch HW, Nelson DC, Wirsen CO. (1989). Massive natural occurrence of unusually large bacteria (Beggiatoa sp.) at a hydrothermal deep-sea vent site. Nature 342: 834–836. [Google Scholar]
- Karl DM, Wirsen CO, Jannasch HW. (1980). Deep-sea primary production a Galapagos hydrothermal vents. Science 207: 1345–1347. [Google Scholar]
- Lesniewski RA, Jain S, Anantharaman K, Schloss PD, Dick GJ. (2012). The metatranscriptome of a deep-sea hydrothermal plume is dominated by water column methanotrophs and lithotrophs. ISME J 6: 2257–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Baker BJ, Anantharaman K, Jain S, Breier JA, Dick GJ. (2015). Genomic and transcriptomic evidence for scavenging of diverse organic compounds by widespread deep-sea archaea. Nat Commun 6: 8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueders T, Wagner B, Claus P, Friedrich MW. (2004). Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ Microbiol 6: 60–72. [DOI] [PubMed] [Google Scholar]
- Lueders T. (2010) Stable isotope probing of hydrocarbon-degraders In: Timmis K (ed) Handbook of Hydrocarbon and Lipid Microbiology. Springer: Berlin Heidelberg, pp 4011–4026. [Google Scholar]
- McCollom TM. (2005). Energetic constraints on subsurface biomass production within the igneous ocean crust. Geochim Cosmochim Acta 69: A200–A200. [Google Scholar]
- Mehta MP, Baross JA. (2006). Nitrogen fixation at 92 degrees C by a hydrothermal vent archaeon. Science 314: 1783–1786. [DOI] [PubMed] [Google Scholar]
- Meyer JL, Akerman NH, Proskurowski G, Huber JA. (2013). Microbiological characterization of post-eruption ‘snowblower' vents at Axial Seamount Juan de Fuca Ridge. Front Microbiol 4: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JL, Huber JA. (2014). Strain-level genomic variation in natural populations of Lebetimonas from an erupting deep-sea volcano. ISME J 8: 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld JD, Dumont MG, Vohra J, Murrell JC. (2007). Methodological considerations for the use of stable isotope probing in microbial ecology. Microb Ecol 53: 435–442. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Chen Y, Dumont MG, Murrell JC. (2008). Marine methylotrophs revealed by stable-isotope probing, multiple displacement amplification and metagenomics. Environ Microbiol 10: 1526–1535. [DOI] [PubMed] [Google Scholar]
- Opatkiewicz AD, Butterfield DA, Baross JA. (2009). Individual hydrothermal vents at Axial Seamount harbor distinct subseafloor microbial communities. FEMS Microbiol Ecol 70: 413–424. [DOI] [PubMed] [Google Scholar]
- Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ. (2011). Microbial ecology of the Dark Ocean above, at, and below the Seafloor. Microbiol Mol Biol Rev 75: 361–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner M, Bach W, Hentscher M, Koschinsky A, Garbe-Schonberg D, Streit WR et al. (2009). Short-term microbial and physico-chemical variability in low-temperature hydrothermal fluids near 5 degrees S on the Mid-Atlantic Ridge. Environ Microbiol 11: 2526–2541. [DOI] [PubMed] [Google Scholar]
- Perner M, Petersen JM, Zielinski F, Gennerich H-H, Seifert R. (2010). Geochemical constraints on the diversity and activity of H-2-oxidizing microorganisms in diffuse hydrothermal fluids from a basalt- and an ultramafic-hosted vent. FEMS Microbiol Ecol 74: 55–71. [DOI] [PubMed] [Google Scholar]
- Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R et al. (2011). Hydrogen is an energy source for hydrothermal vent symbioses. Nature 476: 176–180. [DOI] [PubMed] [Google Scholar]
- Potter LC, Millington P, Griffiths L, Thomas GH, Cole JA. (1999). Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem J 344: 77–84. [PMC free article] [PubMed] [Google Scholar]
- Radajewski S, McDonald IR, Murrell JC. (2003). Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr Opin Biotechnol 14: 296–302. [DOI] [PubMed] [Google Scholar]
- Reysenbach AL, Shock E. (2002). Merging genomes with geochemistry in hydrothermal ecosystems. Science 296: 1077–1082. [DOI] [PubMed] [Google Scholar]
- Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert SM, Vetriani C. (2012). Chemoautotrophy at deep-sea vents past, present, and future. Oceanography 25: 218–233. [Google Scholar]
- Stewart FJ, Ulloa O, DeLong EF. (2012). Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol 14: 23–40. [DOI] [PubMed] [Google Scholar]
- Takai K, Inagaki F, Nakagawa S, Hirayama H, Nunoura T, Sako Y et al. (2003). Isolation and phylogenetic diversity of members of previously uncultivated epsilon-proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol Lett 218: 167–174. [DOI] [PubMed] [Google Scholar]
- Takai K, Campbell BJ, Cary SC, Suzuki M, Oida H, Nunoura T et al. (2005). Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl Environ Microbiol 71: 7310–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Suzuki M, Nakagawa S, Miyazaki M, Suzuki Y, Inagaki F et al. (2006). Sulfurimonas paralvinellae sp nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteo-bacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas 'denitrificans comb. nov and emended description of the genus Sulfurimonas. Int J Syst Evol Microbiol 56: 1725–1733. [DOI] [PubMed] [Google Scholar]
- Urich T, Lanzen A, Stokke R, Pedersen RB, Bayer C, Thorseth IH et al. (2014). Microbial community structure and functioning in marine sediments associated with diffuse hydrothermal venting assessed by integrated meta-omics. Environ Microbiol 16: 2699–2710. [DOI] [PubMed] [Google Scholar]
- Ver Eecke HC, Butterfield DA, Huber JA, Lilley MD, Olson EJ, Roe KK et al. (2012). Hydrogen-limited growth of hyperthermophilic methanogens at deep-sea hydrothermal vents. Proc Natl Acad Sci USA 109: 13674–13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ver Eecke HC, Akerman NH, Huber JA, Butterfield DA, Holden JF. (2013). Growth kinetics and energetics of a deep-sea hyperthermophilic methanogen under varying environmental conditions. Environ Microbiol Rep 5: 665–671. [DOI] [PubMed] [Google Scholar]
- Vetriani C, Voordeckers JW, Crespo-Medina M, O'Brien CE, Giovannelli D, Lutz RA. (2014). Deep-sea hydrothermal vent Epsilonproteobacteria encode a conserved and widespread nitrate reduction pathway (Nap). ISME J 8: 1510–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG et al. (2009). Metagenome of a versatile Chemolithoautotroph from expanding oceanic dead zones. Science 326: 578–582. [DOI] [PubMed] [Google Scholar]
- Wankel SD, Germanovich LN, Lilley MD, Genc G, DiPerna CJ, Bradley AS et al. (2011). Influence of subsurface biosphere on geochemical fluxes from diffuse hydrothermal fluids. Nat Geosci 4: 461–468. [Google Scholar]
- Wirsen CO, Jannasch HW, Molyneaux SJ. (1993). Chemosynthetic microbial activity at Mid-Atlantic Ridge hydrothermal vent sites. J Geophys Res Solid Earth 98: 9693–9703. [Google Scholar]
- Wommack KE, Williamson SJ, Sundbergh A, Helton RR, Glazer BT, Portune K et al. (2004). An instrument for collecting discrete large-volume water samples suitable for ecological studies of microorganisms. Deep-Sea Res Pt I-Oceanograph Res Papers 51: 1781–1792. [Google Scholar]
- Yakimov MM, La Cono V, Smedile F, Crisafi F, Arcadi E, Leonardi M et al. (2014). Heterotrophic bicarbonate assimilation is the main process of de novo organic carbon synthesis in hadal zone of the Hellenic Trench, the deepest part of Mediterranean Sea. Environ Microbiol Rep 6: 709–722. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Takai K. (2011). Sulfur metabolisms in epsilon- and gamma-proteobacteria in deep-sea hydrothermal fields. Front Microbiol 2: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.