Abstract
Regulating DNA replication is essential for all living cells. The DNA replication initiation factor DnaA is highly conserved in prokaryotes and is required for accurate initiation of chromosomal replication at oriC. DnaA-independent free-living bacteria have not been identified. The dnaA gene is absent in plastids and some symbiotic bacteria, although it is not known when or how DnaA-independent mechanisms were acquired. Here, we show that the degree of dependency of DNA replication on DnaA varies among cyanobacterial species. Deletion of the dnaA gene in Synechococcus elongatus PCC 7942 shifted DNA replication from oriC to a different site as a result of the integration of an episomal plasmid. Moreover, viability during the stationary phase was higher in dnaA disruptants than in wild-type cells. Deletion of dnaA did not affect DNA replication or cell growth in Synechocystis sp. PCC 6803 or Anabaena sp. PCC 7120, indicating that functional dependency on DnaA was already lost in some nonsymbiotic cyanobacterial lineages during diversification. Therefore, we proposed that cyanobacteria acquired DnaA-independent replication mechanisms before symbiosis and such an ancestral cyanobacterium was the sole primary endosymbiont to form a plastid precursor.
Introduction
Chromosomal DNA replication is a fundamental process required to inherit the genetic information to progeny in both prokaryotes and eukaryotes. Replication in most bacteria, which have single copy genome, is initiated at an oriC region, whereas in eukaryotes and some archaea, the process is initiated at multiple origins. Initiation of bacterial DNA replication is regulated by the DnaA initiator protein. DnaA binds to the DnaA box located at oriC to facilitate the unwinding of duplex strands (Katayama et al., 2010; Scholefield et al., 2011); consequently, replisome complexes are recruited to the unwinded oriC. As such, DnaA is essential for DNA replication initiation in most bacteria. Indeed, with the exception of certain symbiotic species, there are no known free-living, DnaA-independent bacteria (Akman et al., 2002; Ran et al., 2010; Nakayama et al., 2014).
Cyanobacteria are prokaryotes that utilize an oxygen-producing photosynthetic system and share a common ancestor with chloroplasts. Cyanobacteria display morphological and ecological diversity relating to structural characteristics (for example, rod shaped, coccus and filamentous) and environmental habitat (for example, ocean, fresh water, desert) (Herrero and Flores, 2008). Among many cyanobacteria, three typical freshwater cyanobacteria, Synechococcus elongatus PCC 7942 (rod shape), Synechocystis sp. PCC 6803 (coccus) and Anabaena sp. PCC 7120 (filamentous) (hereafter S. elongatus, Synechocystis and Anabaena), have been used as model organisms for the study of cyanobacterial physiology such as photosynthesis, circadian rhythm, nitrate fixation and development (Herrero and Flores, 2008). Molecular phylogenetic analysis of DNA sequence converge to a monophyletic origin for plastid (Criscuolo and Gribaldo, 2011; Shih et al., 2013; Li et al., 2014; Ochoa de Alda et al., 2014), meaning that the plastid originated from a single primary endosymbiosis in which a heterotrophic protist engulfed and retained a cyanobacterium in its cytoplasm. However, the identification of the nearest current cyanobacterial species remains controversial (Deusch et al., 2008; Criscuolo and Gribaldo, 2011; Shih et al., 2013; Li et al., 2014; Ochoa de Alda et al., 2014) and then characteristics of the ancestral cyanobacterium are largely unclear.
Few studies have focused on the mechanism of cyanobacterial DNA replication, as it was believed to be similar to the bacterial mechanism. Indeed, the dnaA gene and several replication-related factors are conserved in many cyanobacteria. Our recent studies in S. elongatus revealed that replication is initiated at the DnaA box-containing oriC region (Watanabe et al., 2012) by a light-dependent mechanism (Ohbayashi et al., 2013); this suggests that replication is DnaA dependent, as in typical bacteria. In contrast, the dnaA gene is not essential for cell viability in Synechocystis (Richter et al., 1998), although additional characteristics were not addressed. The dnaA gene is inactivated by a transposon insertion in the symbiotic cyanobacterium Nostoc azolla (Akman et al., 2002) and is not conserved in the diatom endosymbiont that is of cyanobacterial origin (termed a spheroid body) with a 2.7-Mb genome (Ran et al., 2010). Moreover, recent study suggested that most of the DNA replication enzymes in chloroplast originated from cyanobacteria and others (Moriyama and Sato, 2014), and the vestige of symbiotic evolution was observed. However, the only dnaA ortholog is not conserved in chloroplasts of plants and algae for which the genomes have been sequenced (Supplementary Table S1), except for Cucumis sativus. It is possible that the dnaA gene of C. sativus is derived from horizontal gene transfer or genome sequence data of C. sativus include some contaminations because this dnaA gene matches that of Enterobacter cloacae ECNIH5, ECR091 and ECNIH3 at 100% according to blast search. These suggest that dependence on DnaA varies among cyanobacteria and was lost before the emergence of symbiosis and when or how DnaA-independent mechanisms were acquired in chloroplast has been mysterious.
In this study, we examined DnaA dependency on DNA replication in three model freshwater cyanobacteria. We demonstrate the diversification of DnaA dependency in the cyanobacterial lineage. We observed that S. elongatus was DnaA dependent, but could adapt to dnaA deletion by plasmid integration into the genome; in addition, dnaA deletion conferred an advantage in long-term culturing conditions. Conversely, Synechocystis and Anabaena were DnaA independent, and replication-sequence analysis suggested multiple replication origins, unlike many other bacteria.
Materials and methods
Bacterial strains and culture conditions
Wild-type (WT) and mutant strains of S. elongatus PCC 7942, Synechocystis sp. PCC 6803 and Anabaena sp. PCC 7120 were grown at 30 °C in BG-11 liquid medium under white light (50 μmol m−2 s−1) with 2% CO2 aeration. To synchronize cell proliferation, cells were cultured for 10 days until they reached the stationary phase, and were then diluted with fresh BG-11 medium, such that the optical density at 750 nm (OD750) was 0.2. After culture for 18 h in the dark, the culture was transferred to the light condition (time 0) to restart cell growth; OD750 and cell number were measured 3 h later. To assess cell growth, synchronized cells were labeled with bromodeoxyuridine (BrdU) for 1–4 h and examined by immunocytochemistry using an antibody against BrdU (Roche Diagnostics, Tokyo, Japan); BrdU incorporation was assessed using microscopy and western blot analysis (100 ng DNA). Cell viability was evaluated using SYTOX Green (Invitrogen, Carlsbad, CA, USA) staining.
Antiserum against SyfDnaA
Mouse anti-SyfDnaA antiserum was generated as follows: the full-length dnaA gene (Synpcc7942_1100) was PCR-amplified using primers dnaA-F-BamHI and dnaA-R-SalI (Supplementary Table S3) and the fragment was cloned into pDEST-cold-TF (Imamura et al., 2008). The resulting vector was transformed into Escherichia coli strain Rosetta 2(DE3)pLys (Takara Bio Inc., Otsu, Japan). Protein purification and polyclonal antibody generation (Japan Lamb Co., Ltd, Hiroshima, Japan) were performed as previously described (Watanabe et al., 2013).
Replication-sequencing (Repli-seq) analysis
BrdU-labeled DNA was isolated by immunoprecipitation, and a library was prepared for next-generation sequencing using an Illumina genome analyzer (San Diego, CA, USA), as previously described (Watanabe et al., 2012).
Pulsed-field gel electrophoresis analysis and Southern blot hybridization
Cells were embedded in agarose, lysed with lysozyme and proteinase K (Bio-Rad Laboratories, Inc., Hercules, CA, USA), digested with NdeI (Takara Bio Inc.) and subjected to pulsed-field gel electrophoresis under conditions recommended by the manufacturer (Bio-Rad: switch time, 1–10 s; run time, 20 h; angle, 120° voltage gradient, 6 V cm−1 in 0.5 × Tris-buffered EDTA at 14 °C). The gel was then transferred to a Hybond-N+ nylon membrane (GE Healthcare Japan, Tokyo, Japan), hybridized with digoxigenin-labeled probes generated by PCR using primers specific to each region (Supplementary Table S3, Southern blotting), and detected using a digoxigenin High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics, Tokyo, Japan) according to the manufacturer's instructions.
RNA sequencing analysis
Total RNA was extracted from each strain as previously described; rRNA was removed from 1 μg of total RNA using the Ribo-Zero Magnetic Kit for Gram-Negative Bacteria (Epicentre, Madison, WI, USA), and a library was constructed with the remaining RNA using the NEBNext mRNA Library Prep Master Mix Set for Illumina (New England Biolabs, Herts, UK). In total, 17 791 908, 22 260 758 and 16 294 140 sequence reads were obtained for WT and ΔdnaA-1 and -2, respectively, that were mapped to the S. elongatus reference genome. Reads per kilobase (kb) per million mapped reads were calculated using CLC Genomics Workbench software version 7.0.4 (CLC Bio, Germantown, MD, USA).
Phylogenetic analysis
Phylogenetic relationship between cyanobacteria was analyzed by 16S ribosomal RNA sequences using MEGA ver. 6.06 (Tamura et al., 2013). The sequence acquired from Cyanobase (http://genome.microbedb.jp/cyanobase) were aligned by MUSCLE, and phylogenetic tree was generated by the neighbor-joining method based on the alignment file, from which probability was confirmed by the 1000-time trial using the bootstrap method. The graphics of phylogenetic tree was arranged by Fig Tree version 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
Results
Functional role of DnaA in DNA replication and effect of dnaA deletion in S. elongatus
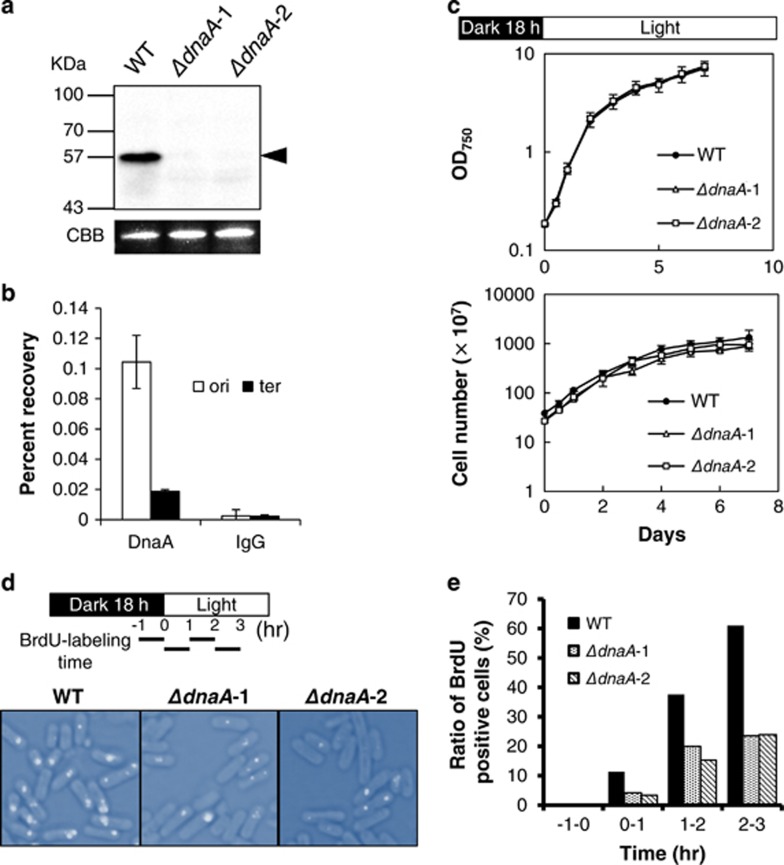
We analyzed DnaA protein expression and evaluated the protein's ability to bind oriC in S. elongatus. DnaA was constitutively expressed (Figure 1a and Supplementary Figure S1a) and bound to oriC but not to the opposite region (ter) (Figure 1b), indicating that it acts as an initiator protein in S. elongatus. To determine whether replication initiation can occur in a DnaA-independent manner as in Synechocystis (Richter et al., 1998), we constructed a dnaA deletion mutant. After two rounds of selection, 18 colonies were isolated during the stationary growth phase with complete disruption of the dnaA gene (Supplementary Figures S1b and c), from which two mutants (ΔdnaA-1 and -2) were selected. Expression of the DnaA protein was undetectable in these disruptants (Figure 1a), but cell growth was comparable to that of the WT strain (Figure 1c). Replication was assayed by pulse-labeling cells with BrdU using thymidine kinase transductants (Watanabe et al., 2012), followed by immunocytochemical analysis of BrdU incorporation (Figure 1d). The ratio of BrdU-positive cells was reduced by >50% in dnaA disruptants compared with that of the WT strain at each time point (Figure 1e). These results suggest that dnaA deficiency decreases the replication initiation frequency and that DnaA plays an important role in the initiation of DNA replication in S. elongatus.
Figure 1.
Deletion of dnaA reduces DNA replication initiation frequency but not growth in S. elongatus. (a) Analysis of DnaA expression by western blotting. (b) DnaA binding at oriC and ter was evaluated by chromatin immunoprecipitation using an anti-DnaA antibody and quantitative PCR to amplify oriC and ter regions. (c) Cell growth at the stationary phase; OD750 (upper) and cell number (lower) were measured. (d) Schematic representation of culture conditions and the BrdU labeling period. Cells were labeled with BrdU for 1 h at each time point, as indicated by bars (BrdU labeling time). BrdU-labeled cells at 2–3 h after synchronization are shown in merged bright-field and immunofluorescence images. (e) Quantitative analysis of BrdU-positive cells. Total and BrdU-positive cell numbers are as follows: WT: −1 to 0 h=376, 0 to 1 h=451, 1 to 2 h=362, 2 to 3 h=471; dnaA1: −1 to 0 h=453, 0 to 1 h=441, 1 to 2 h=429, 2 to 3 h=422; dnaA2: −1 to 0 h=482, 0 to 1 h=391, 1 to 2 h=423, 2 to 3 h=438, respectively.
Elucidation of the origin of DNA replication in dnaA disruptants
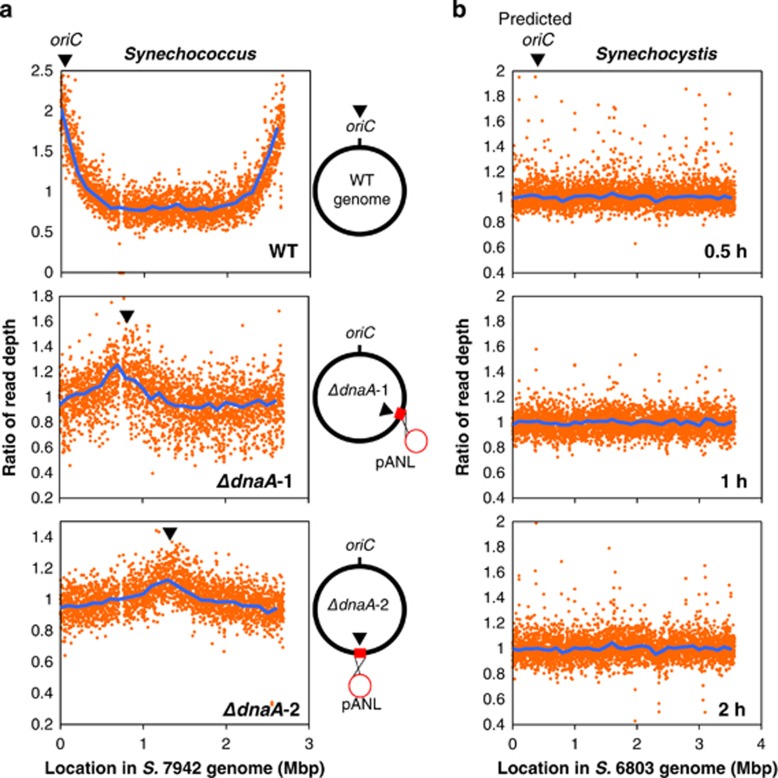
Given that ΔdnaA-1 and -2 exhibited DNA replication activity, we performed Repli-seq analysis to identify the replication initiation site. BrdU-labeled DNA was immunoprecipitated using an anti-BrdU antibody and analyzed using next-generation sequencing. In the WT strain, a large number of sequencing reads was mapped only at oriC, as shown in our previous study (Watanabe et al., 2012) (Figure 2a). Interestingly, in dnaA disruptants, the peak was observed in two different regions (Figure 2a), indicating that DNA replication was initiated from sites other than oriC.
Figure 2.
Replication origin in S. elongatus and Synechocystis. Ratios of read depth at each genomic position analyzed by Repli-seq in WT and two dnaA disruptants of S. elongatus (a) and WT Synechocystis (b). (a) Synchronized cells were labeled with BrdU for 4 h; libraries of BrdU-labeled DNA were analyzed by next-generation sequencing and mapped using the S. elongatus (S. 7942) genome as a reference. Genome maps are shown to the right of the depth plot. Inverted triangles denote replication initiation points. (b) Synchronized Synechocystis cultures were labeled with BrdU for 0.5, 1 and 2 h. Sequence reads were mapped using the Synechocystis (S. 6803) genome as a reference.
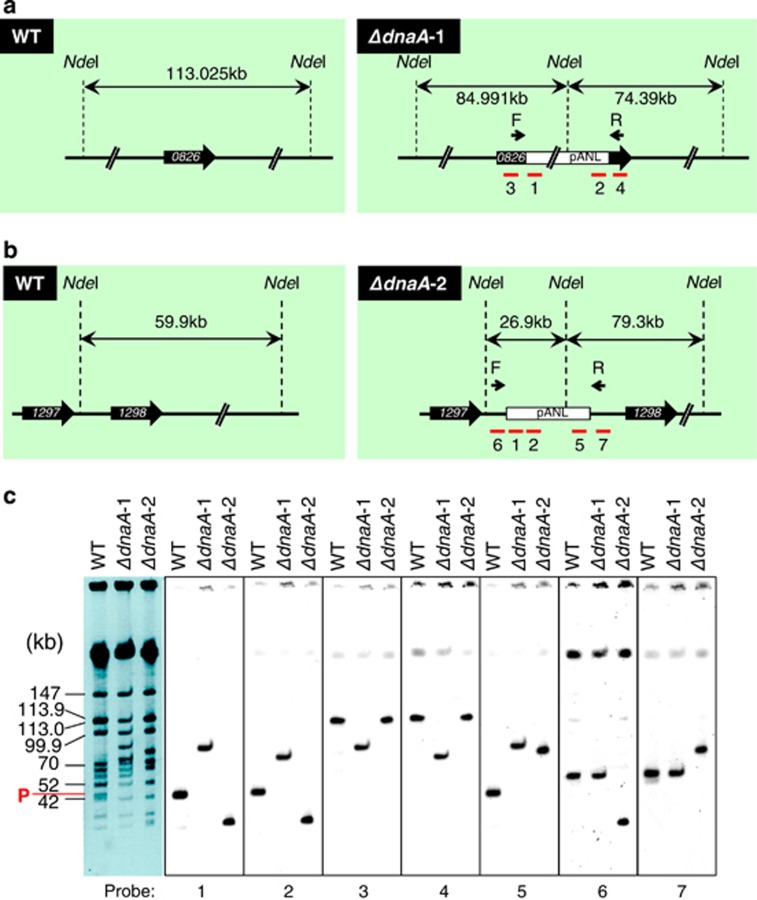
To test whether a DnaA-independent replication mechanism was acquired by a suppressor mutation, we performed whole-genome sequencing of dnaA disruptants. Surprisingly, the 46 kb pANL plasmid, which is extra-chromosomal in WT S. elongatus (Chen et al., 2008), was integrated into the ΔdnaA-1 and -2 chromosome at different sites, although there were no other mutations. Moreover, Southern blotting and capillary sequencing revealed that all pANL copies were integrated in the middle of the Syn7942_0826 gene in ΔdnaA-1 and in the intergenic region between Syn7942_1297 and Syn7942_1298 in ΔdnaA-2 (Figure 3 and Supplementary Figure S2), respectively. These integration sites were identified as new replication origins by Repli-seq analysis (Figures 2a and 3, and Supplementary Figure S2). Interestingly, capillary sequencing also revealed that the chromosome sequence was shifted to the plasmid sequence at 7 or 3 bp (GAAAATC or ACC) homologous regions in ΔdnaA-1 and -2, respectively (Supplementary Figure S2). These findings indicate that plasmids were integrated by homologous recombination via single crossovers at short homologous sequences. We also investigated the involvement of the DNA helicase DnaB, which forms the pre-priming complex with DnaA at oriC, and then DnaB binds to the oriC region related to the other region. Preferential binding at this site was not detected in dnaA disruptants, although DnaB was found to bind the oriC region rather than other regions in WT (Supplementary Figure S3), indicating that dnaA disruptants could not recruit DnaB to the oriC region. These results revealed that the DNA replication origin in dnaA disruptants was shifted from oriC to a plasmid integration site, strongly suggesting that the DnaA/oriC system shifted to a plasmid-based DNA replication initiation system.
Figure 3.
Chromosomal insertion of plasmid pANL in dnaA disruptants of S. elongatus. Schematic representation of the region surrounding the plasmid insertion site in ΔdnaA-1 (a) and ΔdnaA-2 (b). Sizes of fragments digested with NdeI are indicated for WT (left) and dnaA disruptants (right) strains. Red bars with numbers indicate probes used for Southern blot hybridization in (c). The pANL plasmid was integrated into the middle of the Synpcc7942_0826 gene and downstream of the Synpcc7942_1297 gene in ΔdnaA-1 and ΔdnaA-2, respectively, and corresponded to new replication origins, as determined by Repli-seq. (c) Pulsed-field gel electrophoresis (PFGE) pattern (left) and results of Southern hybridization with DNA probes specific to the plasmid (1, 2 and 5), Synpcc7942_0826 gene (3 and 4) and intergenic region between Synpcc7942_1297 and Synpcc7942_1298 (6 and 7), as shown in (a) and (b). Chromosome fragment sizes and plasmid fragments (P; 46 kb) are indicated to the left of PFGE image.
Increased viability of dnaA disruptants under long culture conditions
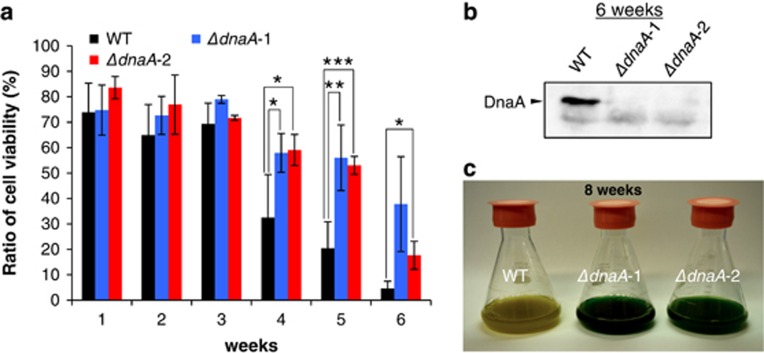
The reason why diversification of DnaA dependency occurred in cyanobacteria is unclear. The functional significance of dnaA deficiency was investigated by assessing cell growth in dnaA disruptants. The loss of DnaA function could have been advantageous on survival in certain environmental conditions if cyanobacteria had evolved a variety of DnaA dependence mechanisms. Although we tested the growth and viability under some stress conditions, including high light, low and high temperature and oxidative stress, significant differences were not observed. However, after long culture periods, viability was notably higher in ΔdnaA-1 and -2 compared with that of WT cells (Figures 4a and c). A viability assay was performed using SYTOX Green that exclusively stains dead cells. At 4 and 5 weeks after inoculation, ∼60% viability was observed in both ΔdnaA-1 or -2 disruptants, in contrast with respective viabilities of 32% and 20% in WT cells. DnaA protein was expressed until 6 weeks (Figure 4b) in WT cells, indicating that DnaA was very stable in long culture conditions. These results indicate that deletion of the dnaA gene conferred a survival advantage to S. elongatus; indeed, the disruptants were isolated from stationary phase cultures (Supplementary Figure S1b). Thus, these culture conditions could be necessary to obtain complete dnaA disruptants. The mechanistic basis for the enhanced survivability of ΔdnaA-1 and -2 was investigated by performing a transcriptome analysis. In stationary phase cultures of dnaA disruptants, transcription of many metabolism-related genes, including the gene encoding adenosine triphosphate synthase and genes involved in photosynthesis and carbon metabolism, was downregulated compared with that of the WT strain (Supplementary Figure S5 and Supplementary Table S2). These data suggest that disruptants maintained a low level of metabolic activity during the stationary phase to preserve cellular energy and enhance viability. DnaA could also directly regulate expression of these genes, given its function as a transcription factor in many bacteria (Messer and Weigel, 1997; Ishikawa et al., 2007; McAdams and Shapiro, 2009).
Figure 4.
DnaA deficiency confers a long-term survival advantage. (a) Cell viability in WT S. elongatus and dnaA disruptants. Total (bright-field) and viable (with autofluorescence and lacking green fluorescence) cells were counted by fluorescence microscopy (Supplementary Figure S4), and the percentage of viable cells (%) was calculated. Data represent the mean of three biological replicates; error bars indicate s.d. *0.05<P<0.1, **0.01<P<0.05, ***P<0.01 (Student's t-test). (b) DnaA expression after 6 weeks of culture. (c) WT and dnaA disruptant cultures after 8 weeks of culture.
DnaA is not essential for DNA replication and cell growth in Synechocystis and Anabaena
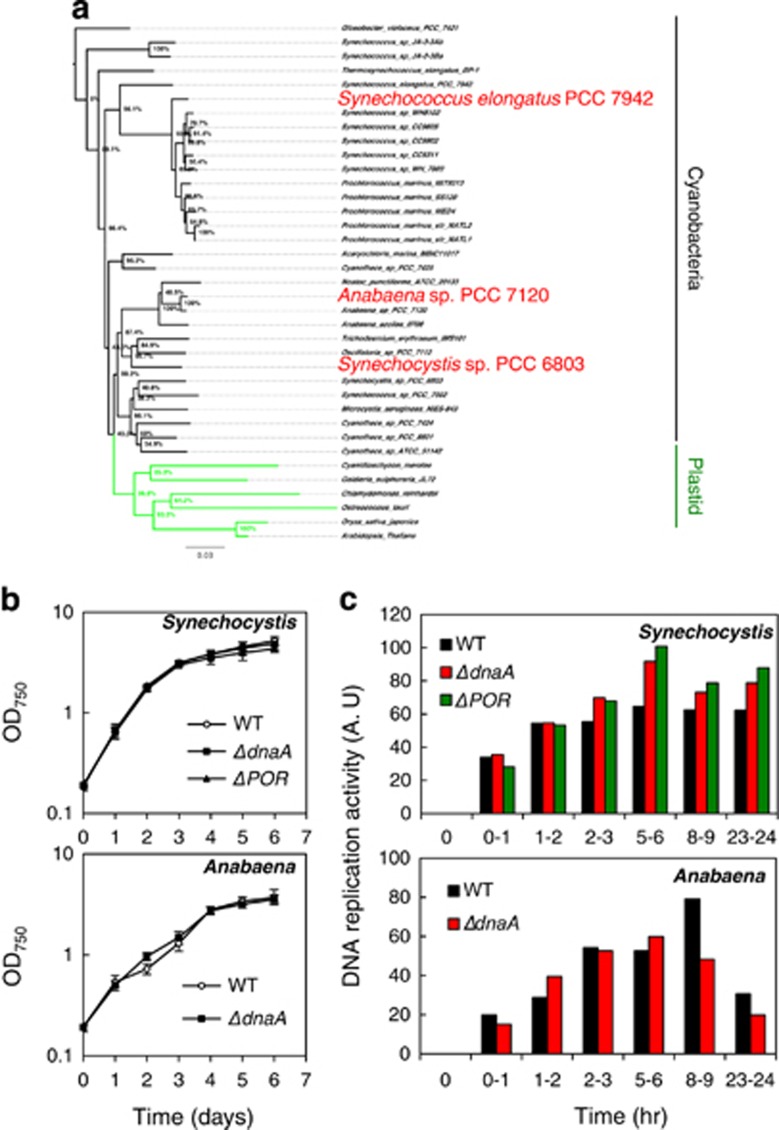
To determine whether dnaA is universally required for DNA replication, we constructed dnaA deletion mutants in model cyanobacteria Synechocystis and Anabaena that are more closely related to chloroplasts than S. elongatus, as shown in a phylogenetic tree (Turner et al., 1999; Falcon et al., 2010; Shih et al., 2013; Ochoa de Alda et al., 2014) (Figure 5a and Supplementary Figure S6). We readily obtained dnaA deletion mutants using Synechocystis and Anabaena, in contrast to S. elongatus. In these genera, growth and DNA replication in the disruptants were indistinguishable from those in WT cells (Figures 5b and c). Deletion of the dnaA box-containing predicted oriC region (POR) could also be obtained in Synechocystis, and replication and cell growth were unaffected in the Synechocystis POR-deletion mutant. We verified whether these mutants possessed a suppressor mutation such as plasmid integration, as detected in S. elongatus using a next-generation sequencing approach. No mutations or plasmid integrations were detected in Synechocystis and Anabaena, indicating that these utilized a DnaA/oriC-independent system of DNA replication.
Figure 5.
DnaA dependence is variable among cyanobacteria. (a) Phylogenetic tree of the phylum Cyanobacteria (black bar) and plastids (green bar) based on 16S rRNA alignment. (b) Cell growth in dnaA and POR disruptants of Synechocystis (upper) and a dnaA disruptant of Anabaena (lower). Data represent the mean of three biological replicates; error bars indicate s.d. (c) DNA replication activity as determined by western blot analysis of BrdU-labeled DNA in WT cells and Synechocystis (upper) and Anabaena (lower) deletion mutants. Signal intensity was quantified as arbitrary units (AU). The x axis shows periods of BrdU pulse labeling.
To identify alternative replication initiation sites, a Repli-seq analysis was performed using a WT Synechocystis strain. There were no peaks observed at either POR or other genomic regions (Figure 2b), although a replication origin could be detected in the case of a single origin. This finding suggests that there are multiple replication origins that fire asynchronously, as in eukaryotic nuclear chromosomal replication. Leading and lagging strand replication are subjected to different mutational pressures, resulting in an asymmetric genomic composition (Lobry and Sueoka, 2002; Lobry and Louarn, 2003), including GC skew, that has been used to predict the DNA replication origin at the shift point of GC skew (Frank and Lobry, 1999; Arakawa and Tomita, 2007). However, in Synechocystis, the replication origin was not predicted from GC skew, as there are many shift points (Watanabe et al., 2012). Moreover, these genomic compositions suggest the multiple replication origin in some prokaryote (Gao, 2014, 2015). These also support our hypothesis that Synechocystis has multiple replication origins.
Discussion
Freshwater cyanobacteria such as S. elongatus have evolved and diversified while maintaining a dependence on DnaA for the initiation of DNA replication, with dnaA disruptants acquiring a survival advantage in the stationary phase. In contrast, DNA replication in cyanobacteria that are closely related to chloroplasts, such as Synechocystis and Anabaena, is DnaA independent. Thus, DnaA dependency corresponds to the phylogenetic relationship and then we propose that ancestral cyanobacteria acquired DnaA-independent mechanism before symbiosis as a fundamental biological phenomenon and such a cyanobacterium facilitated the evolution of symbiosis.
In the S. elongatus dnaA disruptants, an episomal plasmid was integrated into the chromosome, and chromosomal replication was initiated using the plasmid initiation system. In E. coli, it is known that the replication origin of a plasmid or phage is integrated into the chromosome in oriC- and dnaA-defective mutants (Lindahl et al., 1971; Nishimura et al., 1971; Louarn et al., 1982; Gowrishankar, 2015). These strains, which depend on exogenous elements for their replication, displayed no advantageous compared with WT in both E. coli and Bacillus subtilis. In contrast, the S. elongatus dnaA disruptant not only exhibited normal growth, but also displayed increasing cell viability in long culturing conditions. Interestingly, polyploid archaea tolerate the deletion of all replication origins and mutants exhibit a faster growth rate compared with WT counterparts (Hawkins et al., 2013). Freshwater cyanobacteria are also polyploid and their chromosomes are replicated asynchronously; that is, they may not initiate replication only once per chromosome per cell cycle (Chen et al., 2012; Jain et al., 2012; Watanabe et al., 2012) in contrast to bacteria that have a single chromosome copy and therefore tightly regulate replication initiation via the DnaA/oriC system. Thus, polyploidy and asynchronous replication could have enabled the acquisition of DnaA-independent replication control and facilitated DnaA-independent growth under various environmental conditions in freshwater cyanobacteria.
The S. elongatus dnaA disruptants were viable under conditions of nutrient starvation (Figure 4), suggesting that DnaA dependence not only affected DNA replication, but also environmental adaptation during cyanobacterial evolution. In fresh water, changes in environmental conditions such as nutrient availability are common. Therefore, freshwater cyanobacteria can survive for long periods without nutrients. In contrast, nutrients are constant in the ocean owing to the occurrence of waves. Thus, considering DnaA dependence with respect to the natural habitat of cyanobacteria, it can be concluded that marine and freshwater species are DnaA dependent and DnaA independent, respectively. Marine cyanobacterial dnaA gene expression oscillates with the cell cycle (Holtzendorff et al., 2001), and a GC skew pattern is consistent with that of the oriC region (Watanabe et al., 2012), suggesting that these species use DnaA for DNA replication, similar to other well-characterized bacteria. In this respect, the freshwater cyanobacterium S. elongatus is closely related phylogenetically to marine cyanobacteria (Figure 5a).
Among photosynthetic microorganisms, cyanobacteria are the only primary symbionts that have evolved into plastids (Reyes-Prieto et al., 2007). Subsequently, the reorganization of genetic material and regulatory mechanisms, including those pertaining to DNA replication, occurred between the nucleus and the symbiont (Kleine et al., 2009; Moriyama et al., 2014). Twinkle, an organelle DNA helicase and primase, is shared by plastids and mitochondria in green plants, whereas cyanobacterial dnaB helicase and dnaG primase are conserved in red algae. Thus, the evolutionary history of DNA replication from a cyanobacterial to a plastid system can be discerned from genetic evidence. Interestingly, dnaA is the only gene that is not conserved between red algae, the cyanobacterial symbiont Nostoc azollae (Ran et al., 2010) and the spheroid bodies (Nakayama et al., 2014) of diatoms. We have shown that cyanobacteria have the capacity to shift the DNA replication initiation system from chromosomal to plasmid type by dnaA deletion. In red algae plastids, the substitution of cyanobacteria-type DNA polymerase (Pol III) with a plasmid-type (Pol I-like) (Moriyama et al., 2011, 2014) suggests that polymerase reorganization occurred at early stages of symbiosis. We propose that the DnaA-independent replication initiation system in cyanobacteria accelerated the reorganization of replication components and that this flexibility in DNA replication yielded a preadaptive genotype that enabled symbiosis.
Acknowledgments
We thank Drs Kazuharu Arakawa and Motohiro Akashi for advice on bioinformatics analyses; Dr Yuh Shiwa for advice on the analysis of next-generation sequencing data; and Tomoko Kojima for comments on the manuscript. This study was funded by the Sasagawa Scientific Research Grant from the Japan Science Society (to RO), by the MEXT-supported Program for Strategic Research Foundation at Private Universities, 2013–2017 (S1311017), by Grant-in-Aid for Scientific Research (C) to YK (15K07368), and by Grants-in-Aid for Young Scientists (B) to SW (25850056).
Author contributions
RO, SW and HY designed the experiments. RO performed the experiments. SE constructed Anabaena mutant strains. RO and YK analyzed Repli-seq and RNA sequencing data. RO, SW, YK, SE, TC and HY wrote the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M et al. (2002). Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet 32: 402–407. [DOI] [PubMed] [Google Scholar]
- Arakawa K, Tomita M. (2007). The GC skew index: a measure of genomic compositional asymmetry and the degree of replicational selection. Evol Bioinform Online 3: 159–168. [PMC free article] [PubMed] [Google Scholar]
- Chen AH, Afonso B, Silver PA, Savage DF. (2012). Spatial and temporal organization of chromosome duplication and segregation in the cyanobacterium Synechococcus elongatus PCC 7942. PLoS One 7: e47837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Holtman CK, Magnuson RD, Youderian PA, Golden SS. (2008). The complete sequence and functional analysis of pANL, the large plasmid of the unicellular freshwater cyanobacterium Synechococcus elongatus PCC 7942. Plasmid 59: 176–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. (2011). Large-scale phylogenomic analyses indicate a deep origin of primary plastids within cyanobacteria. Mol Biol Evol 28: 3019–3032. [DOI] [PubMed] [Google Scholar]
- Deusch O, Landan G, Roettger M, Gruenheit N, Kowallik KV, Allen JF et al. (2008). Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol Biol Evol 25: 748–761. [DOI] [PubMed] [Google Scholar]
- Falcon LI, Magallon S,, Castillo A. (2010). Dating the cyanobacterial ancestor of the chloroplast. ISME J 4: 777–783. [DOI] [PubMed] [Google Scholar]
- Frank AC, Lobry JR. (1999). Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene 238: 65–77. [DOI] [PubMed] [Google Scholar]
- Gao F. (2014). Recent advances in the identification of replication origins based on the Z-curve method. Curr Genomics 15: 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F. (2015). Bacteria may have multiple replication origins. Front Microbiol 6: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar J. (2015). End of the beginning: elongation and termination features of alternative modes of chromosomal replication initiation in bacteria. PLoS Genet 11: e1004909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins M, Malla S, Blythe MJ, Nieduszynski CA, Allers T. (2013). Accelerated growth in the absence of DNA replication origins. Nature 503: 544–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A, Flores FG. (2008) The Cyanobacteria: Molecular Biology, Genomics, and Evolution. Caister Academic Press: Norfolk, UK. [Google Scholar]
- Holtzendorff J, Partensky F, Jacquet S, Bruyant F, Marie D, Garczarek L et al. (2001). Diel expression of cell cycle-related genes in synchronized cultures of Prochlorococcus sp. strain PCC 9511. J Bacteriol 183: 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura S, Hanaoka M, Tanaka K. (2008). The plant-specific TFIIB-related protein, pBrp, is a general transcription factor for RNA polymerase I. EMBO J 27: 2317–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Ogura Y, Yoshimura M, Okumura H, Cho E, Kawai Y et al. (2007). Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res 14: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain IH, Vijayan V, O'Shea EK. (2012). Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc Natl Acad Sci USA 109: 13638–13643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Ozaki S, Keyamura K, Fujimitsu K. (2010). Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol 8: 163–170. [DOI] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D. (2009). DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol 60: 115–138. [DOI] [PubMed] [Google Scholar]
- Li B, Lopes JS, Foster PG, Embley TM, Cox CJ. (2014). Compositional biases among synonymous substitutions cause conflict between gene and protein trees for plastid origins. Mol Biol Evol 31: 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl G, Hirota Y, Jacob F. (1971). On the process of cellular division in Escherichia coli: replication of the bacterial chromosome under control of prophage P2. Proc Natl Acad Sci USA 68: 2407–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR, Sueoka N. (2002). Asymmetric directional mutation pressures in bacteria. Genome Biol 3: RESEARCH0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR, Louarn JM. (2003). Polarisation of prokaryotic chromosomes. Curr Opin Microbiol 6: 101–108. [DOI] [PubMed] [Google Scholar]
- Louarn J, Patte J, Louarn JM. (1982). Suppression of Escherichia coli dnaA46 mutations by integration of plasmid R100.1. derivatives: constraints imposed by the replication terminus. J Bacteriol 151: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams HH, Shapiro L. (2009). System-level design of bacterial cell cycle control. FEBS Lett 583: 3984–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W, Weigel C. (1997). DnaA initiator—also a transcription factor. Mol Microbiol 24: 1–6. [DOI] [PubMed] [Google Scholar]
- Moriyama T, Sato N. (2014). Enzymes involved in organellar DNA replication in photosynthetic eukaryotes. Front Plant Sci 5: 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama T, Terasawa K, Sato N. (2011). Conservation of POPs, the plant organellar DNA polymerases, in eukaryotes. Protist 162: 177–187. [DOI] [PubMed] [Google Scholar]
- Moriyama T, Tajima N, Sekine K, Sato N. (2014). Localization and phylogenetic analysis of enzymes related to organellar genome replication in the unicellular rhodophyte Cyanidioschyzon merolae. Genome Biol Evol 6: 228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Kamikawa R, Tanifuji G, Kashiyama Y, Ohkouchi N, Archibald JM et al. (2014). Complete genome of a nonphotosynthetic cyanobacterium in a diatom reveals recent adaptations to an intracellular lifestyle. Proc Natl Acad Sci USA 111: 11407–11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Caro L, Berg CM, Hirota Y. (1971). Chromosome replication in Escherichia coli. IV. Control of chromosome replication and cell division by an integrated episome. J Mol Biol 55: 441–456. [DOI] [PubMed] [Google Scholar]
- Ochoa de Alda JA, Esteban R, Diago ML, Houmard J. (2014). The plastid ancestor originated among one of the major cyanobacterial lineages. Nat Commun 5: 4937. [DOI] [PubMed] [Google Scholar]
- Ohbayashi R, Watanabe S, Kanesaki Y, Narikawa R, Chibazakura T, Ikeuchi M et al. (2013). DNA replication depends on photosynthetic electron transport in cyanobacteria. FEMS Microbiol Lett 344: 138–144. [DOI] [PubMed] [Google Scholar]
- Ran L, Larsson J, Vigil-Stenman T, Nylander JA, Ininbergs K, Zheng WW et al. (2010). Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS One 5: e11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber AP, Bhattacharya D. (2007). The origin and establishment of the plastid in algae and plants. Annu Rev Genet 41: 147–168. [DOI] [PubMed] [Google Scholar]
- Richter S, Hagemann M, Messer W. (1998). Transcriptional analysis and mutation of a dnaA-like gene in Synechocystis sp. strain PCC 6803. J Bacteriol 180: 4946–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield G, Veening JW, Murray H. (2011). DnaA and ORC: more than DNA replication initiators. Trends Cell Biol 21: 188–194. [DOI] [PubMed] [Google Scholar]
- Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E et al. (2013). Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA 110: 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S, Pryer KM, Miao VP, Palmer JD. (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol 46: 327–338. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Ohbayashi R, Shiwa Y, Noda A, Kanesaki Y, Chibazakura T et al. (2012). Light-dependent and asynchronous replication of cyanobacterial multi-copy chromosomes. Mol Microbiol 83: 856–865. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Hanaoka M, Ohba Y, Ono T, Ohnuma M, Yoshikawa H et al. (2013). Mitochondrial localization of ferrochelatase in a red alga Cyanidioschyzon merolae. Plant Cell Physiol 54: 1289–1295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.