Abstract
Nitrite-oxidizing bacteria (NOB) of the genus Nitrospina have exclusively been found in marine environments. In the brine–seawater interface layer of Atlantis II Deep (Red Sea), Nitrospina-like bacteria constitute up to one-third of the bacterial 16S ribosomal RNA (rRNA) gene sequences. This is much higher compared with that reported in other marine habitats (~10% of all bacteria), and was unexpected because no NOB culture has been observed to grow above 4.0% salinity, presumably due to the low net energy gained from their metabolism that is insufficient for both growth and osmoregulation. Using phylogenetics, single-cell genomics and metagenomic fragment recruitment approaches, we document here that these Nitrospina-like bacteria, designated as Candidatus Nitromaritima RS, are not only highly diverged from the type species Nitrospina gracilis (pairwise genome identity of 69%) but are also ubiquitous in the deeper, highly saline interface layers (up to 11.2% salinity) with temperatures of up to 52 °C. Comparative pan-genome analyses revealed that less than half of the predicted proteome of Ca. Nitromaritima RS is shared with N. gracilis. Interestingly, the capacity for nitrite oxidation is also conserved in both genomes. Although both lack acidic proteomes synonymous with extreme halophiles, the pangenome of Ca. Nitromaritima RS specifically encodes enzymes with osmoregulatory and thermoprotective roles (i.e., ectoine/hydroxyectoine biosynthesis) and of thermodynamic importance (i.e., nitrate and nitrite reductases). Ca. Nitromaritima RS also possesses many hallmark traits of microaerophiles and high-affinity NOB. The abundance of the uncultured Ca. Nitromaritima lineage in marine oxyclines suggests their unrecognized ecological significance in deoxygenated areas of the global ocean.
Introduction
The genus Nitrospina encompasses chemolithoautotrophic nitrite-oxidizing bacteria (NOB), which catalyse the oxidation of nitrite to nitrate, to meet the energy demands for their general metabolism and the fixation of carbon (Lücker and Daims, 2014 and references therein). Ribosomal RNA (rRNA) gene sequences related to members of this genus have exclusively been detected in marine environments, where they are abundant below the euphotic zone and in mesopelagic waters (~10% of all bacteria), and exhibit a preference for marine sediments and oxygen minimum zones (Fuchs et al., 2005; Mincer et al., 2007; Santoro et al., 2010; Zaikova et al., 2010; Füssel et al., 2012; Beman et al., 2013; Nunoura et al., 2015). Recent phylogenomic-based analyses of the draft genome of Nitrospina gracilis 3/211 (Lücker et al., 2013) placed this type species, which was isolated from the Atlantic Ocean (200 miles off the mouth of the Amazon River (Watson and Waterbury, 1971), into a novel bacterial phylum (Nitrospinae). Besides N. gracilis, the only other so far cultivated species of this genus is N. watsonii, which was isolated from the suboxic zone of the Black Sea (Spieck et al., 2014). However, both are phylogenetically closely related and represent only a minor fraction of the vast Nitrospinae 16S rRNA gene sequence diversity in marine environments (Lücker and Daims, 2014).
A recent 16S rRNA gene-based high-throughput survey of bacterioplankton in the brine–seawater interface (BSI) of several brine pools in the Red Sea revealed that Nitrospina-like bacteria constituted up to one-third of the bacterial community in the BSI of Atlantis II Deep (Ngugi et al., 2015). In this oxic–anoxic transition layer, nitrite was undetectable (<0.1 μM), and extremely low concentrations of dissolved oxygen (DO2; ~2.2 μM; Ngugi et al., 2015) and inorganic phosphate (<1 μM; Bougouffa et al., 2013) were encountered. Also, temperature (32 °C), salinity (5.6%) and heavy metal concentrations were higher compared with that in the normal deep-seawater (Ngugi and Stingl, 2012; Ngugi et al., 2015). In comparison, putative NOB have been found to constitute only 2–4% of the overall bacterioplankton community in the overlying oxygenated water column (200–1500 m) that is characterized by isothermal (22 °C) and isohaline (~4.1%) conditions (Qian et al., 2011; Bougouffa et al., 2013). We therefore speculated that the oxic–anoxic interface layers of deep hypersaline anoxic basins (Antunes et al., 2011) could be potential ‘hotspots' for Nitrospinae. How such NOB thrive in the polyextreme BSI remains unclear as no NOB culture has been observed to grow at above 40 g NaCl l–1 (Oren, 2011), presumably due to the low net energy gained from their metabolism (ΔG0′=–74 kJ mol–1 NO2–), which is insufficient for both growth and osmoregulation (Oren, 1999).
Considering the significant divergence of 16S rRNA gene sequences of diverse Nitrospinae representatives (Lücker and Daims, 2014), and the stark physicochemical differences between the BSI of Atlantis II Deep (high salinity and temperature, deeper depth) and the natural habitats of both Nitrospina species described to date (less salinity, shallow water depth; (Watson and Waterbury, 1971; Spieck et al., 2014), we were prompted to investigate the phylogenetic diversity of Nitrospina-like NOB in the BSI of Atlantis II Deep in detail. We also explored the potential genetic basis for their abundance and probable success in the polyextreme BSI environment using single-cell genomics (Lasken, 2012; Stepanauskas, 2012).
Materials and methods
Single-cell sorting, whole-genome amplification and genome annotation
BSI samples were collected from Atlantis II Deep during the Third KAUST-WHOI Red Sea sampling expedition (October/November 2011) as described in (Ngugi et al., 2015). The metadata and sampling details are provided in Supplementary Table S1. Samples were sent to the Single Cell Genomics Center at the Bigelow Laboratory for Ocean Sciences (East Boothbay, ME, USA; http://www.bigelow.org/scgc), where single-cell sorting, whole-genome amplification, sequencing using the Illumina HiSeq 2000 platform (San Diego, CA, USA), assembly using SPAdes v.2.5.1 (Bankevich et al., 2012) and quality control to detect and remove potential contaminants in the assembled contigs were all performed following previously described protocols (Rinke et al., 2013, 2014).
Two single-cell-amplified genomes (SAGs) from the BSI of Atlantis II Deep in the Red Sea—that is, SCGC AAA799-A02 and SCGC AAA799-C22 (in short, RS-SAGs), out of 40 SAGs were identified as being Nitrospina-like, based on the phylogeny of their 16S rRNA genes (Figure 1a and Supplementary Table S2; see Supplementary Data for details regarding the Methods). Their contigs were subsequently annotated using an automated annotation pipeline for microbial genomes, INDIGO (INtegrated Data Warehouse of MIcrobial GenOmes) (Alam et al., 2013) as described recently (Ngugi et al., 2015). Genome completeness and fidelity was estimated using a suite of 104 single-copy marker genes universally present in genome-sequenced bacteria as implemented in CheckM v.0.9.7 (Parks et al., 2015; Supplementary Table S3), whereas pairwise average nucleotide identities (ANIs) between the RS-SAGs and other published Nitrospinae genomes/SAGs were calculated using JSpecies (Richter and Rosselló-Móra, 2009). The average amino-acid identity (AAI) of orthologous protein-coding genes and the degree of gene order conservation (synteny) were determined according to the method of Yelton et al. (2011), using one of the RS-SAGs with the largest assembly (SCGC AAA799-C22; Table 1) as the query. The isoelectric point (pI) of the predicted protein-coding genes were calculated using the ‘iep' script in the EMBOSS v.6.5.1 software package (Rice et al., 2000) as described in Ngugi et al. (2015). SMART was used to predict the modular structures of protein-coding genes (http://smart.embl-heidelberg.de), whereas MAPLE was used to characterize the completeness of KEGG (Kyoto Encyclopedia of Genes and Genomes) functional modules (Takami et al., 2012) and to provide a taxonomical breakdown of the predicted (pan)-genomes. Transport proteins were deduced via the web-based transporter (TransAAP) annotation tool (http://www.membranetransport.org/).
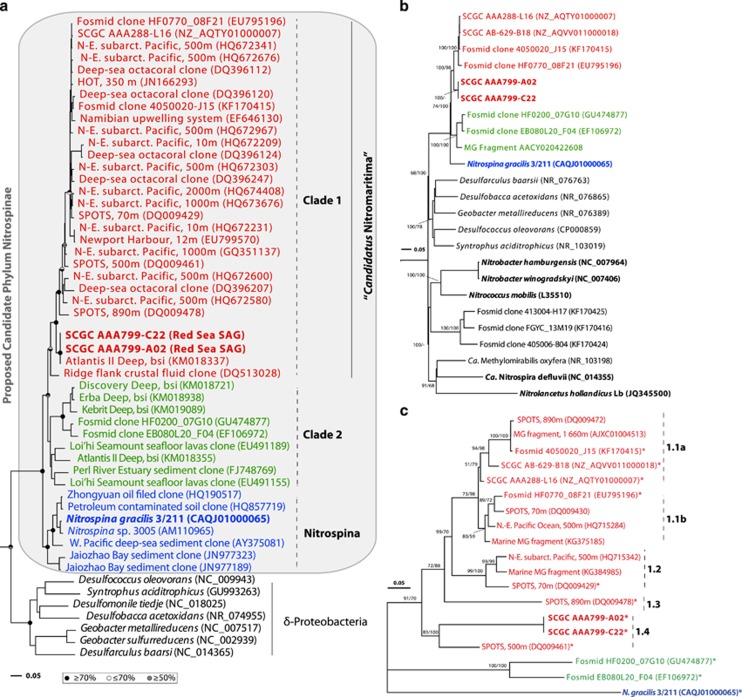
Figure 1.
Phylogenetic placement of the Red Sea Nitrospina-like SAGs (in bold red) inferred using the full-length 16S rRNA (a) and 23S rRNA (b) genes, and the internal transcribed spacer (ITS) region (c). Bootstrap values for maximum-likelihood (ML) and posterior probabilities for Bayesian analysis (BA) are symbolized by an upper (ML) and lower (BA) half circle (in a) or indicated as values above the tree nodes (in b and c). Strains with validated nitrite-oxidizing activity are shown in bold. Bars indicate 5% substitutions per nucleotide position. Flexistipes sinusarabici (NC_015672) and Denitrovibrio acetiphilus (NC_013943) were used as outgroup for the 16S and 23S rRNA gene trees, whereas the ITS tree is unrooted. ITS sequences with asterisk denote those with a corresponding 16S and/or 23S rRNA gene. Although not shown in the 16S rRNA gene tree, SCGC AB-629-B18 carries a partial gene (849 bp), which is 99.5% identical to that of SCGC AAA288-L16.
Table 1. General features of SAGs from this study relative to the type species (Nitrospina gracilis) and related Candidatus Nitromaritima SAGs.
| SAGs/strain | SCGC AAA799-A02a | SCGC AAA799-C22a | SCGC AB-629-B06b,c | SCGC AB-629-B18b,c | SCGC AAA288-L16b,d | N. gracilisb |
|---|---|---|---|---|---|---|
| Draft size (bp) | 1 404 272 | 1 678 166 | 708 914 | 2 139 866 | 2 077 614 | 3 067 213 |
| No. of contigs/N50 (bp) | 97/708 347 | 88/864 303 | 90/354 767 | 73/1 137 313 | 136/1 084 788 | 109/1 571 461 |
| % GC content | 49.4 | 50.5 | 39.6 | 39.7 | 39.5 | 56.2 |
| % Coding regions | 87.6 | 88.2 | 83.1 | 85.8 | 87.5 | 89.6 |
| Predicted ORFs | 1379 | 1639 | 766 | 2206 | 2182 | 2965 |
| % ORFs predicted functional | 74.2 | 76.7 | 61.9 | 60.7 | 72.5 | 78.8 |
| ORF density (ORF/kb) | 1.14 | 1.13 | 1.20 | 1.17 | 1.14 | 1.12 |
| Average ORF size | 892 | 903 | 769 | 832 | 833 | 927 |
| No. of tRNAse | 24 (13) | 18 (14) | 4 (3) | 37 (19) | 32 (15) | 45 (20) |
| No. of rRNA operons (16S-23S-5S) | 1 | 1 | None | 1f | 1 | 1 |
| % Genome overlap with N. gracilisg | 11 | 14 | 5 | 15 | 13 | 100 |
| % ANI of overlapping genome with N. gracilisg | 67 | 67 | 63 | 64 | 63 | 100 |
| % ORFs shared with N. gracilish | 51 | 57 | 44 | 56 | 58 | 100 |
| % AAI of shared ORFs with N. gracilish | 58 | 59 | 58 | 58 | 58 | 100 |
| % Genome completenessi | 20 | 40 | 30 | 73 | 75 | 97 |
Abbreviations: AAI, average amino-acid identity; ANI, average nucleotide identity; INDIGO, INtegrated Data Warehouse of MIcrobial GenOmes; ORF, open-reading frames; SAGs, single-cell-amplified genomes.
Assembly statistics: longest contig size are 66 004 bp (AA799-A02) and 123 650 bp (AAA799-C22), whereas the minimum size is 2 kbp for both.
Based on annotations via our INDIGO pipeline (Alam et al., 2013) using the published draft assemblies.
SAGs from the North Altantic Ocean (Station Archimedes 4, 511 m).
SAG from the North Pacific Ocean (Station ALOHA, 770 m; Swan et al., 2011).
Brackets indicate the number of unique amino acids encoded by the tRNAs.
This SAGs carries an rRNA operon with a partial 16S rRNA gene (849 bp) that is 99.5%, 96.7% and 92% identical to AAA288-L16, AAA799-A02/C22 and N. gracilis, respectively.
Based on whole-genome blastn alignment of the assembled nucleotide bases.
Based on reciprocal best blast hits of their protein-coding genes.
Estimated using 104 single-copy genes common in bacterial genomes using CheckM (Parks et al., 2015). For more details see Supplementary Table S3.
Comparative genomics
Comparative genomics was performed via the phylogenomic pipeline EDGAR (Blom et al., 2009) as described in Ngugi et al. (2015) using the draft genome of N. gracilis 3/211 (accession no. CAQJ00000000) as our reference (Lücker et al., 2013). To make results of the gene-calling and functional prediction steps more coherent for comparative genomics, we reannotated the reference genome and three available Nitrospina-like SAGs that we retrieved from the IMG database under SAG names SCGC AB-629-B06, SCGC AB-629-B18 and SCGC AAA288-L16 using the INDIGO pipeline (Alam et al., 2013). Because of the incompleteness of the RS-SAGs (Supplementary Table S3), we subsequently chose to compare their pan-genome against N. gracilis, and where mentioned, also to the (pan)-genomes of the above Nitrospina-like SAGs from the North Atlantic (SCGC AB629-B06 and SCGC AB629-B18, in short ‘NA-SAGs') and North Pacific (SCGC AAA288-L16, in short ‘NP-SAG' Swan et al., 2011) oceans. The pan-genome is defined here as the non-redundant gene inventory of genome sets, which consists of the ‘core genome' as well as the ‘variable genome'. The variable genome encompasses singleton genes that are specific to each pan-genome, and a flexible gene set that is found in more than one but not all pan-genomes. Hereafter, we will refer to the pan-genome of the RS-SAGs (from the Red Sea) and the two NA-SAGs as Candidatus Nitromaritima RS and Ca. Nitromaritima NA, respectively. For consistency, the single NP-SAG (from the North Pacific) will be referred to as Ca. Nitromaritima sp. NP.
Phylogenetic and fragment recruitment analyses
Phylogenetic analyses were conducted using the 16S (⩾1400 bp) and 23S (⩾2600 bp) rRNA genes, the 16S and 23S internal transcribed spacer region (ITS) and the functional marker genes encoding for nitrite oxidoreductase (NXR) and putative oxygen-based terminal oxidases as described in supporting materials using Geneious Pro v.7.1.2 (http://www.geneious.com).
To estimate the relative abundance of genotypes represented by RS-SAGs in metagenomic data sets of their natural environment and to validate their occurrence (relative to N. gracilis) in other marine environments (Supplementary Table S4), we used comparative fragment recruitment and coverage-based analyses (Rusch et al., 2007; Konstantinidis and DeLong, 2008), as described in Ngugi et al. (2015). These data sets included the water column and brine pools of the Red Sea (Abdallah et al., 2014; Ferreira et al., 2014; Ngugi et al., 2015; Lotaief et al., in preparation) and the water column of the Eastern Tropical South Pacific Oxygen Minimum Zone (ETSP-OMZ; Stewart et al., 2011; Bryant et al., 2012).
Frequency of Nitrospina-like NXR genes in marine habitats
The fact that the NXR enzyme occurs in two phylogenetically distinct forms (Sorokin et al., 2012; Lücker et al., 2010; 2013; Figure 5), which have different nitrite affinities, allowed us to examine the potential abundance of low- and high-affinity NOB (or LNOB and HNOB, respectively) by querying the above metagenomic data sets (Supplementary Table S4) for related gene homologues. Homologues of nrxA genes were searched using tblastx (e-vlaue cutoff score of 10–20) and their relative abundance was estimated by determining the frequency of the retrieved homologues relative to hits of housekeeping genes, which were similarly obtained from the same data sets, as described by Howard et al. (2008). These housekeeping genes were assumed to occur as single-copy genes per cell. Homologues encoding for the archaeal ammonia monooxygenase (AmoA), and respiratory nitrate reductases (NarG; Figure 5) were similarly quantified using customized databases of each gene. Six housekeeping genes retrieved from the genome of Escherichia coli O145:H28 (i.e., recA, dnaK, gyrB, rpoB, atpD and tufA; accession no. CP006027) were used for the normalization. Only metagenomic data sets with a coefficient of variation <20% in the average counts of the housekeeping genes were used.
Statistical analyses
Heatmaps, boxplots and statistical tests were performed within the R software platform (http://www.r-project.org/) using the ggplot2 (http://had.co.nz/ggplot2/) and vegan (http://vegan.r-forge.r-project.org/) packages. Data are expressed as mean±s.d., and differences are considered significant at P<0.05.
Data availability and nucleotide accession numbers
The SGAs and annotations reported in this study can be accessed at http://www.cbrc.kaust.edu.sa/indigo/dataCategories.do. The whole-genome sequences have also been deposited at NCBI under BioProject number PRJNA276499 with accession numbers JZKI00000000 (SCGC AAA799-A02) and JZKJ00000000 (SCGC AAA799-C22).
Results and discussion
Phylogenetic placement of our single-cell genomes
A recent survey of bacterioplankton communities in the BSI of Atlantis II Deep (~2.0 km below the central Red Sea) demonstrated that Nitrospina-like bacteria are relatively abundant in this brine pool (Ngugi et al., 2015). Based on phylogenetic analyses of rRNA gene sequences from two SAGs, which were obtained from the BSI of the same brine pool, we now show that these abundant Nitrospina-like bacteria are highly diverged from N. gracilis (Watson and Waterbury, 1971). Although the rRNA gene sequences of our SAGs are 99.9% identical, their 16S and 23S rRNA genes have identities of only 92% and 85%, respectively, to that of N. gracilis (Figures 1a and b). Phylogenetic analyses also indicate that the rRNA gene sequences of both N. gracilis and our SAGs form distinct lineages within the newly proposed candidate Phylum Nitrospinae (Lücker et al., 2013; Figure 1). The distant affiliation of RS-SAGs to cultivated Nitrospina species at the rRNA gene level implies that they represent a novel uncultured lineage within this phylum. Phylogenetic analyses based on 16S rRNA genes also indicate that the sequence groups of this novel lineage consist of two phylogenetically discrete groups (Clades 1 and 2) that have an average between-clade sequence dissimilarity of 8.4% (Supplementary Table S2). The intraclade average distances range from 3.5% (Clade 1) to 7.3% (Clade 2) at the 16S rRNA gene level, which suggests that the sequences within these clades most likely encompass two separate genera based on the 95% sequence-identity threshold for this taxonomic rank (Hugenholtz et al., 1998). A robust phylogenetic analysis carried out using the ITS region between the 16S and 23S rRNA genes (Figure 1c) indicated further that there are at least five subclusters within Clade 1 (mean pairwise ITS distance of 24.7%), which are distinct from lineages represented by the uncultured Clade 2 and the type species N. gracilis.
Clade 1 encompasses sequences from our single cells and also a representative of the most predominant Nitrospina-like sequence cluster in the BSI of Atlantis II Deep and Discovery Deep (accession no.: KM018337; Guan et al., 2015); representatives of this cluster have an abundance of 80–89% relative to all Nitrospina-like sequences in a recent 454 data set of the same locations (Ngugi et al., 2015; Supplementary Figure S1). It also contains two large-insert DNA fragments (accession nos. EU795196 and KF170415) from the Saanich Inlet (125–500 m depth; (Wright et al., 2013). Interestingly, two publicly available single-cell genomes from the mesopelagic waters of the North Pacific (Station ALOHA, 770 m; SCGC AAA288-L16; Swan et al., 2011) and the North Atlantic (Station Archimedes 4, 511 m; SCGC AB-629-B18) that are 99.5% identical at the 16S rRNA gene level, also affiliate with Clade 1. Our SAGs show identities of only 95% (16S), 93% (23S), and 76% (ITS) to these SAGs from the Atlantic and Pacific Oceans, suggesting that they might consititute a different species. Similarly, Clade 2 is comprised of two fosmid clones from the Montery Bay (Station M1; EB080L20_F04; Mincer et al., 2007) and HOT (station ALOHA; HF0200_07G10; Rich et al., 2011), plus three low-abundance, Nitrospina-like sequences (<1% of all bacterial clones; Guan et al., 2015) from the BSI of three brines in the Red Sea (Erba, Nereus and Kebrit). Because Clade 1 forms a separate cluster from Clade 2 and that of N. gracilis, and considering the average interclade 16S sequence identity of ~91% (Supplementary Table S2) as well as the prominence of Clade 1 sequences in marine environments, we accordingly propose a new provisional genus for Clade 1, ‘Candidatus Nitromaritima' (latin, maritima; belonging to the sea).
Genome features of Nitrospinae SAGs
To get a broader insight into the genomic traits of the two Ca. Nitromaritima SAGs from Atlantis II Deep, we compared their genomic features with those of N. gracilis 3/211 (Lücker et al., 2013) and the Ca. Nitromaritima SAGs from the oxygenated mesopelagic zone of the North Atlantic and North Pacific oceans (Table 1). The draft genome assemblies of SAGs from the Red Sea (RS-SAGs) consist of 88 (SCGC AAA799-C22) and 97 (SCGC AAA799-A02) contigs with a total size of 1.40–1.68 Mbp. Although GC biases cannot be ruled out completely from the MDA and sequencing procedures (Yilmaz et al., 2010), the GC content of RS-SAGs (~50%) is higher compared with that in Ca. Nitromaritima SAGs from the Atlantic (SCGC AB-629-B06 and SCGC AB-629-B18) and the Pacific (SCGC AAA288-L16) of ~40% both are also lower compared with that of N. gracilis (56% GC). Based on a proposed set of 104 single-copy genes found universally in sequenced bacterial genomes (Parks et al., 2015; Supplementary Table S3), we estimated that our SAGs are 20–40% complete, whereas the Ca. Nitromaritima SAGs from the Atlantic and Pacific Oceans (0.71–2.04 Mbp in size) have an estimated completeness of 30–75%. Thus, the complete Ca. Nitromaritima genome would be around 2.71 Mbp (Supplementary Table S3), which is slightly smaller than the 3.2-Mbp genome of N. gracilis (Lücker et al., 2013). Analogous to N. gracilis, >70% of the protein-coding genes in RS-SAGs were predicted functional, with 64% of their pan-proteomes having homologues in the phyla δ-Proteobacteria, Firmicutes, γ-Proteobacteria and Nitrospirae (Supplementary Figure S2).
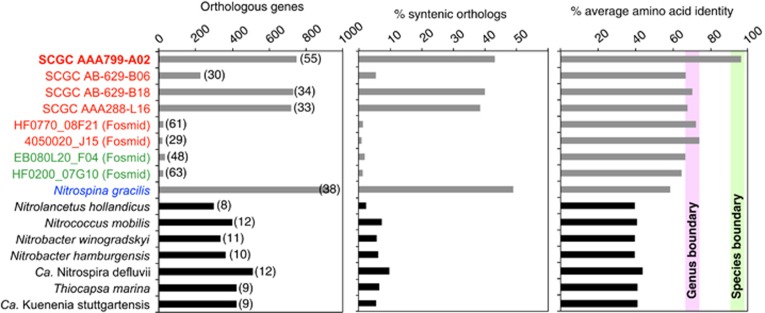
Results of blast-based multiple alignments of genome pairs indicate that the assembled RS-SAGs overlap by 43–52%, with a corresponding ANI of 99% (Supplementary Table S5). These two share 46–55% of their predicted proteins and the AAI of orthologues is high at ~97% (Figure 2). Thus, as expected, over 94% of the 723-shared orthologues were predicted to be syntenic in RS-SAGs, and therefore potentially functionally conserved. However, when the RS-SAGs were compared with the mesopelagic NP- and NA-SAGs, and to N. gracilis, the overlapping genomic fraction was only 11–20%, whereas the ANI values ranged from ~67% to 69%. Only 30–38% of the predicted proteins of N. gracilis and the Nitrospina-like SAGs from the Atlantic and Pacific oceans are predicted to have orthologues in RS-SAGs, and at relatively low identity levels (58.6–69.6% AAI; Figure 2). Despite this, 49% of the orthologues shared by RS-SAGs and N. gracilis are predicted to be syntenic, which suggests that many functions are still conserved between these divergent Nitrospinae lineages. Considering the currently accepted genome-based operational boundaries for species circumscription at the nucleotide (>95% ANI) and proteome (66–72% AAI) levels (Konstantinidis and Tiedje, 2007, and references therein), this additionally supports a novel genus-level placement of the Nitrospina-like RS-SAGs within the proposed phylum Nitrospinae (Lücker et al., 2013).
Figure 2.
Gene conservation between Red Sea SAGs (in bold) and Nitrospina-like bacteria (grey bars) and canonical NOB (black bars). Orthologues were identified using one of the RS-SAGs (SCG A799-C22) as a reference based on the synteny script of Yelton et al. (2011). Nitrospina-like genomes are colour-coded based on their phylogenetic placement in Figure 1. The percentage fraction of orthologous genes in the respective genome (left panel) is indicated in parentheses, whereas the operational genus and species boundaries (bars in the right panel) are as reported by Konstantinidis and Tiedje (2007).
Capturing the genotypic diversity of marine Nitrospinae bacteria
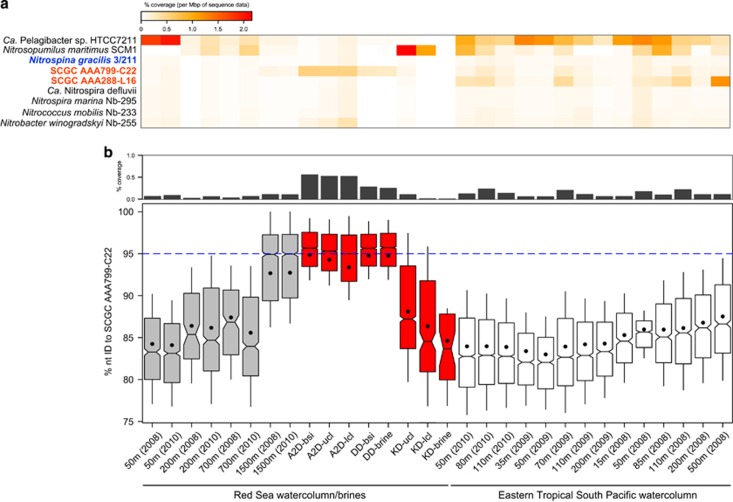
Given the above phylogenomic scenario, we next evaluated the suitability of cultivated Nitrospinae members (i.e., N. gracilis) for capturing the diversity of environmental marine clades through comparative fragment recruitments (Rusch et al., 2007). Based on the coverage and average identity of recruited metagenomics reads against the genome of N. gracilis (Lücker et al., 2013) and SAGs (Figure 3), our analyses show that close relatives of Ca. Nitromaritima but not N. gracilis, are abundant in the BSI of Atlantis II Deep and the ETSP-OMZ. Here, both Ca. Nitromaritima SAGs from the Red Sea (SCGC AAA799-C22) and the Pacific Ocean (SCGC AAA288-L16) recruited significantly more reads than N. gracilis (Figure 3a). The level of recruitment by N. gracilis was surprisingly low (<0.1% of sequence data) even in marine environments, which are known to harbour an abundant (up to 9% of the bacterioplankton) and active ‘Nitrospina' community (Beman and Carolan, 2012; Allers et al., 2013; Beman et al., 2013). These results not only corroborate the ubiquity of Nitrospina-like bacteria in OMZs (Füssel et al., 2012; Beman et al., 2013) but also place representatives of the Ca. Nitromaritima clade as the numerically dominant ‘Nitrospina'-like NOB in the BSI (Red Sea) and in the OMZ (ETSP) habitats.
Figure 3.
Metagenomic fragment recruitment results (a) showing a high degree of coverage when queried against Ca. Nitromaritima SAGs (in bold red) from the Red Sea (SCGC AAA799-C22) and the Pacific Ocean (SCGC AA288-L16) relative to the genomes of N. gracilis (in blue) and other NOB and marine bacterioplankton (in black). (b) Highlights the greater coverage (upper panel) and higher nucleotide identities (lower panel) of recruited reads against the Red Sea SAG both in the overlying 1500-m depth water column of Atlantis II Deep (A2D) and the next three transition layers (bsi, brine–seawater interface; ulc and lcl, upper and lower convective layers) of this brine pool. The blue dotted line demarcates the operational species-based nucleotide identity level for denoting similar genotypes as the reference. DD, Discovery Deep; KD, Kebrit Deep.
An extremely low degree of recruitment (<0.1% coverage) was observed also from genomes of other marine-associated NOB (Figure 3a), including the unpublished genomes of Nitrococcus mobilis (Watson and Waterbury, 1971) and Nitrospira marina (Watson et al., 1986), which were obtained from the IMG database (Markowitz et al., 2014). This implies that the genotypes related to these cultured marine NOB are numerically insignificant in the photic and mesopelagic zones of the Red Sea and the ETSP-OMZ, which is consistent with the niche separation observed between Nitrospina and Nitrococcus species (Füssel, 2014), and between Nitrospina and Nitrospira species (Nunoura et al., 2015). It also reflects their different metabolic strategies, for example, Nitrospina sustains growth at low oxygen and nitrite concentrations (Lücker et al., 2013; Nowka et al., 2015), whereas Nitrococcus adopts an organoheterotrophic lifestyle similar to Nitrobacter (Spieck and Bock, 2005; Füssel, 2014).
Although high salinity is generally recognized to constrain the success of NOB, with >40 g NaCl l–1 (or 4.0% salinity) reported as being inhibitory (Oren, 2011), our results suggest that Ca. Nitromaritima species possibly have a broader salinity (and temperature) range in contrast to this previous assumption. This is evidenced from the increasingly higher recruitment coverage (from ~0.1% to 0.6%) and constantly higher ANI (94±4%) of recruited reads from the overlying 1500-m deep-sea layer towards the first three convective layers of Atlantis II Deep (Figure 3b). In these environments, salinity varies from 4.1% to 15.4% and temperature ranges from 22 to 65 °C (Ngugi et al., 2015). Moreover, Ca. Nitromaritima sequences are also relatively more abundant in the BSI of the adjacent Discovery Deep (~0.3% coverage, 95±4% identity; 13.8% salinity) than in the sulphide-rich BSI and brine of Kebrit Deep (<0.1% coverage; 18.2% salinity). In both of these cases, however, ‘Nitrospina'-like 16S rRNA genes constitute only about 2% of the bacterial community. Hence, the differential recruitment success observed between the two Ca. Nitromaritima SAGs from the Red Sea and the North Pacific —both being highest in their geographical locations where they were isolated from (Figure 3a) —is possibly due to habitat-specific physicochemical differences, which in turn reiterates the phylogenotypic divergence of this group (Lücker and Daims, 2014; this study).
The pan-genome of Ca. Nitromaritima RS
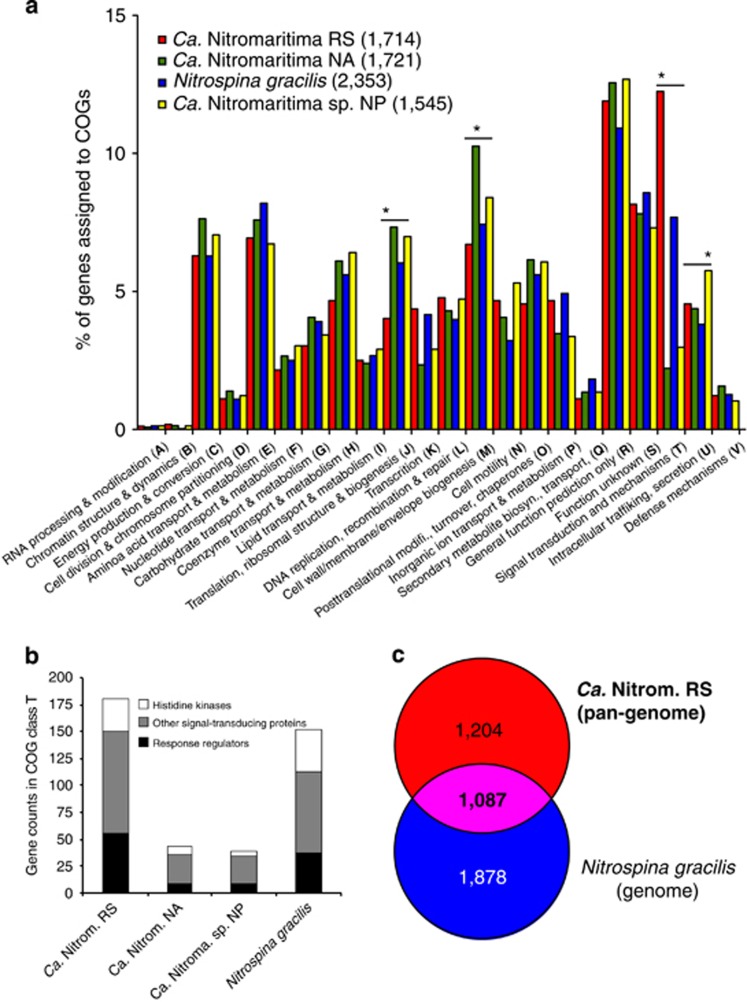
As our main objective was to elucidate potential metabolic attributes differentiating RS-SAGs from their closest cultivated sibling (N. gracilis), we next conducted comparative (pan)-genome analyses through the phylogenomic platform EDGAR (Blom et al., 2009). For clarity, the pan-genome of RS-SAGs, which we simply refer to as Ca. Nitromaritima RS, as they represent cells of the same species (99% ANI and 97% AAI) that are derived from the same sample and location, is defined here as the total non-redundant gene inventory of their ‘core' and ‘variable' genome sets. The pan-genome of Ca. Nitromaritima RS encompasses 2291 genes with a core set of 723 genes, whereas that estimated for the North Atlantic SAGs (or Ca. Nitromaritima NA) comprises of 2492 genes and a core geneset of 482. Hence, the pan-genome size of Ca. Nitromaritima RS is also in the same range as the individual genomes of Ca. Nitromaritima sp. NP (2182 genes) and N. gracilis (2965 genes), which should allow for meaningful comparitive (pan)-genomics.
The COG categories J (translation, ribosomal structure and biogenesis) and T (signal transduction mechanisms) are, respectively, under- and over-represented in Ca. Nitromaritima RS, as compared with the Ca. Nitromaritima SAGs from the Atlantic and Pacific oceans (Figure 4a). Notably, the number of signal-transducing histidine kinases (30 vs 4–8) and response regulators (56 vs 9) are higher in Ca. Nitromaritima RS and N. gracilis relative to the other Ca. Nitromaritima SAGs (Figure 4b). Although these two-component systems may not solely be required for adapting to the polyextreme environments of Atlantis II Deep as they are abundant also in N. gracilis, their unequal distribution among closely related Ca. Nitromaritima species, however, implies that Ca. Nitromaritima RS uses an extensive, albeit unknown array of interactions that are varied from its mesopelagic counterparts.
Figure 4.
Comparative (pan)-genome analysis of the three geographical separated Ca. Nitromaritima SAGs and Nitrospina gracilis. (a) Enriched or under-represented clusters of orthologous genes (COGs) in each (pan)-genome with significantly different COGs denoted by an asterisk above the bars. The total gene counts assigned to COGs in each (pan)-genome are shown in brackets. (b) Illustrates the over-representation of COGs in class T in Ca. Nitromaritima RS and N. gracilis. (c) The size of the ‘core' genome (in bold) and unique gene sets of the RS-SAGs pan-genome (Ca. Nitromaritima RS) and the genome of N. gracilis.
To fully characterize the genomic content and metabolic (dis)similarities between Ca. Nitromaritima RS and N. gracilis, we performed further comparisons between these two data sets. Using the N. gacilis genome as a reference, we found that it shared 1087 genes with Ca. Nitromaritima RS, and that 1204 and 1878 genes were unique to Ca. Nitromaritima RS and N. gracilis, respectively (Figure 4c). Although the core genome is relatively large and accounts for 36.7–47.4% of their protein-coding genes, we also observed that this proportion drops to ~23% when the Atlantic and Pacific Ocean SAGs are included (Supplementary Figure S3), implying an even greater diversity among the marine Nitrospinae lineages.
Besides housekeeping, defence and transport functions, the core genome of N. gracilis and Ca. Nitromaritima RS is predicted to encode the genetic repertoire for nitrite oxidation as detailed below. Additionally, with the exception of some key enzymes of the tricarboxylic acid cycle (TCA; Hügler and Sievert, 2011), namely pyruvate:ferredoxin oxidoreductase (PFOR), 2-oxoglutarate:ferredoxin oxidoreductase (OGFOR), succinyl-CoA synthase, succinate dehydrogenase and phosphoenolpyruvate carboxykinase, all other enzymes are missing in Ca. Nitromaritima RS, presumably due to the low coverage of the RS-SAGs. The SAGs from the Atlantic and Pacific oceans possess (in addition to PFOR and OGFOR) the ATP-dependent citrate lyase that is key for the reductive tricarboxylic acid pathway, and which is present also in N. gracilis (Lücker et al., 2013). Enzymes for anaplerotic TCA reactions are also present in the core genome of N. gracilis and Ca. Nitromaritima RS (Supplementary Table S6), including those for replenishing pyruvate (via alanine dehydrogenase), oxaloacetate (via oxaloacetate decarboxylase or aspartate oxidase) and fumarate (via adenylosuccinate synthetase and adenylosuccinate lyase). Although the functionality of reductive TCA enzymes requires experimental confirmation, it is probable that this pathway represents the universal CO2 fixation mode for members of this phylum.
Interestingly, the predicted PFOR of both Nitrospina and Ca. Nitromaritima RS—and not the Atlantic or Pacific SAGs—colocalizes with a rubrerythrin-like protein upstream of the PFOR operon (Supplementary Figure S4). This indicates that the putative rubrerythrin could be important for protection against oxidative damage (Sztukowska et al., 2002) as all Nitrospinae genomes lack classical reactive oxygen defence mechanisms (e.g., catalase and superoxide dismutase; Lücker et al., 2013; this study); alternatively, the peroxidases in their genomes might also serve this purpose.
Interestingly, unlike the almost complete genome of N. gracilis (and the other three Nitrospina-like SAGs), Ca. Nitromaritima RS is predicted to encode for a pyruvate-water dikinase that could convert phosphoenol pyruvate to pyruvate with concomitant phosphorylation of ADP to ATP. However, N. gracilis, Ca. Nitromaritima RS and NA also encode for a pyruvate dehydrogenase, which could decarboxylate pyruvate to acetyl-CoA, thereby regenerating NAD(P)+. Coupling this fact with the potential dual capacity to regenerate pyruvate (and succinate) and oxaloacetate via the complete methylcitrate cycle and the predicted sodium-translocating oxaloacetate decarboxylase present in all Nitrospinae genomes investigated here, and the possession of a predicted NAD+-dependent malic enzyme (EC. 1.1.1.38) in Ca. Nitromaritima RS (see Figure 6 below), implies that these organisms use similar and also alternative routes for replenishing these central metabolic intermediates.
Surprisingly, although motility and flagella have never been observed in cultured Nitrospina species (Watson and Waterbury, 1971; Spieck et al., 2014), the Ca. Nitromaritima–Nitrospina core genome is predicted to encode genes for flagellar assembly and bacterial chemotaxis. This suggests that motility is an unseen but common trait among members of this novel phylum. Additionally, their core genome also harbours a vitamin B12-uptake system and a few genes of the last steps of the vitamin B12 biosynthesis pathway—that is, adenosylation to the nucleotide loop assembly (Moore et al., 2013), as well as several cobalamin-requiring enzymes (e.g., methionine synthase, methylmalonyl-CoA and ribonucleotide reductase). Because N. gracilis has an almost complete genome and also appears to lack the initial genes for the cobalamin biosynthesis pathway, we speculate that Nitrospinae are vitamin B12 scavengers, which potentially rely on their cobalamin-producing partners (e.g., ammonia-oxidizing archaea; Doxey et al., 2014), for this essential nutrient. It may also partly explain why N. watsonii grows poorly in a minimal synthetic medium without seawater (Spieck et al., 2014), which reportedly contains vitamin B12 of up to 30 pM (Sañudo-Wilhelmy et al., 2012).
Genome-based evidences for nitrite oxidation capacity
As mentioned earlier, Ca. Nitromaritima RS and NA are predicted to possess the capacity for nitrite oxidation. The key signature enzyme for chemolithotrophic nitrite oxidation is the membrane-anchored molybdeterin-binding NXR (Meincke et al., 1992), which occurs in two phylogenetically distinct forms: a cytoplasmic-oriented type (Spieck and Bock, 2005) and a periplasmatic type (Lücker et al., 2010; 2013). Both RS-SAGs carry a single periplasmatic-type NXR operon (Figure 5a), which harbours the substrate-binding α-subunit (NxrA) with a predicted signal peptide on the N terminus, an Fe-S cluster-containing β-subunit (NxrB), a γ-subunit (NxrC) and a putative TorD-like chaperone subunit (NxrD) inserted between NxrB and NxrC (Meincke et al., 1992). NxrD probably orchestrates cofactor acquisition and subunit assembly. The nxrABDC operon structure of Ca. Nitromaritima species is therefore dissimilar to N. gracilis, where the nxrD gene is isolated from the two nxrABC operons (Lücker et al., 2013). However, like in N. gracilis (Lücker et al., 2013), the NxrC subunit of the two Ca. Nitromarita SAGs is also predicted to possess a signal peptide that overlaps with a transmembrane-spanning region. Altogether, these evidences imply that the NXR complex of Ca. Nitromaritima is oriented similarly as that of N. gracilis (Lücker et al., 2013; Spieck et al., 2014), in that the NxrC subunit is anchored to the cytoplasmic membrane, whereas the catalytic subunit (NxrA) faces the periplasm. The NxrABDC from Ca. Nitromaritima species is closely related to the N. gracilis enzymes by 91% (NxrA), 97% (NxrB), 75% (NxrD) and 64–69% (NxrC) amino-acid sequence identities, suggesting that they are functionally equivalent. Interestingly, downstream of the NXR gene cluster are two genes coding for a putative nitrate/nitrite sensor-like histidine kinase (NarX) and its cognate response regulator, which is not the case in N. gracilis.
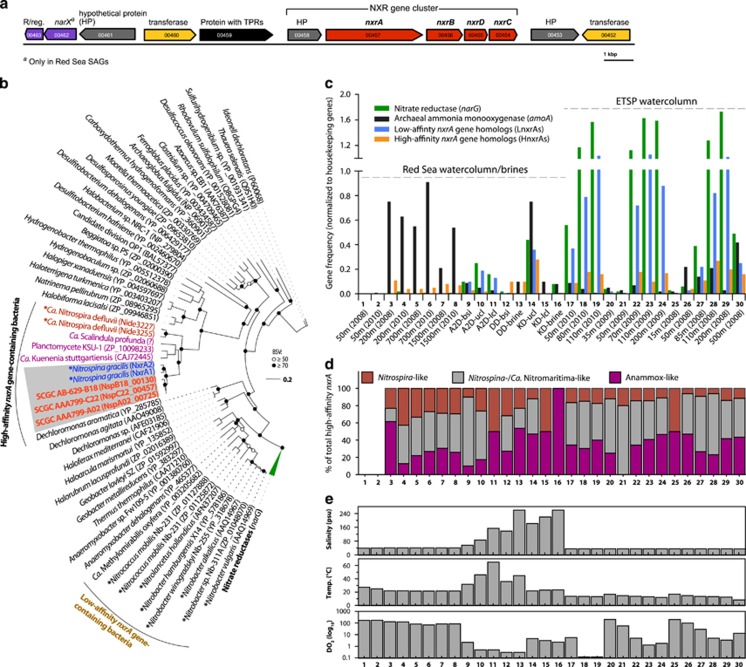
Figure 5.
Organization (a), phylogeny (b) and distribution (c and d) of genes coding for nitrite oxidoreductase (NXR) in the Red Sea SAGs. (a) Shows the NXR gene cluster (in bold) in one of the RS-SAGs (SCGC AAA799-C22), whereas (b) illustrates the phylogenetic position of the nxrA genes among other selected members of the type II dimethyl sulphoxide reductase enzyme family. Coloured leaves of the maximum-likelihood tree show the placement of the two distinct forms of nxrA genes of low- and high-affinity NOB discussed in the main text. Validated aerobic nitrite oxidizers are marked with an asterisk, whereas Ca. Nitromaritima SAGs are shown in bold. (c and d) The frequency of key genes for nitrite dissimilation (relative to housekeeping genes) and the taxonomy of nxrA genes in metagenomic data sets, whereas (e) highlights the variability in three key environmental variables among these data sets. ‘Anammox-like' refers to anaerobic ammonia-oxidizing Planctomycetes. Note that the frequency of nxrA gene types of anammox bacteria is combined together with those of high-affinity NOB in panel c.
Phylogenetically, the nxrA and nxrB encoding genes of RS-SAGs cluster with those of NOB (Figure 5b and Supplementary Figure S5), which harbour periplasmic-oriented NXRs (e.g., Nitrospira and Nitrospina; Lücker et al., 2010, 2013) and have high nitrite affinities (Km=~10–29 μM; (Schramm et al., 1999; Maixner et al., 2006; Nowka et al., 2015). This suggests that the local NOB in the BSI probably operate at low nitrite concentrations. High-affinity NOB (HNOB) are known to be repressed by high nitrite concentrations (>20 mM) and achieve growth at lower concentrations (10- to 100-fold; Spieck and Lipski, 2011) compared with LNOB harbouring the cytoplasmic-oriented NXR (e.g., Nitrobacter and Nitrococcus). The latter require 2–30 mM nitrite (Spieck and Bock, 2005) and have a high Km for NO2– of ~0.41 to 1 mM (Hunik et al., 1993; Sorokin et al., 2012; Nowka et al., 2015).
Based on the relative abundance of the high-affinity nxrA gene (i.e., HnxrA) homologues comprising those of HNOB (Nitrospina- and Nitrospira-like) and anaerobic ammonia-oxidizing (anammox) bacteria, as well as low-affinity nxrA-like genes from LNOB in metagenomic data sets (Figures 5c and d), we can deduce that the organisms harbouring HnxrA are generally abundant at suboxic zones (<20 μM DO2) and are less prominent in the photic (oxygenated) marine zones. At the specific level of nxrA gene counts from LNOB and HNOB only—that is, excluding nxrA gene homologues of anammox bacteria that have a different physiology from NOB, we found that the LNOB were up to 10 times more frequent than HNOB, especially in the brine habitats and the water column of the ETSP (Supplementary Figure S6). These differences, however, might not be directly correlated to their population structure because of biases in genome copies of nxrA genes between these NOB, which is highest among LNOB (e.g., 2–4 in Nitrobacter and Nitrolancentus species; Starkenburg et al., 2006, 2008; Sorokin et al., 2012) and lowest in HNOB (e.g., 2 copies in Nitrospira and Nitrospina species; Lücker et al., 2010, 2013). Based on the taxonomy of the best blastx hits (e-value <10–20 and >90% coverage), we observed that relative to all HnxrAs in the BSI and ETSP-OMZ, most sequences were closely related with Nitrospina/Ca. Nitromaritima-like bacteria (18–60%), decreasing in the order of relative abundance to anammox-like bacteria (10–50%), and Nitrospira (6–50% Figure 5d), which underscores the role of Nitrospinae in deoxygenated marine environments (Levipan et al., 2014).
Despite the potential copy number differences, the abundance of nxrA gene type of HNOB covaried positively with that of LNOB (Spearman's rank correlation r=0.53, P<0.001; Supplementary Table S7), suggesting that the same environmental factors probably influence their ecology. Our data also show that the frequency of both nxrA gene types is strongly and positively correlated with that of narG (r=0.53–0.97, P<0.001), and that HNOB positively correlate with amoA gene frequencies (r=0.40, P<0.05). These results corroborate molecular data that indicate considerable correlations of Nitrospina-like 16S rRNA gene abundances with thaumarchaeal amoA and 16S rRNA gene frequencies (Mincer et al., 2007; Santoro et al., 2010), and findings from tracer experiments in the ESTP-OMZ (Beman and Carolan, 2012), which showed that the activities of NOB are most likely sustained by nitrate reducers and ammonia oxidizers.
Furthermore, taking into account three key environmental variables known to influence the population structure and activities of NOB, namely temperature, salinity and DO2 (Focht and Verstraete, 1977; Schramm et al., 1999; Alawi et al., 2009; Lebedeva et al., 2010; Oren, 2011), we found that the frequency of all the above genes is significantly negatively correlated with DO2 levels (and temperature; Table 2). Surprisingly, the occurence of both nxrA gene types of HNOB and LNOB showed no significant correlation with salinity (P>0.05), suggesting that it could be less important than DO2 and temperature for shaping the ecology of marine NOB.
Table 2. Spearman's correlation coefficient (r) between gene frequency and three environmental variables.
| Genea,b |
Parameter |
||
|---|---|---|---|
| Dissolved O2 | Temperature | Salinity | |
| nxrA (low-affinity type) | −0.42** | −0.50** | −0.30 |
| All nxrA (high-affinity type)c | −0.58*** | −0.33* | 0.14 |
| HNOB-only nxrA (high-affinity type)d | −0.66*** | −0.19 | 0.17 |
| narG | −0.46** | −0.53*** | −0.25 |
| amoA | 0.01 | −0.57*** | 0.29 |
Abbreviations: HNOB, high-affinity nitrite-oxidizing bacteria.
*P<0.05; **P<0.01; ***P<0.001.
Gene frequencies are based on data presented in Figure 5c.
See Figure 5b for phylogeny of nxrA genes.
Also includes nxrA genes from anaerobic ammonia-oxidizing (anammox) bacteria.
Excludes anammox-like nxrA genes.
Niche adaptation traits of Ca. Nitromaritima RS
Oxic–anoxic environments are considered as suitable habitats for chemolithoautotrophs, principally due to the opposing steep gradients of chemical reactants (Brune et al., 2000). The polyextreme BSI poses additional challenges such as the high salinity, increasing temperatures (at least in Atlantis II Deep) and elevated concentration of heavy metals (Antunes et al., 2011; Ngugi et al., 2015). Salinity in particular imposes thermodynamic constraints for chemolithoautotrophic NOB (Oren, 2011), as very little energy is gained from their metabolism (ΔG0′=–74 kJ mol–1 NO2–) to support both growth and osmoregulation. Accordingly, the variable and unique (Supplementary Table S8) genome of Ca. Nitromaritima RS should reveal traits for its adaptation to the polyextreme BSI environment of Atlantis II Deep.
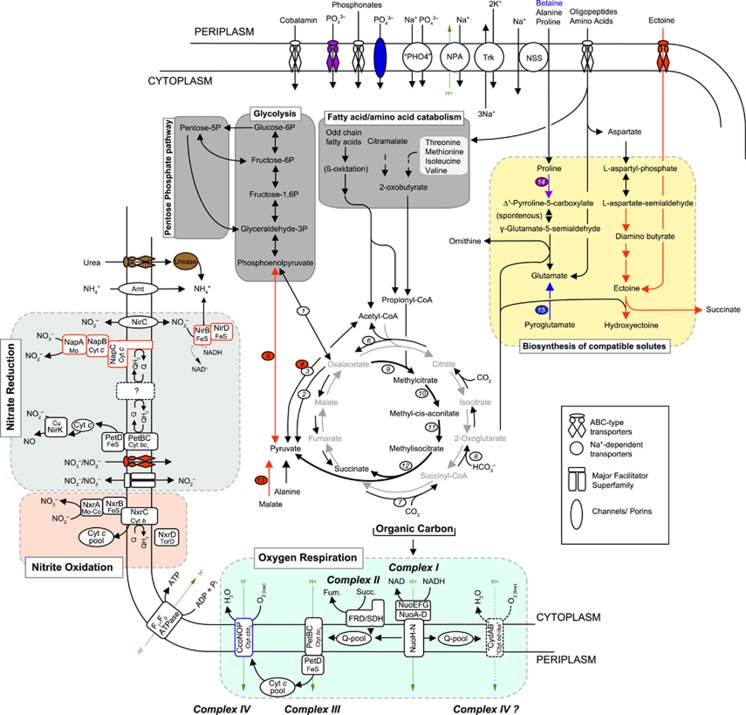
Among all currently genome-sequenced NOB, only Nitrococcus mobilis Nb-231 and Ca. Nitromaritima RS harbour the potential to synthesize the osmoprotectant ectoine (Supplementary Figure S7), and in the latter case, also hydroxyectoine. The occurrence of a putative ABC-type transporter (EhuACBD) for ectoine in Ca. Nitromaritima RS and its localization next to EctABCD operon (Supplementary Figure S7) implies that Ca. Nitromaritima RS can use extracellular ectoine. In view of the thermoprotective properties of hydroxyectoine (Tanne et al., 2014) and the estimated lower energy costs for biosynthesis of osmolytes with five to six carbons (Oren, 1999), this genomic trait possibly represents a crucial adaptation for such NOB in the hypersaline and relatively hot Atlantis II brine pool. The fact that the genome also encodes transporters and enzymes involved in the synthesis and interconversion of other small compatible solutes (proline, glutamate and glycerol; Figure 6 and Supplementary Table S9B) implies that Ca. Nitromaritima RS additionally uses mixtures of osmolytes for osmoregulation. Interestingly, although Ca. Nitromaritima RS and N. gracilis harbour a predicted Trk-type system (Figure 6) that is used by many bacteria, including extreme halophiles for maintaining high internal concentrations of K+ (Corratgé-Faillie et al., 2010), their predicted proteomes do not show the characteristic acid-shifted isoelectric point of typical halophiles (Supplementary Figure S8). This implies that these energy-limited organisms heavily depend on compatible solutes for osmoprotection.
Figure 6.
Metabolic scheme of predicted pathways that are either unique to the pan-genome of Ca. Nitromaritima RS (in red), encoded only in the genome of Nitrospina gracilis (in blue), shared by both (in purple), present in all Nitrospina-like genomes (in black), or only in the other Ca. Nitromaritima SAGs (in brown). Dashed black arrows and a question mark symbol denote uncertain reactions, whereas grey and thick black arrows show the oxidative tricarboxylic acid cycle and the methylcitrate (anaplerotic) pathways, respectively. Important enzymes discussed in the main text are indicated with an oval shape: (1) phosphoenolpyruvate carboxykinase, (2) oxaloacetate decarboxylase, (3) pyruvate:ferredoxin oxidoreductase, (4) pyruvate dehydrogenase, (5) pyruvate-water dikinase, (6) ATP-dependent citrate lyase, (7) succinyl-CoA synthase, (8) oxoglutarate:ferredoxin oxidoreductase, (9) methylcitrate lyase, (10) methylcitrate dehydratase, (11) aconitase, (12) 2-methylisocitrate synthase, (13) NAD+-dependent malic enzyme, (14) proline dehydrogenase, (15) 5-oxoprolinase. NPA, sodium/proton antiporter; NSS, sodium:solute symporter; Trk, a potassium uptake system.
Chemolithoautotrophic nitrite oxidizers require oxygen as a terminal acceptor for their metabolism. Because very low DO2 concentrations are a permanent feature of the BSI environment (~0.3–5 μM in the BSI; Ngugi et al., 2015), there is also a selective pressure for microorganisms to remain metabolically active under low ambient DO2 levels. Unsurprisingly, all Ca. Nitromaritima species (and N. gracilis) lack homologues of the low-affinity aa3-type haeme–copper oxidases (complex IV) that are present in Nitrobacter species (Starkenburg et al., 2008) but encode for an energy-converting electron transport chain that shuttles electrons to terminal oxidase systems capable of using critically low DO2 levels (Figures 6 and 7). Although they also lack a high-affinity cbb3-type haeme–copper oxidases that is present in N. gracilis (Lücker et al., 2013), presumably due to their low coverage, they appear to encode two putatively different and phylogenetically distinct oxygen-based terminal oxidases (Supplementary Figure S9).
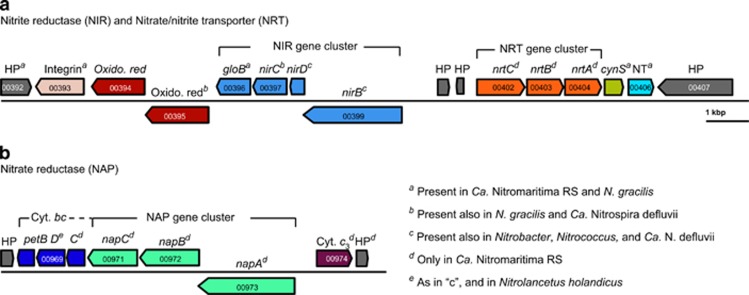
Figure 7.
The organization of key enzymes in the pan-genome of Ca. Nitromaritima RS predicted to encode for (a) a cytoplasmic nitrite reductase (NIR) and an ABC-type nitrate/nitrite transporter (NRT), and (b) a putative nitrate reductase (NAP). Superscripts ‘a' to ‘e' following the gene symbols highlight the presence or absence of the respective gene in other NOB as indicated in the legend. Gene loci IDs are based on annotations of the Red Sea SAG, SCGC AAA799-C22. cynS, cyanate hydratase; HP, hypothetical protein; NT, a high-affinity nickel transporter.
The predicted subunit I of these two putative enzymes have a pairwise average identity of only 32% at the amino acid-level. The first enzyme exhibits properties similar to haeme–copper oxidases, namely presence of four haeme and two copper-binding sites (Supplementary Figure S9B and Pereira et al., 2001), and also shows considerable similarity (40% AAI) to the uncharacterized but functionally transcribed ‘cyt. bd-like oxidase' (NIDE0901) of Ca. Nitrospira defluvii (Lücker et al., 2010). The α-subunit has a signal peptide and four to five transmembrane domains, which implies localization in the membrane, and is colocalized with other additional subunits with di-haeme residues. The second predicted enzyme (designated here as ‘bd-like enzymes' Supplementary Figure S9A) has an unclear function as it is only 26% identical to canonical bd-type oxidases and has neither copper-binding sites nor residues implicated in quinol binding (Yang et al., 2007). The predicted subunit I possesses between 13 and 15 transmembrane domains, which contrasts with canonical bd-type quinol oxidases (Bertsova et al., 1997; Yang et al., 2007). Although functional equivalence cannot be ascertained by homology, we speculate that the ‘cyt. bd-like oxidases' might be involved in electron transfer, which necessitates an assessment of their proton-pumping potential and roles in the membrane energetics of Nitrospina and its relatives.
Ca. Nitromaritima RS appears also to have another energy production mode. Unlike all other so-far sequenced NOB, it is predicted to encode a putative periplasmic-oriented nitrate reductase (NAP) colocalized together with three putative cytochrome bc-like subunits that potentially function as electron carrier units (Figure 7 and Supplementary Table S8). This implies the possible capacity to use nitrate as an electron acceptor, which should be advantageous because it is energetically more favourable, for example, when coupled to the oxidation of formate (ΔG0′=–369 kJ mol−1 NO3–) or H2 (ΔG0′=–123 kJ mol−1 NO3–) compared with growth solely dependent on the oxidation of nitrite with O2 (ΔG0′=–74 kJ mol−1 NO2–). Similar to other NOB, Ca. Nitromaritima RS harbours also a periplasmic copper-containing nitrite reductase (NirK), which typically catalyses the reduction of nitrite to nitric oxide (NO). However, both formate dehydrogenase and/or the catalytic subunit of a hydrogenase (see below), which would enable the generation of a proton-motive force, assuming that both are membrane-bound and cytoplasmatically oriented, are missing in these low-coverage SAGs. The physiological role of the NAP system therefore remains unclear. Experimentally, only Nitrobacter species, Nitrospira moscoviensis and N. mobilis have been shown to reduce nitrate with low-potential electron donors (i.e., H2, formate, acetate and pyruvate; Smith and Hoare, 1968; Bock et al., 1987; Freitag et al., 1987; Füssel, 2014; Koch et al., 2015), producing nitrite and NO, and also ammonium (Nitrobacter). These NOB, however, only possess NIR-type nitrite reductases (NirK, NirA and NADH-dependent nitrite reductase, NirBD) but lack nitrate reductases; here, NXR is proposed to function in the reverse direction under anoxic conditions (Starkenburg et al., 2008; Füssel, 2014).
Similar to all sequenced NOB, Ca. Nitromatima RS is also equipped with a mechanism for reducing nitrite to ammonia based on the presence of a cytoplasmic NirBD. However, it remains unclear whether NirBD is repressed by the high ammonium concentrations (2–387 μM) in the interfaces of Atlantis II Deep (Ngugi et al., 2015). Intriguingly, N. gracilis appears to use a ferredoxin-dependent nitrite reductase (NirA) instead of NirBD for nitrite assimilation (Lücker et al., 2013), which is similar to some Nitrospira species (Koch et al., 2015). All Nitrospinae species also possess an Amt-type transporter for taking up ammonium, which can alternatively be supplied from the hydrolysis of urea and cyanate as Nitrospinae encode, albeit differentially (Figure 6), a urease (absent in N. gracilis) and a cyanate hydratase (present in all Nitrospinae genomes).
Interestingly, Nitrospina and Nitrospira species appear to have adopted other electron-scavenging pathways also with higher net energy yields such as the Knallgas metabolism (ΔG0′=–237 kJ mol−1 H2; Koch et al., 2014) and anaerobic nitrate/nitrite reduction coupled to H2 oxidation (Ehrich et al., 1995). However, none of the Ca. Nitromaritima SAGs encode the putative cytoplasmic type 3b (bidirectional) hydrogenase of N. gracilis (Lücker et al., 2013), although one of the RS-SAGs (SCGC AAA799-C22) harbours a partial operon encoding homologues of proteins known to be essential for the maturation of hydrogenases, namely an endopeptidase and the so-called hyp genes, HypD and HypE (Forzi and Sawers, 2007). The presence of a predicted Na+-proton antiporter in the Ca. Nitromaritima–Nitrospina core genome (Supplementary Table S6) also implies the potential to generate an electrochemical sodium gradient (Padan and Schuldiner, 1994), which is likely coupled to the proton-motive force generated by nitrite oxidation (or via nitrate reduction), to drive the uptake of extracellular nutrients (including osmolytes such as proline and glycine betaine) using several putative Na+-dependent solute uptake systems encoded in their genomes (Figure 6).
Although present also in N. gracilis, the potential capacity to detoxify heavy metals (e.g., mercury, zinc, cobalt, copper and cadmium) and the possession of alternative mechanisms to sequester scarce nutrients, for example, phosphate using a high-affinity phosphate-specific transporter (PstSCABU) or the phosphonates uptake (PhnDEC) and degradation system, are all probably also necessary in the polyextreme habitat of Ca. Nitromaritima RS.
Conclusions
Although NOB have an important role in oceanic nitrification and have great phylogenetic diversity and distribution, our understanding of their functions and ecology in the global ocean has generally lagged behind that of their ammonia-oxidizing counterparts, mostly due to the paucity of cultivated marine representatives. This is unfortunately true for members of the proposed phylum Nitrospinae, despite of their prevalence in large swaths of the suboxic ocean. Using single-cell genomics, we show here that the abundant Nitrospina-like bacteria in the BSI of Atlantis II Deep encompass a novel lineage within this phylum (referred to here as Ca. Nitromaritima RS), which is divergent from all cultivated Nitrospina species. Most importantly, we show that these single-cell genomes possess the genetic repertoire for nitrite oxidation, which implies that the uncultured lineages of the phylum Nitrospinae are probably all capable of nitrite oxidation. Using metagenomic fragment recruitment, we also show that Ca. Nitromaritima-like but not Nitrospina-like genotypes are abundant in the anoxic and hypersaline, deeper convective layers of Atlantis II Deep. Genomic data indicate that Ca. Nitromaritima RS possess machineries for taking up a mixture of osmolytes as well as for the biosynthesis ectoine/hydroxyectoine, and are thus capable of combating salt stress. The predicted potential to use nitrate as an alternative electron acceptor coupled putatively to the oxidation formate or hydrogen might be crucial in adapting to the microoxic, polyextreme brine environment.
Acknowledgments
We thank Dr Sebastian Lücker for providing the nitrite reductase (NxrA/B) gene alignments and Dr Andre Antunes for help with sample collection. Financial support to RS was provided by the US National Science Foundation Grant OCE-1232982. This study was supported by King Abdullah University of Science and Technology through the SEDCO Research Excellence award and baseline funds to US.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abdallah RZ, Adel M, Ouf A, Sayed A, Ghazy MA, Alam I et al. (2014). Aerobic methanotrophic communities at the Red Sea brine–seawater interface. Front Microbiol 5: 487–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam I, Antunes A, Kamau AA, Alawi WB, Kalkatawi M, Stingl U et al. (2013). INDIGO—INtegrated Data Warehouse of MIcrobial GenOmes with Examples from the Red Sea Extremophiles. PLoS One 8: e82210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alawi M, Off S, Kaya M, Spieck E. (2009). Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environ Microbiol Rep 1: 184–190. [DOI] [PubMed] [Google Scholar]
- Allers E, Wright JJ, Konwar KM, Howes CG, Beneze E, Hallam SJ et al. (2013). Diversity and population structure of Marine Group A bacteria in the Northeast subarctic Pacific Ocean. ISME J 7: 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes A, Ngugi DK, Stingl U. (2011). Microbiology of the Red Sea (and other) deep-sea anoxic brine lakes. Environ Microbiol Rep 3: 416–433. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beman JM, Carolan MT. (2012). Deoxygenation alters bacterial diversity and community composition in the ocean's largest oxygen minimum zone. Nat Commun 4: 2705–2705. [DOI] [PubMed] [Google Scholar]
- Beman JM, Leilei Shih J, Popp BN. (2013). Nitrite oxidation in the upper water column and oxygen minimum zone of the eastern tropical North Pacific Ocean. ISMEJ 7: 2192–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsova YV, Bogachev AV, Skulachev VP. (1997). Generation of protonic potential by the bd-type quinol oxidase of Azotobacter vinelandii. FEBS Lett 414: 369–372. [DOI] [PubMed] [Google Scholar]
- Blom J, Albaum SP, Doppmeier D, Pühler A, Vorhölter F-J, Zakrzewski M et al. (2009). EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinform 10: 154. [DOI] [PMC free article] [PubMed]
- Bock E, Wilderer PA, Freitag A. (1987). Growth of Nitrobacter in the absence of dissolved oxygen. Water Res 22: 245–250. [Google Scholar]
- Bougouffa S, Yang JK, Lee OO, Wang Y, Batang Z, Al-Suwailem A et al. (2013). Distinctive microbial community structure in highly stratified deep-sea brine water columns. Appl Environ Microbiol 79: 3425–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune A, Frenzel P, Cypionka H. (2000). Life at the oxic–anoxic interface: microbial activities and adaptations. FEMS Microbiol Rev 24: 691–710. [DOI] [PubMed] [Google Scholar]
- Bryant JA, Stewart FJ, Eppley JM, DeLong EF. (2012). Microbial community phylogenetic and trait diversity declines with depth in a marine oxygen minimum zone. Ecology 93: 1659–1673. [DOI] [PubMed] [Google Scholar]
- Corratgé-Faillie C, Jabnoune M, Zimmermann S, Véry AA, Fizames C, Sentenac H. (2010). Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family. Cell Mol Life Sci 67: 2511–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxey AC, Kurtz DA, Lynch MD, Sauder LA, Neufeld JD. (2014). Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISMEJ 9: 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. (1995). A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol 164: 16–23. [DOI] [PubMed] [Google Scholar]
- Ferreira AJS, Siam R, Setubal JC, Moustafa A, Sayed A, Chambergo FS et al. (2014). Core microbial functional activities in ocean environments revealed by global metagenomic profiling analyses. PLoS One 9: e97338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focht DD, Verstraete W. (1977). Biochemical ecology of nitrification and denitrification. In: Alexander M (ed), Advances in Microbial Ecology. Springer-Verlag: New York, NY, USA, pp 135–214. [Google Scholar]
- Forzi L, Sawers RG. (2007). Maturation of [NiFe]-hydrogenases in Escherichia coli. Biometals 20: 565–578. [DOI] [PubMed] [Google Scholar]
- Freitag A, Rudert M, Bock E. (1987). Growth of Nitrobacter by dissimilatoric nitrate reduction. FEMS Microbiol Lett 48: 105–109. [Google Scholar]
- Fuchs BM, Woebken D, Zubkov MV, Burkill P, Amann R. (2005). Molecular identification of picoplankton populations in contrasting waters of the Arabian Sea. Aquat Microb Ecol 39: 145–157. [Google Scholar]
- Füssel J. (2014). Impacts and importance of ammonia and nitrite oxidation in the marine nitrogen cycle. PhD thesis, Max Planck Institute for Microbial Ecology, Bremen, Germany.
- Füssel J, Lam P, Lavik G, Jensen MM, Holtappels M, Günter M et al. (2012). Nitrite oxidation in the Namibian oxygen minimum zone. ISME J 6: 1200–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Hikmawan T, Antunes A, Ngugi DK, Stingl U. (2015). Diversity of methanogens and sulfate-reducing bacteria in the interfaces of five deep-sea anoxic brines of the Red Sea. Res Microbiol 166: 688–699. [DOI] [PubMed] [Google Scholar]
- Howard EC, Sun S, Biers EJ, Moran MA. (2008). Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ Microbiol 10: 2397–2410. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR. (1998). Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180: 4765–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunik JH, Meijer H, Tramper J. (1993). Kinetics of Nitrobacter agilis at extreme substrate, product and salt concentrations. Appl Microbiol Biotechnol 40: 442–448. [Google Scholar]
- Hügler M, Sievert SM. (2011). Beyond the Calvin Cycle: autotrophic carbon fixation in the ocean. Annu Rev Mar Sci 3: 261–289. [DOI] [PubMed] [Google Scholar]
- Koch H, Galushko A, Albertsen M, Schintlmeister A, Gruber-Dorninger C, Lücker S et al. (2014). Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science 345: 1052–1054. [DOI] [PubMed] [Google Scholar]
- Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E et al. (2015). Expanded metabolic versatility of ubiquitous nitrite-oxidzing bacteria from the genus Nitrospira. Proc Natl Acad Sci USA 112: 11371–11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, DeLong EF. (2008). Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J 2: 1052–1065. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. (2007). Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin in Microbiol 10: 504–509. [DOI] [PubMed] [Google Scholar]
- Lasken RS. (2012). Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol 10: 631–640. [DOI] [PubMed] [Google Scholar]
- Lebedeva EV, Off S, Zumbrägel S, Kruse M, Shagzhina A, Lücker S et al. (2010). Isolation and characterization of a moderately thermophilic nitrite-oxidizing bacterium from a geothermal spring. FEMS Microbiol Ecol 75: 195–204. [DOI] [PubMed] [Google Scholar]
- Levipan HA, Molina V, Fernandez C. (2014). Nitrospina-like bacteria are the main drivers of nitrite oxidation in the seasonal upwelling area of the Eastern South Pacific (Central Chile ∼36°S). Environ Microbiol Rep 6: 565–573. [DOI] [PubMed] [Google Scholar]
- Lücker S, Daims H. (2014). The Family Nitrospinaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F. (eds), The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria, vol. XIX. Springer-Verlag: Berlin, Germany, pp 231–237.
- Lücker S, Nowka B, Rattei T, Spieck E, Daims H. (2013). The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B et al. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA 107: 13479–13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M et al. (2006). Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol 8: 1487–1495. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen I-MA, Chu K, Szeto E, Palaniappan K, Pillay M et al. (2014). IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res 42: D568–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meincke M, Bock E, Kastrau D, Kroneck P. (1992). Nitrite oxidoreductase from Nitrobacter hamburgensis: redox centers and their catalytic role. Arch Microbiol 158: 127–131. [Google Scholar]
- Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF. (2007). Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol 9: 1162–1175. [DOI] [PubMed] [Google Scholar]
- Moore SJ, Lawrence AD, Biedendieck R, Deery E, Frank S, Howard MJ et al. (2013). Elucidation of the anaerobic pathway for the corrin component of cobalamin (vitamin B12). Proc Natl Acad Sci USA 110: 14906–14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngugi DK, Blom J, Alam I, Rashid M, Ba-Alawi W, Zhang G et al. (2015). Comparative genomics reveals adaptations of a halotolerant thaumarchaeon in the interfaces of brine pools in the Red Sea. ISME J 9: 396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngugi DK, Stingl U. (2012). Combined analyses of the ITS loci and the corresponding 16S rRNA genes reveal high micro- and macrodiversity of SAR11 populations in the Red Sea. PLoS One 7: e50274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowka B, Daims H, Spieck E. (2015). Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl Environ Microbiol 81: 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunoura T, Takaki Y, Hirai M, Shimamura S, Makabe A, Koide O et al. (2015). Hadal biosphere: insight into the microbial ecosystem in the deepest ocean on Earth. Proc Natl Acad Sci USA 112: E1230–E1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. (1999). Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev 63: 334–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. (2011). Thermodynamic limits to microbial life at high salt concentrations. Environ Microbiol 13: 1908–1923. [DOI] [PubMed] [Google Scholar]
- Padan E, Schuldiner S. (1994). Molecular physiology of the Na+/H+ antiporter in Escherichia coli. Exp Biol 196: 443–456. [DOI] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MM, Santana M, Teixeira M. (2001). A novel scenario for the evolution of haem–copper oxygen reductases. Biochim Biophys Acta 1505: 185–208. [DOI] [PubMed] [Google Scholar]
- Qian P-Y, Wang Y, Lee OO, Lau SCK, Yang J, Lafi FF et al. (2011). Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J 5: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Rich VI, Pham VD, Eppley J, Shi Y, DeLong EF. (2011). Time-series analyses of Monterey Bay coastal microbial picoplankton using a ‘genome proxy' microarray. Environ Microbiol 13: 116–134. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Lee J, Nath N, Goudeau D, Thompson B, Poulton N et al. (2014). Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat Protoc 9: 1038–1048. [DOI] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol 5: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro AE, Casciotti KL, Francis CA. (2010). Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol 12: 1989–2006. [DOI] [PubMed] [Google Scholar]
- Sañudo-Wilhelmy SA, Cutter LS, Durazo R, Smail EA, Gómez-Consarnau L, Webb EA et al. (2012). Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci 109: 14041–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm AA, de Beer DD, van den Heuvel JCJ, Ottengraf SS, Amann RR. (1999). Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol 65: 3690–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJ, Hoare DS. (1968). Acetate assimilation by Nitrobacter agilis in relation to its ‘obligate autotrophy'. J Bacteriol 95: 844–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin DY, Lücker S, Vejmelkova D, Kostrikina NA, Kleerebezem R, Rijpstra WIC et al. (2012). Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J 6: 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieck E, Bock E. (2005). The lithoautotrophic nitrite-oxidizing bacteria. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds), Bergey's Manual of Systematic Bacteriology. Springer-Verlag: New York, NY, USA, pp 149–153. [Google Scholar]
- Spieck E, Keuter S, Wenzel T, Bock E, Ludwig W. (2014). Characterization of a new marine nitrite-oxidizing bacterium, Nitrospina watsonii sp. nov., a member of the newly proposed phylum ‘Nitrospinae'. Syst Appl Microbiol 37: 170–176. [DOI] [PubMed] [Google Scholar]
- Spieck E, Lipski A. (2011). Cultivation, growth physiology, and chemotaxonomy of nitrite-oxidizing bacteria. Meth Enzymol 486: 109–130. [DOI] [PubMed] [Google Scholar]
- Starkenburg SR, Chain PSG, Sayavedra-Soto LA, Hauser L, Land ML, Larimer FW et al. (2006). Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl Environ Microbiol 72: 2050–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkenburg SR, Larimer FW, Stein LY, Klotz MG, Chain PSG, Sayavedra-Soto LA et al. (2008). Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter. Appl Environ Microbiol 74: 2852–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanauskas R. (2012). Single cell genomics: an individual look at microbes. Curr Opin Microbiol 15: 613–620. [DOI] [PubMed] [Google Scholar]
- Stewart FJ, Sharma AK, Bryant JA, Eppley JM, DeLong EF. (2011). Community transcriptomics reveals universal patterns of protein sequence conservation in natural microbial communities. Genome Biol 12: R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, LAMY D et al. (2011). Potential for chemolithoautotrophy among ubiquitous Bacteria lineages in the dark ocean. Science 333: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Sztukowska M, Bugno M, Potempa J, Travis J, Kurtz DM. (2002). Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol Microbiol 44: 479–488. [DOI] [PubMed] [Google Scholar]
- Takami H, Taniguchi T, Moriya Y, Kuwahara T, Kanehisa M, Goto S. (2012). Evaluation method for the potential functionome harbored in the genome and metagenome. BMC Genomics 13: 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanne C, Golovina EA, Hoekstra FA, Meffert A, Galinski EA. (2014). Glass-forming property of hydroxyectoine is the cause of its superior function as a desiccation protectant. Front Microbiol 5: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U. (1986). Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing acterium. Arch Microbiol 144: 1–7. [Google Scholar]
- Watson SW, Waterbury JB. (1971). Characteristics of two marine nitrite-oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Archiv Mikrobiol 77: 203–230. [Google Scholar]
- Wright JJ, Mewis K, Hanson NW, Konwar KM, Maas KR, Hallam SJ. (2013). Genomic properties of Marine Group A bacteria indicate a role in the marine sulfur cycle. ISME J 8: 455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Zhang J, Vakkasoglu AS, Hielscher R, Osborne JP, Hemp J et al. (2007). Glutamate 107 in subunit I of the cytochrome bd quinol oxidase from Escherichia coli is protonated and near the heme d/heme b595 binuclear center. Biochemistry 46: 3270–3278. [DOI] [PubMed] [Google Scholar]
- Yelton AP, Thomas BC, Simmons SL, Wilmes P, Zemla A, Thelen MP et al. (2011). A semi-quantitative, synteny-based method to improve functional predictions for hypothetical and poorly annotated bacterial and archaeal genes. PLoS Comput Biol 7: e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz S, Allgaier M, Hugenholtz P. (2010). Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat Methods 7: 943–944. [DOI] [PubMed] [Google Scholar]
- Zaikova E, Walsh DA, Stilwell CP, Mohn WW, Tortell PD, Hallam SJ. (2010). Microbial community dynamics in a seasonally anoxic fjord: Saanich Inlet, British Columbia. Environ Microbiol 12: 172–191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SGAs and annotations reported in this study can be accessed at http://www.cbrc.kaust.edu.sa/indigo/dataCategories.do. The whole-genome sequences have also been deposited at NCBI under BioProject number PRJNA276499 with accession numbers JZKI00000000 (SCGC AAA799-A02) and JZKJ00000000 (SCGC AAA799-C22).