Abstract
Few microbial time-series studies have been conducted in open ocean habitats having low seasonal variability such as the North Pacific Subtropical Gyre (NPSG), where surface waters experience comparatively mild seasonal variation. To better describe microbial seasonal variability in this habitat, we analyzed rRNA amplicon and shotgun metagenomic data over two years at the Hawaii Ocean Time-series Station ALOHA. We postulated that this relatively stable habitat might reveal different environmental factors that influence planktonic microbial community diversity than those previously observed in more seasonally dynamic habitats. Unexpectedly, the data showed that microbial diversity at 25 m was positively correlated with average wind speed 3 to 10 days prior to sampling. In addition, microbial community composition at 25 m exhibited significant correlations with solar irradiance. Many bacterial groups whose relative abundances varied with solar radiation corresponded to taxa known to exhibit strong seasonality in other oceanic regions. Network co-correlation analysis of 25 m communities showed seasonal transitions in composition, and distinct successional cohorts of co-occurring phylogenetic groups. Similar network analyses of metagenomic data also indicated distinct seasonality in genes originating from cyanophage, and several bacterial clades including SAR116 and SAR324. At 500 m, microbial community diversity and composition did not vary significantly with any measured environmental parameters. The minimal seasonal variability in the NPSG facilitated detection of more subtle environmental influences, such as episodic wind variation, on surface water microbial diversity. Community composition in NPSG surface waters varied in response to solar irradiance, but less dramatically than reported in other ocean provinces.
Introduction
Microbial community structure and function have pivotal roles in the biogeochemical dynamics of marine ecosystems, yet the microbial ocean remains largely undersampled. Coordinated time-series studies are a key strategy for addressing this undersampling, and improve understanding of the complex interplay between environmental variability and microbial community diversity and dynamics. Several recent time-series efforts focusing on marine surface waters have observed dramatic seasonality in microbial communities, including studies in the Western English Channel (Gilbert et al., 2012), the Sargasso Sea (Morris et al., 2005; Treusch et al., 2009), coastal waters near southern California (Fuhrman et al., 2006) and coastal waters in Antarctica (Murray et al., 1998). Seasonal variability at these locations has been attributed to changes in the physical habitat, including solar irradiance, stratification and mixing. For example, Gilbert et al. (2012), observed dramatic shifts in microbial richness and community composition in the English Channel that correlated with changing day lengths that vary by as much as 8 h between seasons. Clear seasonal patterns in community composition were also observed at the oligotrophic Bermuda Atlantic Time-series Study (BATS, Treusch et al., 2009), where fluctuations in microbial populations varied with the annual cycle of deep convective mixing in the winter, a predictable spring bloom and late summer/early autumn stratification of the upper ocean (Giovannoni and Vergin, 2012).
Compared with other oceanic regions, the physicochemical environment of the North Pacific Subtropical Gyre (NPSG) exhibits relatively low seasonality (Bingham and Lukas, 1996). For example, at the Hawaii Ocean Time-series (HOT) Station ALOHA, a well-studied site representative of the NPSG, there is only a 3.09 h time difference between the longest and shortest days of the year, and sea surface water temperatures vary <4 °C annually (HOT Data Organization and Graphical Systems (DOGS)-see Methods). Additionally, predominantly stratified surface waters create oligotrophic conditions at Station ALOHA year round, unlike the more seasonally oligotrophic waters at BATS. Currently, it is unknown whether the milder climatic and hydrographic seasonal variability at Station ALOHA results in differences in microbial seasonality compared with other oceanic regions. Since the NPSG represents the largest circulation feature on Earth and substantially impacts major global biogeochemical cycles, better understanding its biological dynamics remains an important endeavor (Karl and Lukas, 1996; Karl and Church, 2014).
To investigate the potential seasonality in microbial dynamics at Station ALOHA and identify possible physical and biogeochemical drivers, we examined changes in microbial communities at two discrete depths, 25 m and 500 m, for near-monthly time intervals over a 2-year period. We used bacterial small subunit SSU ribosomal RNA (SSU rRNA) amplicon and shotgun metagenomic sequences to follow changes in microbial taxonomic and functional gene diversity and representation. Amplicon sequencing was used to identify differences between microbial communities by comparing bacterial small subunit ribosomal RNA gene sequences within and between samples directly. Metagenomic shotgun sequencing was used to capture genes from a broader array of cells from all domains, as well as their viruses, and provide broader insight into microbial community composition and variability. Two fundamental dimensions of biodiversity were investigated; alpha diversity, defined as the diversity within individual time points, and beta diversity, defined as the dissimilarity in community composition between pairs of time points. We also used a weighted co-occurrence network analyses to identify clusters of co-varying organisms and protein-coding genes in our samples. We postulated that microbial community dynamics analyzed using these diversity metrics and analytical approaches would reveal clear but potentially muted seasonal trends via correlations with biotic and abiotic seasonal changes. We further hypothesized that the comparatively low seasonal variability in the NPSG might reveal the influence of different, potentially more subtle environmental factors on microbial diversity that have not been reported in previous studies.
Materials and methods
Site and sample collection
The HOT program has been conducting research cruises at approximately monthly intervals at Station ALOHA (22°45'N, 158°00'W) to make physical, chemical and biotic observations since 1988 (Karl and Lukas, 1996). Water samples for the current study were collected on HOT cruises between August 2007 and September 2009 (HOT cruises #194–215). Sampling dates and times are listed in Supplementary Table S1. Corresponding HOT environmental observations were downloaded from the HOT-DOGS website http://hahana.soest.hawaii.edu/hot/hot-dogs/. Environmental measurement protocols are available on the HOT-DOGS site. Environmental measurements were collected during the same cruise and where possible the same day as microbial sampling. HOT program measurements sampled from between 20 and 30 m were utilized for contextual information on microbial community dynamics occurring at 25 m, with the exception of silicate, which was measured between 5 and 25 m. Temperature and salinity measurements collected between 495 and 505 m were averaged for 500 m analyses. Other 500 m environmental measurements (for example, nutrient and dissolved oxygen concentrations) sampled between 470 and 530 m were utilized for the analyses of microbial community structure at 500 m. Curated environmental measurements are available in Supplementary Tables S3 and S4. Pigments were measured using high-performance liquid chromatography and total picoplankton cell numbers were estimated using epi-fluorescence microscopy. The depth of the deep chlorophyll maximum was identified by visual inspection of water column fluorescence data. Mixed layer depth values were based on a 0.125-unit potential density criterion.
In addition, we downloaded daily mean sea level height anomaly data from the Integrated Climate Data Center (http://icdc.zmaw.de/ssh_aviso.html?&L=1). We obtained wind velocity and solar radiation data (incident light energy from 0.28 to 2.8 μm wavelengths in watts m−2) collected by the Upper Oceans Processes Group at the Woods Hole Oceanographic Institution with the WHOTS buoy located at Station ALOHA (retrieved from http://uop.whoi.edu/projects/WHOTS/whots.html). For comparisons between wind velocity and surface chlorophyll concentrations across a longer time scale (1989–2009), wind velocity measurements were retrieved from the NOAA National Data Buoy #51001 located 450 km away from Station ALOHA (retrieved from http://www.ndbc.noaa.gov/station_page.php?station=51001). Theoretical hours of daylight were calculated using the Lammi's Online-Photoperiod Calculator V1.94L (http://www.sci.fi/~benefon/sol.html).
Microbial cells were sampled during HOT cruises by filtering 20 L of seawater, collected with a CTD rosette sampler, through an inline 47 mm diameter, 1.6 μm pore-size GF/A pre-filter (Whatman, Piscataway, NJ, USA) followed by collection on a 0.22 μm pore-size Sterivex GV filter (Millipore, Billerica, MA, USA) using a peristaltic pump. Immediately after filtering was completed, 2 ml of sterile DNA storage buffer (50 mm Tris-HCl, 40 mm EDTA and 0.75 m sucrose) was added to the Sterivex cartridges, and the filters were flash frozen in liquid nitrogen and stored at −80 °C until DNA extraction.
DNA extraction and sequencing
Cells were lysed directly in Sterivex filter units and DNA in the crude lysate was purified on a Quick-Gene 6101 system (Fujifilm, Tokyo, Japan) using DNA Tissue Kit L (Autogen, Holliston, MA, USA). Modifications made to the manufacturer's cell lysis protocol are described by Sharma et al. (2013). Shotgun pyrosequencing was performed using either FLX or Titanium series chemistry (Supplementary Table S1) on a Roche Genome Sequencer FLX instrument according to manufacturer's recommendations (Roche, Indianapolis, IN, USA). FLX or Titanium Rapid Library Preparation protocols were used for library construction. Libraries were quantified using the Titanium Slingshot kit (Fluidigm, San Francisco, CA, USA) and added to emulsion PCR reactions at 0.1 molecules per bead.
Bacterial amplicon libraries targeting the V1-V3 region of bacterial SSU rRNA genes were generated with 27F (5′ AGAGTTTGATCCTGGCTCAG 3′) and 534R (5′ ATTACCGCGGCTGCTGG 3′) primers using PCR amplification protocols established for the Human Microbiome Project (Jumpstart Consortium Human Microbiome Project Data Generation Working Group, 2012). To increase yield while keeping the number of amplification cycles low, triplicate PCR reactions using 20 amplification cycles were run for each sample then pooled. Amplicons were purified using the QiaQuick PCR Clean-Up kit (Qiagen, Valencia, CA, USA) and their size was verified with agarose gel electrophoresis. Following this step, equal quantities of all PCR reactions from the same ocean depth were pooled together, and sequenced using a 454 Genome Sequencer. Sequencing of amplicons was carried out using the Titanium Rapid Library Preparation protocol. We empirically determined the optimal conditions for library preparation since the amplicon DNA fragments were shorter than the fragment length targeted by the library preparation kit. The manufacturer's protocol was followed except adaptor-ligated libraries were not diluted before size selection with AMPure XP beads and 1/4 of the recommended volume of amplification primers was used in emulsion PCR reactions. All metagenomic and amplicon sequencing data is available in the NCBI SRA database (accession numbers in Supplementary Tables S1 and S2).
Metagenomic sequence analysis and annotation
Before analysis of sequencing data, duplicate identical DNA sequences, which were likely 454 sequencing protocol artifacts were removed from metagenomic data sets using previously described computational methods (Stewart et al., 2010). BLASTX searches were conducted against an in-house database comprised of NCBI RefSeq plant and microbe peptide databases (release 51) combined with peptide sequences from marine taxa whose genomes were sequenced using single-cell sequencing (Swan et al., 2011; Swan et al., 2013). Reads were assigned to the taxonomy of their best match, provided the match had a bit score of 50 or greater. Reads matching multiple hits equally well were assigned to the lowest common ancestor of all equally scored top hits. A table containing the number of reads in each sample assigned to each genome or taxonomic group is available in the supplement (Supplementary Tables S6 and S7).
SSU rRNA genes were identified in the metagenomic data using BLASTN searches against the ARB-Silva non-redundant SSU rRNA reference database with a minimum bit score cutoff of 50 (Pruesse et al., 2007, release 102). Reads identified as SSU genes were assigned to their top hit in the database and corresponding leaf on the ARB-Silva non-redundant SSU reference guide phylogeny. By using the ARB software program, the reference phylogeny was then pruned to only include sequences that matched SSU sequences in our metagenomic libraries (Ludwig et al., 2004). This phylogeny was used for subsequent phylogenetic analyses.
Metagenomic reads were also binned into de novo protein-coding ‘functional gene' clusters by first using the gene-finding program MetaGene to identify amino acid sequences within reads (Noguchi et al., 2006). Amino acid sequences originating from the same ocean depth were then pooled, clustered first to a 90% identity threshold, followed by 60% identity threshold, both with a 70% minimum overlap, using cd-hit (Li and Godzik, 2006).
Amplicon sequence analysis and annotation
Amplicons were analyzed within the software package QIIME (Caporaso et al., 2010). First PCR and 454 sequencing artifacts were removed using the AmpliconNoise and Perseus algorithms (Quince et al., 2011). Next, de-multiplexed sequences were binned into de novo operational taxonomic units (OTUs) at 97% identity using the UCLUST algorithm (Edgar, 2010). Reference sequences from each OTU were aligned using PyNAST, uninformative base positions based on the default lanemask were removed and a phylogeny was constructed using the FastTree algorithm (Price et al., 2009; Caporaso et al., 2010b). This phylogeny, with OTUs as leaves was used to calculate subsequent amplicon diversity metrics. Aggressive taxonomic assignments were made outside of the QIIME package, by comparing the OTU reference sequences to the ARB-Silva SSU database using BLASTN (release 108). Reads were assigned to the lowest common ancestor of all database hits with a score within 5% of the top score, provided that bit scores were 50 or higher and the database sequences spanned at least 95% of the amplicon sequence. On occasion, all but a small number of the top hits were to the same taxonomic assignment and the few incongruous hits had low pintail values indicating a high probability the hits were chimeras. In such cases the taxonomy of the majority was assigned to the reference sequence.
Statistical analyses
We estimated alpha diversity with SSU rRNA genes from both amplicon and metagenomic samples, using the metric termed phylogenetic diversity (PD), which is similar to taxonomic richness, but incorporates the phylogenetic relatedness of organisms (Faith, 1992). We estimated beta diversity within SSU rRNA genes using the UniFrac Metric, which is similar to the Jaccard Index that quantifies dissimilarity in taxonomic composition between pairs of samples but unifrac also incorporates the phylogenetic relatedness of organisms (Lozupone and Knight, 2005). We used the protein-coding gene clusters to calculate functional alpha diversity with richness, and to calculate functional beta diversity with the Jaccard Index, parallel to our rRNA phylogenetic metrics PD and Unifrac. Metrics described thus far all depend on the presence, but not abundance of the organisms in the samples. To investigate the additional influence of varying taxon abundances, we also calculated parallel abundance-based metrics (see Supplementary Material).
Alpha diversity within the amplicon data and SSU reads extracted from the metagenomic libraries were calculated in R using the packages vegan and picante (Faith, 1992; Allen et al., 2009; Kembel et al., 2010; Oksanen et al., 2013). PD rarefaction curves were generated in QIIME, as were Unifrac distances (Caporaso et al., 2010). Functional alpha and beta diversity measures were calculated using Vegan and the Python package SciPy (Jones et al., 2001; Oksanen et al., 2013). To account for differing sequencing depths between samples, rarefaction re-sampling was conducted for all indices by averaging the diversity values generated from 100 random subsamples of each community. Communities were subsampled to the read depth of the smallest of the samples being compared. See figure captions for subsampling levels used for each analysis (Figures 2–4). Spearman's correlation coefficients and corresponding two-sided P-values were calculated between alpha diversity and environmental measurements using the R function cor.test. Mantel tests using Spearman's coefficient implemented in the vegan package were used to test the significance of correlations between beta diversity measures and Euclidean distances between environmental measurements. P-values were adjusted to account for multiple tests using the Benjamini–Hochberg (BH) procedure.
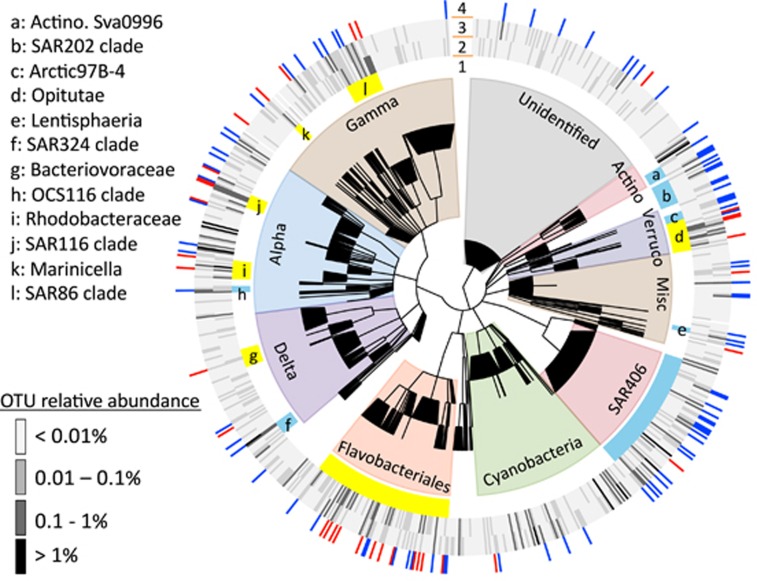
Because of cruise schedules and the nature of solar radiation at HOT, most of our microbial sampling took place when Station ALOHA was experiencing annual extremes in incoming solar irradiance. Therefore to identify microorganisms whose relative abundances may change with variation in incoming solar irradiance, we divided samples into high- and low-light samples. Samples collected in October 2007, February 2008, October 2008 and April 2009 (cruises 196, 200, 205 and 210) that did not fall during annual extremes in incoming shortwave solar radiation were removed. A DESeq2 enrichment analysis was used to test the null hypothesis that the Log2 fold change in the number reads mapping to a given taxonomic group or OTU between high- and low-light times of the year is zero (Love et al., 2014). A detailed description is available elsewhere (Love et al., 2014), but in brief, DESeq2, using count data, tests for differential abundances using negative binomial generalized linear models and estimates size and dispersion factors to control for differences in sequencing depth between libraries and dispersion between taxa. P-values are adjusted to account for multiple tests using the BH procedure after an independent filtering criterion is applied to remove tests that have little chance of showing significance. We included taxa that DESeq2 flagged as potential outliers, because they appeared biologically reasonable. To test for differential abundance in clades within the amplicon data, we collapsed OTU counts into clade counts based on Arb-Silva taxonomic assignments (see above). We choose clades that corresponded to roughly the family level and had relative abundances that summed >1% across all samples. The cladogram in Figure 5 displaying these results was built using the program GraPhlAn (https://bitbucket.org/nsegata/graphlan/).
Weighted co-occurrence network analyses
The R package WGCNA (Langfelder and Horvath, 2008) was used for weighted co-occurrence network analyses of both SSU amplicon and community metagenomic data. For the 25 and 500 m SSU amplicon data sets, count matrices of OTUs were constructed whereby counts represented the number of sequences identified as belonging to a particular OTU with the total number of counts in each sample used for normalization. Low abundance OTUs (<50 counts across all time points or 0 values for at least 15 time points) were excluded on the grounds that they were not likely to yield robust seasonal patterns. To corroborate seasonal patterns identified in the 25 m OTU data, count matrices were also constructed for the 25 m community metagenomic data by mapping sequence reads onto ortholog clusters using methods described previously (Aylward et al., 2015). Briefly, metagenomic reads were first mapped against RefSeq v. 62 using LAST (Frith et al., 2010) with parameters ‘-b 1 -x 15 -y 7 -z 25 -e 80 -F 15 -u 2 -Q 0', with bit scores calculated afterwards and only hits having scores ⩾ 50 retained. Ortholog clusters were constructed for phylogenetic groups of interest (Prochlorococcus, SAR11, SAR116, SAR86, SAR324, SAR406, Roseobacter, pelagiphage and cyanophage) with ProteinOrtho (Lechner et al., 2011) using select genomes that had a high number of reads mapping across metagenomes. For annotations a representative protein was selected from each ortholog cluster (the longest protein in the cluster, or a randomly selected protein in case of ties) and queried against the Kyoto Encyclopedia of Genes and Genomes (Kanehisa and Goto, 2000) using LAST (default parameters, bit score cutoff of 50). Count tables in which columns represented metagenomes and rows represented ortholog clusters were then constructed, in which counts were given based on the number of reads mapping to all of the proteins comprising each of the ortholog clusters. Ortholog clusters having <50 total reads mapping were excluded from subsequent analysis. Ortholog cluster count tables were normalized using the DESeq function ‘varianceStabilizingTransformation'.
Pairwise Pearson correlations were calculated for all OTUs or ortholog clusters in the count tables and a matrix of pairwise adjacency scores was subsequently generated using the equation:
where a is the adjacency, s is the Pearson correlation, i and j are the rows and columns of the correlation and adjacency matrix, respectively, and β is the soft threshold calculated using the scale-free topology index with the guideline that a soft threshold yielding an R2>0.8 for scale-free topology fit is suitable (Zhang and Horvath, 2005). The ‘blockwiseModules' command in WGCNA was used to identify subgroups of OTUs (termed ‘modules') with co-varying temporal profiles (minimum group size >5 specified for OTUs, 30 for ortholog clusters). Clustering was performed using the average linkage hierarchical clustering algorithm, and the Dynamic Tree Cut package (Langfelder and Horvath, 2008) was used to determine module delineations. The first principle components (‘eigengenes') of modules were calculated using the ‘moduleEigengenes' command in WGCNA, with default parameters. OTUs or ortholog clusters were classified as ‘unassigned' if their correlation to the eigengene of the module to which they were assigned was <0.3. Modules having eigengene dissimilarities <0.25 were subsequently merged. Because these methods do not distinguish between positive and negative correlations, we separated modules post facto by calculating the Pearson correlation of each OTU or ortholog cluster's temporal profile with the eigengene of the subgroup to which it belonged; positively and negatively correlated OTUs or ortholog clusters were then denoted with ‘pos' or ‘neg'. Full information for weighted network analysis of OTUs and metagenomes can be found in the Supplementary Material entitled ‘network data sets'.
To further examine co-correlation patterns and corroborate weighted network analysis findings, we also employed a more commonly used unweighted network analysis to examine seasonal patterns in the 25 m data set. In this approach, significantly correlated OTUs were identified independently of weighted co-occurrence analyses using the ‘CorAndPvalue' function in the WGCNA package, which uses Student's t-test to calculate P-values from pairwise Pearson values while accounting for the total number of observations. OTUs found to be significantly correlated (BH adjusted P<0.05) were then used to construct unweighted networks in which edges were drawn between OTUs with significantly correlated or anticorrelated temporal profiles (Figure 6). Network construction was performed using the R package igraph (Csardi and Nepusz, 2006). The adjacency matrix used for unweighted network construction can be found in Supplementary Table S4.
Results and discussion
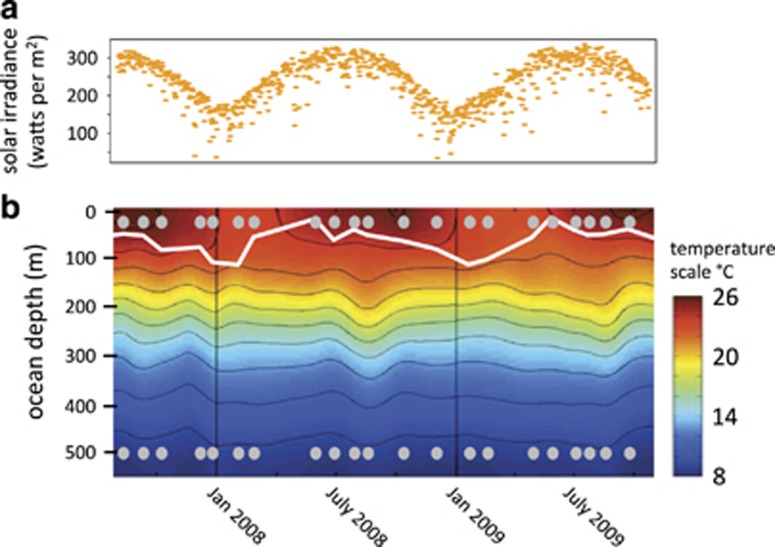
Microbial plankton samples were collected at roughly monthly intervals from August 2007 through September 2009, at 25 and 500 m depths during HOT program cruises to Station ALOHA in the NPSG. As is typical for this region, we observed a persistently stratified upper ocean and only mild seasonal variation in surface waters (Figure 1). At the ocean's surface, mixing depths ranged from 13 to 111 m and water temperatures and bacterial cell numbers tended to peak in the late summer and fall (Figure 1 and Supplementary Figure S1). In contrast, there was little evidence of seasonality in physical or biogeochemical dynamics at 500 m, with conditions being relatively homogenous throughout the year, relative to near-surface waters (Figure 1). For example, maximum and minimum temperature values differed by 3.8 versus 1.4 °C, salinity values differed by 0.52 versus 0.08 (Practical Salinity Scale 1978) and dissolved organic carbon differed by 12.0 versus 7.4 μmol kg−1 at 25 and 500 m, respectively. In addition, nitrate+nitrite and phosphate concentrations were all at least an order of magnitude higher at 500 m compared with 25 m (Supplementary Tables S3 and S4).
Figure 1.
Environmental variation at Station ALOHA during our study period. (a) Orange ovals display the 24-hour daily average solar irradiance (light energy in watts from 0.28 to 2.8 μm wavelengths (shortwave) per m2) at the ocean's surface measured at the nearby WHOTS buoy. The color gradient in (b) signifies water temperature across time and depth. The white line outlines the bottom of the mixed layer measured during HOT cruises. Dots mark sampling points.
We characterized microbial samples using both shotgun metagenomic pyrosequencing of whole community DNA (hereafter referred to as metagenomic data sets or samples), and bacterial SSU rDNA amplicon sequencing. Amplicon-based methods are the most commonly used method for studying microbial diversity. However, shotgun metagenomic approaches are becoming more common as sequencing costs decrease (Logares et al., 2009; Bryant et al., 2012). Comparing functional gene (protein-coding gene) diversity with SSU rRNA-based measures is also useful and relevant, since gene composition ultimately dictates how microbes can potentially interact with each other and their environment (Gilbert et al., 2010; Raes et al., 2011; Barberán et al., 2012; Bryant et al., 2012; Fierer et al., 2012). For each of the 21 discrete time points and two depths studied, we generated metagenomic libraries with an average of 1.1 million reads of an average length of 360 base pairs (bps) (Supplementary Table S1). We generated 21 amplicon libraries at 25 m and 19 at 500 m, each with an average of 12 000 reads of ~350 bps in length (Supplementary Table S2).
As anticipated, Prochlorococcus and SAR11, the dominant microorganisms in surface waters year round at Station ALOHA, comprised roughly 50% and 10% of metagenomic reads across all samples, respectively (Supplementary Figure S2). This was consistent with the corresponding amplicon data, as well as with previous reports in this same oceanic region (Supplementary Figure S3; Schmidt et al., 1991; Campbell and Vaulot, 1993; Eiler et al., 2011). At 500 m, SAR11, SAR324, SAR406, Nitrospina, SAR202 and Thaumarchaeota were consistently the most abundant taxa (Supplementary Figures S4 and S5).
We compared SSU rRNA alpha and beta diversity metrics generated by the two different sequencing approaches (SSU rRNA amplicons versus shotgun metagenomic sequencing) and also compared diversity metrics generated from amplicon SSU rRNA to diversity metrics generated from metagenomic functional genes to investigate whether the different methods were consistent with one another (Supplementary Material). At 25 m, all the diversity metrics were generally well correlated. In contrast, diversity metrics at 500 m were not consistently correlated. We propose that lower correspondence between amplicon verses metagenomic data sets in 500 m samples likely reflects lower variability between samples at 500 m (see next section). Detailed results and additional discussion are available in the supplement.
Microbial communities at 25 versus 500 m in the NPSG
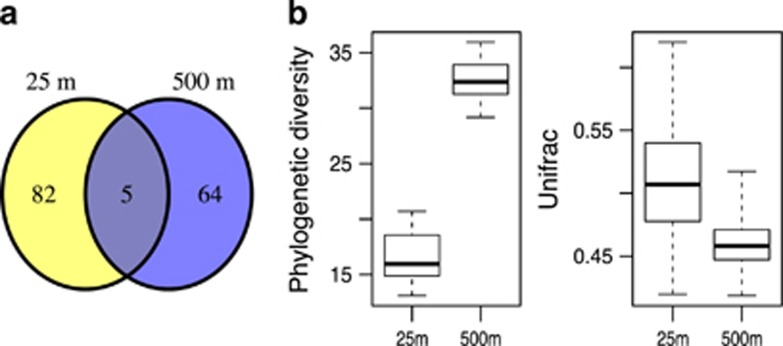
An emerging pattern in pelagic systems is an increase in alpha diversity at intermediate water depths compared with surface waters (Treusch et al., 2009; Jing et al., 2013, Sunagawa et al., 2015). Discrepancies in this richness pattern can usually be explained by the taxonomic resolution of the study (Brown et al., 2009; Kembel et al., 2011). Consistent with these findings, we observed a broad array of dominant taxa at 500 m, which yielded higher phylogenetic and functional richness compared with that found at 25 m (Figure 2, Supplementary Figures S10 and S11). Explanations for higher richness in the mesopelagic are not immediately obvious, since productivity is highest in the photic zone. The distance from sunlight-driven energy and productivity, however, likely causes intense resource competition at depth. This may promote a more diverse and even community containing a broader range of heterotrophic and chemolithotrophic lifestyles and taxa compared with shallower waters. Evidence for more prevalent chemolithotrophic associated activities, including ammonia oxidation, sulfur oxidation and CO2 fixation have all been reported in the mesopelagic at the NPSG (Karl et al., 1984; Ingalls et al., 2006; Hansman et al., 2009; Swan et al., 2011; Giovannoni and Vergin, 2012).
Figure 2.
Comparison of microbial communities at 25 and 500 m. (a) Venn diagram displaying the number of OTUs from each depth with relative abundances >0.1 % at either one or both depths. All amplicon data within each depth were combined to calculate relative abundances. (b) Boxplots comparing the distribution of phylogenetic diversity and Unifrac values between 25 and 500 m samples based on amplicon data. Whiskers delineate the full range of values. The variance of Unifrac values was significantly smaller in 500 m samples compared with 25 m samples (Levene's test, P<0.05). In b, amplicon data was resampled to 6906 reads per sample.
Previous studies have demonstrated that in the NPSG, steep physical and chemical gradients and perennial stratification are reflected in strong vertical structure in microbial community composition (DeLong et al., 2006). Moreover, previous studies have observed that microbial communities in the near-surface ocean tend to be more variable in time compared with communities in deeper water, likely because the physical environment below the euphotic zone is more stable (for example, Lee and Fuhrman, 1991; Treusch et al., 2009; Rich et al., 2011; Ghiglione et al., 2012). Results derived from our metagenomic and amplicon data were consistent with these previously described depth-dependent trends.
When combining all the amplicon data from both 25 and 500 m together, we only observed five OTUs with relative abundances >0.1% at both depths studied (Figure 2, Supplementary Table S5). These OTUs mapped to SAR11 and SAR406. Other shared OTUs that were abundant at one depth were at least an order of magnitude less abundant at the other depth (Supplementary Table S5).
Variance between time points was significantly higher among 25 m samples than among 500 m samples, for the majority of alpha and beta diversity metrics including Unifrac (Levene's test P<0.05, Figure 2 and Supplementary Figure S10). In addition, we observed that microbial communities at 500 m had lower average beta diversity indices compared with 25 m, indicating that the composition of microbial communities was more similar across time points at 500 m (Figure 2, and Supplementary Figure S10). However, when incorporating abundance information into Unifrac values (weighted-Unifrac, Lozupone et al., 2007), values were higher at 500 m compared with 25 m. This is likely because two groups, Prochlorococcus and Pelagibacter, dominated all the 25 m time points.
At 500 m, no consistent correlations between microbial alpha and beta diversity metrics and environmental measurements were found (BH adjusted P<0.10, Supplementary Tables S10 and S11). Also weighted gene co-occurrence network analyses revealed that only a minority of 500 m OTUs could be clustered into modules and these modules showed no significant correlations with environmental parameters (Supplementary Figure S16, Supplementary Table S14). Mesopelagic communities may of course have responded to environmental variables we did not measure. The minimal variability in 500 m samples also suggests that at this depth the temporal frequency we sampled was not optimal for detecting variability in microbial communities. Our 500 m results are consistent with previous work at Station ALOHA, and at global scales, that showed that in the bathypelagic, individual environmental variables appear to have only a small effect on free-living communities, but perhaps a stronger influence on particulate associated communities (Eiler et al., 2011; Salazar et al., 2015). Microbes in the mesopelagic have been shown to be very active on particulate matter, which is rapidly transported from surface water to the ocean floor (Karl et al., 1984, 2012). Including particulate associated microbial communities in mesopelagic studies, as well as incorporating longer sampling periods might reveal more variability and perhaps seasonality in mesopelagic microbial communities than we could observe with this data set.
Wind correlates with alpha diversity in NPSG surface waters
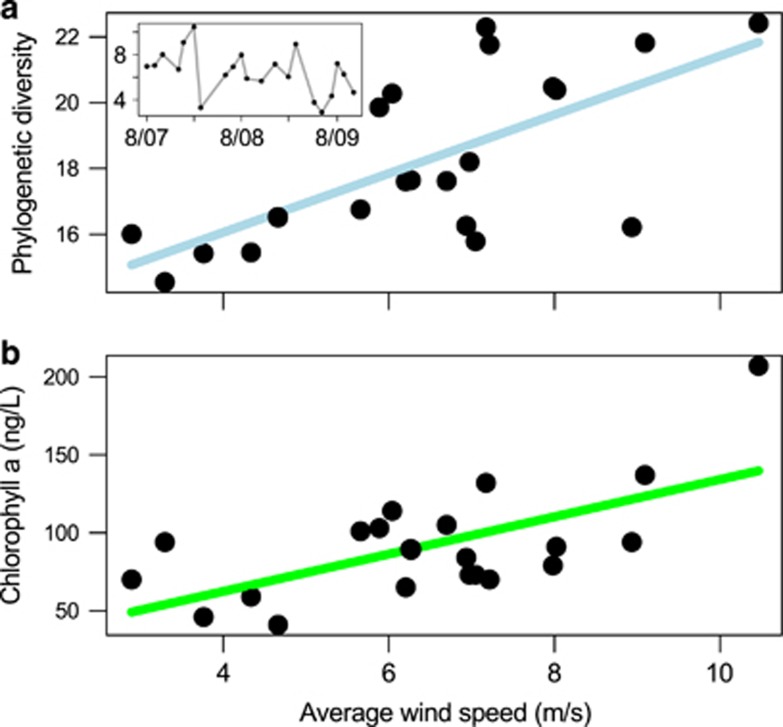
To reveal potential environmental factors influencing variability in alpha diversity across our 25 m time series, we looked for correlates between the alpha diversity measures and the physical, chemical and biotic environmental parameters measured during HOT cruises and at the nearby WHOTS meteorological buoy. Unlike previous studies, we did not observe significant correlations between alpha diversity and seasonally driven environmental parameters such as temperature and mixed layer depth (Gilbert et al., 2012; Ladau et al., 2013). Instead all alpha diversity metrics were consistently most strongly correlated with the average wind speed of the days leading up to sample collection (Figure 3, Supplementary Table S8). The correlation between wind and alpha diversity was statistically significant for amplicon phylogenetic diversity (PD) and functional richness (Spearman r=0.7, BH adjusted P<0.10). This trend held when averaging the wind speeds from 3 to 10 days before the sampling date (Supplementary Figure S12). The alpha diversity metric phylogenetic entropy, which incorporates taxon abundance information, is less influenced by rare taxa than PD. The lower correlation between wind and phylogenetic entropy (Spearman r< 0.50) combined with an OTU abundance distribution demonstrates that low abundance taxa largely contributed to the wind-associated increase in alpha diversity (Supplementary Table S8, Supplementary Figure S13).
Figure 3.
Relationship between wind speed and phylogenetic diversity at 25 m. Inset in a shows the average wind speed (m s-1) of the 4 days before sample collection across time. (a) Relationship between average wind speed and phylogenetic diversity of samples characterized using amplicon data (BH adjusted P<0.01, Spearman r=0.7). The relationship was consistent when considering other alpha diversity measures and average wind speeds derived from a narrower or broader range of days leading up to sampling (Supplementary Table S8, Supplementary Figure S12). Each amplicon data set was resampled to 9661 reads (see Methods). (b) Relationship between average wind speed and chlorophyll a concentrations (P<0.05, Spearman r=0.46).
Chlorophyll a concentrations (chl a) and several other phytoplankton pigments were also correlated with amplicon PD (BH adjusted P<0.10 Supplementary Table S8b) and chl a was reciprocally correlated with average wind speeds (Spearman r=0.46, P<0.05, Figure 3). The relationship between wind speed and chlorophyll concentrations at HOT appears to be a long-term trend, consistent with findings from global-scale studies (Kahru et al., 2010). We compared 20 years of HOT near-surface ocean chl a measurements from 1989 to 2009 to wind speed measurements collected from the NOAA Nation Data Buoy #51001. Chl a was significantly correlated with wind speeds over the 20-year period (Spearman r=0.26, P<0.001). Surface chlorophyll concentrations are also impacted by phytoplankton photoadaption, where per cell pigment concentrations rapidly adjust to current light levels (Ohman et al., 1982; Letelier et al., 1993; Winn et al., 1995). The wind-chl a relationship across the 20-year period remained significant after statistically accounting for phytoplankton photoadaption by modeling chl a and day length (Supplementary Figure S14, linear regression P<0.001, r2=0.13).
Strong wind-driven mixing events and wind/eddy interactions can spur phytoplankton blooms in the open ocean (Winn et al., 1992; Letelier et al., 2000; McGillicuddy et al., 2007). Wind may also be fertilizing surface waters by transporting nutrients including fixed nitrogen, phosphorus and iron through both wet and dry dust deposition (DiTullio and Laws, 1991; Young et al., 1991; Karl and Tien, 1997; Jickells et al., 2005; Fitzsimmons et al., 2014). We did not observe significant increases in autotrophic biomass or cyanobacteria cell concentrations with increasing wind speeds (Supplementary Figure S14). These data likely reflect a wind-driven increase in chlorophyll per cell, as opposed to increased cell numbers, suggesting that alpha diversity is not a product of wind-driven increases in oxygenic photoautotroph abundance.
Photoautotrophs deeper in the water column have higher per cell chlorophyll concentrations to adjust for lower light levels (Winn et al., 1995). Thus the increases in microbial alpha diversity and chlorophyll may also result from entrainment of microorganisms located deeper in the water column via wind-driven mixing. In addition, wind-driven mixing increases the rate at which organisms cycle through the extreme light conditions at the top of the mixed layer and poorly lit conditions at the bottom of the mixed layer, and so may influence microbial communities and chl a concentrations by modifying light exposure. Incoming solar irradiance and wind speed were not significantly correlated (Spearman r=−0.23, P=0.32), albeit the three windiest sampling periods occurred in the winter. Additional studies with high-resolution sampling of strong wind events in the NPSG will help improve the mechanistic understanding of our observations.
Solar irradiance correlates with beta diversity in surface water
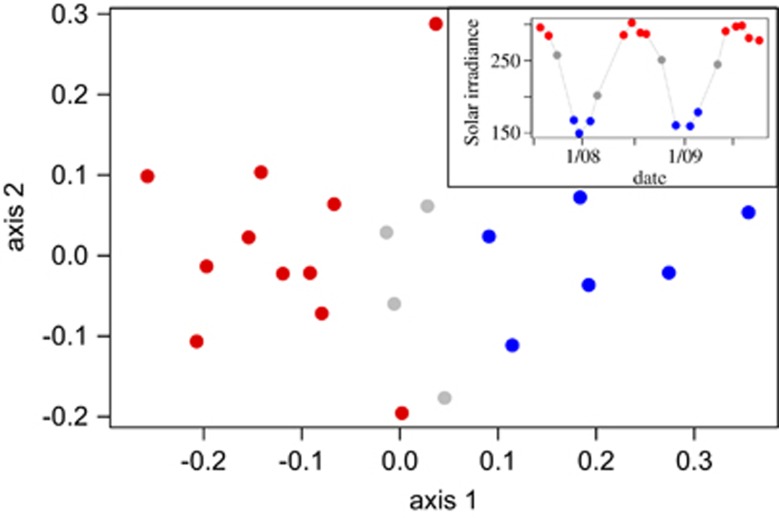
To uncover potential environmental factors influencing beta diversity across our 25 m time series, we looked for correlates among the beta diversity measures and changes in physical, chemical and biotic environmental properties (Supplementary Table S9). Unlike alpha diversity, the beta diversity measures demonstrated a significant seasonal trend. All the non-abundance-based beta diversity indices consistently correlated with the average solar irradiance reaching the ocean's surface derived from light energy from 0.28 to 2.8 μm wavelengths (Crescenti et al., 1989, Figure 4, Mantel test, BH adjusted P<0.05, Supplementary Table S9). The lower correlation between solar irradiance and the abundance-based measures demonstrates that lower abundance taxa largely contributed to this pattern, similar to our alpha diversity observations. The seasonal variables temperature and mixed layer depth were also correlated with some beta diversity measures, but the relationship was not as strong, nor as consistent as with solar irradiance (Supplementary Table S9). Chl a also correlated with most beta diversity measures, presumably reflecting the phytoplankton response to changing light levels via photoadaption. Our results are consistent with recent global-scale studies that found seasonally fluctuating variables including light and temperature strongly impact marine surface communities (Raes et al., 2011; Sunagawa et al., 2015). The stronger impact of light compared with temperature we observed at Station ALOHA contrasted with findings from the Tara Oceans Expedition, and may reflect differing impacts of these variables at different spatial scales (Sunagawa et al., 2015).
Figure 4.
Nonmetric multidimensional scaling plot to visualize Unifrac distances between amplicon data sets at 25 m. Stress=0.15. Inset shows the solar irradiance (Wm−2) averaged across the 30 days leading up to sample collection. Red indicates high-incident solar irradiance data points that were >260 W m−2. Blue indicates low-incident solar irradiance data points that were <175 W m−2. Colors in inset correspond to colors in main figure. Each amplicon data set was resampled to 9661 reads (see Methods).
Serial sampling date did weakly correlate with some of the beta diversity indices at 25 m suggesting there may have been some autocorrelation among our samples. Ocean currents and water masses in the open ocean are very dynamic and therefore it is unlikely that any such autocorrelation we observed comes from sampling the same microbial population each month. Instead, regional-scale processes likely drive the observed temporal autocorrelation. Seasonal trends, including solar irradiance fall under this description. Regardless, the significant relationships between incoming solar irradiance and compositional dissimilarity were still significant after taking the time separating sampling dates into account (partial Mantel tests, P<0.05) demonstrating that temporal autocorrelation is not an overarching driver in the patterns we observed.
Because Station ALOHA is located between the Tropic of Cancer and the equator, it experiences longer periods of maximum incoming solar irradiance in the summer followed by sharp dips during the winter (Figure 1). We compared beta diversity measures to the average incoming solar radiation using 24 h measurements across the 30 days before sampling. Therefore our solar irradiance values incorporate the influence of hours of daylight, cloud cover and seasonal variability in light intensity. Community composition significantly varied with hours of daylight alone as well, but the correlation was not as strong as that derived from the daily average over the preceding 30 days (Supplementary Table S9). This suggests that the quantity of incoming solar irradiance is impacting microbial community composition rather than members of the community exhibiting photoperiodism.
The observed seasonal changes in microbial communities at Station ALOHA are more subtle than what has been previously reported at other marine microbial time-series stations. This likely reflects the less extreme environmental variability at Station ALOHA compared with coastal sites (Murray et al., 1998; Morris et al., 2005; Fuhrman et al., 2006; Gilbert et al., 2012). For example, Rickettsiales and Rhodobacterales alternate being the most abundant clade in surface water in the Western English Channel depending on season (Gilbert et al., 2012). At BATS, Prochlorococcus cell densities are an order of magnitude higher than other picophytoplankton until the spring water column mixing when Synechococcus bloom, Prochlorococcus cells decline and cell densities of these two groups of cyanobacteria become comparable (DuRand et al., 2001). At Station ALOHA, changes in community composition are less pronounced. Prochlorococcus and Pelagibacter remain the dominant organisms year round (Supplementary Figure S2 and S3). This is consistent with a 5-year study showing weak and inconsistent seasonality in total Prochlorococcus cell counts at Station ALOHA (Malmstrom et al., 2010). However these dominant groups (for example, Prochlorococcus, Pelagibacter) undoubtedly harbor microdiverse subpopulations that may also fluctuate with light (Kashtan et al., 2014).
The changes we observed in the relative abundance of some taxa at Station ALOHA between annual high and low solar irradiance periods, however, were consistent with seasonal studies in other oceanic regions (Figure 5). The clades that were more abundant in the amplicon data during high irradiance periods, SAR86, SAR116 and Rhodobacteraceae, were also reported as being more abundant during low nutrient, peak summer water stratification periods at BATS and/or were more abundant in the summer than winter in the Western English Channel (Mary et al., 2006; Treusch et al., 2009; Gilbert et al., 2012; Giovannoni and Vergin, 2012; Figure 5). This was consistent with the genomes we identified in the metagenomic data whose abundance varied with solar radiation when annotating metagenomic reads with the NCBI RefSeq database (Supplementary Figure S15). We also observed a small but statistically significant increase in the relative abundance of some Pelagibacter strains during high solar irradiance periods (Supplementary Figure S15). Similarly, Carlson et al. (2009) found that SAR11 cell densities distinctly increase when the mixed layer depth shoals at BATS in the spring.
Figure 5.
Cladogram highlighting bacterial OTUs and clades with differential relative abundances between samples experiencing high- and low-incident solar irradiance at 25 m (as defined in Figure 4). Shaded areas of branches delineate broad taxonomic groups. The innermost ring (ring #1) delineates clades that are more abundant during high light (>260 W m −2 average solar irradiance of the preceding 30 days; yellow) or low light (< 200 W m−2 average solar irradiance of the preceding 30 days; light blue) sampling points (DESeq2 with BH correction, P<0.1). Letters correspond to listed clade names. The next two rings display the average relative abundance to within an order of magnitude of each OTU during high (ring # 2) and low light (ring # 3) sampling points. Red and blue bars in the outermost ring (ring # 4) mark individual OTUs that are significantly more abundant during high (red) and low (dark blue) light sampling points (DESeq2 with BH correction, P<0.1). Only OTUs with relative abundances across all samples summing to greater than 0.01% are shown. Abbreviations: Alpha, Gamma and Delta, denote corresponding Proteobacteria classes; Verruco, Verrucomicrobia; Actino: Actinobacteria.
The clades we observed to be more abundant during low-light periods of the year, including OCS116, SAR324, SAR202, Synechococcus and SAR406, were either reported as more abundant in surface water at BATS during spring mixing periods or as having peak abundances deeper in the water column at Station ALOHA (Gordon and Giovannoni, 1996; Giovannoni et al., 1996; Wright et al., 1997; Treusch et al., 2009; Giovannoni and Vergin, 2012; Figure 5 and Supplementary Figure S15). Consistent with our observations, previous studies have reported that Synechococcus cell counts at Station ALOHA peak in the winter (Malmstrom et al., 2010). We also observed that low light adapted Prochlorococcus marinus ecotypes NATL1A and NATL2A reached maximal abundances in our study at 25 m during low-light sampling periods (Supplementary Figure S15). Prochlorococcus strains NATL1A and NATL2A reach their highest abundances deeper in the water column, although they can tolerate short periods of more intense light exposure (Moore et al., 1998; Johnson et al., 2006; Kettler et al., 2007; Zinser et al., 2007). Similar seasonal trends have also been observed in NATL and other Prochlorococcus ecotypes (Zinser et al., 2007; Malmstrom et al., 2010; Kashtan et al., 2014).
Solar radiation can influence microbes in a variety of ways including direct impacts on cell physiology, indirect impacts through food web dynamics and chemical transformation of organic matter (Moran and Miller, 2007; Ruiz-González et al., 2013). Incident solar radiation also impacts ocean hydrology. For example, the deepening of the mixed layer in the winter may increase transport of microbes from the lower photic zone to 25 m. Our data indicated however, that solar irradiance was more strongly correlated with beta diversity than mixed layer depth.
Solar radiation and temperature also exert top-down controls on microbial community structure, through changes in bacterivory and viral infection (Tsai et al., 2005; An-Yi et al., 2009; Tsai et al., 2012; Ruiz-González et al., 2013). Over the 2-year period examined, we observed an increase in the relative abundance of several cyanophage genomes in low-light samples (Supplementary Figure S15). Similarly, Parsons et al. (2012) observed that viral particle concentrations in the mixed layer at BATS are lowest during periods of summer water stratification. Our observations, however, likely reflect an increase either in phage intracellular production or extracellular attachment, rather than an increase in planktonic phage particles, since free viral particles should not be efficiently retained on the sampling filters we used.
Co-occurrence network analyses of OTUs and orthologs
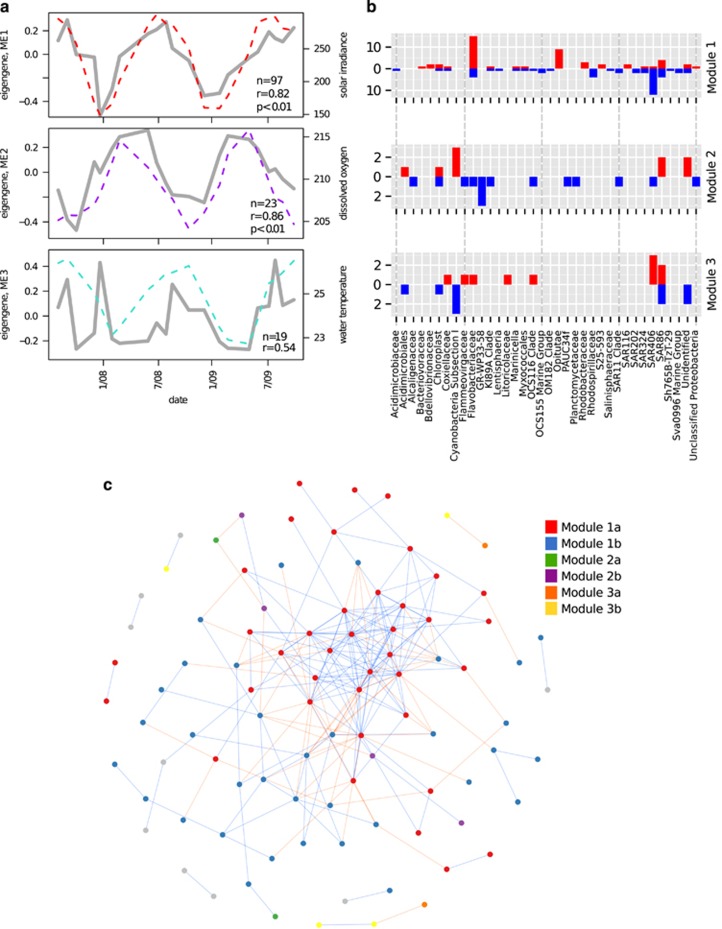
A weighted co-occurrence network analysis (Langfelder and Horvath, 2008) was used to identify potential subgroups of co-varying organisms in our samples that might be responding to seasonal or other measured environmental parameters not captured by the community level analysis. Taxa within co-varying subgroups may be directly or indirectly interacting or sharing a similar niche space. Using the 25 m amplicon data, we identified six clusters of co-correlated OTUs ('modules') that contained over five OTUs (Figure 6 and Supplementary Figure S16). Consistent with our previous findings, the first principal component ('eigengene' Langfelder and Horvath, 2008) of these modules most strongly correlated with seasonally fluctuating environmental measurements or average wind speed (Supplementary Table S12). The eigengenes of the modules containing the largest number of OTUs (96 and 23 total) significantly correlated with the seasonally fluctuating measurements solar irradiance and dissolved oxygen concentrations (Spearman r, BH adjusted P<0.1, Figure 6, Supplementary Table S12). Interestingly the peak of module 1 and 2's eigengenes are slightly offset, suggesting a potential seasonal succession. Several phylogenetic groups had OTUs with both positive and negative correlations to individual modules, or OTUs that were present in multiple modules, suggesting that there are distinct sets of ecotypes from different clades that appear to be co-varying. For example, there are OTUs belonging to Flavobacteriaceae and clade SAR406 that either positively or negatively correlated with module 1, which peaked in early summer. OTUs belonging to clade SAR86 showed a similar pattern in module 3, which most strongly co-varied with water temperature (Figure 6).
Figure 6.
Weighted co-occurrence network analysis at 25 m. (a) OTU trends in the three largest modules with module eigengenes (gray solid lines, left axis) overlaid with their most strongly correlated environmental measurement (dashed lines, right axis). n and r correspond to the total number of OTUs in the module and the Spearman's correlation between the module and overlaid environmental measurement, respectively. P-values (p) are shown where significant (BH adjusted P<0.1). Note that modules contain both positively and negatively correlating OTUs, resulting in sets of OTUs whose relative abundances either positively or negatively correlate with eigengene trends. The bar charts (b) display the number of OTUs that are positively (red) or negatively (blue) correlated with the eigengene trend displayed to the left (a). (c) An unweighted network of OTUs identified in the 25 m data set. Nodes represent OTUs that are colored according to the module to which they were assigned in the weighted network analysis, with ‘a' and ‘b' signifiers representing OTUs that are either positively or negatively associated with the eigengene of that module, respectively (gray nodes were either assigned to another module or not assigned to any module). Edges are colored based on whether the OTUs were found to be correlated (blue) or anticorrelated (red). Note that although the network is unweighted similar patterns identified in the weighted network analysis are recapitulated, such as tight clustering of correlated and anticorrelated nodes assigned to module 1 in the weighted network analysis.
These co-correlation patterns were not unique to the weighted network analysis, as an unweighted network analysis yielded similar clustering patterns for the 25 m data (Figure 6c). Particularly noteworthy in the unweighted network analysis was the tight clustering of OTUs assigned to module 1 of the weighted network (both correlated and anticorrelated to the eigengene of this module), demonstrating that the patterns of seasonal succession identified in this module are particularly prominent.
To corroborate the patterns at 25 m and uncover additional information as to how the genomic content of taxonomic groups may vary over time, we used weighted co-occurrence network analysis to explore the temporal patterns of protein-coding ortholog clusters within the abundant taxa in our 25 m metagenomic samples (Supplementary Figure S17). As expected, we observed that ortholog clusters from the same taxonomic group tended to cluster together into the same module, consistent with read abundance for ortholog clusters being influenced mainly by microbial abundances. Some ortholog clusters, however, were placed in different modules than the majority of clusters from the same taxa, suggesting the presence of microdiversity within these clades that might not co-vary uniformly with clade abundance. These orthologs may belong to genomic islands present only in certain ecotypes within a clade, or to mobile elements present only in some microbial groups. Confirmation of these observations awaits more complete reference data sets (from single-cell genomes or larger metagenomes), to allow mapping of these genes on specific ecotypes over time.
Analysis of the ortholog cluster temporal profiles at 25 m corroborated and extended our other analytical approaches. For example, the eigengenes of modules containing most cyanophage and clade SAR324 ortholog clusters, co-varied significantly with seasonally variable parameters (Supplementary Table S13 and Supplementary Figure S17). SAR324 in particular was positively associated with a deeper mixed layer, reflecting its greater predominance in deeper waters. In addition, eigengenes of some of the smaller modules significantly correlated with total autotrophic biomass, nitrate+nitrite and particulate carbon concentrations. Module 1, containing the largest number of ortholog clusters, did not significantly correlate with any measured environmental parameters, being primarily composed of core genes in phylogenetic groups that are abundant throughout the year (Prochlorococcus, SAR11) (see Supplementary Table S3).
The majority of SAR116 genes were found in modules 1 and 6 suggesting that although many SAR116 types are present throughout the year, some of its variants change in abundance. Several SAR116 ortholog clusters present in module 6 were associated with cofactor and vitamin metabolism, flagellar biosynthesis, several transporters and many housekeeping genes (most positively correlated with module 6 and peaking in abundance in spring and summer months). Similarly, module 10 was positively associated with solar radiation and contained a number of SAR11 ortholog clusters annotated as transporters and genes involved in central carbon and amino acid metabolism (all positively correlated with module 10 and peaking in abundance in the summer) (see Supplementary Table S3). Interestingly, module 10 also included two Prochlorococcus ortholog clusters annotated as PhoB and PhoR, which are involved in phosphate acquisition during phosphate-limiting conditions, and were more abundant in the summer.
Conclusion
The results of this study support the hypothesis that the dampened seasonal variation in the physical environment at the NPSG may provide greater sensitivity for the detection of microbial community responses to non-seasonal phenomena. Research at Station ALOHA conducted by the HOT program over the past 25 years has shown that episodic (for example, mesoscale eddies) and inter-annual-scale (Pacific Decadal Oscillation and El Niño-Southern Oscillation), as well as seasonal processes, all may be important in carbon cycling and plankton dynamics (Letelier et al., 2000; Fong et al., 2008; Bidigare et al., 2009; Karl et al., 2012). Over a 2-year period, in the absence of strong interannual variation we observed that episodic variation in wind speed was the predominant correlate of variability in microbial alpha diversity, but that seasonal variation, in particular solar irradiation, was key to microbial beta diversity. The former feature may be unique to the NPSG, but the latter appears to be a common feature in other marine surface water systems around the globe.
Future work that leverages long-term time-series analyses on a variety of spatial and temporal scales has significant potential to further enhance our understanding of the temporal dynamics and variability of microbial community diversity. Advances in technologies that increase the number of available reference genomes from indigenous ecotypes (Swan et al., 2013), that allow greater sequencing depth to facilitate more complete metagenomic assemblies (Sharon and Banfield, 2013), and that enable more automated, highly resolved sampling over a broader array of nested temporal scales (Robidart et al., 2014), all promise to improve our ability to observe the microbial ocean over increasingly resolved and relevant scales of space and time.
Acknowledgments
We thank Dr Robert R. Bidigare, Yoshimi M. Rii and members of the DeLong Lab for valuable discussions, Craig Nosse for assistance with WHOTS Buoy data, Lance Fujieki for assistance with HOT environmental data, Rachel Barry and Tsultrim Palden for preparing samples for pyrosequencing and Ramunas Stepanauskas for providing single-cell genomes before publication. We are indebted to the HOT scientists and staff and to the captains and crews of the research vessel R/V Kilo Moana and R/V KOK and for logistical support. This work was supported by grants from the Gordon and Betty Moore Foundation #492.01 and 3777 (EFD), #3794 (DMK), and by the US Environmental Protection Agency STAR Fellowship (JAB). In addition, We acknowledge NSF for support of the HOT program (including the most recent OCE1260164 to MJC and DMK) the Center for Microbial Oceanography: Research and Education (C-MORE; EF0424599 to DMK and EFD) and the Simons Collaboration on Ocean Processes and Ecology (SCOPE; #329108 to DMK, EFD). Data from the WHOTS surface mooring are gratefully acknowledged; the NOAA Climate Observation Division provides funding to Robert A Weller and Albert J Plueddemann at WHOI to support the long-term deployment of the surface mooring. This work is a contribution of the Center for Microbial Oceanography: Research and Education, and the Simons Collaboration on Ocean Processes and Ecology.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allen B, Kon M, Bar-Yam Y. (2009). A new phylogenetic diversity measure generalizing the Shannon index and its application phyllostomid bats. Am Nat 174: 236–243. [DOI] [PubMed] [Google Scholar]
- An-Yi T, Chin W-M, Chiang K-P. (2009). Diel patterns of grazing by pigmented nanoflagellates on Synechococcus spp. in the coastal ecosystem of subtropical western Pacific. Hydrobiologia 636: 249–256. [DOI] [PubMed] [Google Scholar]
- Aylward FO, Eppley JM, Smith JM, Chavez FP, Scholin CA, DeLong EF. (2015). Microbial community transcriptional networks are conserved in three domains at the ocean basin scales. Proc Natl Acad Sci USA 17: 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Fernández-Guerra A, Bohannan BJM, Casamayor EO. (2012). Exploration of community traits as ecological markers in microbial metagenomics. Mol Ecol 8: 1909–1917. [DOI] [PubMed] [Google Scholar]
- Bidigare RR, Chai F, Landry MR, Lukas R, Hannides CCS, Christensen SJ et al. (2009). Subtropical ocean ecosystem structure changes forced by North Pacific climate variations. J Plankton Res 31: 1131–1139. [Google Scholar]
- Bingham FM, Lukas R. (1996). Seasonal cycles of temperature, salinity and dissolved oxygen observed in the Hawaii Ocean Time-series. Deep Sea Res Part 2 Top Stud Oceanogr 43: 199–213. [Google Scholar]
- Brown MV, Philip GK, Bunge JA, Smith MC, Bissett A, Lauro FM et al. (2009). Microbial community structure in the North Pacific ocean. ISME J 3: 1374–1386. [DOI] [PubMed] [Google Scholar]
- Bryant JA, Stewart FJ, Eppley JM, DeLong EF. (2012). Microbial community phylogenetic and trait diversity declines with depth in a marine oxygen minimum zone. Ecology 93: 1659–1673. [DOI] [PubMed] [Google Scholar]
- Campbell L, Vaulot D. (1993). Photosynthetic picoplankton community structure in the subtropical North Pacific Ocean near Hawaii (Station ALOHA). Deep Sea Res Part 1 Oceanogr Res Pap 40: 2043–2060. [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. (2010. b). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. (2009). Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISMEJ 3: 283–295. [DOI] [PubMed] [Google Scholar]
- Crescenti GH, Payne RE, Weller RA. (1989) Improved meteorological measurements from buoys and ships (IMET): preliminary comparison of pyranometers WHOI Tech Reports. WHOI-89-47, IMETTR-89-04 Woods Hole Oceanographic Institution: Woods Hole, MA, USA. [Google Scholar]
- Csardi G, Nepusz T. (2006) The igraph software package for complex network research. InterJournal Complex Systems, 1695.
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U et al. (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496–503. [DOI] [PubMed] [Google Scholar]
- DiTullio GR, Laws EA. (1991). Impact of an atmospheric-oceanic disturbance on phytoplankton community dynamics in the North Pacific Central Gyre. Deep Sea Res A 38: 1305–1329. [Google Scholar]
- DuRand MD, Olson RJ, Chisholm SW. (2001). Phytoplankton population dynamics at the Bermuda Atlantic Time-series station in the Sargasso Sea. Deep Sea Res Part 2 Top Stud Oceanogr 48: 1983–2003. [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Eiler A, Hayakawa DH, Rappé MS. (2011). Non-random assembly of bacterioplankton communities in the subtropical North Pacific Ocean. Front Microbiol 2: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP. (1992). Conservation evaluation and phylogenetic diversity. Biol Conserv 61: 1–10. [Google Scholar]
- Fierer N, Leff J, Adams B, Nielsen UN, Bates ST, Lauber CL et al. (2012). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci USA 109: 21390–21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimmons JN, Hayes CT, Al-Subiai SN, Zhang R, Morton PL. et al. (2015). Daily to decadal variability of size-fractionated iron and iron-binding ligand at the Hawaii Ocean Time-series Station ALOHA. Geochim Cosmochim Acta 171: 303–324. [Google Scholar]
- Fong AA, Karl DM, Lukas R, Letelier RM, Zehr JP, Church MJ. (2008). Nitrogen fixation in an anticyclonic eddy in the oligotrophic North Pacific Ocean. ISME J 2: 663–676. [DOI] [PubMed] [Google Scholar]
- Frith MC, Hamada M, Horton P. (2010). Parameters for accurate genome alignment. BMC Bioinformatics 11: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA 103: 13104–13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione J-F, Galand PE, Pommier T, Pedrós-Alió C, Maas EW, Bakker K et al. (2012). Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc Natl Acad Sci USA 109: 17633–17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Swift P, Thomas S, Cummings D, Temperton B et al. (2010). The taxonomic and functional diversity of microbes at a temperate coastal site: a ‘multi-omic' study of seasonal and diel temporal variation. PLoS One 5: e15545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Rappe MS, Vergin KL, Adair NL. (1996). 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the Green Non-Sulfur bacteria. Proc Natl Acad Sci USA 93: 7979–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Vergin KL. (2012). Seasonality in ocean microbial communities. Science 335: 671–676. [DOI] [PubMed] [Google Scholar]
- Gordon D, Giovannoni S. (1996). Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl Environ Microbiol 62: 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman RL, Griffin S, Watson JT, Druffel ERM, Ingalls AE, Pearson A et al. (2009). The radiocarbon signature of microorganisms in the mesopelagic ocean. Proc Natl Acad Sci USA 106: 6513–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ERM et al. (2006). Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA 103: 6442–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jickells TD, An ZS, Andersen KK, Baker a R, Bergametti G, Brooks N et al. (2005). Global iron connections between desert dust, ocean, biogeochemistry and climate. Science 308: 67–71. [DOI] [PubMed] [Google Scholar]
- Jing H, Xia X, Suzuki K, Liu H. (2013). Vertical profiles of bacteria in the tropical and subarctic oceans revealed by pyrosequencing. PLoS One 8: e79423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311: 1737–1740. [DOI] [PubMed] [Google Scholar]
- Jones E, Oliphant T, Peterson P, Others A. (2001), SciPy: Open source scientific tools for python. Available at: http://www.scripy.org/ (accesse 15 September 2013).
- Jumpstart Consortium Human Microbiome Project Data Generation Working Group. (2012). Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One 7: e39315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahru M, Gille ST, Murtugudde R, Strutton PG, Manzano-Sarabia M, Wang H, Mitchell BG. (2010). Global correlations between winds and ocean chlorophyll. J Geophys Res 115: C12040. [Google Scholar]
- Kanehisa M, Goto S. (2000). KEGG: Kyoto encyclopedia of genomes and genomes. Nucl Acids Res 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl DM, Church MJ, Dore JE, Letelier RM, Mahaffey C. (2012). Predictable and efficient carbon sequestration in the North Pacific Ocean supported by symbiotic nitrogen fixation. Proc Natl Acad Sci USA 109: 1842–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl DM, Church MJ. (2014). Microbial oceanography and the Hawaii Ocean Time-series programme. Nature Rev Micro 12: 699–713. [DOI] [PubMed] [Google Scholar]
- Karl DM, Knauer GA, Martin JH, Ward BB. (1984). Bacterial chemolithotrophy in the ocean is associated with sinking particles. Nature 309: 54–56. [Google Scholar]
- Karl DM, Lukas R. (1996). The Hawaii Ocean Time-series (HOT) program: Background, rationale and field implementation. Deep-Sea Res Part 2 Top Stud Oceanogr 43: 129–156. [Google Scholar]
- Karl DM, Tien G. (1997). Temporal variability in dissolved phosphorus concentrations in the subtropical North Pacific Ocean. Mar Chem 56: 77–96. [Google Scholar]
- Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A et al. (2014). Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344: 416–420. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Eisen JA, Pollard KS, Green JL. (2011). The phylogenetic diversity of metagenomes. PLoS One 6: e23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S et al. (2007). Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3: 2515–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladau J, Sharpton TJ, Finucane MM, Jospin G, Kembel SW, O'Dwyer J et al. (2013). Global marine bacterial diversity peaks at high latitudes in winter. ISMEJ 7: 1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, Findeiβ S, Steiner L, Marz M, Stadler PF, Prohaska SJ. (2011). Proteinortho: detection of (Co-)orthologs in large-scale analysis. BMC Bioinformatics 12: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Fuhrman JA. (1991). Spatial and temporal variation of natural bacterioplankton assemblages studied by total genomic DNA cross-hybridization. Limnol Oceanogr 36: 1277–1287. [Google Scholar]
- Letelier R, Karl DM, Abbott MR, Flament P, Freilich M, Lukas R et al. (2000). Role of late winter mesoscale events in the biogeochemical variability of the upper water column of the North Pacific Subtropical Gyre. J Geophys Res 105: 28723–28739. [Google Scholar]
- Letelier RM, Bidigare RR, Hebel DV, Ondrusek M, Winn CD, Karl DM. (1993). Temporal variability of phytoplankton community structure based on pigment analysis. Limnol Oceanogr 38: 1420–1437. [Google Scholar]
- Li W, Godzik A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- Logares R, Sunagawa S, Salazar G, Cornejo-Castillo FM, Ferrera I, Sarmenta H, Hingamp P et al. (2009). Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environ Microbiol 16: 2659–2671. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R. (2007). Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73: 1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Buchner A et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom RR, Coe A, Kettler GC, Martiny AC, Frias-Lopez J, Zinser ER et al. (2010). Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. ISME J 4: 1751–7362. [DOI] [PubMed] [Google Scholar]
- Mary I, Cummings D, Biegala I. (2006). Seasonal dynamics of bacterioplankton community structure at a coastal station in the western English Channel. Aquat Microb Ecol 42: 119–126. [Google Scholar]
- McGillicuddy DJ, Anderson LA, Bates NR, Bibby T, Buesseler KO, Carlson CA et al. (2007). Eddy/wind interactions stimulate extraordinary mid-ocean plankton blooms. Science 316: 1021–1026. [DOI] [PubMed] [Google Scholar]
- Moore LR, Rocap G, Chisholm SW. (1998). Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393: 464–467. [DOI] [PubMed] [Google Scholar]
- Moran MA, Miller WL. (2007). Resourceful heterotrophs make the most of light in the coastal ocean. Nat Rev Microbiol 5: 792–800. [DOI] [PubMed] [Google Scholar]
- Morris RM, Vergin KL, Cho J-C, Rappé MS, Carlson CA, Giovannoni SJ. (2005). Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr 50: 1687–1696. [Google Scholar]
- Murray AE, Preston CM, Massana R, Taylor LT, Blakis A, Wu K et al. (1998). Seasonal and spatial variability of Bacterial and archaeal assemblages in the Coastal Waters near Anvers Island, Antarctica. Appl Environ Microbiol 64: 2585–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H, Parker J, Takagi T. (2006). MetaGene: prokayrotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 34: 5623–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman MD, Anderson GC, Ozturgut E. (1982). A multivariate analysis of planktonic interactions in the eastern tropical North Pacific. Deep Sea Res A 29: 1451–1469. [Google Scholar]
- Oksanen J, Kindt R, Pierre L, O'Hara B, Simpson GL, Solymos P et al. (2013), vegan: Community Ecology Package, R package version 2.0. Available at: http://vegan.r-forge.r-project.org.
- Parsons RJ, Breitbart M, Lomas MW, Carlson CA. (2012). Ocean time-series reveals recurring seasonal patterns of virioplankton dynamics in the northwestern Sargasso Sea. ISME J 6: 273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. (2011). Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Letunic I, Yamada T, Jensen LJ, Bork P. (2011). Toward molecular trait-based ecology through integration of biogeochemical, geographical and metagenomic data. Mol Syst Biol 7: 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich VI, Pham VD, Eppley J, Shi Y, DeLong EF. (2011). Time-series analyses of Monterey Bay coastal microbial picoplankton using a ‘genome proxy' microarray. Environ Microbiol 13: 116–134. [DOI] [PubMed] [Google Scholar]
- Robidart JC, Church MJ, Ryan JP, Ascani F, Wilson ST, Bombar D. (2014). Ecogenomic sensor reveals controls on N2-fixing microorganisms in the North Pacific Ocean. ISME J 8: 1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-González C, Simó R, Sommaruga R, Gasol JM. (2013). Away from darkness: a review on the effects of solar radiation on heterotrophic bacterioplankton activity. Front Microbiol 4: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G, Cornejo-Castillo FM, Benítez-Barrios V, Fraile-Nuez E, Álvarez-Salgado XA, Duarte CM et al. (2015). Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J e-pub ahead of print 7 August 2015 doi:10.1038/ismej.2015.137. [DOI] [PMC free article] [PubMed]
- Schmidt T, DeLong E, Pace N. (1991). Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol 173: 4371–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Becker JW, Ottesen EA, Bryant JA, Duhamel S, Karl DM et al. (2013). Distinct dissolved organic matter sources induce rapid transcriptional responses in coexisting populations of ProchlorococcusPelagibacter and the OM60 clade. Environ Microbiol 16: 2815–2830. [DOI] [PubMed] [Google Scholar]
- Sharon I, Banfield JF. (2013). Genomes from metagenomics. Science 342: 1057–1058. [DOI] [PubMed] [Google Scholar]
- Stewart FJ, Ottesen EA, DeLong EF. (2010). Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J 4: 896–907. [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G et al. (2015). Structure and function of the global ocean microbiome. Science 348: 1261359. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, González JM et al. (2013). Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA 110: 11463–11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA et al. (2009). Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J 3: 1148–1163. [DOI] [PubMed] [Google Scholar]
- Tsai A-Y, Chiang P-P, Chang J, Gong GC. (2005). Seasonal diel variation of picoplankton and nanoplankton in a subtropical western Pacific coastal ecosystem. Limnol Oceanogr 50: 1221–1231. [Google Scholar]
- Tsai A-Y, Gong GC, Sanders RW, Chiang KP, Huang JK, Chan YF. (2012). Viral lysis and nanoflagellate grazing as factors controlling diel variations of Synechococcus spp. summer abundance in coastal waters of Taiwan. Aquat Microb Ecol 66: 159–167. [Google Scholar]
- Winn C, Chiswell S, Firing E, Karl DM, Lukas R. (1992) Hawaii Ocean Time-series data report 2, 1990. University of Hawaii, HI, USA. SOEST Tech Rep 92-1.
- Winn CD, Campbell L, Christian JR, Letelier RM, Hebel D V, Dore JE et al. (1995). Seasonal variability in the phytoplankton community of the North Pacific Subtropical Gyre. Global Biogeochem Cycles 9: 605–620. [Google Scholar]
- Wright T, Vergin K, Boyd P, Giovannoni S. (1997). A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Appl Environ Microbiol 63: 1441–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW, Carder KL, Betzer PR, Costello DK, Duce RA, DiTullio GR et al. (1991). Atmospheric iron inputs and primary productivity: Phytoplankton responses in the North Pacific. Global Biogeochem Cycles 5: 119–134. [Google Scholar]
- Zhang B, Horvath S. (2005). A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: Article 17. [DOI] [PubMed] [Google Scholar]
- Zinser ER, Johnson ZI, Veneziano D, Chisholm SW. (2007). Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol Oceanogr 52: 2205–2220. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.