Abstract
Endogenous intestinal microbiota have wide-ranging and largely uncharacterized effects on host physiology. Here, we used reverse-phase liquid chromatography-coupled tandem mass spectrometry to define the mouse intestinal proteome in the stomach, jejunum, ileum, cecum and proximal colon under three colonization states: germ-free (GF), monocolonized with Bacteroides thetaiotaomicron and conventionally raised (CR). Our analysis revealed distinct proteomic abundance profiles along the gastrointestinal (GI) tract. Unsupervised clustering showed that host protein abundance primarily depended on GI location rather than colonization state and specific proteins and functions that defined these locations were identified by random forest classifications. K-means clustering of protein abundance across locations revealed substantial differences in host protein production between CR mice relative to GF and monocolonized mice. Finally, comparison with fecal proteomic data sets suggested that the identities of stool proteins are not biased to any region of the GI tract, but are substantially impacted by the microbiota in the distal colon.
Introduction
The human gut harbors a complex ecosystem of microbes, comprising as many as 100 trillion bacterial cells (Whitman et al., 1998). Collectively, these bacteria constitute our microbiota, which outnumbers human cells in the body by a factor of 10 (Savage, 1977). The gut microbiota modulates various aspects of host physiology, including nutritional status, metabolism and immune-system maturation (Gill et al., 2006; Chow et al., 2010). Furthermore, the composition of the gut microbiota has been implicated in several human diseases, including type 1 diabetes (Wen et al., 2008), obesity (Ley et al., 2006; Turnbaugh et al., 2006), asthma (Penders et al., 2007) and inflammatory bowel disease (Frank et al., 2007; Penders et al., 2007; Maslowski and Mackay, 2011). DNA sequencing technologies have enabled the profiling of microbial communities and their association with human health and disease (Eckburg et al., 2005; Gill et al., 2006; Ley et al., 2006). Although these studies yielded key insights into host–microbe relationships, the direct effects that microbes have on host proteomes have only begun to be measured (Verberkmoes et al., 2009; Erickson et al., 2012; Lichtman et al., 2015; Muth et al., 2015). New tools to discover host-derived factors within the intestinal lumen hold tremendous potential for discovering molecular mediators of host–microbe interactions and deriving mechanistic understandings of this complex, biologically important system.
Recently, we described the first mass spectrometry-based proteomics approach for specifically interrogating host proteins that are secreted into the gastrointestinal (GI) lumen and shed into stool (Lichtman et al., 2013). Although host responses to changes in microbial composition were previously investigated in a targeted manner (Cash et al., 2006; Martens et al., 2008; Suzuki and Fagarasan, 2008), this novel technique allows the discovery of unanticipated host proteins that respond to and shape changes to the microbiota. One powerful aspect of this approach is that the stool host proteome could integrate interactions occurring throughout the length of the GI tract. Considering the diversity in structure, function and cell types throughout the gut, understanding where proteins are produced and the extent to which feces can serve as a gut-wide survey is of utmost importance for future investigations.
Here, we investigate the effects of microbial colonization on host protein expression as a function of location along the GI tract. Examination of the spatial distribution of host proteins makes it possible to associate them with discrete intestinal niches. Using gnotobiotic mice, we determined that the GI tract region had a larger impact on host protein abundances than did the microbial colonization state, and that changes in microbial colonization affected patterns of protein abundance and diversity in a location-specific manner. Comparison of these data with fecal proteome data sets allowed us to measure the extent to which the stool proteome serves as a proxy for characterizing the proteins found along the entire GI tract. We conclude that stool proteins provide a reasonable global snapshot of host protein expression throughout the length of the GI tract, but the host proteome is continually manipulated by microbes as they transit the lower GI tract. This regional approach to examine host proteomics fills an important gap in our understanding of the interdependence between the commensal microbiota and host physiology and responses to colonization.
Results
Regional protein composition varies among colonization states and across the length of the GI tract
To investigate the expression of host proteins along the GI tract, we compared mice harboring one of three gut microbial colonization states (n=3 per state): (1) germ-free (GF), (2) colonized with a single commensal microbe, Bacteroides thetaiotaomicron (BT) and (3) conventionally raised (CR) with a normal, pathogen-free mouse microbiota. BT was chosen based on its status as a common and abundant member of the human microbiota that has been extensively characterised (Hooper et al., 2000; Martens et al., 2008; Mahowald et al., 2009; Sonnenburg et al., 2010). We analyzed the luminal contents of five GI regions dissected from each experimental animal: (1) stomach, (2) jejunum, (3) ileum, (4) cecum and (5) proximal colon. Luminal material was processed as previously described (Lichtman et al., 2013) (Materials and methods), and analyzed via LC-MS/MS (liquid chromatography-coupled tandem mass spectrometry). We identified 853 unique proteins (Supplementary Table S1) across the three colonization states and five locations after filtering the results to a 1% peptide false discovery rate (FDR) for each mass spectrometry run and an experiment-wide 5% protein FDR. Proteins were quantified by spectral counts normalized by protein length and total spectral assignments.
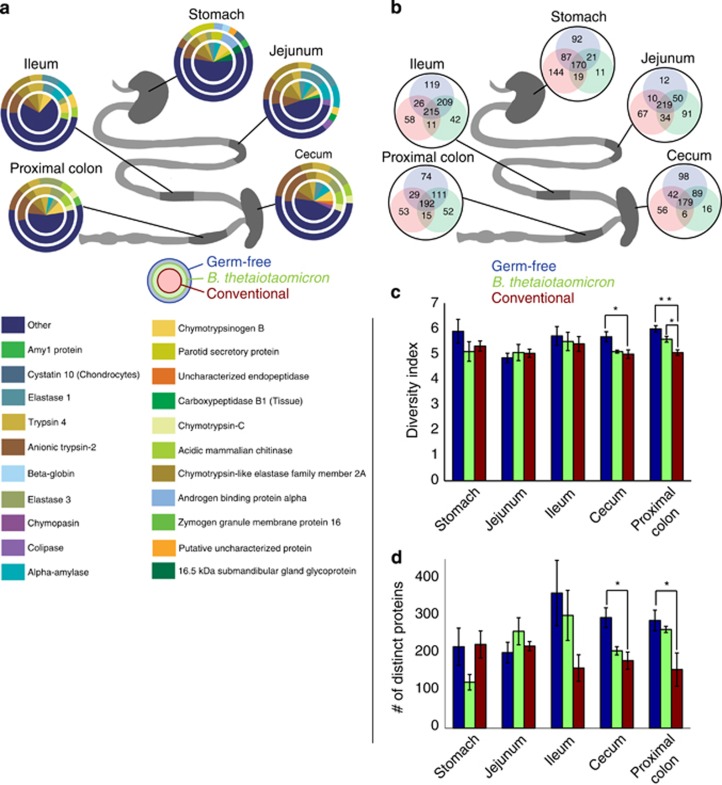
As many as 11 proteins (average of 7.0±1.6) identified from any of the five luminal regions individually accounted for at least 3% of the total host-protein mass from that region, as estimated by spectral counts (Figure 1a, Supplementary Table S2). These high-abundance proteins in aggregate constituted an average of 52±11% of the total host protein abundance estimated in this way, ranging from 36±12% in stomach samples from GF mice to 63±4% in jejunum samples from GF mice (Figure 1a). Anionic trypsin-2 and chymotrypsin-like elastase family member 2A were the highest abundance proteins across all locations and colonization states (Figure 1a). The ubiquitous presence of these proteases throughout the GI tract and the high jejunal abundance of co-lipase, a lipase coenzyme secreted by the pancreas (Lowe, 2002), supports our data set's validity due to their previous description in numerous studies. Notably, the fifth most abundant protein was an uncharacterized protein with predicted alpha-amylase activity (Figure 1a), suggesting that this approach has utility in functionally annotating in silico-predicted open reading frames.
Figure 1.
Protein abundance varies by location along the gut as well as by colonization state. (a) Each slice corresponds to the abundance of a protein; proteins with <3% of the total abundance in a given location and colonization state were grouped into the ‘other' slice (dark blue). (b) The number of unique proteins in the indicated colonization state. All overlaps between colonization states were significant (hypergeometric P<0.0004). (c) Mean Shannon–Weiner index, a metric of protein diversity (*P<0.03, **P<0.003, n=3). (d) Mean number of proteins identified (*P<0.05, n=3). Error bars represent standard error of the means.
GI host-protein expression is primarily driven by location rather than colonization state
Visual inspection of abundance-weighted protein representation suggested greater similarity across colonization states than GI locations (Figure 1a). To quantify this similarity, we pooled each set of triplicate analyses and counted the number of proteins shared across each colonization state at each GI location (Figure 1b). Although the degree of overlap varied between colonization states, the number of proteins shared between any two colonization states always outnumbered the proteins specific to any particular state (Figure 1b). Moreover, GF and BT mice were generally much more similar to each other than either one was to CR mice, with the exception of the stomach, where GF and CR samples had the greatest overlap (Figure 1b).
We next counted the total number of proteins shared across each GI region regardless of colonization state (Table 1). These data suggest that ~65% of proteins occur in every examined GI region. We further note that the cecum and proximal colon were most similar in the proteins they express (Table 1), in accordance with their spatial proximity along the GI tract. Eighteen proteins were identified in every sample, independent of location, colonization state and replicate (Supplementary Table S3), consistent with their high abundance throughout the GI tract. The observation that host proteins are observed throughout the GI is consistent with a model in which proteins produced in a proximal region of the GI tract are not degraded as they transit to distal regions. Alternatively, these proteins could be expressed ubiquitously throughout the gut.
Table 1. Proteins persist throughout the GI tract.
| Percentage of proteins found in {region}… |
…that were also found in {region} |
||||
|---|---|---|---|---|---|
| Stomach | Jejunum | Ileum | Cecum | Prox col | |
| Stomach | 100 | 77.8 | 68.7 | 88.3 | 73.4 |
| Jejunum | 65.1 | 100 | 64.6 | 74.1 | 71.5 |
| Ileum | 65.1 | 77.6 | 100 | 73.3 | 74 |
| Cecum | 67.5 | 74.5 | 62.8 | 100 | 81.6 |
| Prox col | 71 | 77.8 | 68.7 | 88.3 | 100 |
Abbreviation: GI, gastrointestinal. Identified proteins were pooled from all three colonization states. Each entry is the percentage of proteins found in the region indicated in the row which were also found in the region indicated in the column.
To further assess the similarities between the protein profiles of each region and colonization state in each sample, we used spectral count-estimated abundances to calculate the Shannon–Weiner diversity index, an information-theoretic measure in which higher values indicate higher diversity (Magurran, 2004). Host-protein diversity was lower in contents from CR mice than from BT mice (proximal colon) and GF mice (proximal colon and cecum) (Figure 1c). To address whether the differences in diversity were due to host proteome richness (number of proteins) or evenness (expression level of the proteins), we compared the number of proteins found in each region and colonization state. Significantly more proteins were identified from GF mice than from CR mice in both the cecum and the proximal colon (Figure 1d). This suggests that the GF host-protein populations we were able to observe by LC-MS/MS are richer than those of CR mice. Additionally, in GF samples, highly abundant proteins (each constituting at least 3% of total protein abundance) represented 46% and 51% of the total protein abundance in the cecum and colon, respectively, compared with 64% and 67% in CR mouse samples (Figure 1a). These data suggest that similar numbers of proteins are present in all colonization states, but increased bacterial load in the large intestines of CR-colonized mice resulted in a commensurate decrease in the number of identifiable host proteins. This notion is supported by our observation that of the 518 718 and 509 276 MS/MS spectra acquired from all GF and CR specimens, respectively, a substantially larger proportion (17% vs 12%) was confidently assigned to host proteins derived from GF animals. Thus, the absence of microbial proteins gives the mass spectrometer more opportunities to measure host proteins in proportion to their underlying abundances. We conclude that since decreased microbial complexity can enhance the identification of low-abundance host proteins, caution should be exercised when comparing low-abundance proteins across multiple colonization states.
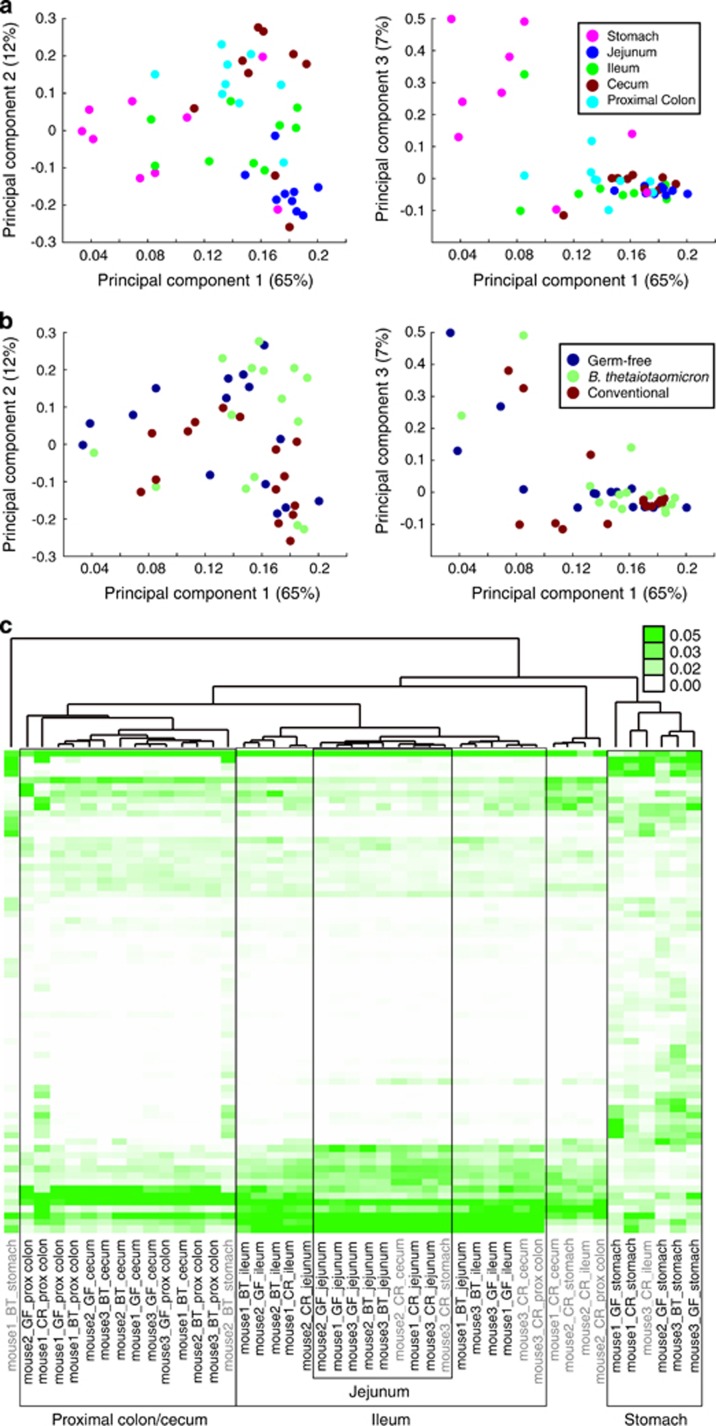
Our observations suggested that both GI region and colonization state affect the host proteome. To identify the major driver of proteome variation, we applied unsupervised clustering methods to our abundance profiles (Materials and methods). Principal component analysis (PCA) revealed that luminal contents clustered predominantly based on GI location rather than on colonization state (Figures 2a and b). Hierarchical clustering of the protein profiles further demonstrated the similarity between connected regions of the gut; we observed five prominent clusters, each consisting mainly of samples from one or two adjacent locations in the GI tract (Supplementary Figure S1). To minimize the effects of stochastic noise contributed by low-abundance proteins, further clustering was performed with the 72 most abundant proteins across all samples (Figure 2c); the resulting groupings closely matched those generated by clustering the entire protein data set (Supplementary Figure S1). Samples from the stomach clustered farthest from the rest of the data set, consistent with our observation that the third principal component primarily distinguished stomach-derived material (Figure 2a). In contrast, samples from the proximal colon and the cecum clustered together (Figure 2c, Supplementary Figure S1), although PCA identified greater distinction between these two contiguous GI regions (Figure 2a). Samples from the ileum and the jejunum also co-clustered; the jejunum samples formed a sub-cluster within the larger ileum cluster (Figure 2c, Supplementary Figure S1). Four samples did not cluster according to location, colonization state or mouse replicate (Figure 2c). These samples and samples that clustered away from their corresponding replicates were from CR mice, indicative of the greater variability in the CR mouse proteome compared with GF and BT states as we previously observed (Lichtman et al., 2013).
Figure 2.
Unsupervised clustering shows that variation in the host-secreted proteome is driven by GI location rather than by colonization state. (a, b) PCA of the normalized spectral counts for the 853 proteins identified in any of the 45 experimental samples, plotting principal component 1 against principal components 2 (left) or 3 (right). (c) Hierarchical clustering of the abundances of the 72 most abundant proteins using Euclidean distance and average linkage metrics. The five clusters denoted by boxes are comprised mainly of samples from one or two adjacent locations along the GI tract. Samples are labelled with the mouse, colonization state and region with the labels of samples that clustered correctly based on location colored black.
Whether considering all proteins identified here, or just those with the greatest abundance, hierarchical clustering demonstrated that luminal protein abundances generally corresponded to GI location. To demonstrate the feasibility of identifying region-specific proteins, we used random forests to generate a classifier of samples into locations (stomach, jejunum/ileum and cecum/proximal colon), based on 247 proteins with total relative abundances greater than 0.01. Across all mice, the out-of-bag (OOB) error rate was only 24%, indicating a high level of classification accuracy. Consistent with hierarchical clustering (Figure 2c), the high variability of CR host proteomes contributed to these animals' misclassification. Based on this, we recalculated random forest classifiers for two subgroups consisting of the BT/GF and the CR mice. These classifiers resulted in OOB error rates of 0% and 93%, respectively, further demonstrating the negative impact of CR variability on classification. These results further illustrate the variability that exists in the conventional mouse proteome.
Since hierarchically clustering the most abundant proteins (Figure 2c) re-capitulated sample relationships derived from whole-proteome data set clustering (Supplementary Figure S1), we explored how few proteins could be used to obtain the same clusters. Restricted protein sets that best distinguished these regions would be inherently valuable for understanding biological functions specific to each region. We created new random forest classifiers, now based only on the top 10% of proteins with high importance in the initial random forest analysis (Supplementary Table S4), rather than abundance alone. The error rates for these new reduced classifiers performed at least as well as the entire set of proteins (OOB error rates of 22%, 0% and 60% for all, BT/GF and CR mice, respectively). Additional cross-validation was performed by creating classifiers leaving out one mouse at a time for each colonization state and predicting the performance based on the remaining mice. This resulted in error rates of 22%, 0% and 53%, very similar to the OOB rates. Thus, the small subsets of proteins designated here could classify gut locations with comparable effectiveness to the entire proteome. Furthermore, this smaller set out-performed the proteome-wide classification for the variable CR data sets. Most interestingly, only one protein, chymopasin, was shared between the GF/BT classifier (22 proteins) and the CR classifier (12 proteins). Together, these data support the role of the microbiota in expanding the phenotypic variability in the host intestinal mucosa.
Ontological classes predict functional differences across GI locations and colonization states
To assess the functional relevance of location and colonization state-specific protein representation in the GI, we assigned gene ontologies (GOs) to each of the 853 proteins in our data set (Materials and methods) and repeated PCA and hierarchical clustering on the resulting 1520 unique functional annotations. Strikingly, the primary principal component accounted for >86% of observed variation, and promoted region-specific sample segregation (Supplementary Figure S2). Accordingly, hierarchical clustering of GO assignments grouped GI regions more tightly than individual animals or colonization state (Supplementary Figure S3), consistent with clustered protein abundances (Figure 2,Supplementary Figure S1). Samples from the stomach again differed from the remaining GI regions (Supplementary Figure S3), indicating divergent functions of stomach proteins compared with proteins in the rest of the GI tract. The functions of proteins in the proximal colon and cecum were indistinguishable from each other in this analysis; however, samples from the ileum and jejunum formed distinct groups, indicating a lesser degree of functional equivalency between these regions in comparison with protein abundances (Figure 2c).
To demonstrate the feasibility of identifying region-specific functions, we used random forests to classify samples according to GI location (stomach, jejunum/ileum and cecum/proximal colon), based on the 257 GO terms with total relative abundance greater than 0.1. Across all mice, the OOB error rate was only 24%, similar to classification using protein identities. As expected considering hierarchical clustering analysis (Supplementary Figure S3), when random forests were generated for four locations (stomach, jejunum, ileum and cecum/proximal colon), as opposed to grouping the jejunum and the ileum, the OOB error rate remained low (33%). In both cases, the OOB error rates were much higher for CR than for GF/BT mice (53% vs 7% for 4 locations) as seen previously. We then determined the functions that were most important for differentiating these locations by creating new random forests based on the 10% of GO terms with the greatest importance from the original classifier (Supplementary Table S5). Peptidase activity, inflammatory response, cell adhesion and cell proliferation functions were among the most discriminating ontologies, consistent with location-specific regulation of these GI functions. Taken together, these results indicate that variation in both protein abundance and functionality is more strongly influenced by GI location than by the colonization state of the host.
Protein-abundance patterns change according to GI location
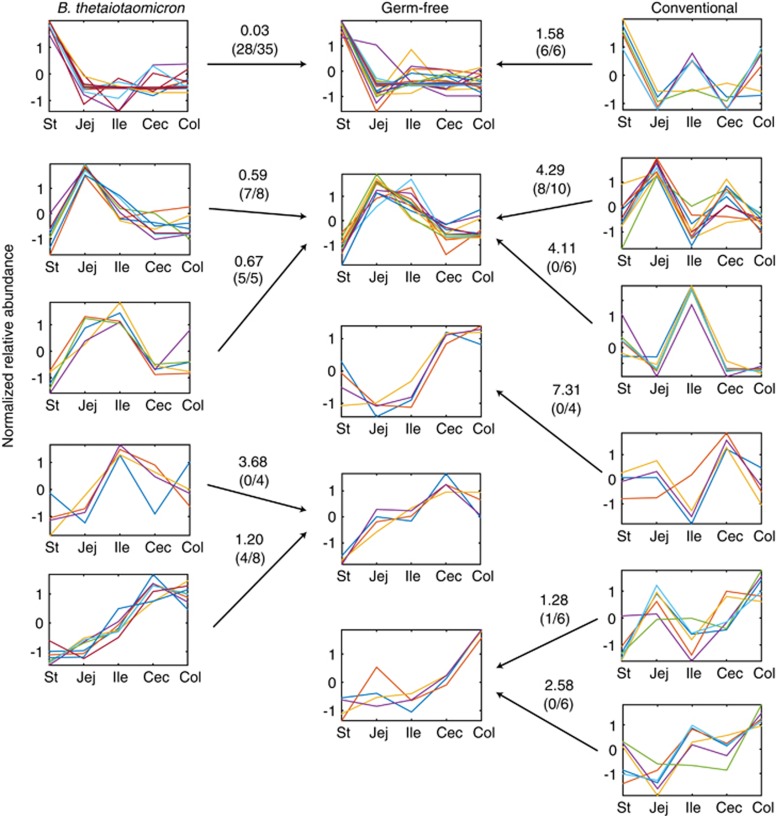
Global evaluations of protein and GO assignments support the notion that host-derived luminal proteins are effective descriptors of GI regions and colonization states. The activities of specific host proteins, however, are expected to drive the global protein differences measured across each GI region. To associate proteins with their most prominent GI regions, we applied k-means clustering to the abundances of the 72 most abundant proteins (Materials and methods). Relative abundances were z-score normalized before conducting k-means clustering on the refined data set, with the most coherent protein groups assigned to five, five and six distinct and unique expression clusters for GF, BT and CR samples, respectively (Figure 3). To measure overlapping abundance patterns between colonization states, we calculated the squared Euclidean distance between cluster centroids for all pairwise comparisons between colonization states. The smallest distances between BT clusters and GF clusters ranged from 0.03 to 3.7 arbitrary units, while the smallest distances between CR clusters and GF clusters ranged from 1.27 to 7.30 arbitrary units (Figure 3). In general, the expression patterns of luminal samples from BT mice were more closely related to those of GF mice than CR mice (Figure 3). This is consistent with our assessment of global protein counts (Figure 1b), further supporting the conclusion that a complex microbiota substantially alters host-protein expression throughout the GI tract, though to a lesser extent than GI location.
Figure 3.
K-means clustering reveals distinct patterns of protein abundance along the GI tract. K-means clustering (Materials and methods) on z-score-normalized, highly abundant proteins for six clusters in data from CR samples and five clusters in data from GF and BT samples. Numbers associated with arrows are the squared Euclidean distance and the number of proteins in common between the non-GF cluster (BT or CR) and the indicated GF cluster. For example, 28 of 35 proteins in the top BT cluster were found in the top GF cluster and collectively indicated a squared Euclidean distance of 0.03. St, stomach; Jej, jejunum; Ile, ileum; Cec, cecum; Col, proximal colon.
The large dynamic range of these clusters indicates that the proteins in each have functional relevance to specific GI regions. We assessed the functional significance of observed protein-abundance distributions (Table 2) from the ontological features of each cluster (Supplementary Table S6). As expected from GI anatomy, salivary (for example, alpha-amylase 1, androgen-binding protein gamma and 16.5 kDa submandibular gland glycoprotein) and stomach-specific (for example, gastrokine-1 and gastricsin) proteins were found in the clusters with high abundance in the stomach and low abundance in other locations (Supplementary Table S6). Proteins known to be secreted into the GI tract in the duodenum (for example, pancreatic triacylglycerol lipase, co-lipase (Lowe, 2002) and chymotrypsinogen B) as well as other proteases (for example, elastase 1 and carboxypeptidase A1) were present in clusters that had the highest expression in the jejunum and the ileum (Supplementary Table S6). Clusters of proteins that were most highly expressed in the cecum and proximal colon included anionic trypsin-2 and acidic mammalian chitinase (Supplementary Table S6), which also have high jejunal expression in conventional mice, indicating microbiota-dependent expression in the proximal gut. Several uncharacterized proteins (Supplementary Table S6) were highly abundant across the five locations (Supplementary Table S4); the abundance patterns identified here may provide novel insight into their function.
Table 2. The fecal host-secreted proteome is representative of global proteome composition across the GI tract.
| Region | % Proteins found in feces that are found in region | % Proteins found in region that are found in feces |
|---|---|---|
| Stomach | 56.8 | 63.9 |
| Jejunum | 61.9 | 54.4 |
| Ileum | 54.0 | 57.4 |
| Cecum | 68.3 | 60.4 |
| Prox col | 66.7 | 63.9 |
Pairwise comparisons between the fecal proteome determined from Lichtman et al. (2013) and the proteome identified at each location along the GI tract.
The host fecal proteome captures the proteome of each region of the GI tract
To test the extent to which the fecal proteome reconstitutes host-protein expression throughout the gut, we compared the proteins previously identified in feces (Lichtman et al., 2013) with those identified at each location along the GI tract (Materials and methods). Although host-centric fecal proteome surveys did not directly represent the full diversity of host-protein expression along the GI (Table 2), the overlap in protein identity between feces and any single region of the gut was statistically significant (hypergeometric P<2 × 10−29). Moreover, proteins found in any single GI region were no more likely to be present in stool than proteins detected in any other region (Table 2). This includes comparisons between fecal proteins and those associated with the distal gut, which by proximity would be expected to demonstrate the greatest degree of similarity.
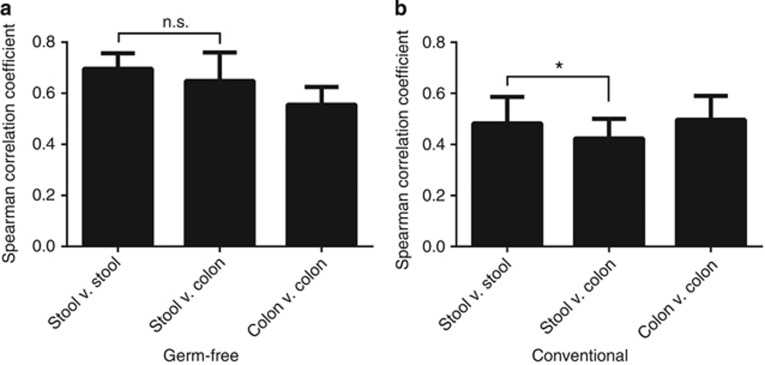
Two hypotheses could explain why the stool did not demonstrate more similarity to the distal end of the gut: either the host-derived proteome is physically transformed during its transit between the proximal colon and feces, or inter-experiment variability overshadowed relatively subtle correlations between these matrices across different sets of mice. To assess this, we compared the abundances of proteins identified from GF (Supplementary Table S7) and CR (Supplementary Table S8) proximal colon samples with stool samples from two (GF) or three (CR) sets of mice (n=2–4 mice per set). In the case of GF mice, the correlations of protein abundances between any two stool samples were not significantly greater than the correlations between any stool sample and any sample from the proximal colon (Figure 4a). These data support sampling bias as the underlying explanation for differences between the distal gut host proteome and the fecal proteome of GF mice. Conversely, the correlations between any two stool samples collected from CR mice were significantly greater than the correlation between any stool sample and colon sample (Figure 4b). We conclude from this analysis that whereas stochastic sampling contributes to observed differences between the proximal colon and fecal proteomes, the presence of microbiota in the distal colon exacerbates these differences through their continued influence on host proteins as they transit through the remaining GI tract.
Figure 4.
Differences between proximal colon and stool samples are microbiota dependent. Spearman correlation coefficients were calculated for the comparison of any two samples from the proximal colon and stool of (a) GF and (b) CR mice as seen in Supplementary Tables S6 and S7 (*P<0.01, n.s.=not significant by unpaired, two-tailed t-test).
Discussion
Here we have presented a spatially resolved comparison of host proteins detected along the GI tract. We also evaluated how the expression of host proteins is influenced by three microbiota-colonization states. Our major findings are that: (1) the overlap in protein profiles is greatest between very simple microbiota states (BT and GF); (2) variation in protein abundance is driven primarily by location rather than by colonization state; and (3) proteins identified in fecal samples generally represent proteins that originate throughout the GI tract, although they do not directly provide a nuanced representation of any given GI location. Although previous studies monitored the host-secreted proteome of fecal samples (Lichtman et al., 2013) or focused on targeted processes across GI locations (Vaishnava et al., 2011), ours is the first study to generate a global view of host proteins along the GI tract and across different colonization states. This work provides a rich data set that should accelerate future studies of location-specific host-protein expression. Proteins that we have identified as acting as location classifiers could provide GI biomarkers of microbiota-related diseases and host colonization states.
Taken together, our analyses provide a global perspective on the host contribution to the GI luminal proteome and specific candidate proteins that should guide future investigations into the outstanding issues mentioned above. From a broad perspective, our finding that GI-specific GOs (for example, ‘peptidase activity') emerged as being significantly enriched over nondescript ones (for example, cell part) supports the utility of our approach. Considering individual proteins, several detected in our data set are currently uncharacterized and these data may suggest functional roles for these proteins. We also detected, at lower abundance, antimicrobial proteins REG3γ and alpha-defensin as well as inflammatory proteins serotransferrin and S100A9 that are likely to be good markers of increased inflammatory status under conditions that deviate from the ‘healthy' conditions surveyed here. More generally, measuring host-protein expression in intestinal tissues across locations and colonization states (by proteomics or transcript-based measurements) will enable the evaluation of correlations between region-specific protein production and luminal protein abundance. Such a complementary study would provide insight into the extent of protein carryover between GI locations, and how the microbiota changes host gene expression in a global sense. Likewise, microbial community information such as 16S rRNA sequencing would allow the clustering of particular microbial taxa with host-protein expression. By demonstrating the power of host-centric proteomics, this study establishes a roadmap for necessary work to further elucidate this complex aspect of the host–microbe relationship.
One challenge of our approach is the introduction of analytical and biological noise from bacterial proteins. As seen in the number and diversity of proteins identified in CR compared with GF mice (Figures 1c and d), the presence of bacterial proteins greatly increases the abundance threshold at which host proteins can be identified. This limitation reduces the ability to sensitively distinguish biological causes of altered protein representation from technical obstacles. In addition, the experimental methodology implemented here does not account for glycosylated and phosphorylated peptides; it has been estimated that almost 50% of all mammalian proteins are glycosylated and that a third are phosphorylated (Zhang et al., 2010). Thus, future studies that probe this unqueried segment of the host-secreted proteome could help to clarify and extend our results. For example, we predict that there will be significant colonization-dependent differences in immunoglobulin profiles, given that there is a large difference in serum immunoglobulin profiles between GF and CR mice (Meeuwsen et al., 1989). We expect that application of more nuanced enrichment and fractionation protocols, as deployed in other proteome investigations (Zaia, 2008; Wang et al., 2011), hold the potential to provide even greater insight into the host proteins directing and responding to the commensal microbiota.
Materials and methods
Animal handling
In total, 45 protein extract samples were measured. Three Swiss-Webster mice in each of the three colonization states (GF, BT and CR) were killed, and luminal contents were obtained from the following locations of each mouse: stomach, jejunum, ileum, cecum and proximal colon. The small intestine was sectioned into 16 equal segments, of which sections 5–10 were identified as jejunum and sections 11–15 were identified as ileum. The proximal colon was identified as the first 1–2 cm of large intestine distal to the cecum. All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of Stanford University.
Sample preparation
Sample preparation was conducted as previously described (Lichtman et al., 2013). Briefly, luminal contents of dissected intestine sections were extracted, immediately frozen in liquid nitrogen and stored at −80 °C until use. Luminal contents were suspended in 500 μl of solution (8 M urea, 100 mM NaCl, 25 mM Tris, pH 8.2 with cOmplete protease inhibitors (Roche, Indianapolis, IN, USA)), and then thoroughly disrupted by vortexing. Insoluble material was pelleted by centrifugation (2500 g for 8 min at room temperature), followed by ultracentrifugation (35 000 g for 30 min at 4 °C) to pellet bacteria. The final supernatant was reduced and alkylated with iodoacetamide, followed by fractionation using a reverse-phase C-4 cartridge (Grace Vydak, Columbia, MD, USA) as previously described (Lichtman et al., 2013). Proteins in the 60% acetonitrile fraction were digested into peptides using trypsin (Promega, Madison, WI, USA; V5111) overnight at 37 °C and desalted using C-18 Sep-pak cartridges (Waters, Milford, MA, USA).
Mass spectrometry
Desalted, tryptic digests were analyzed by LC-MS/MS on an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, Santa Clara, CA, USA). Briefly, peptides were eluted over a 180-min gradient from a 15-cm C-18 reverse-phase column. The mass spectrometer acquired tandem mass spectra using a top-10, data-dependent acquisition workflow; MS1 was collected in the orbitrap at 60 000 resolution and subsequent MS/MS was acquired in the ion trap. Peak lists were generated with the msConvert algorithm (Chambers et al., 2012) (v. 3.0.45). Spectra were assigned to peptides using the SEQUEST (Eng et al., 1994) algorithm (v. 28.12), and searching a protein sequence database consisting of the mouse proteome (Uniprot, downloaded 30 October 2012), and reversed ‘decoy' versions of these proteins (Elias and Gygi, 2007). Data from each individual sample were filtered to a 1% peptide FDR and subsequently filtered to an experiment-wide 5% protein FDR using a linear discriminant analysis (Huttlin et al., 2010). All raw data are available on PRIDE (Vizcaíno et al., 2013) with the data set identifier PXD002838. Spectral counts for each individual protein within a given sample were divided by the total assigned counts within the same sample and further normalized by protein length.
Protein-abundance comparisons
Each section of the protein-abundance pie charts represents the summed abundance across all replicates for a given experimental condition. The core proteome (independent of colonization state) was determined using the ‘mintersect' function from the MATLAB File Exchange. Proteins were included in the core proteome if they were identified in at least one replicate across all locations and colonization states. The significance of the overlaps was assessed using the cumulative hypergeometric distribution.
Shannon–Weiner diversity index
Shannon–Weiner diversity indices were calculated using the ‘index_SaW' function, available on the MATLAB File Exchange, on normalized abundance data. One-way analysis of variance was conducted on the Shannon–Weiner indices for each location along the GI tract using the ‘anova1' function in MATLAB, and Tukey-Kramer tests were performed using the ‘multcompare' function to determine which colonization states within a location were associated with significant differences in diversity.
Unsupervised clustering methods
PCA was performed on the normalized spectral counts for the 853 identified proteins by first generating a covariance matrix of all 45 samples. We performed hierarchical clustering on the data set of normalized spectral counts using Euclidean distance and average linkage metrics with Cluster (de Hoon et al., 2004; v. 3.0) and Treeview (Saldanha, 2004; v. 1.1.6r4). PCA and hierarchical clustering were also performed on the 2991 GO terms associated with this data set.
Random forest analysis
For each of three groupings (all mice, BT and GF mice, and CR mice), we performed a three-stage classification analysis. First, we generated a random forest classifier using the R package randomForest (Liaw and Wiener, 2002) based on all proteins in each group with total relative abundances >0.01. Each random forest consisted of 10 000 trees and classified samples according to three locations: stomach, small intestine (jejunum/ileum) and large intestine (cecum/proximal colon). Second, we selected the proteins in the top 10th percentile of importance from each random forest, based on the mean decrease in Gini score. These proteins served as the basis for a new random forest classifier with the same parameters as before. Third, we confirmed the OOB error rates by performing leave-one-out cross-validation within each grouping; classifiers based on the proteins selected in the second step were trained based on all mice except one, and locations of the five samples from the remaining mouse were predicted from the classifier.
The same random forest procedure was applied to the 1520 unique GO terms classified into three (stomach, small intestine, large intestine) or four locations (stomach, jejunum, ileum, large intestine). We selected the GO terms in the top 10th percentile of importance from each random forest based on the mean decrease in Gini score.
GO analysis
To facilitate the analysis of the relative abundances of GO terms, the murine gene association file was downloaded from the European Bioinformatics Institute website (Dimmer et al., 2012). Using MATLAB, we generated a binary vector indicating the presence or absence of each GO term for every protein, and multiplied the normalized spectral count for any given sample and protein by the corresponding binary GO vector. Finally, we summed the normalized abundances for each GO term across all proteins within each sample.
K-means clustering
We used mean abundances across the three mouse replicates to perform k-means clustering, with squared Euclidean as the distance metric and >1000 replicates per run. Clustering was carried out separately for each colonization state; cluster numbers of 8, 12 and 16 were tested for each state. Proteins in clusters of low abundance (<8 × 10−3 arbitrary units) were removed from the data set. The remaining high-abundance proteins were normalized across location using the ‘zscore' function in MATLAB. These normalized, high-abundance proteins were then re-clustered using k-means with trials of five, six and seven clusters. To compare clusters across colonization states, we used squared Euclidean distance to compare the centroids of each of the six and five clusters from CR and BT mice, respectively, to the centroids of each of the five clusters from GF mice; the GF cluster that was most similar to each CR cluster and BT cluster was selected based on the shortest distance between centroids.
Determination of correlation between gut and fecal proteomes
Fecal proteome and host-secreted gut proteome samples were compared by identifying the total number of proteins in the overlap between each pair of data sets. The core fecal proteome was generated from previously published data from the stool of mice with the colonization states used here (Lichtman et al., 2013). Raw data from only the 60% protein fraction (used in this study) were re-processed using the steps described above, including peptide and protein FDR filtering and quantification. Proteins were included in the fecal or location-specific proteome if they had been identified in any one of the three colonization states. The significance of overlap between any two regions was calculated using the hypergeometric distribution.
Proximal colon contents were compared with fecal proteomes from several different experiments by calculating the Spearman correlation coefficient for each binary comparison, considering only the proteins that were identified in at least one of the two samples, using the ‘spearmanr' function in the SciPy python package. The results were clustered (Euclidean distance, average linkage) by rows and columns. Significant differences in correlation were calculated using Student's two-tailed, non-paired t-test.
Analysis of relative GO term abundances according to colonization state
The data set containing the normalized GO term abundances across all samples was subdivided into three groups, each containing data from the 15 samples encompassing a given colonization state. These three groups were then condensed by summing the GO term abundances across all locations along the GI tract. To compare the abundances of the three replicates for each colonization state, we used the ‘anova1' function in MATLAB for each GO term to determine the significance of differences between all pairs of the three colonization states. The FDR and the corresponding q-values were calculated using the ‘mafdr' function in MATLAB on the set of all anova1-derived P-values. A q-value threshold of 0.01 was selected and the GO terms that met this threshold were recorded, along with the corresponding mean and standard deviation of the normalized abundance for each GO term in each colonization state. To determine the specific pairs of colonization states that differed significantly for each GO term, we used the ‘multcompare' function in MATLAB on the statistics generated by anova1 using a P-value threshold of 0.01.
Acknowledgments
We would like to thank members of the Elias and Sonnenburg lab members, Carlos Gonzales and Katherine Ng for artistic support; and teaching assistants Alexandre Colavin, Miriam Gutschow and Sam Smits for helpful feedback. This work was supported by a Curriculum Development Award from The MathWorks, Inc. (to KCH and JLS), a grant from the National Institutes of Health (R01-DK085025) (to JLS), the Stanford Systems Biology Center funded by National Institutes of Health grant P50 GM107615 (to KCH) and grant DGE-114747 from the National Science Foundation (to JSL).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Cash H, Whitham C, Behrendt C, Hooper L. (2006). Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313: 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S et al. (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30: 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. (2010). Host-bacterial symbiosis in health and disease. Adv Immunol 107: 243–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. (2004). Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- Dimmer EC, Huntley RP, Alam-Faruque Y, Sawford T, O'Donovan C, Martin MJ et al. (2012). The UniProt-GO Annotation database in 2011. Nucleic Acids Res 40: D565–D570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M et al. (2005). Diversity of the human intestinal microbial flora. Science 308: 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214. [DOI] [PubMed] [Google Scholar]
- 3rd. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a aprotein database. J Am Soc Mass Spectrom 5: 976–989. [DOI] [PubMed] [Google Scholar]
- Erickson AR, Cantarel BL, Lamendella R, Darzi Y, Mongodin EF, Pan C et al. (2012). Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease. PLoS One 7: e49138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS et al. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312: 1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Falk PG, Gordon JI. (2000). Analyzing the molecular foundations of commensalism in the mouse intestine. Curr Opin Microbiol 3: 79–85. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA et al. (2010). A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. (2006). Human gut microbes associated with obesity. Nature 444: 1022–1023. [DOI] [PubMed] [Google Scholar]
- Liaw A, Wiener M. (2002). Classification and regression by randomForest. R News 2: 18–22. [Google Scholar]
- Lichtman JS, Marcobal A, Sonnenburg JL, Elias JE. (2013). Host-centric proteomics of stool: a novel strategy focused on intestinal responses to the gut microbiota. Mol Cell Proteomics 12: 3310–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JS, Sonnenburg JL, Elias JE. (2015). Monitoring host responses to the gut microbiota. ISME J 9: 1908–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe ME. (2002). The triglyceride lipases of the pancreas. J Lipid Res 43: 2007–2016. [DOI] [PubMed] [Google Scholar]
- Magurran AE. (2004) Measuring Biological Diversity 1st edn. Blackwell Sciences Ltd.: Malden, MA, USA. [Google Scholar]
- Mahowald M a, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A et al. (2009). Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA 106: 5859–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski KM, Mackay CR. (2011). Diet, gut microbiota and immune responses. Nat Immunol 12: 5–9. [DOI] [PubMed] [Google Scholar]
- Meeuwsen CG, De Visser H, Hazenberg MP, Wostmannn BS, Pleasantsa JR, Benner R et al. (1989). Serum immunoglobulin levels and naturally occurring antibodies against carbohydrate antigens in germ-free BALB/c mice fed chemically defined ultrafiltered diet. Eur J Immunol 19: 2335–2339. [DOI] [PubMed] [Google Scholar]
- Muth T, Kolmeder CA, Salojärvi J, Keskitalo S, Varjosalo M, Verdam FJ et al. (2015). Navigating through metaproteomics data: a logbook of database searching. Proteomics 15: 3439–3453. [DOI] [PubMed] [Google Scholar]
- Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F et al. (2007). Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut 56: 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. (2004). Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Savage D. (1977). Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31: 107–133. [DOI] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN et al. (2010). Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141: 1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Fagarasan S. (2008). How host-bacterial interactions lead to IgA synthesis in the gut. Trends Immunol 29: 523–531. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O et al. (2011). The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J et al. (2009). Shotgun metaproteomics of the human distal gut microbiota. ISME J 3: 179–189. [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM et al. (2013). The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41: D1063–D1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T et al. (2011). Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11: 2019–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley R, Volchkov P, Stranges P. (2008). Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455: 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. (1998). Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95: 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaia J. (2008). Mass spectrometry and the emerging field of glycomics. Chem Biol 15: 881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Guo T, Li X, Datta A, Park JE, Yang J et al. (2010). Simultaneous characterization of glyco- and phosphoproteomes of mouse brain membrane proteome with electrostatic repulsion hydrophilic interaction chromatography. Mol Cell Proteomics 9: 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.