Abstract
Infantile hemangioma (IH) is the most common vascular tumor of infancy, and it uniquely regresses in response to oral propranolol. MicroRNAs (miRNAs) have emerged as key regulators of vascular development and are dysregulated in many disease processes, but the role of miRNAs in IH growth has not been investigated. We report expression of C19MC, a primate-specific megacluster of miRNAs expressed in placenta with rare expression in postnatal tissues, in glucose transporter 1–expressing (GLUT-1–expressing) IH endothelial cells and in the plasma of children with IH. Tissue or circulating C19MC miRNAs were not detectable in patients having 9 other types of vascular anomalies or unaffected children, identifying C19MC miRNAs as the first circulating biomarkers of IH. Levels of circulating C19MC miRNAs correlated with IH tumor size and propranolol treatment response, and IH tissue from children treated with propranolol or from children with partially involuted tumors contained lower levels of C19MC miRNAs than untreated, proliferative tumors, implicating C19MC miRNAs as potential drivers of IH pathogenesis. Detection of C19MC miRNAs in the circulation of infants with IH may provide a specific and noninvasive means of IH diagnosis and identification of candidates for propranolol therapy as well as a means to monitor treatment response.
Infantile hemangioma endothelial cells express and release placental microRNAs into patient blood where they are detectable as biomarkers and provide clues to the tumor’s pathogenesis.
Introduction
Infantile hemangioma (IH) is a benign vascular tumor that proliferates rapidly after birth and then eventually involutes, often being replaced by fibrofatty tissue (1). IH are potentially disfiguring tumors that occur in 4%–5% of infants and can lead to significant morbidity, including airway obstruction, high-output cardiac failure, hypothyroidism, amblyopia, liver failure, ulceration, and hemorrhage (2–5). IH can resemble other congenital vascular anomalies but is unique in its growth pattern, prominent expression of placental vascular protein markers (6), and accelerated regression in response to the nonselective β-adrenergic antagonist propranolol (7). Early empiric propranolol therapy has emerged as the first-line treatment for IH, often obviating the need for invasive surgical procedures by preventing or reversing the tumor’s growth prior to its reaching a morbid size or causing skin ulceration and scarring. The decision to initiate propranolol therapy is most often based on history and physical examination alone; the definitive diagnosis of IH requires surgical biopsy and immunohistochemical staining demonstrating glucose transporter 1 (GLUT-1) protein (coded for by SLC2A1) expression in endothelial cells (6, 8), a property unique to IH among vascular anomalies and shared by the placental chorionic villous capillaries (8, 9). Currently, the pathogenesis of IH and the mechanism of action of propranolol in IH regression remain under investigation.
MicroRNAs (miRNAs) have emerged as key regulators of vascular development (10) and are dysregulated in many disease processes (11). These short, noncoding RNA molecules silence gene expression by inducing cleavage or blocking translation of target mRNAs. miRNAs are released by cells into the circulation and other body fluids, in which they are stable and detectable by qRT-PCR, making them ideal biomarkers of a wide range of disease processes, including cancer (12). Here, we analyzed miRNA expression in IH and found that the chromosome 19 miRNA cluster (C19MC), known to be expressed in placenta, embryonic stem cells, and some tumors (13–15), is highly expressed in IH and that GLUT-1+ IH endothelial cells are the major C19MC-expressing cell type. C19MC miRNAs were detected in the circulation of infants and young children with IH but not in the blood of unaffected children or those with other vascular anomalies. Validated targets of these miRNAs, which include inhibitors of cell proliferation and angiogenesis (16–20), suggest that C19MC is important in IH pathogenesis.
Results
C19MC miRNA detection in IH tumors.
To investigate the potential role of miRNAs in IH pathogenesis, we profiled the miRNA transcriptomes of tissue from IH or lymphatic malformations (LMs), or from skin adjacent to IH lesions, from a total of 24 patients, using a multiplex hybridization miRNA array (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/jci.insight.88856DS1). Using unsupervised hierarchical clustering, we identified the C19MC miRNA cluster as overexpressed in IH when compared with skin or LMs (Figure 1A).
Figure 1. C19MC miRNAs are specific biomarkers of infantile hemangioma.
(A) Multiplexed tissue miRNA analysis and unsupervised hierarchical clustering of IH, LM, and skin identified C19MC miRNAs as upregulated in IH. (B) Tissue from 22 additional infants and children with the indicated vascular anomalies as well as tissue from 5 healthy placentas were analyzed by qRT-PCR for GLUT-1 mRNA, the endothelial miR-126a-3p, and the C19MC miRNAs miR-517a-3p and miR-518d-5p. Only placenta and IH expressed the C19MC miRNAs miR-517a-3p and 518d-5p. GLUT-1 was expressed in placenta and IH; background signal in other samples is due to erythrocyte GLUT-1 expression. The y axis represents relative mRNA or miRNA compared with levels of GAPDH or RNU48, respectively. (C) Circulating miR-517c-3p and miR-126a-3p were analyzed by qRT-PCR in plasma from 60 additional infants and children with the indicated vascular anomalies and from 5 patients with no vascular anomalies. Only plasma from patients with IH had detectable levels of miR-517c-3p, while miR-126a-3p was detected in all samples. Infants and children with large (>2-cm) and proliferative (in patients younger than 9 mo of age) IH had significantly higher levels of circulating miR-517c-3p compared with infants and children with smaller or partially involuted tumors, while miR-126a-3p levels were not affected by tumor size or patient age. Statistical analysis was performed using single-factor ANOVA, comparing groups with n = 3 or greater samples. For miR-517c-3p, follow-up subgroup analysis was performed using Tukey’s test, following ANOVA (**P < 0.01). For miR-126a-3p, values did not reach significance (P = 0.62, ANOVA). The y axis represents the relative miRNA level compared with the average pooled miRNA from all the samples. All points illustrated on the qRT-PCR graphs represent individual patient samples, and error bars represent standard deviations. Pla, placenta; IH, infantile hemangioma; Skin, healthy skin adjacent to IH; TA, tufted angioma; KH, kaposiform hemangioendothelioma; PG, pyogenic granuloma; VM, venous malformation; NICH, noninvoluting congenital hemangioma; RICH, rapidly involuting congenital hemangioma; LM, lymphatic malformation; AVM, arteriovenous malformation; JNA, juvenile nasopharyngeal angiofibroma; No lesion, plasma from patient without vascular anomaly.
Using qRT-PCR, we confirmed expression of 43 C19MC miRNAs in tumor tissue from 5 additional children with IH and 46 C19MC miRNAs in tissue from 5 healthy placentas, while no C19MC miRNAs were detected in skin adjacent to IH lesions (Supplemental Figure 2). IH, placenta, and skin equivalently expressed several non-C19MC miRNAs, including endothelial-specific miR-126-3p (21) and vascular smooth muscle/pericyte-specific miR-145-5p (22). To determine if C19MC miRNA expression is specific to IH, we selected two representative C19MC sequences (miR-517a-3p and miR-518d-5p) as well as endothelium-specific miR-126-3p and GLUT-1 mRNA and profiled their expression in excised tissue from 22 children with 7 types of vascular anomaly and 5 healthy placentas using qRT-PCR (Figure 1B). All sampled tissues expressed miR-126-3p, but only IH and placenta expressed detectable levels of miR-517a-3p and miR-518d-5p. GLUT-1 was highly expressed in placenta and IH tissue when compared with tissue from other vascular anomalies.
Because C19MC miRNAs are released into maternal circulation from the placenta (23), we performed qRT-PCR on plasma from 65 infants and children with 9 types of vascular anomaly and 5 healthy control patients to determine if C19MC miRNAs were detectable in the circulation of IH patients. While miR-126a-3p was detected in all plasma samples, only plasma from infants and children with IH had detectable circulating miR-517c-3p (Figure 1C). Plasma from patients with larger (>2 cm) or proliferative (in patients younger than 9 mo of age) IH had significantly higher levels of circulating miR-517c-3p than plasma from patients with smaller or involuting tumors.
C19MC expression in IH endothelial cells.
To determine the cellular origin of C19MC miRNAs in IH, we performed in situ hybridizations (ISH) with probes for miR-517a/c-3p and miR-126a-3p on paraffin sections from 3 children with proliferative or involuted IH. Adjacent sections were subjected to immunofluorescent staining for the endothelial cell surface markers CD31 and GLUT-1 (Figure 2, A and B, and Supplemental Figure 4). As expected, miR-126a-3p was detected in all CD31+ endothelium; however, miR-517a/c-3p was detected only in vessels with CD31+GLUT-1+ endothelial cells. We also performed ISH for miR-517a/c-3p and miR-126a-3p on sections from 3 term placentas (Figure 2C). Consistent with others’ observations on primary cells (24), miR-517a/c-3p was detected in the placental trophoblast and was also detected in the vascular endothelium of the chorionic villi and plate. To confirm that IH GLUT-1+ endothelial cells express miR-517a-3p, a single-cell suspension of freshly excised proliferative IH tissue was labeled with an anti-CD31 antibody and the GLUT-1–binding fragment of HTLV-env fused to EGFP (25). CD31+GLUT-1+ endothelial cells were isolated by FACS and analyzed by qRT-PCR (Figure 2D), which demonstrated enriched expression of miR-517a-3p in the CD31+GLUT-1+ cell population.
Figure 2. GLUT-1+ IH endothelial cells contain C19MC miRNAs.
(A and B) Paraffin sections of (A) proliferative IH (from 6-mo-old child) and (B) involuted IH (from 44-mo-old child) were labeled with immunofluorescent antibodies to endothelial CD31 and IH endothelial GLUT-1. Overlaid DAPI images indicate GLUT-1+CD31+ endothelial cells. On adjacent sections, in situ hybridization probes to endothelial miR-126a-3p and C19MC miR-517a/c-3p indicate that normal CD31+GLUT-1– endothelium (white arrows) contains miR-126a-3p but not miR-517a/c-3p, while CD31+GLUT-1+ tumor endothelium (yellow arrows) contains both miR-126a-3p and miR-517a/c-3p. A scrambled miR probe was used as a negative control; nuclei were stained with Fast Red. Scale bar: 100 μm (middle); 50 μm (bottom). (C) In situ hybridization images of sections from placenta demonstrate miR-517a/c-3p in the placental trophoblast (black arrows). miR-517a/c-3p and miR-126a-3p were detected in villous endothelial cells (magenta arrows). Scale bar: 200 μm (top); 50 μm (bottom). (D) FACS and qRT-PCR analysis of a proliferative IH confirmed GLUT-1+CD31+ cells as the miR-517–expressing cell type. A single-cell suspension from proliferative IH was labeled with GLUT-1–EGFP and CD31-PE antibodies and sorted into GLUT-1+CD31+ and GLUT-1–CD31– populations. RNA isolation and qRT-PCR analysis of these sorted populations demonstrates enriched miR-517a-3p expression in the GLUT-1+CD31+ population. The y axis represents relative mRNA or miRNA compared with levels of GAPDH or RNU48, respectively. Data points on the qRT-PCR graphs indicate duplicate samples prepared at the sorting stage from the same lesion, and error bars represent standard deviations.
Distinguishing IH from other vascular anomalies using C19MC detection.
Early and accurate diagnosis of infants with IH is important clinically, because other vascular anomalies, such as noninvoluting congenital hemangioma (NICH), grossly resemble IH but do not regress in response to propranolol. Tissue miRNA profiling of multiple vascular anomalies (Figure 1B) identified 4 children (example photo in Figure 3A) as having NICH by history, physical examination, and H&E staining of their excised tumors (GLUT-1 staining was not initially performed after excision). While 2 of these tumors had no detectable levels of C19MC miRNAs (illustrated in Figure 1B), 2 tumors surprisingly contained C19MC miRNA levels similar to IH (Figure 3B). Retrospective GLUT-1 staining on paraffin sections from all 4 specimens revealed GLUT-1+ endothelial cells in the 2 C19MC-containing lesions but not in the others (Figure 3C), an observation confirmed by qRT-PCR (Figure 3D). These data revised the diagnosis of the C19MC-expressing tumors from NICH to IH.
Figure 3. C19MC detection is diagnostic for GLUT-1+ IH.
(A) Photograph of a child (NICH 264) clinically diagnosed with NICH who underwent surgical excision. (B) qRT-PCR analysis of the C19MC cluster was performed on cryopreserved tissue from the lesion in A as well as 3 other NICHs (NICH 121, 169, and 364) and one GLUT-1+ IH (IH 344). x-axis labels represent individual C19MC miRNA qRT-PCR assays. y-axis (linear scale) values represent ΔCt between the global mean of all tested miRNAs - assayed miRNA. NICH 169 and 264, but not 121 and 364, contained levels of C19MC miRNAs similar to the proliferative IH 344. All miRNAs below the limit of detection were assigned a ΔCt value of –14. (C) Paraffin sections were prepared from blocks preserved at the time of surgery and stained for GLUT-1. For NICH 121 (top row, DAB staining), GLUT-1 positivity is represented in brown. For NICH 169 and 264 (middle and bottom rows), GLUT-1 positivity is represented by green fluorescence. White arrows indicate GLUT-1– endothelial cells; magenta arrows indicate GLUT-1+ endothelial cells. Only the C19MC-expressing NICHs 169 and 264 expressed endothelial GLUT-1. Scale bar: 100 μm. (D) Individual qRT-PCR assays demonstrate GLUT-1 and miR-517c-3p expression in both NICH 169 and 264 (purple bars), which was absent in NICH 121 (black bars, background GLUT-1 is due to presence of erythrocytes). The y axis represents relative mRNA or miRNA compared with levels of GAPDH or RNU48, respectively. Individual data points represent experimental duplicate samples prepared from the same lesion, and error bars represent standard deviations.
C19MC expression in involuted and propranolol-treated IH tissue.
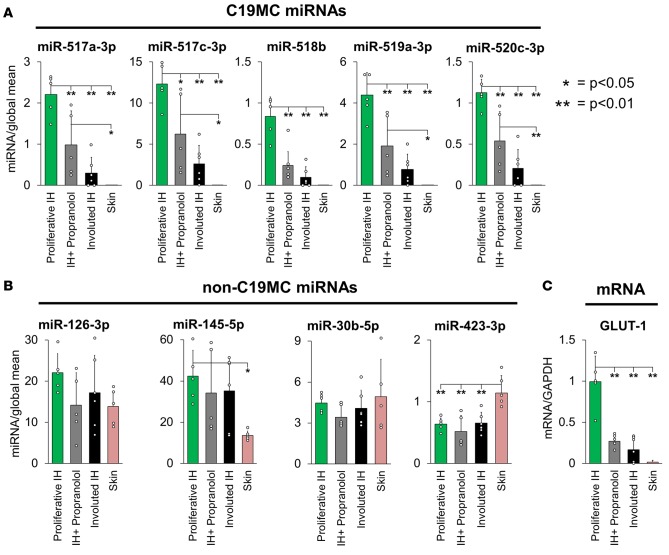
In the majority of cases, both natural involution and propranolol therapy reduce the size and growth of IH tumors and their number of GLUT-1+ endothelial cells (7, 26, 27). To determine whether involution or propranolol treatment affected C19MC miRNA levels in IH, we profiled the expression of 5 representative C19MC miRNAs, 4 non-C19MC miRNAs, and GLUT-1 mRNA using qRT-PCR on tissue excised from infants and children with proliferative IH (age <9 mo, n = 5; mean age 5.34 mo), infants and children with partially involuted IH (age >36 mo, n = 6; mean age 58.9 mo), and infants and children with proliferative IH who were receiving propranolol therapy (n = 5, mean age 16.0 mo) as well as adjacent skin (Figure 4). Both propranolol-treated and involuted IH tissue contained significantly lower levels of all tested C19MC miRNAs as well as GLUT-1 mRNA when compared with proliferative IH, while the non-C19MC miRNAs were unaffected by involution or propranolol therapy.
Figure 4. C19MC miRNA and GLUT-1 mRNA levels are significantly lower in propranolol-treated or partially involuted IH tumors.
Tissue from infants and children with proliferative IH (n = 5, green bars, mean age 5.34 mo), infants and children with proliferative IH who are undergoing propranolol therapy (n = 5, gray bars, mean age 16.0 mo), or infants and children with partially involuted IH (n = 6, black bars, mean age 58.9 mo) or adjacent skin (n = 5, pink bars) was analyzed using qRT-PCR for (A) 5 C19MC miRNAs (miR-517a-3p, miR-517c-3p, miR-518b, miR-519a-3p, and miR-520c-3p), (B) 4 non-C19MC miRNAs (miR-126-3p, miR-145-5p, miR-30b-5p, and miR-423-3p), and (C) GLUT-1 mRNA. Both the propranolol-treated and involuted tumors contained significantly lower levels of C19MC miRNAs, which were undetectable in skin, while non-C19MC miRNAs were detected in all tissues and levels were unaffected by involution or propranolol treatment. ANOVA analysis was used for comparison of all groups, and when deemed significant (P < 0.05) post-hoc Tukey’s test was performed to determine the significance of each group comparison (*P < 0.05; **P < 0.01). The y axis represents relative mRNA or miRNA compared with levels of GAPDH or the global mean of all miRNAs tested, respectively. Indicated data points represent individual patient samples, and error bars represent standard deviations.
Correlation between clinical response to propranolol and circulating C19MC miRNAs.
To determine the correlation of circulating levels of C19MC miRNAs to the clinical response of propranolol therapy in IH patients, we collected plasma from 10 patients with suspected IH at the time of diagnosis and at 1- and 6-month time points of daily oral propranolol therapy (2 mg/kg/d) and profiled the expression of 38 C19MC miRNAs and 23 non-C19MC miRNAs using qRT-PCR. All patients had circulating levels of some C19MC miRNAs at diagnosis, and representative patient photographs and qRT-PCR results for select miRNAs are displayed in Figure 5. Circulating levels of C19MC miRNAs decreased or were undetectable in all patients at 1 month of propranolol treatment, which clinically appeared to correlate with tumor regression. At 6 months, 9 of 10 patients (patients 1010, 1024, and 1053, Figure 5, A–C) showed near complete regression of their tumors and undetectable levels of C19MC miRNAs, while 1 patient (patient 1072, Figure 5D) had minimal tumor regression, followed by tumor regrowth, with a corresponding increase in circulating C19MC levels.
Figure 5. Levels of circulating C19MC miRNAs correlate with IH tumor regression during propranolol treatment and regrowth following propranolol cessation.
Patients (A) 1010, (B) 1024, (C) 1053, and (D) 1072 with suspected IH were photographed and blood was drawn before the initiation of oral propranolol therapy, after 1 month of propranolol, and after 6 months of propranolol. The y axis represents relative miRNA compared with the pooled average of all C19MC miRNAs tested. RNA extracted from plasma was analyzed by qRT-PCR for representative miRNA sequences. Each bar/data point represents one individual patient sample.
Discussion
The posttranscriptional gene regulation by miRNAs occurs in a vast array of cellular processes, including proliferation, differentiation, apoptosis, and angiogenesis (28–35), and alterations in miRNA expression patterns contribute to many pathological processes, including several cancers (36–40). The release of miRNAs into body fluids (41, 42), their stability there (43), their specific uptake by recipient cells (44), and abnormalities in circulating miRNA profiles in disease (45–47) make cell-free miRNAs attractive disease biomarkers and novel therapeutic targets. The identification of C19MC miRNAs in IH endothelial cells and in IH patient plasma establishes them as the first validated circulating biomarker of any vascular anomaly, to our knowledge.
The primate-specific C19MC comprises 46 miRNA genes and is the largest miRNA gene cluster in humans. Validated C19MC targets include inhibitors of cell growth, proliferation, and angiogenesis CDKN1A, PTEN, AKT3, KLF10, TGFBR2, and TIMP2 (each targeted by 4–10 C19MC miRNAs) (16–20), suggesting that C19MC expression may drive the growth of IH through relaxation of normal constraints. Through its lipid phosphatase activity, PTEN inhibits PI3 kinase, which phosphorylates AKT. Our finding of C19MC expression in IH may provide a mechanism for the prior observations that PTEN and AKT3 protein levels are lower, and phospho-AKT levels are higher (presumably through PTEN loss), in IH endothelial cells compared with levels in normal ones (48). In that study, knockdown of AKT3, but not AKT1, was observed to stimulate IH endothelial cell sprouting and migration in vitro, and forced expression of an AKT transgene induced GLUT-1 expression in mouse endothelial cells engrafted on nude mice. Phosphorylated AKT is known to drive SLC2A1 expression in multiple cell types (49–51), and cytoplasmic sequestration of GLUT-1 has been shown to be PTEN dependent (52). Hence, the plasma membrane GLUT-1 expression that is diagnostic of IH may result at least in part from the targeting of PTEN by C19MC miRNAs and resultant AKT activation.
C19MC and related miRNAs are thought to contribute to the etiology of hyperplasias other than IH. miR-519a and miR-519d were both found to be highly expressed in IH. Targeting of RBL2 by overexpressed miR-519a from amplified C19MC activates the DNA methyltransferase DNMT3B, driving the formation of embryonal tumors with multilayered rosettes (53). mir-519d shares its seed sequence with miR-17, miR-20a, and miR-106a, which are associated with cancer and target RB1, among other genes (54). C19MC expression is activated by different mechanisms in different cases of hepatic mesenchymal hamartoma (55, 56), suggesting a causal role.
In placenta, trophoblasts and stromal cells are known to express C19MC miRNAs, which are released into the maternal circulation (13–15, 23). Primary human trophoblasts release exosomes containing C19MC miRNAs, and these are taken up by neighboring cells in vitro and confer resistance to viral infection through autophagy (57), a process which promotes angiogenesis in endothelial cells in vitro (53). Its validated targets and paternal imprinting suggest that, in placenta, C19MC might also drive growth in accord with the competition model of imprinting (19).
Interestingly, IH-derived GLUT-1+CD31+ endothelial cells undergo endothelial-to-mesenchymal transition (EndMT) and become GLUT-1– when grown in culture (27). These cells did not express C19MC (Supplemental Figure 3) and can differentiate into adipocytes, as can IH-derived primary mesenchymal stem cells (58). However, it is not known if the adipose tissue that often appears during IH involution originates from these cells. Future experiments are required to determine if loss of C19MC expression in IH endothelial cells leads to EndMT, loss of the GLUT-1+ phenotype, and subsequent adipogenesis during involution. Whether propranolol, which can modulate miRNA expression in ischemic cardiac tissue (59), directly inhibits C19MC expression in IH remains an open question.
North et al. (6, 60) hypothesized that IH originates from a placental pool of vascular precursor cells, based on the shared expression of 4 chorionic villous capillary endothelial proteins by IH endothelium and the predictable timing of IH presentation. A comparison of the transcriptomes of IH, placenta, and several other normal and diseased tissues found the highest degree of similarity between IH and placenta (61). Further supporting this hypothesis, IH occurs more frequently in the children of mothers with placental anomalies (62) or who have undergone chorionic villous sampling (63). C19MC expression in IH is consistent with a placental origin but does not rule out a stem cell origin for this tumor, as suggested by the Bischoff group (64), as C19MC miRNAS are also expressed in human embryonic stem cells (65).
The specificity of C19MC miRNAs to IH and the correlation between circulating C19MC levels and clinical response to propranolol implies that C19MC profiling may be a useful clinical tool in the diagnosis and treatment of vascular anomalies. As demonstrated in Figure 3A, distinguishing IH from other vascular anomalies and thus determining whether propranolol treatment is appropriate can be challenging. A noninvasive means of confirming IH diagnosis, determining degree of IH proliferation, and following treatment response would significantly refine management of vascular anomalies and may enable earlier diagnoses and initiation of appropriate treatment. Children with visceral or airway IH may benefit significantly, as a circulating biomarker will improve diagnostic accuracy and may lessen the need for invasive endoscopy, high-risk biopsy, and expensive serial imaging.
In summary, we report the expression of C19MC miRNAs, which represent what we believe to be the first identified circulating biomarker of a vascular anomaly, in IH. C19MC miRNAs are unique to IH among all studied vascular anomalies and are released into circulation by GLUT-1+ endothelial cells, making them specific and easily detectable biologic markers of IH. Levels of circulating C19MC miRNAs correlate with tumor size and proliferative state and decrease during propranolol-mediated tumor regression, potentially making circulating C19MC miRNAs useful indicators of proliferative state, responsiveness to propranolol therapy, and residual disease or recurrence following surgical excision. The validated targets of C19MC miRNAs suggest testable hypotheses regarding the mechanisms of IH pathogenesis and the accelerated regression with propranolol therapy.
Methods
Primary human samples.
Healthy placenta samples from full-term pregnancies were obtained from the Global Alliance to Prevent Prematurity and Stillbirth repository at Seattle Children’s Research Institute according to their standard operating procedures (http://gapps.org/docs/Lab_MOP_v2.8.4_Placenta_and_Cord_tissue.pdf). All vascular anomaly tissue specimens and photographs were collected from patients at SCH with informed consent according to institutional review board protocols (SCH IRB no. 11458). Tissue samples were collected intraoperatively during resection, immediately frozen in liquid nitrogen, and cryopreserved in a –80°C freezer. Additional tissue was embedded in paraffin and labeled with the appropriate diagnosis following evaluation by SCH Surgical Pathology. Skin samples were taken directly adjacent to IH lesions and frozen as above. Plasma samples were collected on the day of diagnosis in the SCH Outpatient Laboratory (for vascular anomaly plasma samples), intraoperatively during elective tonsillectomy (for control plasma samples), or at 1- and 6-month follow up visits (for patients on propranolol therapy). Blood from venipuncture was drawn into lavender BD Vacutainer tubes and centrifuged at 1,300 g for 15 minutes at 4°C. The plasma fraction was then aspirated, passed through a 0.8-micron filter, and cryopreserved in 1-ml cryovials at –80°C.
miRNA expression profiling and unsupervised hierarchical clustering.
The nCounter analysis system (Nanostring Technologies) was used to identify miRNAs that were differentially expressed among IH, LM tissue, and skin (66). RNA was isolated from cryopreserved surgical specimens using TRIzol reagent (Life Technologies) and analyzed with the nCounter Human v2 miRNA Expression Assay Kit and nCounter Analysis System (Nanostring) according to the manufacturer’s instructions. Expression data was then log transformed, genes and arrays were centered using the median expression, and unsupervised hierarchical clustering was performed using uncentered correlation symmetry metrics and centroid linkage. The clustered array was visualized using Maple Tree (http://mapletree.sourceforge.net/) and is included in Supplemental Figure 1. Microarray data are accessible to reviewers at NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=inahwuesrvirzix&acc=GSE69136).
Real-time PCR analysis.
Initial qRT-PCR analysis was performed by Exiqon Services (Exiqon) using the miRCURY LNA Universal RT miRNA PCR Custom Pick-&-Mix panel (v3) (Supplemental Figure 2, Figure 3B, and Figure 4, A and B). Briefly, total RNA was extracted from the tissue using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. 2 ng RNA was reverse transcribed in 10-μl reactions using the miRCURY LNA Universal RT microRNA PCR polyadenylation and cDNA synthesis kit (Exiqon). cDNA was diluted 100 fold and assayed in 10-μl PCR reactions according to the protocol for miRCURY LNA Universal RT microRNA PCR; each miRNA was assayed once by qPCR on the microRNA Ready-to-Use PCR, custom Pick-&-Mix panel using ExiLENT SYBR Green mastermix. Negative controls excluding template from the reverse transcription reaction were performed. Amplification was performed in a LightCycler 480 Real-Time PCR System (Roche Diagnostics) in 384-well plates. The amplification curves were analyzed using Roche LC software, both for determination of Ct (by the second derivative method) and for melting curve analysis. The global mean expression of all assays was determined by NormFinder (Aarhus University Hospital) (67). For Supplemental Figure 2 and Figure 4, A and B, results are expressed as the fold change of the target miRNA/global mean. For Figure 3B, results are expressed as ΔCt (Ct global mean – Ct sample). Error bars represent standard deviations between all samples in a given category.
Additional individual qRT-PCR analyses were performed in our laboratory using TaqMan individual miRNA primers (Life Technologies) according to the manufacturer’s instructions. For tissue samples (Figure 1B, Figure 2D, Figure 3D, and Figure 4C), three 50-mg pieces of cryopreserved tissue from each patient were separately homogenized using a TissueRuptor homogenizer (Qiagen), and RNA was extracted using TRIzol Reagent (Life Technologies) according to the manufacturer’s instructions for whole tissue samples. RNA was quantitated on a NanoDrop 2000 spectrophotometer (Thermo Scientific). For miRNA assays, 10 ng of RNA from each sample was reverse transcribed to cDNA using the TaqMan miRNA Reverse Transcriptase Kit (Life Technologies). For mRNA assays, 100 ng of RNA from each sample was reverse transcribed to cDNA using the SuperScript VILO cDNA Synthesis Kit (Life Technologies). cDNA was diluted 1:10 and analyzed by qPCR using TaqMan Universal PCR Mastermix (Life Technologies) on a Fast 7500 qRT-PCR system (Applied Biosystems). Quantitation was performed using the relative standard curve method. For GLUT-1 mRNA qPCR assays, expression was normalized to the relative expression of GAPDH. For all tissue miRNA assays, expression was normalized to the relative expression of the small nucleolar RNA RNU48. In Figure 4, ANOVA analysis was performed across each group for each miRNA assay. Post-hoc Tukey’s test was performed to determine which comparisons were statistically significant (P < 0.05, ANOVA); these significant comparisons are indicated in the figure. ANOVA analysis was performed using Microsoft Excel 2007, and Tukey’s test was performed according to the NIST Engineering Statistics Handbook using the Tukey HSD calculator (68). Error bars represent standard deviations between all samples in a given category.
For cells isolated using flow cytometry (Figure 2E), 50,000 cells were pelleted in 3 separate tubes from each collected fraction, and RNA was isolated using TRIzol Reagent (Life Technologies) according to the manufacturer’s instructions for suspended cells. qRT-PCR was then performed as described above. Error bars represent standard deviations between the 3 pellets collected from each fraction.
For plasma samples (Figure 1C and Figure 5), RNA was isolated from 200 μl of thawed human plasma from each patient using the miRCURY RNA Isolation Kit - Biofluids (Exiqon). For Figure 1C, 9 μl of elutant was then used in the TaqMan miRNA Reverse Transcriptase Kit (Life Technologies), with a 60-minute 42°C step, and qRT-PCR was performed as above, without the 1:10 dilution step. Samples were run in duplicate, and levels of miRNA are expressed using the relative standard curve method. Due to the absence of a reliable circulating miRNA for normalization, a standard curve was generated for each target miRNA using a pool of RNA from the 12 patients with large, proliferative IH, and the abundance of the detected RNA was set to 1. ANOVA analysis was performed across each group (when n ≥ 3) for each miRNA assay. Post-hoc Tukey’s test was performed as described above to determine which comparisons were statistically significant (P < 0.05, ANOVA); these significant comparisons are indicated in the figure. Error bars represent standard deviations between all samples in a given category. For Figure 5, eluted RNA was analyzed using the miRCURY LNA Universal RT miRNA PCR Custom Pick-&-Mix panel (Exiqon) as indicated above.
Flow cytometry.
A 2 × 1 × 1 cm proliferative IH was excised from the right eyebrow of an 8-month-old female patient and transferred to 1X PBS with 1X penicillin-streptomycin-glutamine (1X GPS) (Life Technologies) on ice. The tumor was minced into 2-mm3 pieces with a sterile scalpel blade and rinsed once with cold 1X GPS in PBS to remove surface blood. Tissue pieces were spun down at 1258 x g and transferred to a 50-ml Erlenmeyer flask containing 30 ml of incubation medium (low-glucose DMEM, Life Technologies; 2% fetal bovine serum; 1X GPS; 1X Ca2+Mg2+; 0.1% collagenase A, Roche) and shaken at 37°C at 50 rpm for 30 minutes until turbid. The mixture was transferred to a 50-ml conical centrifuge tube and homogenized with a TissueRuptor homogenizer (Qiagen) at the lowest setting until no large pieces remained. Cells were spun down at 453 x g for 5 minutes and resuspended in 5 ml cold FACS buffer (PBS + 0.5% BSA + 2.2 g/l sodium citrate + 0.8 g/l citric acid). The resuspended cells were sequentially passed through 70-μm and 40-μm strainers and counted on a hemacytometer. Cells were Fc blocked using FcR blocking reagent (Miltenyi Biotech) in FACS buffer (20 μl FcR/80 μl FACS buffer/106 cells) for 15 minutes at room temperature. Cell surface GLUT-1 was then bound using EGFP-conjugated GLUT-1 ligand (Metafora Biosystems) according to the manufacturer’s instructions. Briefly, aliquots of 105 cells were incubated with EGFP-GLUT-1 ligand for 30 minutes at 37°C, centrifuged at 1088 x g at 4°C for 5 minutes, and washed twice with cold FACS buffer. Cells were then labeled with anti-CD31 conjugated to phycoerythrin (CD31-PE, Miltenyi clone AC138) at 4°C for 15 minutes. Cells were washed and resuspended in cold FACS. Cell sorting was performed on a FACSAria II cell sorter (Beckon Dickinson), and cells were collected for qRT-PCR analysis. Cell sorting data are as follows: total cells counted, 5.81 × 106; CD31+GLUT-1+, 1.82 × 105 (6.6%); CD31–GLUT-1–, 1.85 × 105 (6.7%).
ISH.
Formalin-fixed paraffin sections (6-μm thickness) of IH surgical specimens were dewaxed in xylene and rehydrated in an ethanol series followed by PBS. For placenta sections, after dewaxing and rehydration, placenta sections were placed in 15% glacial acetic acid for 15 minutes to inactivate endogenous alkaline phosphatase (69) followed by two 5-minutes washes in PBS, before proteinase K treatment. ISH, with 5′, 3′ digoxigenin-labeled locked nucleic acid probes (LNA) for hsa-miR-126a-3p, hsa-miR-517a/c and a scrambled negative control probe (Exiqon), was performed using an Exiqon ISH optimization kit according to the manufacturer’s instructions. All probes were used at a concentration of 40 nM. Hybridization and posthybridization high-temperature washes were performed at 55°C. Probes were detected using alkaline phosphatase–conjugated sheep anti-digoxigenin Fab fragments and NBT/BCIP (both from Roche). Sections were examined and images were collected using a Leica DM4000B microscope and Leica Application Suite software, v4.5.
Antibodies.
The antibodies used for immunofluorescence were mouse anti–GLUT-1 (NBP1-35926, Novus; used at 1:300 dilution), rabbit anti-CD31 (EPR3094, Abcam; 1:200), and Alexa 488 goat anti-mouse IgG and Alexa 660 goat anti-rabbit IgG (both from Life Technologies; 1:300). For 3,3′-diaminobenzidine (DAB) staining, the primary antibody was a rabbit polyclonal anti–GLUT-1 (355a-16, Cell Marque; 1:400), and the negative control was normal rabbit Ig (760-1029, Ventana; 1:400).
Immunohistochemistry.
For immunofluorescent detection of CD31 and GLUT-1, formalin-fixed paraffin sections (6-μm thickness) of IH surgical specimens were dewaxed in xylene and rehydrated in an ethanol series followed by PBS. Antigen retrieval was performed heating sections in 10 mM citrate, pH 6.0, for 20 minutes in a boiling water bath, followed by 30 minutes of cooling. After washing in PBS, sections were briefly preblocked in 1% BSA in PBS and then blocked in 5% normal goat serum (Jackson ImmunoResearch) in 1% BSA in PBS for 60 minutes. Primary antibodies diluted in 1% BSA in PBS were applied, and the sections were incubated at 4°C overnight and then washed in PBS. Primary antibodies were omitted in negative controls. Secondary antibodies were diluted in 2% normal goat serum, 1% BSA in PBS plus DAPI and applied to the sections for 60 minutes at room temperature. The slides were then washed in PBS and mounted in Prolong Gold Antifade (Life Technologies). Sections were examined and images were collected using a Nikon Ti fluorescence microscope, Nikon A1 confocal system, and Nikon Elements software v. 3.22.09 Build 726.
For DAB detection of GLUT-1 in NICH, slides were stained using a Ventana Benchmark Ultra automated stainer and an iView DAB detection kit according to the manufacturer’s protocols, with the following modifications: the sections were incubated in Cell Conditioner 1 for 64 minutes at 95°C and the primary antibody or negative control Ig was applied to the sections for 32 minutes at 37°C. Red blood cells served as internal positive controls for GLUT-1 detection.
Statistics.
In Figure 1C and Figure 4, ANOVA analysis and Tukey’s post-hoc analysis were conducted according to the NIST Engineering Statistics Handbook (68), using Microsoft Excel 2007 and a commercially available online calculator. P values of less than or equal to 0.05 (ANOVA) were considered significant and prompted subsequent subgroup analysis with Tukey’s test. For Figure 1C, groups with n ≥ 3 were used for comparison.
Study approval.
All human subjects or their guardians provided written informed consent giving permission for the use of their photographs and biologic specimens collected for this study according to institutional review board protocols (SCH IRB no. 11458).
Author contributions
GMS assisted in surgically harvesting tumors, performed qRT-PCR and flow cytometry experiments, and wrote the manuscript. ALK interpreted the array results; performed the ISH, immunostaining, and microscopy; added significant content to the discussion on miRNA target analysis, and GLUT-1 regulation; and edited the manuscript. MEW performed the microarray clustering and statistical analysis and interpreted the results. WPK performed the microarray experiment. RBK and RPK performed qRT-PCR experiments and counting of GLUT-1+ cells. JAP surgically harvested tumors, obtained plasma samples, and recruited participants. JAP and MWM conceived the study. All authors contributed to experimental design.
Supplementary Material
Acknowledgments
We thank Robert Boucek for discussions; Scott C. Manning and Joseph S. Gruss for procurement of tissue samples; Robert Sidbury, Deepti Gupta, Ronna Smith, and Ashley Sapin for procurement of serial plasma samples; Joyce Bischoff and Huang Lan for provision of cell lines and flow cytometry protocols; Eden Palmer for assistance with figure preparation; Rose Goodrich-Jones for maintenance of the tissue databank and patient recruitment; the SCH Pathology Integrative Resource Core laboratory for storage and processing of tissue samples; and Ron Seifert for helpful discussions of microscopy. Confocal microscopy was supported in part by the Mike and Lynn Garvey Cell Imaging Lab at the University of Washington Institute for Stem Cell and Regenerative Medicine. This work was funded by grants from the American Society of Pediatric Otolaryngology (CORE Research grant 314486), the National Institutes of Health (HL-123650), institutional grants from the Seattle Children’s Research Institute Center for Clinical and Translational Research and Center for Developmental Biology and Regenerative Medicine, and the Funding Focus Award from Seattle Children’s Hospital Guild Association
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2016;1(14):e88856. doi:10.1172/jci.insight.88856.
Contributor Information
Andrew L. Kirsh, Email: andrew.kirsh@seattlechildrens.org.
Mark W. Majesky, Email: mwm84@uw.edu.
References
- 1.Enjolras O, Mulliken JB. The current management of vascular birthmarks. Pediatr Dermatol. 1993;10(4):311–313. doi: 10.1111/j.1525-1470.1993.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 2.Metry DW, et al. A prospective study of PHACE syndrome in infantile hemangiomas: demographic features, clinical findings, and complications. Am J Med Genet A. 2006;140(9):975–986. doi: 10.1002/ajmg.a.31189. [DOI] [PubMed] [Google Scholar]
- 3.Huang SA, et al. Severe hypothyroidism caused by type 3 iodothyronine deiodinase in infantile hemangiomas. N Engl J Med. 2000;343(3):185–189. doi: 10.1056/NEJM200007203430305. [DOI] [PubMed] [Google Scholar]
- 4.Frieden IJ, et al. Infantile hemangiomas: current knowledge, future directions. Proceedings of a research workshop on infantile hemangiomas, April 7-9, 2005, Bethesda, Maryland, USA. Pediatr Dermatol. 2005;22(5):383–406. doi: 10.1111/j.1525-1470.2005.00102.x. [DOI] [PubMed] [Google Scholar]
- 5.Kilcline C, Frieden IJ. Infantile hemangiomas: how common are they? A systematic review of the medical literature. Pediatr Dermatol. 2008;25(2):168–173. doi: 10.1111/j.1525-1470.2008.00626.x. [DOI] [PubMed] [Google Scholar]
- 6.North PE, et al. A unique microvascular phenotype shared by juvenile hemangiomas and human placenta. Arch Dermatol. 2001;137(5):559–570. [PubMed] [Google Scholar]
- 7.Léauté-Labrèze C, Dumas de la Roque E, Hubiche T, Boralevi F, Thambo JB, Taïeb A. Propranolol for severe hemangiomas of infancy. N Engl J Med. 2008;358(24):2649–2651. doi: 10.1056/NEJMc0708819. [DOI] [PubMed] [Google Scholar]
- 8.North PE, Waner M, Mizeracki A, Mihm MC. GLUT1: a newly discovered immunohistochemical marker for juvenile hemangiomas. Hum Pathol. 2000;31(1):11–22. doi: 10.1016/S0046-8177(00)80192-6. [DOI] [PubMed] [Google Scholar]
- 9.Harik SI, Kalaria RN, Andersson L, Lundahl P, Perry G. Immunocytochemical localization of the erythroid glucose transporter: abundance in tissues with barrier functions. J Neurosci. 1990;10(12):3862–3872. doi: 10.1523/JNEUROSCI.10-12-03862.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469(7330):336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 13.Flor I, et al. Abundant expression and hemimethylation of C19MC in cell cultures from placenta-derived stromal cells. Biochem Biophys Res Commun. 2012;422(3):411–416. doi: 10.1016/j.bbrc.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Bentwich I, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37(7):766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 15.Noguer-Dance M, et al. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet. 2010;19(18):3566–3582. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- 16.Wu S, et al. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3’ untranslated region. Oncogene. 2010;29(15):2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- 17.Fornari F, et al. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol. 2012;227(3):275–285. doi: 10.1002/path.3995. [DOI] [PubMed] [Google Scholar]
- 18.Kameswaran V, et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014;19(1):135–145. doi: 10.1016/j.cmet.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haecker I, et al. Ago HITS-CLIP expands understanding of Kaposi’s sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog. 2012;8(8):e88856. doi: 10.1371/journal.ppat.1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vlachos IS, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43(Database issue):D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15(2):261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsson E, et al. Discovery of microvascular miRNAs using public gene expression data: miR-145 is expressed in pericytes and is a regulator of Fli1. Genome Med. 2009;1(11):e88856. doi: 10.1186/gm108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo SS, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81(4):717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 24.Donker RB, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod. 2012;18(8):417–424. doi: 10.1093/molehr/gas013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manel N, Kim FJ, Kinet S, Taylor N, Sitbon M, Battini JL. The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell. 2003;115(4):449–459. doi: 10.1016/S0092-8674(03)00881-X. [DOI] [PubMed] [Google Scholar]
- 26.Aboutalebi A, Jessup CJ, North PE, Mihm MC. Histopathology of vascular anomalies. Facial Plast Surg. 2012;28(6):545–553. doi: 10.1055/s-0032-1329929. [DOI] [PubMed] [Google Scholar]
- 27.Huang L, Nakayama H, Klagsbrun M, Mulliken JB, Bischoff J. Glucose transporter 1-positive endothelial cells in infantile hemangioma exhibit features of facultative stem cells. Stem Cells. 2015;33(1):133–145. doi: 10.1002/stem.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113(1):25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 30.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 31.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, et al. Microvesicle-mediated transfer of microRNA-150 from monocytes to endothelial cells promotes angiogenesis. J Biol Chem. 2013;288(32):23586–23596. doi: 10.1074/jbc.M113.489302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada N, et al. Colorectal cancer cell-derived microvesicles containing microRNA-1246 promote angiogenesis by activating Smad 1/5/8 signaling elicited by PML down-regulation in endothelial cells. Biochim Biophys Acta. 2014;1839(11):1256–1272. doi: 10.1016/j.bbagrm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288(15):10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhuang G, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31(17):3513–3523. doi: 10.1038/emboj.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valeri N, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25(4):469–483. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nasser MW, et al. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283(48):33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Gururajan M, et al. miR-154* and miR-379 in the DLK1-DIO3 microRNA mega-cluster regulate epithelial to mesenchymal transition and bone metastasis of prostate cancer. Clin Cancer Res. 2014;20(24):6559–6569. doi: 10.1158/1078-0432.CCR-14-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66(15):7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 41.Weber JA, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Köberle V, et al. Differential stability of cell-free circulating microRNAs: implications for their utilization as biomarkers. PLoS One. 2013;8(9):e88856. doi: 10.1371/journal.pone.0075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 45.Stückrath I, Rack B, Janni W, Jäger B, Pantel K, Schwarzenbach H. Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget. 2015;6(15):13387–13401. doi: 10.18632/oncotarget.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toiyama Y, et al. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg. 2014;259(4):735–743. doi: 10.1097/SLA.0b013e3182a6909d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imaoka H, et al. Circulating microRNA-203 predicts metastases, early recurrence, and poor prognosis in human gastric cancer. Gastric Cancer. 2016;19(3):744–753. doi: 10.1007/s10120-015-0521-0. [DOI] [PubMed] [Google Scholar]
- 48.Phung TL, et al. Akt1 and akt3 exert opposing roles in the regulation of vascular tumor growth. Cancer Res. 2015;75(1):40–50. doi: 10.1158/0008-5472.CAN-13-2961. [DOI] [PubMed] [Google Scholar]
- 49.Barthel A, et al. Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J Biol Chem. 1999;274(29):20281–20286. doi: 10.1074/jbc.274.29.20281. [DOI] [PubMed] [Google Scholar]
- 50.Taha C, Liu Z, Jin J, Al-Hasani H, Sonenberg N, Klip A. Opposite translational control of GLUT1 and GLUT4 glucose transporter mRNAs in response to insulin. Role of mammalian target of rapamycin, protein kinase b, and phosphatidylinositol 3-kinase in GLUT1 mRNA translation. J Biol Chem. 1999;274(46):33085–33091. doi: 10.1074/jbc.274.46.33085. [DOI] [PubMed] [Google Scholar]
- 51.Frauwirth KA, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16(6):769–777. doi: 10.1016/S1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 52.Morani F, et al. PTEN regulates plasma membrane expression of glucose transporter 1 and glucose uptake in thyroid cancer cells. J Mol Endocrinol. 2014;53(2):247–258. doi: 10.1530/JME-14-0118. [DOI] [PubMed] [Google Scholar]
- 53.Kleinman CL, et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet. 2014;46(1):39–44. doi: 10.1038/ng.2849. [DOI] [PubMed] [Google Scholar]
- 54.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapur RP, Berry JE, Tsuchiya KD, Opheim KE. Activation of the chromosome 19q microRNA cluster in sporadic and androgenetic-biparental mosaicism-associated hepatic mesenchymal hamartoma. Pediatr Dev Pathol. 2014;17(2):75–84. doi: 10.2350/13-12-1415-OA.1. [DOI] [PubMed] [Google Scholar]
- 56.Keller RB, Demellawy DE, Quaglia A, Finegold M, Kapur RP. Methylation status of the chromosome arm 19q MicroRNA cluster in sporadic and androgenetic-Biparental mosaicism-associated hepatic mesenchymal hamartoma. Pediatr Dev Pathol. 2015;18(3):218–227. doi: 10.2350/15-01-1600-OA.1. [DOI] [PubMed] [Google Scholar]
- 57.Delorme-Axford E, et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci U S A. 2013;110(29):12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu Y, et al. Mesenchymal stem cells and adipogenesis in hemangioma involution. Stem Cells. 2006;24(6):1605–1612. doi: 10.1634/stemcells.2005-0298. [DOI] [PubMed] [Google Scholar]
- 59.Zhu W, et al. MicroRNA expression analysis: clinical advantage of propranolol reveals key microRNAs in myocardial infarction. PLoS One. 2011;6(2):e88856. doi: 10.1371/journal.pone.0014736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.North PE, Waner M, Brodsky MC. Are infantile hemangiomas of placental origin? Ophthalmology. 2002;109(4):633–634. doi: 10.1016/S0161-6420(02)01071-0. [DOI] [PubMed] [Google Scholar]
- 61.Barnés CM, et al. Evidence by molecular profiling for a placental origin of infantile hemangioma. Proc Natl Acad Sci U S A. 2005;102(52):19097–19102. doi: 10.1073/pnas.0509579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Munden A, et al. Prospective study of infantile haemangiomas: incidence, clinical characteristics and association with placental anomalies. Br J Dermatol. 2014;170(4):907–913. doi: 10.1111/bjd.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bauland CG, Smit JM, Bartelink LR, Zondervan HA, Spauwen PH. Hemangioma in the newborn: increased incidence after chorionic villus sampling. Prenat Diagn. 2010;30(10):913–917. doi: 10.1002/pd.2562. [DOI] [PubMed] [Google Scholar]
- 64.Khan ZA, et al. Multipotential stem cells recapitulate human infantile hemangioma in immunodeficient mice. J Clin Invest. 2008;118(7):2592–2599. doi: 10.1172/JCI33493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laurent LC, et al. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26(6):1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 66.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 67.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 68. NIST/SEMATECH e-Handbook of Statistical Methods. National Institute of Standards and Technology. http://www.itl.nist.gov/div898/handbook Accessed August 29, 2016.
- 69.Gagioti S, Scavone C, Bevilacqua E. Participation of the mouse implanting trophoblast in nitric oxide production during pregnancy. Biol Reprod. 2000;62(2):260–268. doi: 10.1095/biolreprod62.2.260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.