Abstract
Corrinoid auxotrophic organohalide-respiring Dehalococcoides mccartyi (Dhc) strains are keystone bacteria for reductive dechlorination of toxic and carcinogenic chloroorganic contaminants. We demonstrate that the lower base attached to the essential corrinoid cofactor of reductive dehalogenase (RDase) enzyme systems modulates dechlorination activity and affects the vinyl chloride (VC) RDases BvcA and VcrA differently. Amendment of 5,6-dimethylbenzimidazolyl-cobamide (DMB-Cba) to Dhc strain BAV1 and strain GT cultures supported cis-1,2-dichloroethene-to-ethene reductive dechlorination at rates of 107.0 (±12.0) μM and 67.4 (±1.4) μM Cl- released per day, respectively. Strain BAV1, expressing the BvcA RDase, reductively dechlorinated VC to ethene, although at up to fivefold lower rates in cultures amended with cobamides carrying 5-methylbenzimidazole (5-MeBza), 5-methoxybenzimidazole (5-OMeBza) or benzimidazole (Bza) as the lower base. In contrast, strain GT harboring the VcrA RDase failed to grow and dechlorinate VC to ethene in medium amended with 5-OMeBza-Cba or Bza-Cba. The amendment with DMB to inactive strain GT cultures restored the VC-to-ethene-dechlorinating phenotype and intracellular DMB-Cba was produced, demonstrating cobamide uptake and remodeling. The distinct responses of Dhc strains with BvcA versus VcrA RDases to different cobamides implicate that the lower base exerts control over Dhc reductive dechlorination rates and extents (that is, detoxification), and therefore the dynamics of Dhc strains with discrete reductive dechlorination capabilities. These findings emphasize that the role of the corrinoid/lower base synthesizing community must be understood to predict strain-specific Dhc activity and achieve efficacious contaminated site cleanup.
Introduction
The worldwide usage of chlorinated solvents for degreasing, chemical manufacturing and household applications made chlorinated solvents widespread groundwater contaminants (Abelson, 1990). Chlorinated solvents such as tetrachloroethene (PCE), trichloroethene (TCE) and dichloroethenes (DCEs) are toxic and probable human carcinogens, and recently TCE has been implicated in the development of Parkinson disease (ATSDR, 2003, 2004; Goldman et al., 2012). The breakdown of polychlorinated ethenes to the human carcinogen vinyl chloride (VC) is particularly troublesome, and exposure may occur through vapor intrusion into dwellings or consumption of tainted drinking water (Kielhorn et al., 2000; EPA, 2012; NRC, 2013).
In bacterial organohalide respiration, chlorinated compounds serve as the terminal electron acceptors, a cornerstone process for chlorinated solvent bioremediation (Leys et al., 2013). Although diverse bacterial groups contribute to reductive dechlorination of PCE and TCE to cis-1,2-dichloroethene (cDCE), only some Dhc strains dechlorinate DCEs and VC to benign ethene (Maymó-Gatell et al., 1997; He et al., 2003; Löffler et al., 2013a, b). Dhc strain BAV1 and strain GT possess the reductive dehalogenase (RDase) enzyme systems BvcA and VcrA, respectively, that share 54% amino acid similarity (40% identity) and are responsible for reductive dechlorination of DCEs and VC to ethene in the respective Dhc strains (Krajmalnik-Brown et al., 2004; Müller et al., 2004; Sung et al., 2006; McMurdie et al., 2009; Tang et al., 2013; Parthasarathy et al., 2015). Dhc are obligate organohalide respirers and the RDases require a corrinoid cofactor (that is, coenzyme B12) for activity (Yi et al., 2012; Hug et al., 2013). Remarkably, Dhc strains lack the entire set of ~20 genes involved in corrin ring biosynthesis, and growth of Dhc pure cultures strictly depends on exogenous cyanocobalamin (that is, vitamin B12) (Löffler et al., 2013b). Instead of de novo corrinoid biosynthesis, Dhc strains harbor the requisite genes for both the bacterial and the archaeal scavenging pathways that include btuF, btuC and btuD corrinoid transport, cbiZ, cbiB, cobD and cobU cobinamide salvage/activation and cobT, cobC and cobS lower base activation to access growth-supporting cobamides (Yi et al., 2012; Men et al., 2014a, b). In addition, the sequenced Dhc genomes harbor at least one ortholog of cobA that encodes the co(I)rrinoid adenosyltransferase responsible for attaching the upper adenosyl group ligand (Yi et al., 2012).
Recent studies revealed that Dhc strains have specific corrinoid requirements that corrinoid producers such as the methanogen Methanosarcina barkeri strain Fusaro, the acetogen Sporomusa ovata (DSMZ 2662) and the ferric iron reducer Geobacter sulfurreducens cannot fulfill, unless 5,6-dimethylbenzimidazole (DMB) is amended to the medium (Yan et al., 2012, 2013). Studies with Dhc strain 195 revealed that TCE reductive dechlorination was only sustained by DMB-cobamide (Cba), 5-MeBza-Cba and 5-OMeBza-Cba, but not by Bza-Cba, 5-hydroxybenzimidazolyl-cobamide (5-OHBza-Cba or factor III) or phenolic cobamides (Phe-Cba and p-Cresol-Cba) (Yi et al., 2012; Men et al., 2014b). Apparently, the cobamide lower base (Figure 1) determines whether Dhc can or cannot use chlorinated hydrocarbons as electron acceptors. To explore the effects of the lower base on Dhc reductive dechlorination activity in more detail, we prepared naturally occurring cobamides through guided biosynthesis (Allen and Stabler, 2008; Mok and Taga, 2013), and investigated their impact on Dhc growth and reductive dechlorination activity. Pure cultures of Dhc strain BAV1 and strain GT harboring the bvcA and vcrA genes, respectively, and capable of growth with cDCE and VC were amended with the different cobamides and reductive dechlorination rates and extents were analyzed.
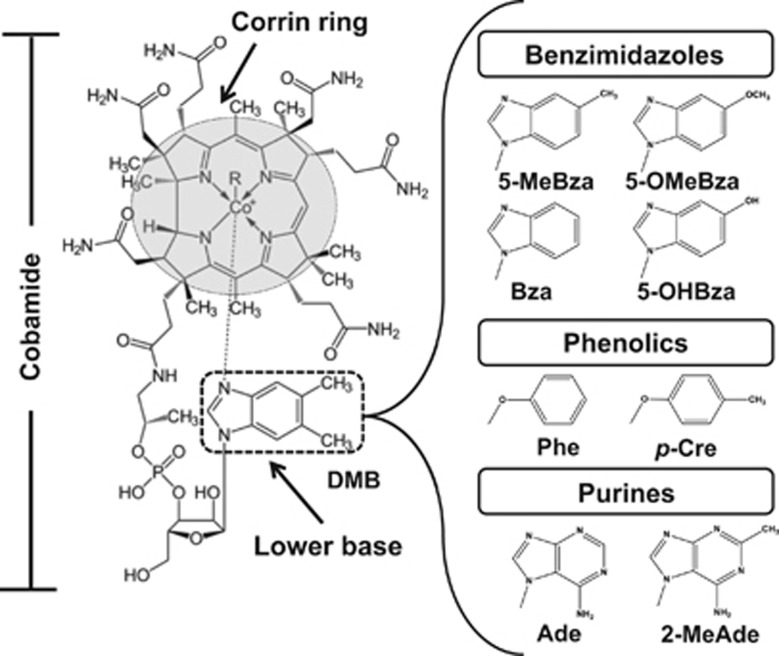
Figure 1.
General structure of the corrin ring system and several naturally occuring lower bases. Ade, adenine; 2-MeAde, 2-methyladenine; p-Cre, 4-methylphenol; Phe, phenol.
Materials and methods
Chemicals
DMB (⩾99%), 5-methylbenzimidazole (5-MeBza; 98%), 5-methoxybenzimidazole (5-OMeBza; 97%), benzimidazole (Bza; 98%), cDCE (⩾99.5%), VC (⩾99.5%), ethene (⩾99.9%) and betaine (⩾99%) were purchased from Sigma-Aldrich (St Louis, MO, USA). Yeast extract and casitone were purchased from Becton, Dickinson and Company (Franklin Lakes, NJ, USA).
Dehalococcoides mccartyi (Dhc) cultures
Dhc strain BAV1 (ATCC BAA-2100) and strain GT (ATCC BAA-2099) were grown in 160 ml serum bottles containing 100 ml defined, completely synthetic mineral salts medium amended with 5 mM acetate as carbon source, 10 ml hydrogen as electron donor, 5 μl neat cDCE (0.60 mM aqueous phase concentration) or 2 ml VC (0.52 mM aqueous phase concentration) as electron acceptor as previously described (Yan et al., 2013). Following equilibration, triplicate bottles were amended with the Wolin vitamin mix (Wolin et al., 1963) not containing cyanocobalamin (vitamin B12). The vessels received 36.9 nM of a purified cobamide. For inoculation, Dhc strain BAV1 or strain GT cell suspensions were centrifuged and the pellets were suspended in cyanocobalamin-free mineral salts medium to avoid cyanocobalamin carryover (Yan et al., 2012). Cultures that failed to dechlorinate cDCE or VC received 10 μM DMB from a filter-sterilized 3 mM aqueous stock solution to restore dechlorination activity. All culture vessels were incubated statically in dark at 30 °C. The cDCE-to-ethene dechlorination rates were calculated based on VC and ethene quantification, each step associated with the release of one chloride ion, and reported as μmol of Cl- released per liter per day.
Cobamide biosynthesis, extraction and purification
DMB-, 5-MeBza-, 5-OMeBza- and Bza-Cba were obtained via guided cobamide biosynthesis using Sporomusa sp. strain KB-1 (GenBank accession no. AY780559.1) cultures supplied with a single lower base compound (200 μM). To achieve higher yields, 800 ml cultures of strain KB-1 were grown in 1.2 l glass vessels using a modified medium (Yan et al., 2013) amended with yeast extract (2 g l−1), casitone (2 g l−1) betaine (50 mM) and a cyanocobalamin-free Wolin vitamin mix (Wolin et al., 1963). After 72–96 h of static incubation at 30 °C in the dark, cells were harvested from 1.6 to 2.4 l culture suspensions by centrifugation at 17 600 g for 15 min at room temperature. Cell pellets were suspended in 15 ml deionized water, briefly incubated in a sonication water bath to achieve homogeneous suspensions and 5 ml aliquots were transferred to sterile 50 ml plastic tubes. Total corrinoids were extracted in the cyano form (that is, a cyanide group as the upper β-ligand) and purified using the potassium cyanide extraction method (Yan et al., 2013). Cobamide-containing fractions were separated by high-performance liquid chromatography (HPLC) and manually collected from the detector outlet according to retention time and diode array detector (DAD) response to remove any remaining traces of native phenolic cobamides (that is, Phe-Cba and p-Cre-Cba). Cobamides were purified as described above. Cobamide absorbance was measured with a Lambda 35 UV/VIS spectrometer (PerkinElmer, Waltham, MA, USA) at 361 nm, and cobamide concentrations were estimated using a molar extinction coefficient of 28 060 mol−1 cm−1 (Pratt, 1972). The purified cyano form of DMB-Cba was indistinguishable from commercial cyanocobalamin. All experiments used purified DMB-Cba in control incubations to verify the absence of any inhibitory compounds introduced during the extraction process.
HPLC and liquid chromatography–mass spectrometry analysis
Cobamide purity and authenticity (for example, distinct singular peaks and m/z values) were analyzed in a combined approach employing an Agilent (Santa Clara, CA, USA) 1200 HPLC system and a Thermo Fisher Scientific (Waltham, MA, USA) Orbitrap Exactive Plus LC/MS system (Supplementary Figures S1 and S2). For HPLC analysis, 20 μl samples were injected onto an Eclipse XDB-C18 column (5 μm, 4.6 × 150 mm) (Agilent) and separated with a flow rate of 1 ml per min at 30 °C using 0.1% (v/v) formic acid (⩾88%, w/v) in water (eluent A) and 0.1% (v/v) formic acid in methanol (eluent B) as mobile phases. The column was equilibrated with 82% eluent A/18% eluent B, and a linear change to 75% A/25% B was applied following sample injection over a 12-min time period. Then, the eluent composition decreased immediately to 25% A/75% B over 3 min followed by a 5-min hold before the column was equilibrated to initial conditions. Cobamides were detected at 361 nm with an Agilent 1260 Infinity DAD and quantified by comparing integrated peak areas with 4-point calibration curves generated with purified cobamides. Liquid chromatography–mass spectrometry analysis was performed using a Dionex Ultimate 3000 system (Thermo Fisher Scientific) with an inline DAD fitted to an Exactive Plus Orbitrap Mass Spectrometer with an electrospray ionization source (Thermo Fisher Scientific). For liquid chromatography–DAD–mass spectrometry analysis, 10 μl aliquots of each sample were injected onto a Kinetex XB-C18 column (2.6 μm, 2.1 × 100 mm) (Phenomenex, Torrance, CA, USA) and separated at a flow rate of 0.2 ml per min at 30 °C using 0.1% formic acid in water (eluent A) and 0.1% formic acid in acetonitrile (eluent B) as mobile phases. The gradient started with 100% A, changed linearly to 85% A after 2.8 min, 75% A after 5.2 min, 90% A after 5.44 min and 100% A after 6.8 min with a 4.2-min hold to achieve column equilibration to starting conditions. The mass spectrometer was operated in full scan mode with a mass range of 750–1800 m/z and a resolution of 140 000. All ion fragmentation was performed on each ion packet in a subsequent event after each full scan using a normalized collision energy of 20 eV with a stepped normalized collision energy of 50%. Electrospray ionization was performed in positive mode with the sheath gas set at 25 arbitrary units, the auxiliary gas at 10 arbitrary units, the spray voltage at 4000 V and a capillary temperature of 350 °C. The DAD was set to detect at 361 nm as well as over a 3D field from 190 to 800 nm.
Corrinoid extraction from Dhc cultures
Dhc cells and culture supernatant were separated by filtration using a 47 mm diameter 0.22 μm pore size membrane (Pall Life Sciences, Port Washington, NY, USA). Intracellular corrinoids were extracted and purified following the KCN extraction protocol (Yan et al., 2013). Supernatant-associated corrinoids were reduced into the upper ligand-free, cobalt(II) form by reductants (for example, HS-, L-cysteine, dithiothreitol) present in the medium (Assaf-Anid et al., 1994; Chiu and Reinhard, 1996; Lesage et al., 1998). To convert cob(II)amides back to the oxidized cyano-cob(III)amide forms, which can be separated and quantified with the established HPLC method, culture supernatants were reacted with ambient air in the presence of KCN for 24 h (Schneider and Stroiński, 1987). This method was validated by extracting a mixture of cobamides with different lower bases from 100 ml of medium, and the average recovery efficiencies from triplicate vessels were 93.4%, 90.6% and 91.1% for DMB-Cba, 5-OMeBza-Cba and 5-Bza-Cba, respectively. Following the 24-h incubation, culture supernatants were loaded onto a C18 Sep-Pak cartridge and washed with 40 ml distilled water. Absorbed corrinoids were eluted with 3 ml methanol, vacuum dried and suspended in 0.5 ml distilled water.
Cobamide uptake in Dhc
Triplicate 100 ml cultures of Dhc strain BAV1 and strain GT were grown with 5 μl cDCE and a cobamide mixture containing equimolar concentrations of DMB-Cba, 5-OMeBza-Cba and 5-Bza-Cba (36.9 nM each). As soon as cDCE was completely dechlorinated to ethene, cobamides remaining in the culture supernatants were recovered, purified and quantified as described above.
Analytical methods
Dhc cells were harvested onto 0.22 μm membrane filters (Merck Millipore Ltd, Darmstadt, Germany) and the genomic DNA was extracted from the filters using the MO BIO Soil DNA Isolation kit (MO BIO, Carlsbad, CA, USA) as previously described (Yan et al., 2012). Dhc 16S rRNA gene copies were enumerated by quantitative PCR with a ViiA 7 real-time PCR system (Life Technologies, Grand Island, NY, USA) using primer pair Dhc1200F/Dhc1271R and probe Dhc1240probe following established protocols (Ritalahti et al., 2006). Chlorinated ethenes and ethene were analyzed with an Agilent 7890 gas chromatograph equipped with a flame ionization detector and a DB-624 capillary column (60 m × 0.32 mm × 1.8 μm) (Sung et al., 2006).
Results
The lower base affects dechlorination rates
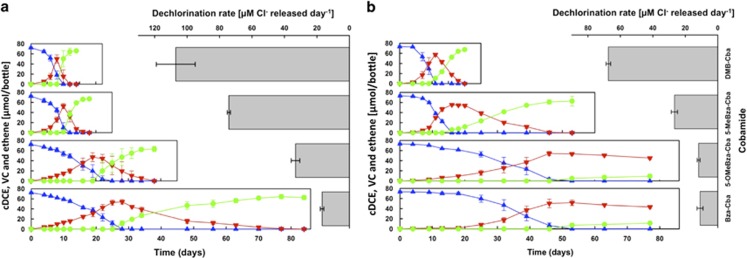
The cobamides DMB-Cba, 5-MeBza-Cba, 5-OMeBza-Cba and Bza-Cba supported complete dechlorination of cDCE to ethene in Dhc strain BAV1 cultures. The highest cDCE-to-ethene dechlorination rates of 107.0±12.0 and 74.3±1.0 μM Cl- released per day were measured in the presence of DMB-Cba and 5-MeBza-Cba, respectively (Figure 2a), and the initial amount of 72.2 (±1.4) μmol cDCE was completely dechlorinated to ethene in 12–18 days. Longer time periods of 38 and 86 days were required to achieve complete cDCE and VC reductive dechlorination to ethene in strain BAV1 cultures amended with 5-OMeBza-Cba or Bza-Cba because of lower cDCE dechlorination rates of 33.2 (±2.6) and 16.8 (±1.1) μM Cl- released per day, respectively (Figure 2a). Strain GT cultures amended with DMB-Cba and 5-MeBza-Cba dechlorinated cDCE to ethene at rates of 67.4 (±1.4) and 26.7 (±1.9) μM Cl- released per day, respectively (Figure 2b). Similar to strain BAV1 cultures, cDCE dechlorination rates decreased in strain GT cultures amended with 5-OMeBza-Cba or Bza-Cba; however, the VC-to-ethene dechlorination step occurred at such low rates that VC, rather than ethene, was formed as dechlorination end product (Figure 2b). In strain GT cultures amended with 5-OMeBza-Cba or Bza-Cba, no more than 23% (17.5 μmol) of the total VC produced from cDCE dechlorination was further dechlorinated to ethene over a 77-day incubation period (Figure 2b).
Figure 2.
Reductive dechlorination of cDCE in (a) Dhc strain BAV1 harboring the BvcA RDase and (b) strain GT harboring the VcrA RDase. Triplicate cultures were amended with cobamides carrying different benzimidazole derivatives as lower bases at initial concentrations of 36.9 nM. Blue triangles, cDCE; red inverted triangles, VC; green circles, ethene. Error bars represent the s.d. of triplicate cultures.
The lower base affects Dhc growth yields
Enumeration of Dhc cell numbers after cDCE-to-ethene dechlorination was complete (that is, in all BAV1 cultures and DMB-Cba- or 5-MeBza-Cba-amended GT cultures) or ceased (that is, in 5-OMeBza- or Bza-Cba-amended GT cultures) revealed that strain BAV1 cultures reached similar growth yields of 1.38 (±0.09) to 1.57 (±0.03) × 108 cells per μmol Cl- released with all cobamides tested (Table 1). For Dhc strain GT, the highest final cell densities of 1.29 (±0.22) × 108 and 1.30 (±0.41) × 108 cells per ml were measured in cultures amended with DMB-Cba and 5-MeBza-Cba, respectively, and these results were consistent with the growth yield expected from complete cDCE-to-ethene dechlorination (Table 1). With 5-OMeBza-Cba or Bza-Cba, strain GT cultures reached cell yields of 6.95 (±1.04) and 7.72 (±0.87) × 107 cells per μmol Cl- released, respectively, that were on average 31% and 23% lower compared with growth yields of 0.92 (±0.15) and 1.10 (±0.24) × 108 cells per μmol Cl- released measured in DMB-Cba or 5-MeBza-Cba amended cultures, respectively. The lower growth yields suggested that the VC-to-ethene reductive dechlorination step was uncoupled from growth in strain GT cultures amended with 5-OMeBza-Cba or Bza-Cba.
Table 1. Growth of Dhc pure cultures amended with different cobamides.
| Dhc strain (Electron acceptor) | Cobamidea | DMBb |
Dhc cell density (16S rRNA gene copies per ml) |
Growth yield (cells (μmol Cl-)-1)c | |
|---|---|---|---|---|---|
| Initial | Final | ||||
| BAV1 | DMB-Cba | − | 4.26±0.87 × 106 | 1.82±0.25 × 108 | 1.38±0.09 × 108 |
| (cDCE) | 5-MeBza-Cba | − | 4.26±0.87 × 106 | 2.09±0.51 × 108 | 1.54±0.39 × 108 |
| 5-OMeBza-Cba | − | 4.26±0.87 × 106 | 1.85±0.39 × 108 | 1.42±0.20 × 108 | |
| Bza-Cba | − | 4.26±0.87 × 106 | 2.07±0.10 × 108 | 1.57±0.03 × 108 | |
| GT | DMB-Cba | − | 5.22±0.54 × 106 | 1.29±0.22 × 108 | 0.92±0.15 × 108 |
| (cDCE) | 5-MeBza-Cba | − | 5.22±0.54 × 106 | 1.30±0.41 × 108 | 1.10±0.24 × 108 |
| 5-OMeBza-Cba | − | 5.22±0.54 × 106 | 4.97±0.99 × 107 | 6.95±1.04 × 107 | |
| Bza-Cba | − | 5.22±0.54 × 106 | 5.72±0.38 × 107 | 7.72±0.87 × 107 | |
| GT | DMB-Cba | − | 2.61±0.27 × 106 | 0.96±0.06 × 108 | 1.17±0.15 × 108 |
| (VC) | 5-MeBza-Cba | − | 2.61±0.27 × 106 | 1.10±0.10 × 108 | 1.39±0.20 × 108 |
| 5-OMeBza-Cba | − | 2.61±0.27 × 106 | 2.08±0.01 × 106 | No growth | |
| 5-OMeBza-Cba | + | 2.61±0.27 × 106 | 1.21±0.34 × 108 | 1.45±0.33 × 108 | |
| Bza-Cba | − | 2.61±0.27 × 106 | 2.37±0.00 × 106 | No growth | |
| Bza-Cba | + | 2.61±0.27 × 106 | 1.22±0.08 × 108 | 1.48±0.11 × 108 | |
Abbreviations: Bza, benzimidazole; Cba, cobamide; cDCE, cis-1,2-dichloroethene; Dhc, Dehalococcoides mccartyi; DMB, 5,6-dimethylbenzimidazole; 5-MeBza, 5-methylbenzimidazole; 5-OHBza, 5-hydroxybenzimidazole; 5-OMeBza, 5-methoxybenzimidazole; VC, vinyl chloride.
Cobamide concentration was 36.9 nM.
DMB was supplied at 10 μM.
Growth yield was estimated from the final cell density after complete or stalled cDCE or VC dechlorination.
Impact of the lower base on dechlorination extent
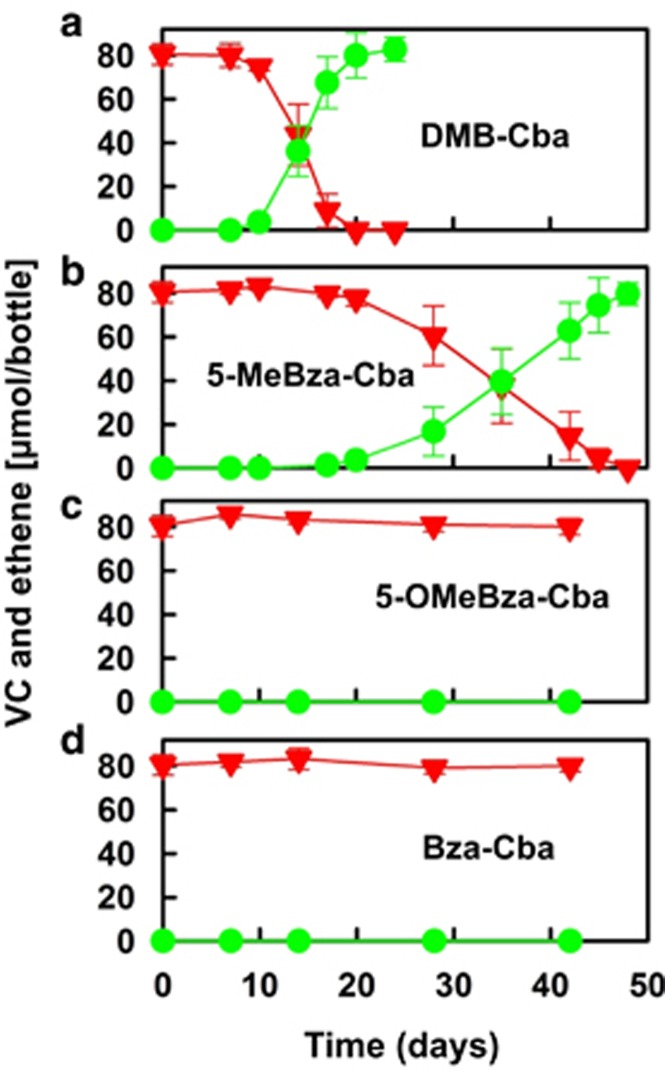
VC stall and lower growth yields suggested that 5-OMeBza-Cba and Bza-Cba were not fully functional in strain GT, and VC was not used as a growth substrate. The key feature distinguishing Dhc strain GT from strain BAV1 is the VC RDase: strain GT uses VcrA whereas strain BAV1 uses BvcA for cDCE and VC reductive dechlorination (Krajmalnik-Brown et al., 2004; Müller et al., 2004; Sung et al., 2006; Tang et al., 2013; Parthasarathy et al., 2015). To further investigate the impact of the lower base on the catalytic activity of the VcrA RDase, growth experiments in medium amended with VC as electron acceptor were performed. Consistent with the observation in cDCE-dechlorinating cultures, only DMB-Cba and 5-MeBza-Cba sustained VC dechlorination in strain GT cultures, and rates of 40.0 (±5.1) and 16.5 (±2.8) μM VC per day, respectively, were observed. Ethene production was negligible (<0.8% of the total VC added) in strain GT cultures amended with 5-OMeBza-Cba or Bza-Cba (Figure 3). In contrast, strain BAV1 cultures that received 5-OMeBza- or Bza-Cba completely dechlorinated VC to ethene. Enumeration of Dhc cells with quantitative PCR confirmed the inability of 5-OMeBza-Cba and Bza-Cba to support growth of strain GT with VC as an electron acceptor. In vessels with initial cell densities of 2.61 (±0.27) × 106 cells per ml (that is, cells introduced with the inoculum), strain GT cell numbers decreased to 2.08 (±0.01) × 106 and 2.37 (±0.09) × 106 cells per ml with 5-OMeBza-Cba or Bza-Cba provided as corrinoid cofactor (Table 1). In contrast, strain GT cell numbers increased to 0.96 (±0.06) × 108 and 1.10 (±0.10) × 108 cells per ml in cultures amended with DMB-Cba and 5-MeBza-Cba, respectively, corresponding to growth yields of 1.17 (±0.15) × 108 and 1.39 (±0.20) × 108 cells per μmol VC dechlorinated (Table 1).
Figure 3.
Reductive dechlorination of VC in Dhc strain GT cultures amended with an initial concentration of 36.9 nM (a) DMB-Cba, (b) 5-MeBza-Cba, (c) 5-OMeBza-Cba or (d) Bza-Cba. Red inverted triangles, VC; green circles, ethene. Error bars represent the s.d. of triplicate cultures.
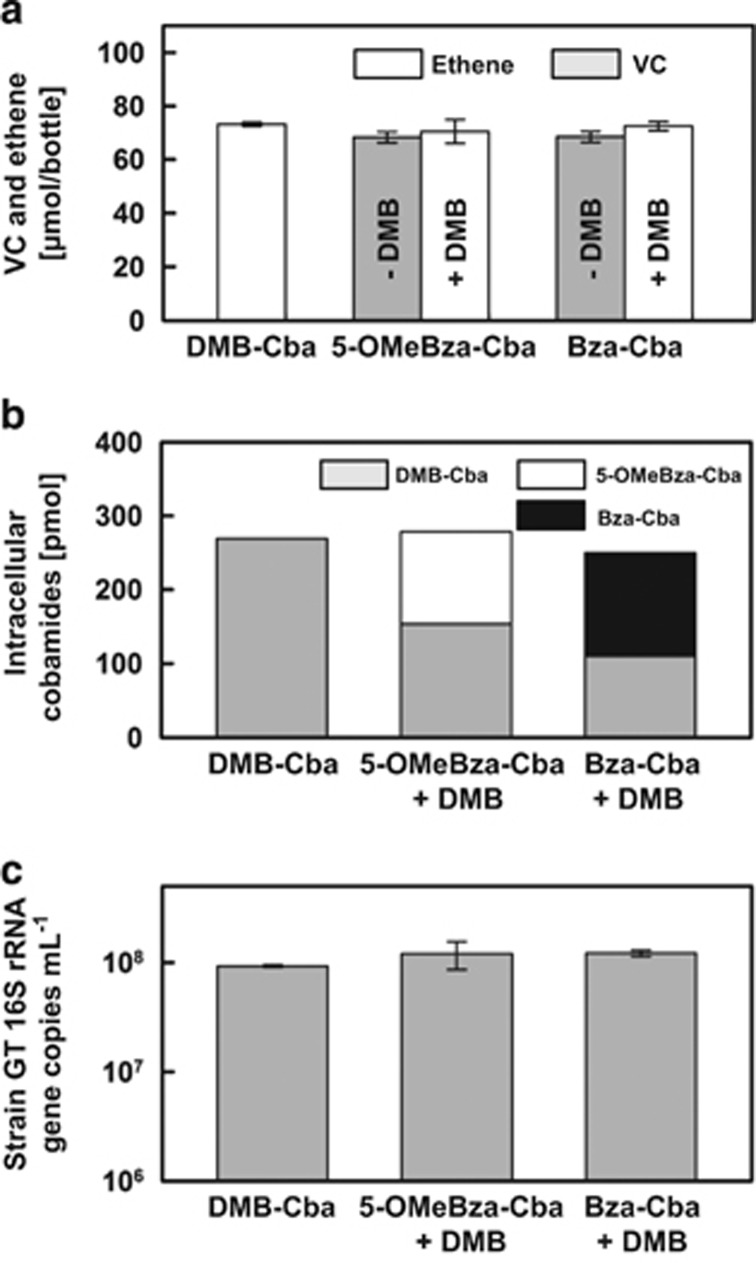
We next tested whether the addition of DMB could rescue the VC-to-ethene dechlorination phenotype in strain GT cultures that had received 5-OMeBza-Cba or Bza-Cba. At 3 weeks following DMB addition to inactive cultures, all VC was dechlorinated to ethene, whereas no ethene formation occurred in cultures without DMB amendment (Figure 4a). To verify whether the restoration of VC dechlorination activity following DMB addition was because of corrinoid remodeling, intracellular corrinoids from strain GT cells were extracted and purified. DMB-Cba was only detected in corrinoid extracts of cells collected from vessels amended with DMB, and DMB-Cba accounted for nearly half of the total intracellular cobamides in strain GT cells grown in medium with 5-OMeBza-Cba or Bza-Cba (Figure 4b). Cell enumeration with quantitative PCR demonstrated that additional growth occurred following restoration of VcrA activity. Cultures that initially received 5-OMeBza-Cba or Bza-Cba and were then amended with DMB to restore the VC dechlorinating phenotype produced 1.21 (±0.34) × 108 and 1.22 (±0.08) × 108 cells per ml culture, respectively (Table 1). The growth yields in 5-OMeBza-Cba and Bza-Cba cultures following DMB addition were 1.45 (±0.33) × 108 and 1.48 (±0.11) × 108 cells per μmol VC dechlorinated, respectively, and are comparable to those observed in strain GT cultures amended with DMB-Cba (1.17 (±0.15) × 108 cells per μmol VC dechlorinated; Figure 4c).
Figure 4.
Restoration of the VC dechlorination phenotype by DMB addition to inactive Dhc strain GT cultures. (a) Comparison of VC-to-ethene dechlorination activity in 5-OMeBza-Cba- or Bza-Cba- (36.9 nM each) amended strain GT cultures in the presence or absence of 10 μM DMB. (b) Quantification of intracellular cobamides extracted from strain GT biomass grown with 5-OMeBza-Cba or Bza-Cba in the presence of DMB and collected from 100 ml cell suspensions. (c) Comparison of the final cell densities in in 5-OMeBza-Cba- or Bza-Cba-amended strain GT cultures with or without DMB (see Table 1).
Cobamide transport in Dhc
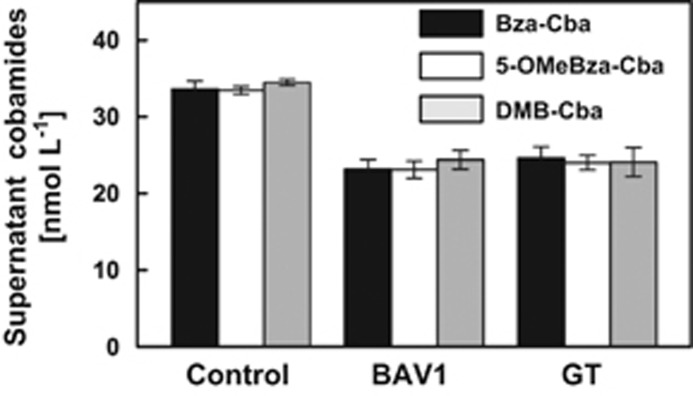
To explore whether differential transport of cobamides with distinct lower bases across the Dhc surface layer and/or cytoplasmic membrane could explain the observations, cDCE-dechlorinating Dhc cultures were supplied with an equimolar mixture of DMB-Cba, 5-OMeBza-Cba and Bza-Cba. Following cDCE conversion to ethene in strain BAV1 and strain GT cultures, the molar concentration ratios of DMB-Cba/5-OMeBza-Cba/Bza-Cba were 1.06:1.00:1.00 and 1.00:1.00:1.02 in strain BAV1 and strain GT culture supernatants, respectively, and therefore not significantly different than the initial conditions. Furthermore, statistically identical concentrations of DMB-Cba (24.4 (±1.2)/24.1 (±1.9) nM), 5-OMeOBza-Cba (23.1 (±1.1)/24.0 (±1.0) nM) and Bza-Cba (23.2 (±1.3)/24.6 (±1.5) nM) were recovered from strain BAV1/strain GT culture supernatants (Figure 5). These findings indicate that surface layer and membrane transport did not discriminate between the three benzimidazole-type cobamides. Intracellular cobamides were also analyzed, and DMB-Cba, 5-OMeBza-Cba and Bza-Cba were present in similar amounts ranging from 2 × 103 to 5 × 103 molecules per strain BAV1 and strain GT cell.
Figure 5.
Quantification of supernatant-associated cobamides recovered from cDCE-dechlorinating Dhc strain BAV1 and strain GT cultures supplied with an equimolar mixture of DMB-Cba, 5-OMeBza-Cba and Bza-Cba (36.9 nM each).
Discussion
Corrinoids in complete form (that is, cobamides) are essential enzyme cofactors exclusively synthesized by a subset of the Archaea and Bacteria, even though these molecules play critical roles for the majority of organisms in all domains of life (Schneider and Stroiński, 1987; Banerjee, 1999; Martens et al., 2002; Gruber et al., 2011; Miles et al., 2011; Parks et al., 2013). Naturally occurring corrinoids carry a variety of lower bases, and as many as 16 benzimidazole, nucleobase and phenol derivatives have been identified (Stupperich et al., 1990; Allen and Stabler, 2008). Microorganisms such as Escherichia coli (ATCC 11105) and Lactobacillus leichmannii (ATCC 4797) are capable of utilizing natural corrinoids with at least 11 different lower bases to fulfill their nutritional requirements (Thompson et al., 1950; Burkholder, 1951; Schneider and Stroiński, 1987). The human gut anaerobe Bacteroides thetaiotaomicron grows with indistinguishable doubling times with DMB-Cba, 5-MeBza-Cba, 5-OMeBza-Cba, Bza-Cba, pseudo-B12 (that is, Cba with adenine as the lower base) or 2-methyladeninyl-Cba as corrinoid cofactor (Degnan et al., 2014). S. ovata and Sporomusa sp. strain KB-1, which natively synthesize phenolic-type cobamides (that is, Phe-Cba and p-Cre-Cba), were also capable of utilizing cobamides with benzimidazole-type lower bases (that is, DMB-Cba, 5-MeBza-Cba, 5-OMeBza-Cba, Bza-Cba) during growth with H2/CO2, betaine or fructose as substrate (Mok and Taga, 2013; Yan et al., 2013). Some inhibitory effects were observed in S. ovata cultures amended with 5-MeBza when methanol or 3,4-dimethoxybenzoate served as growth substrates, suggesting that the structure of the lower base affected cobamide function and essential metabolic pathways (Mok and Taga, 2013; Yan et al., 2013). Reduced versatility in terms of the lower base utilization was observed with the corrinoid-auxotrophic Dhc strains that exhibited a clear preference for cobamides with DMB or 5-MeBza lower bases to perform organohalide respiration with cDCE and VC as electron acceptors.
The PCE-to-cDCE RDase PceA of Sulfurospirillum multivorans and the ortho-dibromophenol RDase NpRdhA of Nitratireductor pacificus strain pht-B shed first light on the structure of RDase enzyme systems and provided details on interactive corrinoid-RDase binding (Bommer et al., 2014; Payne et al., 2014). In both RDases, the corrinoid cofactor (norpseudo-B12 for PceA and DMB-Cba for NpRdhA) is involved in the catalytic cleavage of carbon-chlorine bonds. The lower bases adenine in norpseudo-B12 or DMB in DMB-Cba are uncoordinated and in the base-off conformation to anchor the cofactors deeply inside the RDase scaffold, suggesting that the lower base is not directly involved in catalysis (PDB ID=4UR0, 4RAS). Interestingly, kinetic studies on non-RDase cobamide-dependent enzymes suggested varying affinities of the apoprotein to cobamides with different lower bases (Barker et al., 1960; Lengyel et al., 1960; Tanioka et al., 2010). For example, both the mammalian (sheep) kidney and bacterial (Propionibacterium shermanii) methylmalonyl-CoA mutase have high affinity to DMB-Cba (Km values of 0.021 and 0.024 μM, respectively), whereas the Km values for Bza-Cba were about an order in magnitude greater (Lengyel et al., 1960). In contrast, the glutamate mutase of Clostridium tetanomorphum preferred Bza-Cba over DMB-Cba with Km values of 0.24 and 18 μM, respectively (Barker et al., 1960). Preference for a cobamide with a specific lower base was also observed with the methionine synthase of Spirulina platensis strain NIES-39, which bound pseudo-B12 with a Km of 0.07 μM, and a much higher Km of 16.0 μM was determined for DMB-Cba (Tanioka et al., 2010). These observations suggest that the affinity of the enzyme to cobamides with different lower bases affects the assembly of the functional holoenzyme, which can explain the observed lower base effects on reductive dechlorination rates and extents.
Differential transport of cobamides into the cell can affect RDase maturation and is another possible explanation for the observed decrease in dechlorination rates. However, the experimental data indicated that DMB-, 5-OMeBza- and Bza-Cba were equally transported across the surface layer/cytoplasmic membrane in Dhc strains BAV1 and GT, suggesting that differential cobamide uptake cannot explain the observed decrease in dechlorination performance. Dhc strains lack a peptidoglycan cell wall, and a btuB homolog for controlling cobamide translocation across the outer membrane into the periplasmic space is absent in the sequenced Dhc genomes (Chimento et al., 2003; Yi et al., 2012; Löffler et al., 2013b). Experimental evidence obtained with the human gut anaerobe Bacteroides thetaiotaomicron corroborated that BtuB, rather than the ATP-binding cassette corrinoid transporter system BtuFCD is the necessary component for selective cobamide transport (Degnan et al., 2014).
An unexpected finding was that the activity of the VC RDases BvcA and VcrA was differentially influenced by cobamides with distinct lower bases. Apparently, lower bases can exert post-translational control over corrinoid-dependent enzyme systems, in this example RDases. An observation supporting this hypothesis was made in the PCE-to-cDCE dechlorinator S. multivorans. Following DMB addition, DMB replaced the native adenine lower base to generate nor-B12 that inhibited PceA maturation and export and affected PCE dechlorination and growth of S. multivorans (Keller et al., 2013). This finding further indicated that the lower base affects the binding between the cobamide and the apo-form of the RDase, and the incorporation of a cobamide with an unfavorable lower base affects RDase maturation. In Dhc strains BAV1 and GT, inefficient maturation and impaired export of BvcA and VcrA due to incorporation of a corrinoid cofactor carrying unfavorable 5-OMeBza or Bza lower bases may explain the decreased dechlorination rates, or even the complete loss of VC dechlorination ability in strain GT.
The pure culture studies with cDCE/VC-dechlorinating Dhc strains revealed that methyl substitutions of the benzimidazole backbone at the 5 and 6 positions affect Dhc strains harboring vcrA (strain GT) or bvcA (strain BAV1) differently. Apparently, the growth-supporting benzimidazole-type corrinoids are functionally not equivalent in Dhc strains with distinct RDases. In strain BAV1 (BvcA) and strain GT (VcrA), reductive dechlorination rates decreased in the order DMB-Cba>5-MeBza-Cba>Bza-Cba, and a similar trend was observed in Dhc strain 195 (Yi et al., 2012). DMB-Cba supported the highest TCE dechlorination rates in strain 195 cultures, but no growth occurred with Bza-Cba. Furthermore, Dhc strains are unable to grow with 5-OHBza-Cba in axenic culture or during co-cultivation with a 5-OHBza-Cba-producing methanogen, indicating that substitution of the hydrophobic methyl group with a hydroxyl group in the 5 position and removal of the methyl group in 6 position resulted in a nonfunctional lower base (Yi et al., 2012; Yan et al., 2013). Corrinoid cofactor–protein interactions are governed by hydrogen bonding, as revealed by the crystal structures of PceA and NpRdhA (Bommer et al., 2014; Payne et al., 2014). The findings presented here suggest that the methyl group substitutions in the 5 and 6 positions of the benzimidazole ring have relevant roles for VcrA and BvcA activity, and are possibly involved in stabilizing the mature RDase complex. Such methyl group–protein hydrophobic interactions provide a plausible explanation for the preference of the Dhc RDases BvcA (strain BAV1), VcrA (strain GT) and TceA (strain 195) for a corrinoid with DMB as the lower base.
The effects of the lower base on dechlorination rates and end points have implications for bioremediation practice. A common contaminated site management practice is to supply electron donor when dechlorination rates and extent following initial bioremediation treatment (that is, biostimulation alone or combined with bioaugmentation) decrease (Löffler and Edwards, 2006). The assumption is that hydrogen, the required electron donor for Dhc, is limiting and additions of fermentable carbon substrates increase hydrogen flux. Although repeated electron donor additions can sustain reductive dechlorination activity, evidence that hydrogen flux was actually limiting Dhc activity is difficult to ascertain. A carefully executed long-term study under conditions of sustained low hydrogen flux indicated that vitamin B12 additions, not hydrogen, limited Dhc reductive dechlorination activity and VC to ethene conversion (Fennell et al., 1997). In contaminated aquifers, organohalide-respiring Chloroflexi depend on corrinoid scavenging to acquire this essential cofactor from the environment. Depending on the site biogeochemical conditions and the type of substrate(s) used for biostimulation (that is, electron donors), microorganisms that produce the ‘wrong' lower base(s) may dominate. In such scenarios, continued biostimulation and bioaugmentation will not sustain Dhc activity and lead to efficient contaminant detoxification. Knowledge of the exact lower base requirements of the keystone corrinoid-auxotrophic dechlorinators, as well as the biogeochemical conditions that favor the synthesis of required growth factors by indigenous microbes, will enable strategies to overcome nutritional limitations of organohalide-respiring Chloroflexi and promote faster detoxification rates.
Acknowledgments
This research was supported by the Department of Defense Strategic Environmental Research and Development Program (SERDP project ER-2312) and by the National Institute of Environmental Health Sciences Superfund Research Program (R01ES24294).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abelson PH. (1990). Inefficient remediation of ground-water pollution. Science 250: 733. [DOI] [PubMed] [Google Scholar]
- Allen RH, Stabler SP. (2008). Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am J Clin Nutr 87: 1324–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf-Anid N, Hayes KF, Vogel TM. (1994). Reduction dechlorination of carbon tetrachloride by cobalamin(II) in the presence of dithiothreitol: mechanistic study, effect of redox potential and pH. Environ Sci Technol 28: 246–252. [DOI] [PubMed] [Google Scholar]
- ATSDR. (2003), CERCLA priority list of hazardous substances. Agency for Toxic Substances and Disease Registry.
- ATSDR. (2004), CERCLA priority list of hazardous substances. Agency for Toxic Substances and Disease Registry.
- Banerjee R. (1999) Chemistry and biochemistry of B12. John Wiley & Sons, Inc.: New York, USA. [Google Scholar]
- Barker HA, Smyth RD, Weissbach H, Toohey JI, Ladd JN, Volcani BE. (1960). Isolation and properties of crystalline cobamide coenzymes containing benzimidazole or 5, 6-dimethylbenzimidazole. J Biol Chem 235: 480–488. [PubMed] [Google Scholar]
- Bommer M, Kunze C, Fesseler J, Schubert T, Diekert G, Dobbek H. (2014). Structural basis for organohalide respiration. Science 346: 455–458. [DOI] [PubMed] [Google Scholar]
- Burkholder PR. (1951). Determination of vitamin B12 with a mutant strain of Escherichia coli. Science 114: 459–460. [DOI] [PubMed] [Google Scholar]
- Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. (2003). Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat Struct Biol 10: 394–401. [DOI] [PubMed] [Google Scholar]
- Chiu P-C, Reinhard M. (1996). Transformation of carbon tetrachloride by reduced vitamin B12 in aqueous cysteine solution. Environ Sci Technol 30: 1882–1889. [Google Scholar]
- Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. (2014). Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell Host Microbe 15: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA. (2012), Vapor intrusion database: evaluation and characterization of attenuation factors for chlorinated volatile organic compounds and residential buildings. US Environmental Protection Agency [online]. Office of Solid Waste and Emergency Response, US Environmental Protection Agency, Washington, D.C. 20460, EPA 530-R-10-002 http://www.epa.gov/oswer/vaporintrusion/vi_data.html.
- Fennell DE, Gossett JM, Zinder SH. (1997). Comparison of butyric acid, ethanol, lactic acid, and propionicacid as hydrogen donors for the reductive dechlorination of tetrachloroethene. Environ Sci Technol 31: 918–926. [Google Scholar]
- Goldman SM, Quinlan PJ, Ross GW, Marras C, Meng C, Bhudhikanok GS et al. (2012). Solvent exposures and Parkinson disease risk in twins. Ann Neurol 71: 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber K, Puffer B, Kräutler B. (2011). Vitamin B12-derivatives-enzyme cofactors and ligands of proteins and nucleic acids. Chem Soc Rev 40: 4346–4363. [DOI] [PubMed] [Google Scholar]
- He J, Ritalahti KM, Yang K-L, Koenigsberg SS, Löffler FE. (2003). Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424: 62–65. [DOI] [PubMed] [Google Scholar]
- Hug LA, Maphosa F, Leys D, Löffler FE, Smidt H, Edwards EA et al. (2013). Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Phil Trans R Soc B 368: 20120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S, Ruetz M, Kunze C, Kräutler B, Diekert G, Schubert T. (2013). Exogenous 5,6-dimethylbenzimidazole caused production of a non-functional tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Environ Microbiol 16: 3361–3369. [DOI] [PubMed] [Google Scholar]
- Kielhorn J, Melber C, Wahnschaffe U, Aitio A, Mangelsdorf I. (2000). Vinyl chloride: still a cause for concern. Environ Health Perspect 108: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajmalnik-Brown R, Hölscher T, Thomson IN, Saunders FM, Ritalahti KM, Löffler FE. (2004). Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl Environ Microbiol 70: 6347–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel P, Mazumder R, Ochoa S. (1960). Mammalian methylmalonyl isomerase and vitamin B12 coenzymes. Proc Natl Acad Sci USA 46: 1312–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S, Brown S, Millar K. (1998). A different mechanism for the reductive dechlorination of chlorinated ethenes: kinetic and spectroscopic evidence. Environ Sci Technol 32: 2264–2272. [Google Scholar]
- Leys D, Adrian L, Smidt H. (2013). Organohalide respiration: microbes breathing chlorinated molecules. Phil Trans R Soc B 368: 20120316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler FE, Edwards EA. (2006). Harnessing microbial activities for environmental cleanup. Curr Opin Biotechnol 17: 274–284. [DOI] [PubMed] [Google Scholar]
- Löffler FE, Ritalahti KM, Zinder SH. (2013. a) Dehalococcoides and reductive dechlorination of chlorinated solvents. In: Stroo HF, Leeson A, Ward CH (eds). SERDP ESTCP Environmental Remediation Technology. Springer: New York, NY, pp 39–88. [Google Scholar]
- Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT et al. (2013. b). Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol 63: 11. [DOI] [PubMed] [Google Scholar]
- Martens J-H, Barg H, Warren MJ, Jahn D. (2002). Microbial production of vitamin B12. Appl Microbiol Biotechnol 58: 275–285. [DOI] [PubMed] [Google Scholar]
- Maymó-Gatell X, Chien Y-t, Gossett JM, Zinder SH. (1997). Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276: 1568–1571. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Behrens SF, Müller JA, Goke J, Ritalahti KM, Wagner R et al. (2009). Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet 5: e1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men Y, Seth EC, Yi S, Allen RH, Taga ME, Alvarez-Cohen L. (2014. a). Sustainable growth of Dehalococcoides mccartyi 195 by corrinoid salvaging and remodeling in defined lactate-fermenting consortia. Appl Environ Microbiol 80: 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men Y, Seth EC, Yi S, Crofts TS, Allen RH, Taga ME et al. (2014. b). Identification of specific corrinoids reveals corrinoid modification in dechlorinating microbial communities. Environ Microbiol; e-pub ahead of print 7 May 2014; doi:10.1111/1462-2920.12500. [DOI] [PMC free article] [PubMed]
- Miles ZD, McCarty RM, Molnar G, Bandarian V. (2011). Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc Natl Acad Sci USA 108: 7368–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok KC, Taga ME. (2013). Growth inhibition of Sporomusa ovata by incorporation of benzimidazole bases into cobamides. J Bacteriol 195: 1902–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller JA, Rosner B.M., von Abendroth G, Meshulam-Simon G, McCarty PL, Spormann AM. (2004). Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl Environ Microbiol 70: 4880–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NRC. (2013) National Research Council: alternatives lower case for managing the nation's complex contaminated groundwater sites. The National Academies Press: Washington, DC, http://www.nap.edu. [Google Scholar]
- Parks JM, Johs A, Podar M, Bridou R, Hurt RA Jr, Smith SD et al. (2013). The genetic basis for bacterial mercury methylation. Science 339: 1332–1335. [DOI] [PubMed] [Google Scholar]
- Parthasarathy A, Stich TA, Lohner ST, Lesnefsky A, Britt RD, Spormann AM. (2015). Biochemical and EPR-Spectroscopic investigation into heterologously expressed vinyl chloride reductive dehalogenase (VcrA) from Dehalococcoides mccartyi strain VS. J Am Chem Soc 137: 3525–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne KAP, Quezada CP, Fisher K, Dunstan MS, Collins FA, Sjuts H et al. (2014). Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature 517: 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt JM. (1972) Inorganic Chemistry of Vitamin B12. Academic Press: New York, NY. [Google Scholar]
- Ritalahti KM, Amos BK, Sung Y, Wu Q, Koenigsberg SS, Löffler FE. (2006). Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl Environ Microbiol 72: 2765–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider Z, Stroiński A. (1987) Comprehensive B12: Chemistry, Biochemistry, Nutrition, Ecology, Medicine. Walter de Gruyter & Co.: Berlin, Germany. [Google Scholar]
- Stupperich E, Eisinger H-J, Schurr S. (1990). Corrinoids in anaerobic bacteria. FEMS Microbiol Lett 87: 355–360. [Google Scholar]
- Sung Y, Ritalahti KM, Apkarian RP, Löffler FE. (2006). Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl Environ Microbiol 72: 1980–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Chan WWM, Fletcher KE, Seifert J, Liang X, Löffler FE et al. (2013). Functional characterization of reductive dehalogenases by using blue native polyacrylamide gel electrophoresis. Appl Environ Microbiol 79: 974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanioka Y, Miyamoto E, Yabuta Y, Ohnishi K, Fujita T, Yamaji R et al. (2010). Methyladeninylcobamide functions as the cofactor of methionine synthase in a Cyanobacterium, Spirulina platensis NIES-39. FEBS Lett 584: 3223–3226. [DOI] [PubMed] [Google Scholar]
- Thompson HT, Dietrich LS, Elvehjem CA. (1950). The use of Lactobacillus leichmannii in the estimation of vitamin B12 activity. J Biol Chem 184: 175–180. [PubMed] [Google Scholar]
- Wolin EA, Wolin MG, Wolfe RS. (1963). Formation of methane by bacterial extracts. J Biol Chem 238: 2882–2886. [PubMed] [Google Scholar]
- Yan J, Im J, Yang Y, Löffler FE. (2013). Guided cobalamin biosynthesis supports Dehalococcoides mccartyi reductive dechlorination activity. Phil Trans R Soc B 368: 20120320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Ritalahti KM, Wagner DD, Löffler FE. (2012). Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl Environ Microbiol 78: 6630–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Seth EC, Men YJ, Stabler SP, Allen RH, Alvarez-Cohen L et al. (2012). Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl Environ Microbiol 78: 7745–7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.