Abstract
Most land plants form mutualistic associations with arbuscular mycorrhizal fungi of the Glomeromycota, but recent studies have found that ancient plant lineages form mutualisms with Mucoromycotina fungi. Simultaneous associations with both fungal lineages have now been found in some plants, necessitating studies to understand the functional and evolutionary significance of these tripartite associations for the first time. We investigate the physiology and cytology of dual fungal symbioses in the early-diverging liverworts Allisonia and Neohodgsonia at modern and Palaeozoic-like elevated atmospheric CO2 concentrations under which they are thought to have evolved. We found enhanced carbon cost to liverworts with simultaneous Mucoromycotina and Glomeromycota associations, greater nutrient gain compared with those symbiotic with only one fungal group in previous experiments and contrasting responses to atmospheric CO2 among liverwort–fungal symbioses. In liverwort–Mucoromycotina symbioses, there is increased P-for-C and N-for-C exchange efficiency at 440 p.p.m. compared with 1500 p.p.m. CO2. In liverwort–Glomeromycota symbioses, P-for-C exchange is lower at ambient CO2 compared with elevated CO2. No characteristic cytologies of dual symbiosis were identified. We provide evidence of a distinct physiological niche for plant symbioses with Mucoromycotina fungi, giving novel insight into why dual symbioses with Mucoromycotina and Glomeromycota fungi persist to the present day.
Introduction
Symbioses with soil fungi have existed since plants first began to colonize the Earth's land masses (Redecker et al., 2000; Redecker and Raab, 2006; Smith and Read, 2008) and are thought to have played a key role in establishing terrestrial ecosystems (Pirozynski and Malloch, 1975; Malloch et al., 1980). There are numerous lines of supporting evidence for this view, including plant and fungal fossils (Stubblefield et al., 1987; Remy et al., 1994; Taylor et al., 1995) and molecular data (Simon et al., 1993; Redecker et al., 2000; Redecker and Raab, 2006). Recent studies of ultrastructure (Pressel et al., 2010) and plant–fungal physiology of early-diverging extant land plant lineages (Field et al., 2012, 2015a) provided new insights into the structure–function relationships of non-vascular plants and their symbiotic fungi. Until recently, the fungal associates of the earliest branching plant lineages have been assumed to be members of the arbuscular mycorrhiza-forming clade of fungi, the obligately biotrophic Glomeromycota that lack saprotrophic capabilities.
Application of universal DNA primers, enabling detection of fungi beside Glomeromycota, together with detailed physiological and cytological observations, have now established that the earliest branching lineage of extant liverworts, the Haplomitriopsida (Heinrichs et al., 2005, 2007; Crandall-Stotler et al., 2009; Wikström et al., 2009), often form mutualistic mycorrhiza-like associations exclusively with Mucoromycotina fungi (Bidartondo et al., 2011; Field et al., 2015a). This partially saprotrophic fungal lineage is basal or sister to the Glomeromycota (James et al., 2006; Lin et al., 2014), raising the hypothesis that plant–Mucoromycotina associations represent the ancestral mycorrhizal type for land plants and that these were replaced by the strictly biotrophic Glomeromycota as plants evolved and soil organic matter accumulated (Bidartondo et al., 2011). Although some early branching clades of land plant taxa have been found to associate exclusively with one or other of these fungal groups, representatives of nearly all extant early branching clades of land plants examined thus far host both fungal lineages, sometimes simultaneously (Desirò et al., 2013; Rimington et al., 2015, and a recent report for Haplomitrium mnioides by Yamamoto et al., 2015) (Figure 1a). These discoveries point to more versatile and shifting evolutionary scenarios in early plant–fungus symbioses than hitherto assumed (Field et al., 2015b), suggesting that the ability to engage in simultaneous partnerships with both Mucoromycotina and Glomeromycota fungi may be a basal trait (Desirò et al., 2013; Rimington et al., 2015).
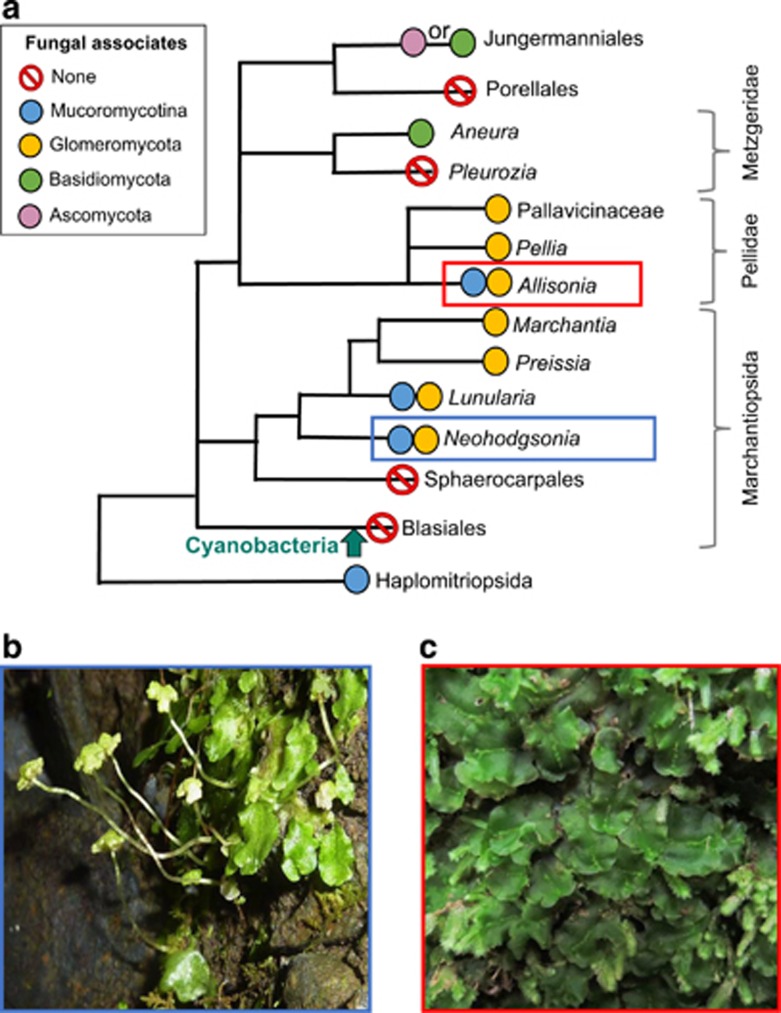
Figure 1.
Liverwort phylogeny and species used in the present study. (a) Liverwort phylogeny (following Wikström et al., 2009) showing key nodes alongside commonly associated fungal symbionts (James et al., 2006; Pressel et al., 2008; Bidartondo and Duckett, 2010; Humphreys et al., 2010; Pressel et al., 2010; Bidartondo et al., 2011; Field et al., 2012; Desirò et al., 2013). Plants of (b) Allisonia cockaynei and (c) Neohodgsonia mirabilis photographed in the field (photo credits: KJ Field and JG Duckett).
The latest evidence (Field et al., 2015a) shows that liverwort–Mucoromycotina symbioses functionally differ from those between liverworts and Glomeromycota fungi (Field et al., 2012) in their ability to maintain efficiency of carbon-for-nutrient exchange between partners across atmospheric CO2 concentrations (a[CO2]). The conditions in these studies simulate the 90% a[CO2] drop coincident with the diversification of terrestrial ecosystems through the Palaeozoic (Berner, 2006; Franks et al., 2014). Although the symbiotic functional efficiency of liverwort–Glomeromycota associations was severely compromised by a simulated Palaeozoic fall in a[CO2], that of Haplomitriopsida liverwort–Mucoromycotina partnerships was unaffected or increased under the modern-day a[CO2] scenario. These findings parallel those in Glomeromycota-associated sporophytes of some vascular plants, which also increased in functional efficiency under lower a[CO2] (Field et al., 2012). Therefore, the hypothesis that Mucoromycotina fungi, switching from saprotrophy to facultative biotrophy, facilitated the evolution and diversification of early land plants under a high a[CO2] and were among the first fungi to form mutualistic symbioses with plants is strengthened (Bidartondo et al., 2011). It remains an open question as to why dual Mucoromycotina/Glomeromycota plant–fungus partnerships today are often restricted to early-branching lineages of land plants (Figure 1a) and thus ‘lost out' to Glomeromycota-specific ones in later-branching plant lineages, such as the angiosperms (Field et al., 2015b).
We investigated the functionality and detailed cytology of the dual fungal associations in wild-collected Neohodgsonia mirabilis, the sister taxon to all other complex thalloid liverworts harbouring mycorrhiza-like associations, and Allisonia cockaynei in the earliest divergent clade of simple thalloid liverworts (Forrest et al., 2006; Crandall-Stotler et al., 2008, 2009; Villarreal et al., 2015) (Figure 1). Using molecular methods, we found that both liverworts hosted simultaneous Glomeromycota and Mucoromycotina fungal partners (see Results). We used a combination of isotope tracers under a modern ambient a[CO2] of 440 p.p.m. and a simulated Palaeozoic (c. 410–390 Ma) atmosphere of 1500 p.p.m. [CO2] (Franks et al., 2014).
We aimed to answer the following questions;
Is there an enhanced carbon cost to liverworts associated simultaneously with Mucoromycotina and Glomeromycota fungi compared with those harbouring single fungal symbionts?
Do plants with dual colonization by Mucoromycotina and Glomeromycota fungi benefit from enhanced nutrient gain in comparison to those harbouring single fungal associations?
Are the costs decreased and benefits increased by elevated a[CO2] for liverworts maintaining dual symbioses with both Mucoromycotina and Glomeromycota fungi?
Are there any characteristic cytological signatures of dual fungal symbiosis as opposed to single fungal associations?
Materials and methods
Plant material and growth conditions
The liverworts Neohodgsonia mirabilis (Perss.) Perss. and Allisonia cockaynei (Steph.) RM Schust. were collected from the South Island of New Zealand in April 2012, and vouchers were deposited in the Natural History Museum, London. We planted the liverworts directly into pots (120 mm diameter × 100 mm depth) soon after collection. Native soil surrounding liverwort rhizoids was left intact to act as a natural inoculum, and pots were carefully weeded regularly to remove any other plant species.
Based on the methods of Johnson et al. (2001), we inserted three mesh-windowed cylindrical cores (Supplementary Figure S1) into each experimental pot. The mesh covering the cores was fine enough to exclude liverwort rhizoids but allows the ingrowth of fungal hyphae. Two of the cores were filled with a homogeneous mixture of acid-washed silica sand (89% core volume), native soil gathered from around the rhizoids of wild plants (10% core volume) and fine-ground tertiary basalt (1% core volume) to act as fungal bait (Field et al., 2012). The third core was filled with glass wool and enabled below-ground respiration sampling throughout the 14C-labelling period.
We maintained plants in controlled environment chambers (BDR16, Conviron, Winnipeg, MB, Canada) with settings chosen according to those of the plant's natural environment (see Supplementary Information). Each species was grown at either 440 p.p.m. a[CO2] (n=10) or at a simulated early-Palaeozoic a[CO2] concentration of 1500 p.p.m. (n=10) (Berner, 2006, Franks et al., 2014). a[CO2] was monitored using CARBOCAP GMP343 CO2 sensors (Vaisala, Birmingham, UK) and maintained through addition of gaseous CO2. Cabinet settings and contents were alternated every 2 weeks, and we regularly rotated all pots within cabinets. Plants were acclimated to chamber/growth regimes for 12 weeks to allow establishment of mycelial networks within pots.
Molecular identification of fungal associates
Wild Neohodgsonia and Allisonia thalli were prepared for molecular analysis within 1 day of collection and immediately following our isotope labelling experiments at the end of the growth period at different a[CO2]. We dissected both plant species in the same way to leave the central part of the thallus and rhizoidal ridge (2–3 mm2) where fungal colonization is the highest. The DNA extraction, amplification and sequencing were performed as per the methods of Gardes and Bruns (1993), Desirò et al. (2013) and Field et al. (2015a) (see Supplementary Information). Sequence identity was inferred from their most closely related BLAST hits (Altschul et al., 1997). Bayesian inference was used to confirm the fungal identity of samples shown to be Glomeromycota or Mucoromycotina by BLAST. Sequences were aligned with reference DNA sequences from GenBank (Benson et al., 2005) using MUSCLE alignment algorithms (Edgar, 2004) within MEGA v. 5.1 (Tamura et al., 2011). We tested evolutionary models in MEGA and selected HKY85 (nst=2) with invgamma rates for Bayesian analysis using MrBayes (Huelsenbeck and Ronquist, 2001).
Quantification of fluxes of C, 33P and 15N between liverworts and fungi
After the 12-week acclimation period, we introduced 100 μl of an aqueous mixture of 33P-labelled orthophosphate (specific activity 148 GBq mmol−1, total 111 ng 33P added) and 15N-ammonium chloride (1 mg ml−1) into one of the soil-filled mesh cores in each pot and 100 μl distilled water into the control core via the installed capillary tubes. Cores in which isotope tracers were introduced were left static in half of the pots to preserve direct hyphal connections with the liverworts. In the remaining half, labelled cores were rotated through 90°, severing the hyphal connections between the plants and core soil immediately prior to addition of isotopes and every other day thereafter (Supplementary Figure S2).
We sealed the top of all soil cores with lanolin and caps 21 days after addition of the isotope tracers. Glass wool-filled cores were sealed with a rubber septum (SubaSeal, Sigma). We then sealed each pot into a 3-l, gas-tight labelling chamber and added 2 ml 10% lactic acid to 15 μl Na14CO3 (specific activity 2.04 TBq mmol−1) in a cuvette within the chamber prior to illumination at 0700 hours. This resulted in the release of a 1.1-MBq pulse of 14CO2 gas. Pots were maintained under growth chamber conditions, and 1 ml of labelling-chamber headspace gas was sampled after 1 h and every 4 h thereafter. Below-ground gas was sampled via the glass-wool filled core after 1 h and every 2 h thereafter to monitor below-ground respiration and 14C flux for around 17 h (see Supplementary Information for further details).
Plant harvest and sample analyses
Plant and soil materials were separated, freeze-dried, weighed and homogenized. In all, 10–30 mg of homogenized samples were digested in 1 ml of concentrated H2SO4. These were heated to 365 °C for 15 min, and 100 μl H2O2 was added to each sample when cool. Samples were reheated to 365 °C, and each clear digest solution was diluted to 10 ml with distilled water. Two ml of each diluted digest were then added to 10 ml of the scintillation cocktail Emulsify-safe (Perkin Elmer, Beaconsfield, UK) and quantified through liquid scintillation. 33P transferred to the plant via fungal mycelium was then calculated as detailed in Supplementary Information (Cameron et al., 2007).
15N abundance was determined using Isotope Ratio Mass Spectrometry (IRMS). Between 2 and 5 mg of freeze-dried, homogenized plant tissue was weighed out into 6 × 4 mm2 tin capsules (Sercon Ltd, Crewe, UK) and analysed using a continuous flow IRMS (PDZ 2020 IRMS, Sercon Ltd). Air was used as the reference standard, and the IRMS detector was regularly calibrated to commercially available reference gases.
14C activity was quantified through sample oxidation and liquid scintillation. Approximately 10–100 mg of freeze-dried sample was placed in Combusto-cones (Perkin Elmer) before oxidation (Model 307 Packard Sample Oxidiser Isotech, Chesterfield, UK). CO2 released through oxidation was trapped in 10 ml Carbosorb prior to mixing with 10 ml Permafluor. Total carbon (12C+14C) fixed by the plant and transferred to the fungal network was calculated as a function of the total volume and CO2 content of the labelling chamber and the proportion of the supplied 14CO2 label fixed by the plants. The difference in carbon between the static and rotated cores is taken as equivalent to the total C transferred from plant to symbiotic fungus within the soil core, noting that a small proportion will be lost through soil microbial respiration. The total carbon budget for each experimental pot was calculated using equations from Cameron et al. (2006), which are detailed in Supplementary Information.
Data from Allisonia and Neohodgsonia are compared in the discussion to published and unpublished data from Haplomitrium and Treubia associated exclusively with Mucoromycotina fungi obtained from experiments using identical conditions within the same controlled environment growth chambers (see Field et al., 2015a). Data are also presented alongside previously published data for Preissia and Marchantia associated only with Glomeromycota fungi from experiments using near-identical experimental conditions within the same controlled environment growth chambers (Field et al., 2012). In these experiments, pots were filled with soil from dune slacks at Aberfraw, Anglesey, UK (Grid Reference: SH 397 648) but were otherwise identical to those of all our other experiments.
Ultrastructural analyses
We processed plants that were wild-collected and from experiments where they were grown at two a[CO2] for transmission and scanning electron microscopy as described previously (Duckett et al., 2006). For transmission electron microscopy, thalli were fixed in 3% glutaraldehyde, 1% fresh formaldehyde and 0.75% tannic acid in 0.05 m Na-cacodylate buffer, pH 7, for 3 h at room temperature. After rinses in 0.1 m buffer, samples were postfixed in buffered (0.1 m, pH 6.8) 1% osmium tetroxide overnight at 4 °C, dehydrated in an ethanol series and embedded in TAAB low viscosity resin via ethanol. Thin sections were cut with a diamond knife, stained with methanolic uranyl acetate for 15 min and in Reynolds' lead citrate for 10 min and observed with a Hitachi H-7100 transmission electron microscope (Hitachi High-Technologies Europe, Maidenhead, UK) at 100 kV. For scanning electron microscopy, we fixed thalli in 3% glutaraldehyde, dehydrated through an ethanol series, critical-point dried using CO2 as transfusion fluid, sputter coated with 390 nm palladium-gold and viewed them using a FEI Quanta scanning electron microscope (FEI, Hillsboro, OR, USA).
Statistics
Effects of plant species, a[CO2] and the interaction between these factors on the C, 33P and 15N fluxes between plants and fungi from this and previous studies (Field et al., 2012, 2015a) were tested using analysis of variance with additional post-hoc Tukey's tests where indicated. Data were checked for homogeneity of variance and normality. Where assumptions for analysis of variance were not met, data were transformed using log10 or arcsine-square-root as indicated in Table 1. Different letters in the figures denote statistical difference (P<0.05) in all the figures. All statistics were carried out using the statistical software package R 3.1.2 (R Core Team, 2012).
Table 1. Summary of differences in mycorrhizal functionality (F ratio from ANOVA) between Neohodgsonia, Alisonia, Haplomitrium, Treubia, Preissia and Marchantia at elevated a[CO2] (1500 p.p.m.) and ambient a[CO2] (440 p.p.m.).
| df | Plant species | CO2 treatment | Species × CO2 | |
|---|---|---|---|---|
| Biomass (g) | 1, 30 | 16.276*** | 18.911*** | 1.937 |
| Fungal carbon in cores (ng)a | 1, 54 | 14.042*** | 31.334*** | 5.087*** |
| Percentage of carbon allocationb | 1, 54 | 5.756*** | 13.900*** | 3.278* |
| Total 33P uptake (ng)b | 1, 30 | 4.498** | 5.714* | 6.483*** |
| [33P] in plant tissue (ng g−1) | 1, 36 | 6.259*** | 3.142 | 3.857* |
| Total 15N uptake (ng) | 1, 20 | 1.889 | 0.953 | 0.822 |
| [15N] in plant tissue (ng g−1)a | 1, 20 | 0.235 | 1.147 | 1.304 |
| 33P-for-C efficiency (ng ng−1) | 1, 36 | 46.220*** | 0.885 | 31.747*** |
| 15N-for-C efficiency (ng ng−1) | 1, 20 | 0.413 | 13.523** | 1.913 |
Abbreviations: ANOVA, analysis of variance; p.p.m., parts per million. *P<0.05, **P<0.01, ***P<0.001; post-hoc Tukey's test.
Data have been log10 transformed to meet the assumptions for ANOVA.
Data have been arcsine-square-root transformed to meet the assumptions for ANOVA.
Results
Molecular identification of fungi
Molecular analyses of fungal partners (n=6) showed that Allisonia and Neohodgsonia plants freshly collected from the field and after our isotope tracing experiments are colonized by both Mucoromycotina and Glomeromycota fungi (Supplementary Figure S3). The Mucoromycotina fungi identified here were the same as those found previously in wild populations of both species (Bidartondo et al., 2011) belonging to groups I and H (sensu, Desirò et al., 2013) in Neohodgsonia and Allisonia, respectively. The Glomeromycota fungal associates were exclusively Glomerales in Allisonia while Neohodgsonia harboured members of both Glomerales and Archaeosporales. Sequences are deposited in GenBank (KR779272-KR7792784).
Plant biomass
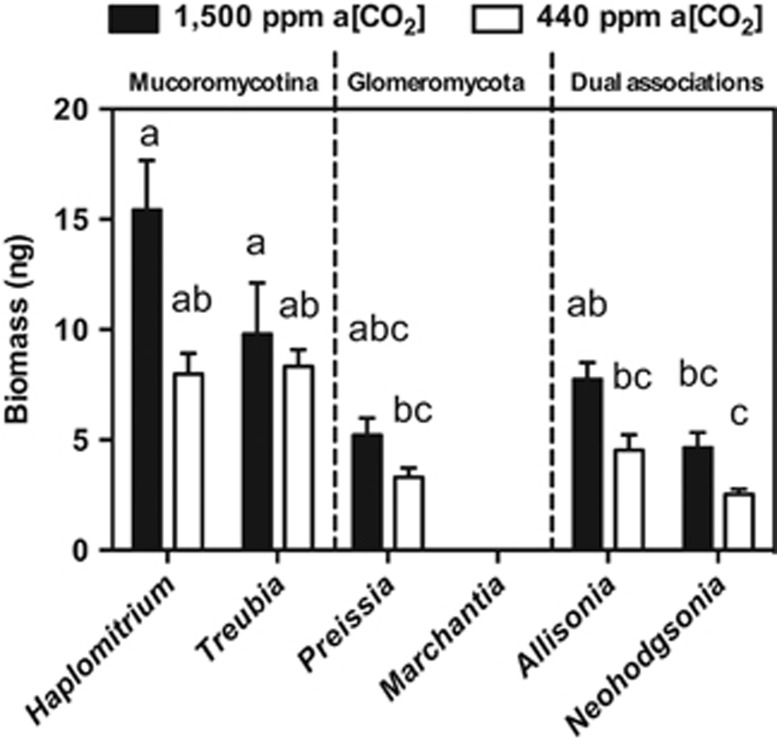
Overall, there was a consistent trend of liverworts achieving greater biomass when grown at a[CO2] of 1500 p.p.m. compared with 440 p.p.m. a[CO2] (Figure 2). We found greater biomass of both Allisonia (41%) and Neohodgsonia (45%) grown at 1500 p.p.m. a[CO2] compared with those grown at a[CO2] of 440 p.p.m.
Figure 2.
Mean total plant biomass (dry) at the end of experimental period in five liverwort species (Field et al., 2012, 2015a) at both 1500 p.p.m. a[CO2] (black bars) and 440 p.p.m. a[CO2] (white bars). Error bars show s.e.m. (n=4 for all species); different letters denote statistical difference where P<0.05 (Tukey's post hoc).
Liverwort-to-fungus carbon transfer
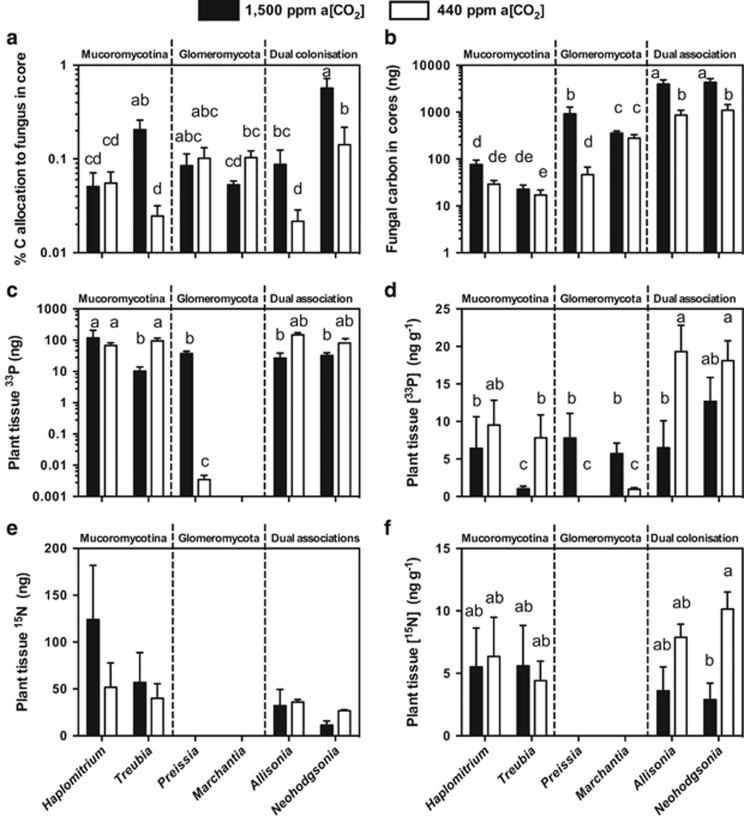
Both Allisonia and Neohodgsonia allocated around four times more photosynthate to their fungal symbionts under the simulated Palaeozoic a[CO2] (Figure 3a) compared with the lower [CO2] (Figure 3a). In terms of total carbon transferred from plants to fungal partners (Figure 3b), each liverwort species transferred more carbon to their fungal symbionts at 1500 p.p.m. a[CO2] than at 440 p.p.m. a[CO2], this difference being significant in Allisonia and Neohodgsonia. As such, the dual fungal symbioses of Neohodgsonia and Allisonia have a greater total carbon ‘cost' at both a[CO2] than any of the other Glomeromycota– or Mucoromycotina–liverwort symbioses (Figure 3b).
Figure 3.
Carbon-for-nutrient exchange between liverworts and their fungal partners. (a) Percentage allocation of plant-derived carbon to fungi within soil cores, (b) total measured plant-fixed carbon transferred to fungi in soil for liverworts with different fungal associations (Mucoromycotina-only, Glomeromycota-only and dual fungal associations; Field et al., 2012, 2015a); (c) total plant tissue 33P content (ng) and (d) tissue concentration (ng g−1) of fungal-acquired 33P in six liverwort species with different fungal associations under 1500 p.p.m. (black bars) a[CO2] and 440 p.p.m. (white bars) a[CO2] (Field et al., 2012, 2015a); (e) total tissue 15N content (ng) and (f) concentration (ng g−1) of fungal-acquired 15N in four liverwort species with different fungal associations (Field et al., 2015a) at both 1500 p.p.m. (black bars) and 440 p.p.m. (white bars) a[CO2]. In all panels, error bars show ±s.e.m. Different letters represent where P<0.05 (analysis of variance, Tukey's post hoc; see Table 1). In panels (a) and (b), n=6 for all species apart from Marchantia, where n=4; (c–e) n=4, where data are available.
Fungal transfer of 33P and 15N to host liverworts
Allisonia and Neohodgsonia acquire 78% and 67% more 33P, respectively, at 440 p.p.m. compared with 1500 p.p.m. a[CO2], also reflected in plant tissue [33P] (Figures 3c and d). When grown at the 1500 p.p.m. a[CO2], the liverworts with dual fungal symbionts showed reduced total 33P uptake (Figure 3c), resulting in greatly reduced 33P concentrations in their tissues (Figure 3d).
The total uptake and assimilation of 15N is reduced by 11% in Allisonia and 57% in Neohodgsonia at 1500 p.p.m. a[CO2] compared with 440 p.p.m. a[CO2] (Figure 3e). In terms of tissue concentration, the same trend is amplified with [15N] being far greater by 250% and 119% in Allisonia and Neohodgsonia, respectively, at 440 p.p.m. a[CO2] compared with when plants are grown at 1500 p.p.m. a[CO2] (Figure 3f).
Nutrient-for-carbon exchange efficiency
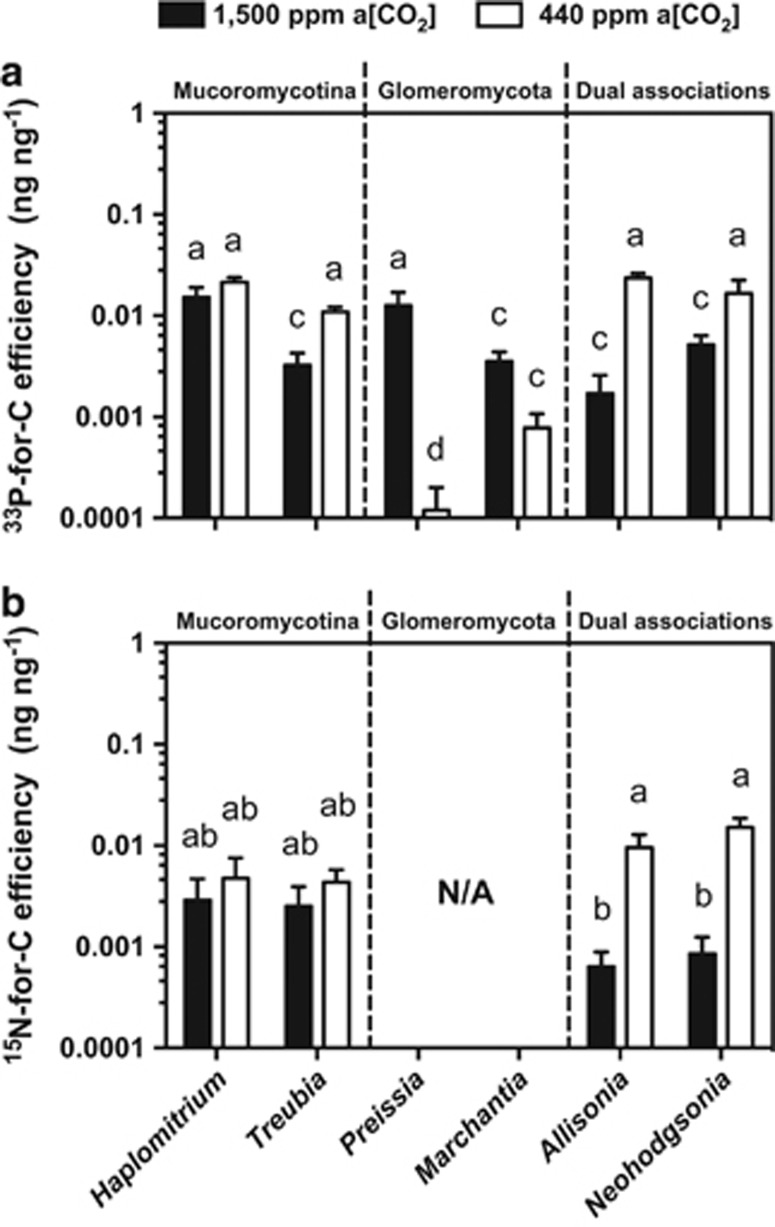
33P-for-C exchange efficiency in Allisonia was >13 times greater at 440 p.p.m. a[CO2] than it was at 1500 p.p.m. a[CO2] (Figure 4a). The same pattern was true in Neohodgsonia, with three times greater 33P-for-C exchange efficiency at the lower a[CO2] (Figure 4a). 15N-for-C exchange was an order of magnitude greater in both Allisonia and Neohodgsonia at the lower a[CO2] compared with the elevated a[CO2] (Figure 4b).
Figure 4.
Nutrient-for-carbon exchange efficiencies between liverworts and their fungal partners. (a) 33P-for-carbon and (b) 15N-for-carbon efficiency for different liverwort species with different fungal associations under both 1500 p.p.m. (black bars) and 440 p.p.m. (white bars) a[CO2] (Field et al., 2012, 2015a,b). Error bars show s.e.m. (n=4 for all species). Different letters indicate where P<0.05 (analysis of variance, Tukey's post hoc).
Cytology of colonization
The cytology of dual colonization by Mucoromycotina and Glomeromycota fungi in wild plants of Neohodgsonia and Allisonia is described here for the first time. As our detailed electron microscopic analyses revealed no major differences, only a minor one in Allisonia (detailed below), between wild and experimental plants grown at contrasting a[CO2] (440 and 1500 p.p.m.), the results are presented together unless otherwise stated.
Neohodgsonia mirabilis
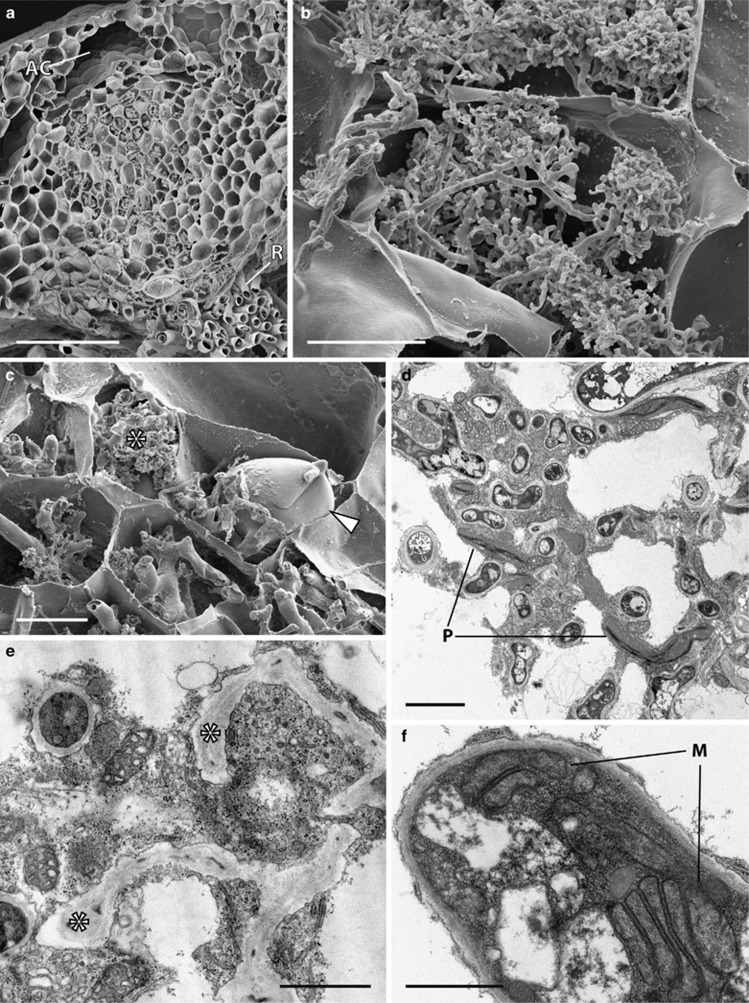
Fungal colonization occupies the central thallus midrib, extending all the way from the rhizoid-bearing ventral surface, the point of fungal entry (see Supplementary Information), to just below the large dorsal air chambers (Figure 5a). Fungal structures comprise numerous arbuscules at various stages of development, from young (Figure 5b) to collapsed and large vesicles occupying a significant proportion of the host cell (Figure 5c). Healthy (Figure 5d) and degenerated arbuscules (Figure 5e), large living hyphae, vesicles and active host cytoplasm are most often present in the same host cell. Fungal trunk hyphae and arbuscular hyphae are surrounded by the host plasma membrane, and the cytoplasm of the host cells comprises numerous Golgi bodies, mitochondria, plastids and microbodies (Figures 5d and e). The latter have a well-developed thylakoid system but are largely devoid of starch deposits (Figure 5d). Fungal hyphae are aseptate and often contain multiple mitochondrial stacks, each comprising of 5–6 mitochondria (Figure 5f).
Figure 5.
Cytology of Neohodgsonia mirabilis grown at 440 and 1500 p.p.m. a[CO2]. Scanning (a–c) and transmission (d–f) electron micrographs (TEM). Both the distribution and cytology of the association remained the same between a[CO2] treatments and are illustrated here in plants grown at 440 p.p.m. a[CO2]. (a) Fungal colonization zone extending from the rhizoid (R) bearing ventral surface of the thallus to just below the dorsal air chambers (AC). (b, c) Young arbuscules (b) and collapsed ones (c) (*) adjacent to a large vesicle (arrowed). (d) fungal hyphae surrounded by active host cytoplasm. Note the plastids (P) with well-developed thylakoid systems but largely devoid of starch. (e) Degenerated arbuscular hyphae (*) surrounded by healthy host cytoplasm. (f) Fungal hyphae typically contain multiple mitochondrial stacks (M). Scale bars: (a) 200 μm; (c) 50 μm; (b) 20 μm; (d) 3 μm; (e, f) 1 μm.
Allisonia cockaynei
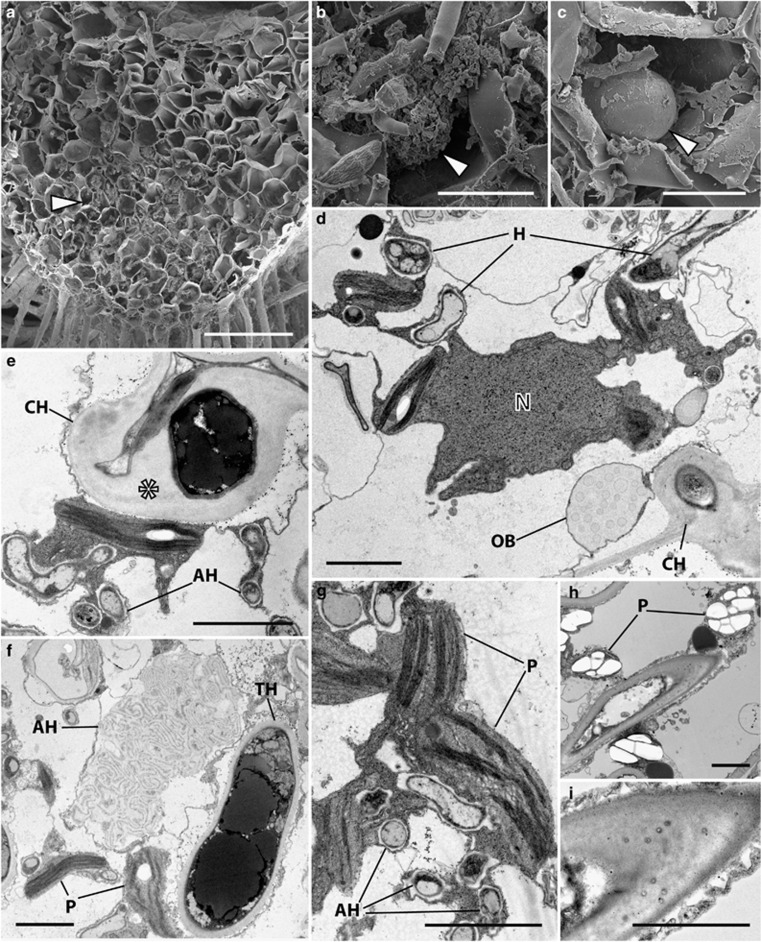
Fungal entry is via the rhizoids (Supplementary Figure S4) with the fungal zone occupying the central region of the thallus, generally the first 10 cell layers from the rhizoid-bearing ventral side with approximately 1/3 of the thallus midrib remaining free of fungal structures (Figure 6a). These comprise large hyphae, arbuscules (Figure 6b) and prominent vesicles (Figure 6c). Host cells are characterised by active cytoplasm, including numerous mitochondria and plastids in close association with the fungus (Figure 6d). Colonizing hyphae traverse the walls of adjacent host cells and have a thick layer of fibrillar material in between the fungus cell wall and the host plasma membrane that surrounds them (Figure 6d) while arbuscular hyphae are characterized by thin cell walls (Figure 6e). These are often collapsed while the colonizing hyphae and host cytoplasm surrounding them persist (Figure 6f). Whereas the plastids of wild plants and those grown at 440 p.p.m. a[CO2] contain little or no starch deposits (Figure 6g), those of plants grown at 1500 p.p.m. a[CO2] have prominent starch grains (Figure 6h). The large colonizing hyphae of wild and experimental plants grown under contrasting a[CO2] regimes are all characterized by plasmodesmata-like channels in the fibrillar material that surrounds them (Figure 6i).
Figure 6.
Cytology of Allisonia cockaynei grown at 440 and 1500 p.p.m. a[CO2]. Scanning (a–c) and transmission (e–i) electron micrographs (TEM). There was no change in the overall distribution of fungal colonization and in the cytology of the fungus between [CO2] treatments, both illustrated here in plants grown at 440 p.p.m. a[CO2] except for panels (h and i). (a) Fungal colonization zone (arrowed) occupying the first 10 cell layers from the rhizoid-bearing ventral surface. (b) Collapsed arbuscules (arrowed) and (c) large vesicle (arrowed). (d) Host cell with active cytoplasm in close association with fungal hyphae (H). Note the colonizing hypha (CH) traversing the host cell wall. N, nucleus; OB, oil body. (e) colonizing hypha with thick layer of fibrillar material (*) in between the fungus cell wall and the host plasma membrane (arrowed) and thin-walled arbuscular hyphae (AH) in close proximity to plastids (P). (g) Arbuscular hyphae in close association with starch-free plastids. (h) In plants grown at 1500 p.p.m. a[CO2] plastids have prominent starch deposits. (i) Plasmodesmata-like channels are present in the fibrillar material that surrounds the colonizing hyphae. Scale bars: (a) 200 μm; (b, c) 20 μm; (d–i) 3 μm.
Discussion
The currently emerging paradigm considers the Mucoromycotina symbiosis with plants to have evolved prior to the emergence of plant–Glomeromycota fungal symbioses (Bidartondo et al., 2011). Moreover, until very recently it has been assumed that early diverging lineages of plants associate with only Glomeromycota (Wang and Qui, 2006). In direct contrast to this, our work shows that basal liverwort lineages (Figure 1a) form simultaneous mutualistic symbioses with both Mucoromycotina and Glomeromycota fungi. This raises novel questions regarding mycorrhizal evolution; given the global radiation and dominance of glomeromycotean symbioses, why have associations with Mucoromycotina fungi persisted? We can now begin to answer this question with the present demonstration that dual associations are significantly more efficient at modern day atmospheric CO2 compared with Palaeozoic CO2, whereas single fungal group partnerships are either unaffected by a[CO2] (Mucoromycotina fungi) or are less efficient under modern day a[CO2] (Glomeromycota fungi). This trade-off provides a physiological niche facilitating the persistence of plant symbioses with Mucoromycotina fungi, singly and in dual partnerships with Glomeromycota fungi to the present day.
Physiological costs and benefits
Our experiments reveal that Neohodgsonia and Allisonia with dual Glomeromycota and Mucoromycotina fungal associations allocated greater percentages and total amounts of photosynthate to their fungal partners at 1500 p.p.m. a[CO2] than at 440 p.p.m. a[CO2] (Figures 3a and b). Our previous studies show that in terms of percentage of carbon allocation, Mucoromycotina partners of Treubia receive seven times greater percentage allocation of plant-fixed carbon at 1500 p.p.m. a[CO2] compared with at 440 p.p.m. a[CO2]. There is little difference in percentage of C allocation in Haplomitrium while in Marchantia and Preissia the percentage of C allocation to Glomeromycota fungi is 1.9 and 1.2 times greater, respectively. This likely resulted in the greater biomasses recorded in all liverworts at elevated a[CO2] (Figure 2).
In all of the combinations of liverwort–fungal symbioses examined thus far, partnerships in which there is a Mucoromycotina fungal symbiont (that is, in Haplomitrium, Treubia, Allisonia and Neohodgsonia) display increased 33P-for-C and 15N-for-C exchange efficiency at 440 p.p.m. a[CO2] compared with at 1500 p.p.m. a[CO2] (Figure 4). In liverwort–Glomeromycota symbioses, the opposite trend is apparent, with 33P-for-C being several orders of magnitude lower in both Marchantia and Preissia at 440 p.p.m. compared with at 1500 p.p.m. a[CO2] (Figure 4a).
Decreased fungal-acquired nutrient uptake in liverworts with Mucoromycotina fungal partners (either single or dual colonizations) at elevated a[CO2] seems at first counter-intuitive, particularly given their larger biomass (Figure 2) and increased photosynthate allocation to fungal partners (Figures 3a and b) in those conditions. However, it is possible that the plants in our experimental pots experienced nutrient limitation (for P, N or both). This seems likely considering the lack of plant-available nutrients in the surrounding sand and its limited accessibility within the soil cores. As such, when a[CO2] is at 1500 p.p.m., the liverworts likely produced excess photosynthates that they might have been unable to utilize for growth or reproduction owing to nutrient limitation. As liverworts are structurally simple plants, with no vasculature or specialized storage organs to provide transport and storage of excess carbohydrates (Kenrick and Crane, 1997), surplus sugars must be either stored as insoluble starch granules within the thallus (observed here in Allisonia; Figure 6h), supplied directly to fungal partner(s) (see Figures 3a and b), or be released into the surrounding soil as exudates.
It is likely that the greater C allocation we observed from liverworts to Mucoromycotina fungal partners in our experiments allows increased hyphal proliferation and fungal sporulation. Given that these processes are demanding in terms of energy and resources (Denison and Kiers, 2011), the fungus would have greater N and P requirements and therefore may assimilate more of the nutrients acquired from its surroundings, rather than surrender them in return for plant carbohydrate. This may provide a mechanism to explain our observations of reduced fungal-acquired nutrient uptake in Mucoromycotina-exclusive and dual Mucoromycotina/Glomeromycota-partnered liverworts at elevated a[CO2], even with enhanced C allocation to fungal partners (Figures 4 and 5). It is also possible that there are further non-nutritional benefits for liverworts in symbiosis with Mucoromycotina fungi that have not been explored here, such as enhanced disease and/or herbivore resistance (Cameron et al., 2013).
In contrast, the liverworts partnered exclusively with obligately biotrophic Glomeromycota fungi in previous experiments (that is, Marchantia and Preissia in Field et al., 2012) operated a more linear exchange of nutrients-for carbon. In this scenario, more photosynthate is supplied to the fungal mycelium at elevated a[CO2], which in turn supplies more 33P to the host plant. At 440 p.p.m. a[CO2], the plant does not maintain the same supply of photosynthates to the fungus, and so the fungus does not return as much nutrient to its host. This pattern of ‘tit-for-tat' reciprocity in plant–Glomeromycota symbiosis has previously been demonstrated in various vascular plant species, both in root-organ culture systems (Kiers et al., 2011) and in whole-plant experiments (Hammer et al., 2011; Fellbaum et al., 2014). Here we demonstrate that this model does not apply in our case of a plant symbioses involving more than one fungal partner and involving Mucoromycotina fungi.
It is possible that by allocating excess photosynthates directly to Mucoromycotina fungal partners, rather than releasing them as C-rich plant exudates, the liverworts avoid providing excess carbohydrate resources to surrounding saprotrophic microbes. This may help to reduce nutrient immobilization by free-living saprotrophic microorganisms and damage or toxicity caused by potential microbial pathogens (Otten et al., 2004). These potential benefits to the plants may contribute to the maintenance of Mucoromycotina fungal partnerships even in plants that can form symbiotic associations with Glomeromycota fungi and may explain why these have not been lost entirely from extant plants through evolutionary time (Rimington et al., 2015). If excess photosynthates are released as exudates from the liverworts, they are likely to enhance nutrient immobilization and increase their nutrient limitation.
Cytological characteristics
Our investigation reveals that the cytology of fungal colonization in both Neohodgsonia and Allisonia is typical of mycorrhizal associations involving Glomeromycota fungi; in both it comprises prominent vesicles and well-developed, short-lived arbuscules and/or fine hyphae surrounded by active host cytoplasm. The last feature is also typical of the intracellular phase in Mucoromycotina associations (Desirò et al., 2013; Strullu-Derrien et al., 2014; Rimington et al., 2015; Field et al., 2015a). However, the key feature of intracellular colonization in Haplomitriopsida–Mucoromycotina symbiosis—hyphal coils with terminal swellings (‘lumps') (Carafa et al., 2003; Duckett et al., 2006)—seems to be unique and has not been observed in any other liverwort–fungus partnerships, including those in Neohodgsonia and Allisonia.
Another diagnostic feature of Mucoromycotina colonization, intercellular fungal proliferation with the production of thick-walled spores in mucilaginous spaces, does occur across plant lineages, including hornworts and lycopods, but neither Neohodgsonia nor Allisonia develop mucilage-filled schizogenous intercellular spaces in their thalli. It is unsurprising therefore that in these two species we did not observe any of the major cytological differences between ambient and elevated a[CO2]-grown plants reported in the Haplomitriopsida (Field et al., 2015a) as the latter were exclusively associated with the intercellular phase of fungal colonization. The single minor cytological difference observed between wild and experimental plants grown at contrasting a[CO2], and restricted to Allisonia, was the presence of far more starch granules within thalli of this species when grown at high a[CO2] (Figure 6h).
The only cytological features that may potentially be indicative of fungal identity in these dual symbioses are the fine/arbuscular and trunk/colonizing hyphal diameters (Strullu-Derrien et al., 2014). In Mucoromycotina–liverwort symbiosis, the fine hyphae range from 0.5 to 1.0 μm and the larger classes are 3–4 μm (Haplomitrium, Treubia), but in Glomeromycota–liverwort symbiosis (Marchantia, Preissia, Pellia) the corresponding dimensions are from 1 to 3 μm and from 4 to 8 μm. Measurements of Neohodgsonia and Allisonia reveal that the fine hyphae range from 0.6 to 1.2 μm, that is, mostly in the Mucoromycotina range, whereas the trunk hyphae, ranging from 3 to 8 μm, are more typical of Glomeromycota. In contrast, vesicles are consistently diagnostic of Glomeromycota. Thus, although the identification of the two different fungi in Neohodgsonia and Allisonia largely rests with the molecular evidence, there are indications from cytology for the presence of both Mucoromycotina and Glomeromycota fungi that could be further explored by cytochemical and cytogenetic techniques. Consequently, regarding the large number of previous studies, particularly in early-diverging plant lineages, in which electron microscopy has been used to describe mycorrhizal associations as ‘glomeromycotean', our findings suggest that cryptic Mucoromycotina associations may sometimes also be occurring simultaneously.
In vitro isolation and resynthesis experiments with liverworts known to engage in dual symbioses and whereby either of the two mycobionts is reintroduced in the host plant will help to determine cytological similarities and/or differences between the two fungal symbionts. Fluorescence in situ hybridization may allow localization of Glomeromycota and Mucoromycotina fungi co-existing in the same host plant to establish which structures belong to which fungus. In the meantime, it is essential that fungal identification is carried out using appropriately inclusive molecular techniques in any mycorrhizal or mycorrhizal-like symbiosis.
Wider perspectives
Our findings indicate that under a modern near-ambient a[CO2], liverworts in partnership with Mucoromycotina, either in single or dual associations alongside Glomeromycota fungi, benefit from greater nutrient gain for carbon outlay than liverworts that maintain mutualistic symbioses with only a Glomeromycota fungal symbiont. From an evolutionary perspective, the relative increases in nutrient exchange efficiency of plants harbouring both types of symbiont at lower a[CO2] may at least partially explain why declining atmospheric a[CO2] over the course of the Palaeozoic would have favoured the retention of both functional types of symbiosis. However, it is important to note that plants living in 1500 p.p.m. a[CO2] were likely to experience other abiotic factors that changed as plants evolved, including soil mineralogy and nutrient supply.
The question remains whether Mucoromycotina fungal symbioses resemble an ancestral condition that gave way to dual (for example, Neohodgsonia and Allisonia) and then solely Glomeromycota symbiosis (for example, Marchantia, Preissia, Conocephalum) or whether co-evolution of plant and fungal symbioses have been more dynamic than previously thought (Field et al., 2015b). Indeed, the liverwort phylogeny (Figure 1a) is associated with repeated losses and re-acquisitions of the same or different fungal symbionts. That liverwort clades supporting dual fungal partnerships have fungus-free sister groups, for example, the Sphaerocarpales and Blasiales (Pressel et al., 2010), points to shifting fungal associations during liverwort evolution. Exclusive plant–Mucoromycotina fungal symbiosis being a basal trait is only supported by these associations being present in liverworts of the Haplomitriopsida (Bidartondo et al., 2011), the sister group to all other liverworts (Forrest et al., 2006; Crandall-Stotler et al., 2008, 2009), with liverworts themselves being the earliest diverging land plant lineage (Alaba et al., 2014; Cox et al., 2014; Qiu et al., 1998, 2006, 2007).
Mounting evidence that a large proportion of taxa in all extant early-diverging plant lineages (Desirò et al., 2013; Rimington et al., 2015; Field et al., 2015a), and likely some Rhynie Chert fossil plants (Strullu-Derrien et al., 2014), form dual symbiosis with both Mucoromycotina and Glomeromycota fungi now corroborates these simultaneous fungal partnerships as being an extremely ancient condition, coincident with the early evolution of land plants. Why some Haplomitriopsida liverworts do not engage in symbiosis with the ubiquitous Glomeromycota fungi remains enigmatic given the clear advantages of dual partnerships demonstrated here. Even less comprehensible are the obligate Glomeromycota relationships in thalloid liverworts such as Marchantia and Pressia, given that recent functional studies clearly demonstrated that the symbiotic functional efficiency of these partnerships is severely compromised by the fall in a[CO2] that occurred through land plant diversification (Field et al., 2012). In this context, it is interesting to note that liverwort clades harbouring exclusively Glomeromycota fungi have much later divergence times than those able to associate with both fungal symbionts (Cooper et al., 2012; Feldberg et al., 2013). Marchantia, Conocephalum and Preissia most likely diverged during the Cretaceous (Wikström et al., 2009; Villarreal et al., 2015), a period of rapid angiosperm and polypodiaceous fern radiation (Schneider et al., 2004). We hypothesize that during this period major changes in abiotic and biotic dynamics, both below ground and above-ground, led to the predominance of the biotrophic Glomeromycota fungi in land plant–fungal interactions. It is possible therefore that these Glomeromycota-specific liverworts evolved in Glomeromycota-dominated environments and never engaged in associations with Mucoromycotina fungi.
In this first assessment of the functionality and cytology of the dual symbiosis of plants with Mucoromycotina and Glomeromycota fungi, we were not able to distinguish between fungal partners using microscopical techniques nor relative carbon allocation to each fungal symbiont. Future research using axenic cultures of plants and symbiotic fungi may enable such comparisons to be made and is an area for future development. More targeted cytological techniques, such as fluorescence in situ hybridization, may provide further novel insights into the associations and should be pursued in the future. With the discovery of dual Mucoromycotina-Glomeromycota symbioses in early branching lineages of living vascular plants (Rimington et al., 2015), it is now critical that we explore how far these might extend into seed plants.
Acknowledgments
We gratefully acknowledge funding from NERC (NE/1024089/1), a Leverhulme Emeritus Fellowship to JGD and a Royal Society University Research Fellowship to DDC. We thank Irene Johnson and Dr Heather Walker for technical assistance and stable isotope analyses. We thank the New Zealand Department of Conservation for collecting permits. We thank the anonymous referees and the editor for their constructive comments on our manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alaba S, Piszczalka P, Pietrykowska H, Pacak AM, Sierocka I, Nuc PW et al. (2014). The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants. New Phytol 206: 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. (2005). GenBank. Nucleic Acids Res 33(suppl 1): D34–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berner RA. (2006). GEOCARBSULF: a combined model for Phanerozoic atmospheric O2 and CO2. Geochim Cosmochim Acta 70: 5653–5664. [Google Scholar]
- Bidartondo MI, Duckett JG. (2010). Conservative ecological and evolutionary patterns in liverwort-fungal symbioses. Proc R Soc B 277: 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidartondo MI, Read DJ, Trappe JM, Merckx V, Ligrone R, Duckett JG. (2011). The dawn of symbiosis between plants and fungi. Biol Lett 7: 574–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DD, Johnson I, Leake JR, Read DJ. (2007). Mycorrhizal acquisition of inorganic phosphorus by the green-leaved terrestrial orchid Goodyera repens. Ann Bot (London) 99: 831–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DD, Leake JR, Read DJ. (2006). Mutualistic mycorrhiza in orchids: evidence from plant-fungus carbon and nitrogen transfers in the green-leaved terrestrial orchid Goodyera repens. New Phytol 171: 405–416. [DOI] [PubMed] [Google Scholar]
- Cameron DD, Neal AL, van Wees SCM, Ton J. (2013). Mycorrhiza-induced resistance: more than the sum of its parts? Trends Plant Sci 18: 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafa A, Duckett JG, Ligrone R. (2003). Subterranean gametophytic axes in the primitive liverwort Haplomitrium harbour a unique type of endophytic association with aseptate fungi. New Phytol 160: 185–197. [DOI] [PubMed] [Google Scholar]
- Cooper ED, Henwood MJ, Brown EA. (2012). Are the liverworts really that old? Cretaceous origins and Cenozoic diversifications in Lepidoziaceae reflect a recurrent theme in liverwort evolution. Biol J Linn Soc 107: 425–441. [Google Scholar]
- Cox CJ, Blaise L, Foster PJ, Embley TM, Civáň P. (2014). Conflicting phylogenies for early land plants are caused by composition biases among synonymous substitutions. Syst Biol 63: 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandall-Stotler B, Stotler R, Long DG. (2008) Morphology and classification of the Marchantiophyta. In: Goffinet B, Shaw AJ (eds). Bryophyte Biology, 2nd edn, Cambridge University Press: Cambridge, UK, pp 1–54. [Google Scholar]
- Crandall-Stotler B, Stotler RE, Long DG. (2009). Phylogeny and classifications of the Marchantiophyta. Edinburgh J Bot 66: 155–198. [Google Scholar]
- Denison RF, Kiers ET. (2011). Life histories of symbiotic rhizobia and mycorrhizal fungi. Curr Biol 21: R775–R785. [DOI] [PubMed] [Google Scholar]
- Desirò A, Duckett JG, Pressel S, Villarreal JC, Bidartondo MI. (2013). Fungal symbioses in hornworts: a chequered history. Proc Roy Soc B 280: 20130207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett JG, Carafa A, Ligrone R. (2006). A highly differentiated glomeromycotean association with the mucilage-secreting, primitive antipodean liverwort Treubia (Treubiaceae): clues to the origins of mycorrhizas. Am J Bot 93: 797–813. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldberg K, Heinrichs JA, Schmidt AR, Váňa J, Schneider H. (2013). Exploring the impact of fossil constraints on the divergence time estimates of derived liverworts. Plant Syst Evol 299: 585–601. [Google Scholar]
- Fellbaum CR, Mensah JA, Cloos AJ, Strahan GE, Pfeffer PE, Kiers ET et al. (2014). Fungal nutrient allocation in common mycorrhizal networks is regulated by the carbon source strength of individual host plants. New Phytol 203: 646–656. [DOI] [PubMed] [Google Scholar]
- Field KJ, Cameron DD, Leake JR, Tille S, Bidartondo MI, Beerling DJ. (2012). Contrasting arbuscular mycorrhizal responses of vascular and non-vascular plants to a simulated Palaeozoic CO2 decline. Nat Commun 3: 835. [DOI] [PubMed] [Google Scholar]
- Field KJ, Pressel S, Duckett JG, Rimington WR, Bidartondo MI. (2015. b). Symbiotic options for the conquest of land. Trends Ecol Evol 30: 477–486. [DOI] [PubMed] [Google Scholar]
- Field KJ, Rimington WR, Bidartondo MI, Allinson KE, Beerling DJ, Cameron DD et al. (2015. a). First evidence of mutualism between ancient plant lineages (Haplomitriopsida liverworts) and Mucoromycotina fungi and its response to simulated Palaeozoic changes in atmospheric CO2. New Phytol 205: 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest LL, Davis EC, Long DG, Crandall-Stotler BJ, Clark A, Hollingsworth ML. (2006). Unravelling the evolutionary history of the liverworts (Marchantiophyta): multiple taxa, genomes and analyses. Bryologist 109: 303–334. [Google Scholar]
- Franks PJ, Royer DL, Beerling DJ, Van de Water PK, Cantroll DJ, Barbour MM et al. (2014) Geophys Res Lett 41: 4685–4694.
- Gardes M, Bruns TD. (1993). ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol 2: 113–118. [DOI] [PubMed] [Google Scholar]
- Hammer EC, Pallon J, Wallander H, Olsson PA. (2011). Tit for tat? A mycorrhizal fungus accumulates phosphorus under low plant carbon availability. FEMS Microbiology Ecology 76: 236–244. [DOI] [PubMed] [Google Scholar]
- Heinrichs J, Gradstein SR, Wilson R, Schneider H. (2005). Towards a natural classification of liverworts (Marchantiophyta) based on the chloroplast gene rbcL. Cryptogamie Bryol 26: 131–150. [Google Scholar]
- Heinrichs J, Hentschel J, Wilson R, Feldberg K, Schneider H. (2007). Evolution of leafy liverworts (Jungermanniidae, Marchantiophyta): estimating divergence times from chloroplast DNA sequences using penalized likelihood with integrated fossil evidence. Taxon 56: 31–44. [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Humphreys CP, Franks PJ, Rees M, Bidartondo MI, Leake JR, Beerling DJ. (2010). Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat Commun 1: 7. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ et al. (2006). Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443: 818–822. [DOI] [PubMed] [Google Scholar]
- Johnson DJ, Leake JR, Read DJ. (2001). Novel in-growth core system enables functional studies of grassland mycorrhizal mycelial networks. New Phytol 152: 555–562. [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. (1997). The origin and early evolution of plants on land. Nature 389: 33–39. [Google Scholar]
- Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E et al. (2011). Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333: 880–882. [DOI] [PubMed] [Google Scholar]
- Lin K, Limpens E, Zhang Z, Ivanov S, Saunders DG, Mu D et al. (2014). Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLOS Genet 10: e1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloch DW, Pirozynski KA, Raven PH. (1980). Ecological and evolutionary significance of mycorrhizal symbioses in vascular plants (A Review). Proc Natl Acad Sci USA 77: 2113–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten W, Bailey DJ, Gilligan CA. (2004). Empirical evidence of spatial thresholds to control invasion of fungal parasites and saprotrophs. New Phytol 163: 125–132. [DOI] [PubMed] [Google Scholar]
- Pirozynski KA, Malloch DW. (1975). The origin of land plants: a matter of mycotrophism. Biosystems 6: 153–164. [DOI] [PubMed] [Google Scholar]
- Pressel S, Bidartondo MI, Ligrone R, Duckett JG. (2010). Fungal symbioses in bryophytes: new insights in the Twenty First Century. Phytotaxa 9: 238–253. [Google Scholar]
- Pressel S, Ligrone R, Duckett JG, Davis EC. (2008). A novel ascomycetous endophytic association in the rhizoids of the leafy liverwort family, Schistochilaceae (Jungermanniidae, Hepaticopsida). Am J Bot 95: 531–541. [DOI] [PubMed] [Google Scholar]
- Qiu Y-L, Cho Y, Cox JC, Palmer JD. (1998). The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature 394: 671–674. [DOI] [PubMed] [Google Scholar]
- Qiu Y-L, Li L, Wang B, Chen Z, Dombrovska O, Lee J et al. (2007). Nonflowering land plant phylogeny inferred from nucleotide sequences of seven chloroplast, mitochondrial, and nuclear genes. Int J Plant Sci 168: 691–770. [Google Scholar]
- Qiu Y-L, Li L, Wang B, Chen Z, KnoopV, Groth-Malonek M et al. (2006). The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad USA 103: 15511–15516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2012) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, ISBN 3-900051-07-0 (http://www.R-project.org/). [Google Scholar]
- Read DJ, Duckett JG, Francis R, Ligrone R, Russell A. (2000). Symbiotic fungal associations in ‘lower' land plants. Philos T R Soc B 355: 815–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redecker D, Kodner R, Graham LE. (2000). Glomalean fungi from the Ordovician. Science 289: 1920–1921. [DOI] [PubMed] [Google Scholar]
- Redecker D, Raab P. (2006). Phylogeny of the Glomeromycota (arbuscular mycorrhizal fungi): recent developments and new gene markers. Mycologia 98: 885–895. [DOI] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H. (1994). Four-hundred-million-year-old vesicular-arbuscular mycorrhizae. Proc Natl Acad Sci USA 91: 11841–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimington WR, Pressel S, Duckett JG, Bidartondo MI. (2015). Fungal associations of basal vascular plants: reopening a closed book? New Phytol 205: 1394–1398. [DOI] [PubMed] [Google Scholar]
- Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Susana Magallón S, Lupia R. (2004). Ferns diversified in the shadow of angiosperms. Nature 428: 553–557. [DOI] [PubMed] [Google Scholar]
- Simon L, Bousquet J, Lévesque RC, Lalonde M. (1993). Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature 363: 67–69. [Google Scholar]
- Smith SE, Read DJ. (2008) Mycorrhizal Symbiosis, 3rd edn, Academic Press: Cambridge, UK. [Google Scholar]
- Strullu-Derrien C, Kenrick P, Pressel S, Duckett JG, Rioult J-P, Strullu D-G. (2014). Fungal associations in Horneophyton ligneri from the Rhynie Chert (c. 407 million year old) closely resemble those in extant lower land plants: novel insights into ancestral plant–fungus symbioses. New Phytol 203: 964–979. [DOI] [PubMed] [Google Scholar]
- Stubblefield SP, Taylor TN, Trappe JM. (1987). Fossil mycorrhizae: a case for symbiosis. Science 237: 59–60. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TN, Remy W, Hass H, Kerp H. (1995). Fossil arbuscular mycorrhizae from the early Devonian. Mycologia 87: 560–573. [Google Scholar]
- Villarreal AJC, Crandall-Stotler BJ, Hart ML, Long DG, Forrest LL. (2015). Divergence times and the evolution of morphological complexity in an early land plant lineage (Marchantiopsida) with a slow molecular rate. New Phytol; e-pub ahead of print 27 October 2015; doi:10.1111/nph.13716. [DOI] [PubMed]
- Wang B, Qui Y-L. (2006). Phylogenetic distribution and evolution of mycorrhizas. Mycorrhiza 16: 299–363. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds). PCR Protocols: a Guide to Methods and Applications. Academic Press: London, UK, pp 315–322. [Google Scholar]
- Wikström N, He-Nygrén X, Shaw JA. (2009). Liverworts (Marchantiopsida) In: Blair Hodges S, Kumar S (eds). The Timetree of Life. Oxford University Press: Oxford, UK, pp 146–152. [Google Scholar]
- Yamamoto K, Degawa Y, Yamada A. (2015). Diverse lineages of Mucoromycotina and Glomeromycota that colonize in the subterranean axes of the Asian liverwort, Haplomitrium mnioides from Japan. Poster session presented at the Eighth International Conference on Mycorrhiza; 3–7 Aug. Shinshu University and University of Tsukuba: Flagstaff, AZ, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.