Abstract
Recent advances in the field of biodosimetry have shown that the response of biological systems to ionizing radiation is complex and depends on the type and dose of radiation, the tissue(s) exposed, and the time lapsed after exposure. The biological effects of low dose radiation on learning and memory are not well understood. An ion mobility-enhanced data-independent acquisition (MSE) approach in conjunction with the IsoQuant software tool was utilized for label-free quantification of hippocampal proteins with the goal of determining protein alteration associated with low-dose whole body ionizing radiation (X-rays, 1 Gy) of 5.5-month-old male C57BL/6J mice post contextual fear conditioning training. Global proteome analysis revealed deregulation of 73 proteins (out 399 proteins). Deregulated proteins indicated adverse effects of irradiation on myelination and perturbation of energy metabolism pathways involving a shift from the TCA cycle to glutamate oxidation. Our findings also indicate that proteins associated with synaptic activity, including vesicle recycling and neurotransmission, were altered in the irradiated mice. The elevated LTP and decreased LTD suggest improved synaptic transmission and enhanced efficiency of neurotransmitter release which would be consistent with the observed comparable contextual fear memory performance of the mice following post-training whole body or sham-irradiation.
Keywords: radiation, hippocampus, label-free quantification, synaptic activity, TCA, myelination
Graphical Abstract
1. INTRODUCTION
The increasing clinical and public health awareness concerning the health effects of ionizing radiation continues to stimulate studies for determining the impact of ionizing radiation on biological systems. Recent advancements in diverse omics technologies revealed the complexity with which biological systems respond to at cellular, organelle and tissue level to different radiation exposure scenarios, including type and dose of radiation and time elapsed after exposure [1; 2].
Much radiation research has focused on high-dose exposure and associated tissue injury including DNA damage and repair mechanisms. High dose ionizing radiation causes water radiolysis and elevated levels of free radicals and reactive oxygen species (ROS) leading to DNA double strand breakage, and oxidative modifications of DNA, proteins and lipids [3]. Many of the ionizing radiation studies have shown that high dose radiation exposure of brain causes loss of cognitive function and memory [4; 5; 6; 7; 8]. Neurotransmission networks are highly susceptible and sensitive to irradiation [9; 10; 11]. Oligodendrocyte myelination mediates neural plasticity by optimizing the performance of the circuitry [12]. High-dose radiation results in demyelination caused by death of oligodendrocytes [13; 14]. Persistent demyelination reduces the reliability of neurotransmission [15]. Alteration of synaptic plasticity was observed accompanied by myelination deficits [16]. We and other have shown that radiation exposure causes impairment of hippocampal neurogenesis and cognitive deficits [9; 17; 18].
However, the biological effects of low dose radiation have been less studied and the mechanistic consequences are less well understood [19]. For instance, recent studies indicate that low dose (ranging from 0.1 Gy to 0.5 Gy) gamma radiation causes deregulation of mitochondrial and synaptic pathways in murine hippocampus and cortex [9]. However, there is also increasing evidence that low dosage exposure to radiation may elicit certain adaptive responses and resilience including the stimulation of neural stem cell proliferation, the neurogenesis of hippocampus and animal learning [20; 21; 22; 23].
The effects of radiation on hippocampus-dependent memory and learning are not well understood. We and others have shown that the hippocampus is highly sensitive to radiation. Radiation exposure impedes neurogenesis (X-rays, 5 Gy) [24], disrupts pathways essentials for the development of functional dendritic structures (X-rays, 1 Gy) [17] in the hippocampus and synaptic signaling pathways (gamma, 1 Gy) [9]. Exposure to low levels of ionizing radiation causes impairment of hippocampal-dependent functions of learning and memory (56Fe-particle, 1 Gy) [25]. In this context, contextual fear conditioning is frequently used to assess hippocampus-dependent memory [26]. Advantages of contextual fear conditioning include the ease with which rodents can be trained on this test and the use of translational fear conditioning tests in humans [27]. Most radiation studies involve irradiation exposure prior to fear learning. However, we recently reported that post-training whole body irradiation (28Si, 1 Gy) of 9-week-old mice showed effects on synaptic plasticity and unexpectedly increased hippocampus-dependent contextual freezing levels. The molecular mechanisms associated with these functional enhancements remain unclear [28].
This study is an extension of this previous work with the aim to develop and apply a quantitative proteomics workflow that would allow determining the protein networks and pathways that are responsive to low dose (1 Gy) ionizing radiation post contextual fear conditioning training. For this purpose we utilized and evaluated a label-free comparative proteomics approach utilizing a data-independent acquisition (DIA) mode, namely the MSE mode available on Waters Q-TOF instruments [29]. The MSE acquisition technique was chosen over the data-dependent acquisition (DDA) mode due to its superiority in providing accurate and precise quantification of precursor ion signals resulting in an improved dynamic range for label-free proteome quantifications [30]. In the MSE acquisition mode the mass spectrometer alters between a low energy (MS) acquisition mode, which provides exact mass information and ion signal for the precursor ions, and an elevated energy (E) mode of acquisition, which functions as a data-independent fragmentation approach yielding accurate mass and intensity information for fragment ions [29]. In the current study ion mobility separations of the precursor ions was combined with MSE to increase overall peak capacity which ultimately yields higher numbers of peptide/protein identifications [31]. The ion mobility-enhanced MSE workflow in conjunction with the IsoQuant software tool [32] allowed determining estimates of protein levels and enabled determination of relative subtle changes in the composition of the hippocampal proteome of the C57/BL6 mice 24 hr after low-dose radiation (1Gy, X-ray) post contextual fear conditioning training. Proteins that showed protein abundance changes between the sham-radiated and the X-ray radiated mice were functionally annotated. The findings of this study provide insights into the mechanisms of response and injury of the hippocampal proteome to low-dose radiation post contextual fear conditioning training.
2. MATERIALS AND METHODS
2.1. Animals
Five and a halve-month-old male C57BL6/J wild-type mice (n =10) purchased from the Jackson Laboratory (Bar Harbor, ME) were used for this study. The mice were housed under a constant 12 h light: 12 h dark cycle. Food (PicoLab Rodent Diet 20, no. 5053; PMI Nutrition International, St. Louis, MO) and water were provided ad libitum. All procedures were approved by Institutional Animal Care and Use Committee at the Oregon Health & Science University (Portland, Oregon).
2.2. Contextual fear conditioning, irradiation of animals and recall testing of conditioned fear
The mice were cognitively trained in a contextual fear conditioning paradigm, involving a five-shock paradigm, consisting of 2 s 0.7 mA shocks, separated by 2 min inter-shock-intervals (ISI), with the first shock at 118 s from the beginning of the trial. The total length of the training session was 10 min. Two hours after training, all mice were brought to a room within the animal facility containing an X-ray irradiator (Rad Source RS2000 Biological Research Irradiator, Suwanee, GA). Half of the mice (n = 5) were placed in a new mouse cage fitting in the irradiator and received whole body irradiation at a dose of 1 Gy (dose rate: 1.25 Gy/min). This dose of irradiation was selected as it affected fear memory in 4-week-old mice (unpublished observations). The other half of the mice (n = 5) were placed in a new mouse cage and received a sham-irradiation procedure by being placed into the new cage for the same duration of time. The mice were assigned to the experimental group (irradiated or sham-irradiated) by repeated random sorting until all initial variables were equal between the groups. After fear-conditioning training, and prior to irradiation, mice were randomly sorted until all initial values (body-weight, baseline-freezing, freezing levels after acquisition, etc.) were not significantly different between groups. The next day, or 24 h after irradiation, the mice were tested for recall of conditioned fear. All freezing data were analyzed using Med Associates software, as previously described [33]. The software analyzes freezing based on a proprietary algorithm scoring with freezing defined as no movement except respiration. Two hours after the contextual test, the mice were sacrificed by cervical dislocation. The hippocampus was dissected and stored on dry ice till further processing.
2.3. Chemicals
Lysis buffer (7M urea, 2M thiourea, 4% CHAPS, 70mM DTT and protease inhibitor cocktail), dithiothreitol (DTT), SDS, iodoacetamide, ammonium bicarbonate were obtained from Sigma (St Louis, MO). Sequencing-grade trypsin and protease MAX solution were purchased from Promega (Madison, WI). Protease inhibitors cocktails were purchased from Roche (Mannheim, Germany). Coomassie (Bradford) protein assay kit was obtained from Pierce (Rockford, IL). [Glu1]-fibrinopeptide B ([Glu1]-Fib) and Saccharomyces cerevisiae enolase digest were obtained from Waters (Milford, MA). All solutions were prepared using MS-grade water from J.T Baker (Center Valley, PA).
2.4. Sample preparation
Each sample was prepared from the hippocampus of the left brain hemisphere of each mouse. For the proteomics study, hippocampal tissues were sonicated in the lysis buffer using 5 bursts of 7 seconds; each burst was followed by a 30s-suspension in ice. The hippocampal tissue preparations were centrifuged for 30 min at 14,000 g to obtain soluble fractions. Lysis buffers were supplemented with a protease inhibitor cocktail to prevent protein degradation by endogenous proteases. All steps were performed either at 4°C or on ice. Total protein amounts in the soluble fractions were determined using the Bradford assay.[34] Proteins were reduced, alkylated and trypsinized at 1:25 (v/w) ratio for 18 h according to the manufacturer’s protocol (Promega). The reactions were stopped by adding 0.5% trifluoroacetic acid and dried under vacuum. Prior to LC-MS analysis, peptides were dissolved in 3% acetonitrile and 0.1% formic acid, and spiked with an enolase internal standard[35] to minimize technical variability originating from LC injection, spray stability, ionization efficiency, sample matrix and other factors. For each LC-MS analysis, a 1 μL LC injection was performed, resulting in the loading of 1 μg of protein and 100 fmol of enolase onto the column.
2.4. Liquid chromatography–mass spectrometry analysis
Tryptic peptides were analyzed in triplicate using a nanoAcquity UPLC coupled to a Synapt G2 HDMS mass spectrometer (Waters, Milford, MA). Peptides were separated on a BEH 130 C18 100 μm × 100 mm column with a particle size of 1.7 μm (Waters). Water containing 0.1% (v/v) formic acid was used as the mobile phase A and acetonitrile (ACN) containing 0.1% (v/v) formic acid was used as the mobile phase B. Peptides were eluted with a gradient of 3% – 40% mobile phase B over 30 min at a flow rate of 400 nL/min. The composition of B increased up to 90% in another 1 min and remained so for 5 min. Further, the composition of B solution reduced to 3% in 1 min and was maintained so for the last 20 min of the analysis.
The eluting peptides were analyzed in the positive ionization mode using the data-independent MSE mode in combination with ion-mobility separation (a.k.a. ion-mobility enhanced MSE, also referred as high definition mass spectrometry (HDMSE)). The capillary voltage was set at 3.0 kV, and the source temperature was set at 100 °C. The instrument was tuned for resolution of 18,000 and instrument settings are listed in Table S1. Mass spectra were acquired in the MSE mode alternating between a low energy scan (6 eV) to obtain peptide precursor information and a high energy scan (ramping from 27 to 45 eV) to acquire fragment ion information. Scan time was set at 0.9 s. The data were post-acquisition lockmass-corrected based on the doubly charged ion of [Glu1]-Fib ([M+2H]2+, m/z 785.8426). For lockmass acquisition, a [Glu1]-Fib solution of 100 fmol/μL at a flow rate of 5 μL/min was infused and a low-energy scan was acquired every 60 s throughout a run. External calibration of the TOF analyzer was performed using NaI solution over the range of m/z 50 to 2000.
2.5. Processing of label-free quantitative proteomic data
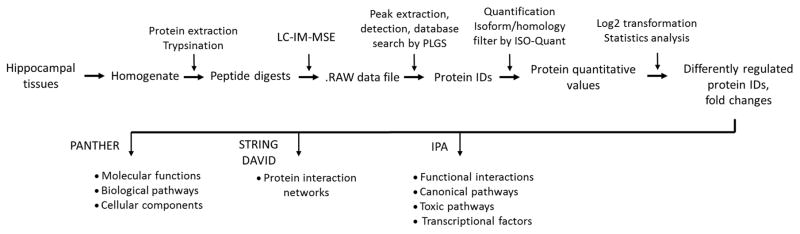
The data analysis strategy is outlined in Figure 1. Ion mobility-enhanced MSE data were processed using the Protein Lynx Global SERVER version 2.5.2 (Waters). The following processing parameters and their respective settings were used: chromatographic peak width and MS TOF resolution, automatic; low-energy threshold, 100 counts; elevated energy threshold, 10 counts; intensity threshold, 750 counts. The following search criteria were set for peptide identification: (i) trypsin as digestion enzyme, (ii) variable carbamidomethylcysteine and methionine oxidation as modifications, and (iii) minimum three identified fragment ions. The “OK filter” was used to obtain high confidence identification results.
Figure 1.
Workflow and data analysis strategy employed in the label-free proteomics study to determine differentially regulated proteins in the sham vs. irradiated group.
Hippocampal protein extracts were trypsinized and subjected to nanoUPLC and analyzed by IMS-enhanced MSE. MassLynx in conjunction with PLGS 2.5 was used for peak extraction, detection and database search; ISO-Quant for protein quantification and for isoform/homology filtering; Perseus for statistical analysis; and other bioinformatics tools for protein interaction analysis and network construction.
Data were post-processed using the software package ISO-Quant by performing retention-time alignment, peak clustering and annotation, peptide FDR filter, protein isoform/homology filter, peak intensity normalization, peptide/protein quantification, and protein FDR filter [32]. Within ISO-Quant, the cluster annotation of the peptide-level FDR was set to 1%, and only proteins identified by at least two peptides (minimum length: six amino acids) were used resulting in an 0.27% FDR on protein levels for all datasets [36]. In the final dataset, only proteins identified in at least two out of three technical replicates as well as three out of five biological replicates were included.
2.6. Western blotting analysis
The hippocampal protein extract was used for SDS PAGE and subsequent Western blotting. Twenty-five microgram of protein were separated on SDS-PAGE (Bis-Tris-Plus, 4–12%) gels and transferred to a PVDF membrane. Membrane blots were blocked in Tris-buffered saline containing 0.1% Tween 20 (TBST) and 5% bovine serum albumin (BSA). The membrane was incubated with primary antibody (CaMKII, Life Technologies, #A14012 and MBP Millipore, #AB980) diluted in 1% BSA-TBST (1:1000) solution overnight at 4°C. The blots were washed and incubated with a donkey anti-rabbit secondary antibody (1:10,000, Santa Cruz, # sc-2004). All membranes were next incubated in ECL reagent according to the manufacturer’s instructions (Thermo Scientific, #32106) and exposed to autoradiography film. Gel and blot images were quantified using the Image J quantification software (National Institutes of Health). Western blotting analyses were conducted using three biological replicates.
2.7. Bioinformatics analysis
To construct the hippocampal proteome network, protein IDs were uploaded to STRING, Search Tool for the Retrieval of Interacting Genes (http://www.string-db.org) [37]. The protein interaction score were extracted and re-visualized by the free open-source platform Cytoscape 3.2.1 (http://www.cytoscape.org/) [38]. The dataset were exported to Perseus (Version 1.4.1.3) and transformed to the logarithmic scale (log2) for statistical analysis. Log transformation is the most popular method used to transform skewed data to normality, increasing the validity of the associated statistical analysis [39]. The regulation of proteins with p value <0.05 were considered as significantly different in this dataset. The gene ontology of the deregulated proteins was analyzed using the PANTHER bioinformatics tools (http://www.pantherdb.org) for functional annotation. A detailed analysis of the functional interaction and biological pathway was performed using the Ingenuity pathway analysis (IPA) software (http://www.ingenuity.com) [40].
3. RESULTS
3.1. Cognitive performance
Figure 2 shows the freezing levels of the sham-irradiated and irradiated mice during the contextual fear memory test. There was no effect of irradiation (2-tailed t-test; p = 0.8606, t = 0.1813) and the freezing levels were comparable in the two groups.
Figure 2.

Comparable contextual fear memory in 5.5-month-old mice one day following post-training sham-irradiation or whole body X-ray irradiation (1 Gy). N = 5 mice/group.
3.2. Radiation-induced changes in the hippocampal proteome
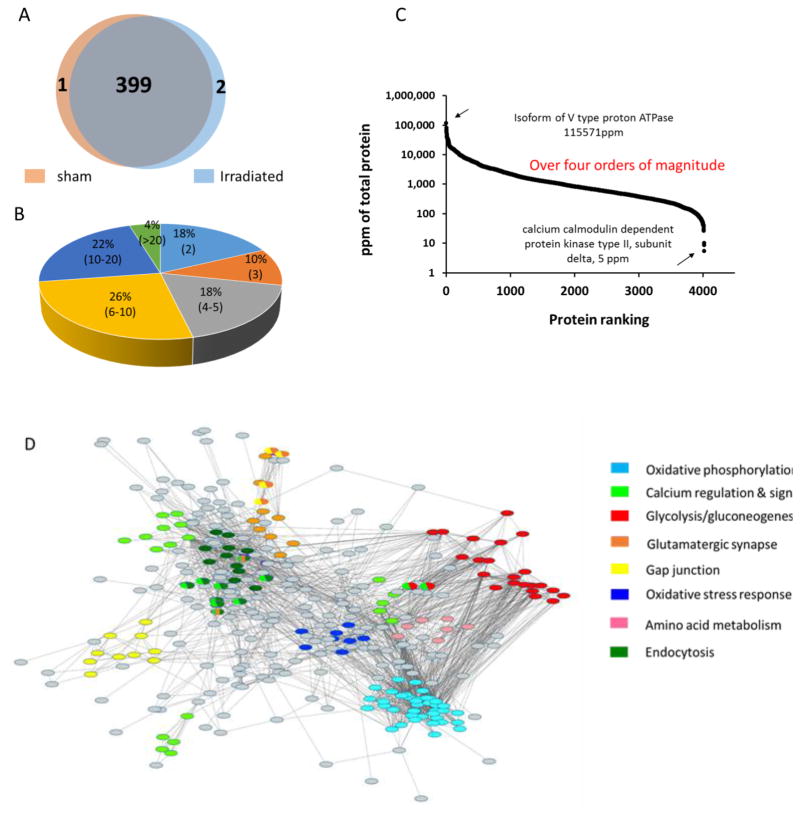
We used a label-free quantitative strategy to investigate the radiation-induced changes in the hippocampal proteome of mice 24 h post exposure to ionizing radiation (1 Gy) applied after training for contextual fear conditioning. Hippocampal tissues from five individual mice were used as biological replicates to account for biological variance. As shown in the Venn diagram (Figure 3A), 400 and 401 proteins were identified in the sham and irradiated group, respectively. There was excellent overlap of proteins between the groups with 399 proteins identified in both groups. We evaluated the overall quality of the label-free proteomics datasets. After log2-transformation of the intensity of each protein, the coefficient of variation (CV) of each protein was calculate and plotted (Figure S1A). The median values of the technical coefficient of variation (CV) obtained from the sham and irradiated groups of all quantified proteins were 0.99%. In comparison, the median CV values for biological replicates were 1.6% and 1.3% for the sham and irradiated group, respectively (Figure S1B). The majority of proteins (over 70%) were identified by at least four peptides, 25% by 10 or more (Figure 3B). The dynamic range of identified proteins in the two groups spanned over four orders of magnitude. The protein with the highest abundance was an isoform of V-type proton ATPase with 115,571 ppm of all the proteins. The protein with the lowest abundance was Ca2+/calmodulin dependent protein kinase type II (CaMKII), subunit delta, with 5 ppm of all proteins (Figure 3C).
Figure 3.
(A) Venn Diagram of the number of hippocampal proteins identified in both groups, sham (buff color) vs. irradiated (light blue color). (B) Pie chart of percentage of proteins according to numbers of peptides used for protein identification (range of numbers of peptides in brackets). (C) Dynamic range of detected mouse hippocampal proteins. The dynamic range of identified proteins across all fractions spanned over four orders of magnitude. (D) Protein network compiling proteins that constitute major functional subgroups of the hippocampal proteome. Nodes with the same color are grouped into one subgroup. Nodes with shared colors are defined as belonging to two or more subgroups. The gray nodes are proteins that did not fall into any of the major subgroups. The proteins that compose the functional subgroups are listed in Table S2.
We used the STRING database to construct a protein interaction network to functionally organize the proteins identified in the proteomics dataset (Figure 3D). The major functional subgroups are highlighted by different colors. The top three subgroups comprised proteins with functions in glycolysis/gluconeogenesis, oxidative stress, and calcium regulation and signaling. Some proteins were shared by two or three groups. An example is mitogen-activated protein kinase (MAPK), which was the hub protein of calcium regulation and signaling, endocytosis and synapse synthesis. Table S2 shows a compilation of the subset of proteins and their functional categorizations.
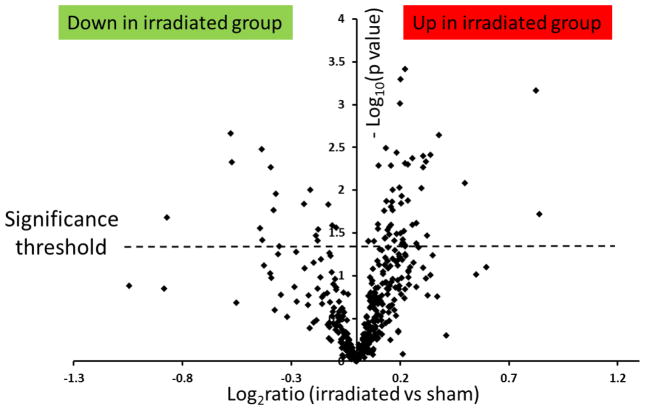
To investigate the proteome changes caused by irradiation, the fold changes of the protein abundance estimates of the irradiated group versus sham group were evaluated. ISOQuant quantification used annotated exact mass retention time (EMRT) tables and unique peptides for TOP3 quantification instead of only peptides identified directly by PLGS in the respective workflow. [32] Figure 4 shows the volcano plot for the distribution of p values versus log2(FC) calculated for the 399 proteins that were common to both groups. The y axis represents the negative logarithm (log10) of p values and the x axis represents the logarithm (log2) of fold changes. The dashed line shows the cut-off of 0.05 to define the significantly differential regulation of proteins between groups. Overall, 73 proteins (shown above the dashed line) were considered as significantly deregulated in irradiated tissues, as compared to sham-irradiated tissues. The differently regulated proteins are compiled in Table 1. The abundance estimates of 17 proteins (23%) were down-regulated and those of 56 (77%) proteins were up-regulated. The fold change distribution of the proteins quantified in this study ranged from 0.56 (down-regulated) to 1.7 (up-regulated).
Figure 4.
Volcano plot of protein abundance changes caused by irradiation using a label-free quantification approach.
The ratios of irradiated versus sham were calculated as log2(fold change) and shown on the x-axis. The y-axis represents the −log10(p-value). Proteins that fall above p <0.05 (above 1.3 on the volcano plot, dashed line) are considered as proteins with significant changes. Among them, 56 proteins were up-regulated and 17 proteins were down-regulated.
Table 1.
List of 73 proteins that showed significant abundance level changes in the sham vs. irradiated group.
| Accession number | Protein name | Gene name | Average (fmol) a | No of peptide b | Fold change c | P-value category d |
|---|---|---|---|---|---|---|
|
| ||||||
| Energy Metabolism | ||||||
|
| ||||||
| Q8BMF4 | Dihydrolipoyllysine residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | Dlat | 84 | 16 | 1.12 | ** |
| G3UVV4 | Hexokinase 1 isoform | Hk1 | 114 | 24 | 1.1 | * |
| Q99KI0 | Aconitate hydratase mitochondrial | Aco2 | 684 | 24 | 0.77 | * |
| A2AKV1 | ATP synthase subunit gamma mitochondrial | Atp5c1 | 129 | 6 | 1.04 | * |
| P08249 | Malate dehydrogenase mitochondrial | Mdh2 | 1046 | 10 | 1.15 | ** |
| P06745 | Glucose 6 phosphate isomerase | Gpi | 401 | 20 | 1.15 | *** |
| Q9D2G2 | Dihydrolipoyllysine residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | Dlst | 52 | 6 | 1.16 | * |
| Q91VA7 | Isocitrate dehydrogenase [NAD+] subunit mitochondrial | Idh3b | 109 | 11 | 0.91 | * |
| P35486 | Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial | Pdha1 | 149 | 5 | 1.07 | * |
| P05202 | Aspartate aminotransferase, mitochondrial | Got2 | 395 | 16 | 1.11 | ** |
|
| ||||||
| Calcium-dependent synaptic plasticity | ||||||
|
| ||||||
| Q9R0N5 | Synaptotagmin-5 | Syt5 | 419 | 7 | 1.12 | * |
| P28663 | Beta-soluble NSF attachment protein | Napb | 94 | 14 | 1.12 | * |
| P63044 | Vesicle-associated membrane protein 2 | Vamp2 | 475 | 4 | 1.19 | * |
| Q3TXX4 | Vesicular glutamate transporter 1 | Slc17a7 | 232 | 6 | 1.77 | *** |
| D3Z7R4 | Synaptotagmin-1 | Syt1 | 71 | 7 | 1.14 | ** |
| O88935 | Synapsin-1 | Syn1 | 580 | 23 | 1.15 | * |
| P63011 | Ras-related protein Rab-3A | Rab3a | 400 | 7 | 1.18 | * |
| P11798 | Calcium/calmodulin-dependent protein kinase type II subunit alpha | Camk2a | 1015 | 18 | 1.41 | ** |
| Q3UKW2 | Calmodulin | Calm1 | 1719 | 11 | 0.89 | * |
| P63328-2e | Isoform 2 of Serine threonine protein phosphatase 2B catalytic subunit alpha isoform (missing residue 448–457) | Ppp3ca | 288 | 12 | 1.21 | * |
| P68510 | 14-3-3 protein eta | Ywhah | 227 | 11 | 1.13 | * |
| P62259 | 14-3-3 protein epsilon | Ywhae | 554 | 16 | 0.79 | * |
| Q61644 | Protein kinase C and casein kinase substrate in neurons protein 1 | Pacsin1 | 137 | 7 | 1.09 | * |
| Q68FD5 | Clathrin heavy chain 1 | Cltc | 376 | 56 | 1.12 | * |
| Q3TWV4 | AP-2 complex subunit mu | Ap2m1 | 92 | 7 | 1.31 | ** |
| P39053 | Dynamin-1 | Dnm1 | 324 | 35 | 1.15 | * |
| A2ALV3 | Endophilin-A1 | Sh3gl2 | 157 | 10 | 0.76 | ** |
| E9Q9A3 | Clathrin coat assembly protein AP180 | Snap91 | 71 | 8 | 1.17 | * |
| Q99PT1 | Rho GDP-dissociation inhibitor 1 | Arhgdia | 105 | 4 | 0.94 | * |
|
| ||||||
| Myelination | ||||||
|
| ||||||
| P20917-2e | Isoform S-MAG (Myelin associated glycoprotein) | Mag | 73 | 6 | 0.67 | ** |
| P04370-14e | Myelin basic protein, isoform 13 | Mbp | 2793 | 11 | 0.88 | * |
| Q61885 | Myelin-oligodendrocyte glycoprotein | Mog | 92 | 4 | 0.74 | * |
| P16330-2e | Isoform CNPI of 2′,3′ cyclic nucleotide 3′ phosphodiesterase | Cnp | 557 | 20 | 0.77 | * |
| P60202 | Myelin proteolipid protein | Plp1 | 1739 | 6 | 0.74 | * |
|
| ||||||
| Others | ||||||
|
| ||||||
| Q9CQZ5 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 | Ndufa6 | 149 | 4 | 1.17 | *** |
| Q3TXX4 | Vesicular glutamate transporter 1 | Slc17a7 | 232 | 6 | 1.77 | *** |
| Q9D0K2 | Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial | Oxct1 | 96 | 8 | 1.15 | *** |
| Q3TWV4 | AP 2 complex subunit mu | Ap2m1 | 92 | 7 | 1.30 | ** |
| A1BN54 | Alpha actinin 1a | Actn1 | 35 | 8 | 1.26 | ** |
| E9PUL5 | Proline rich transmembrane protein 2 | Prrt2 | 67.19 | 3 | 1.19 | ** |
| P57780 | Alpha-actinin-4 | Actn4 | 114 | 12 | 1.25 | ** |
| P97300-3e | Isoform 3 of Neuroplastin | Nptn | 94 | 5 | 1.17 | ** |
| G5E829 | Plasma membrane calcium-transporting ATPase 1 | Atp2b1 | 157 | 19 | 1.18 | ** |
| Q8BH59 | Calcium binding mitochondrial carrier protein Aralar1 | Slc25a12 | 108 | 20 | 1.12 | ** |
| P16858 | Glyceraldehyde-3-phosphate dehydrogenase | Gapdh | 2872 | 13 | 1.07 | ** |
| P43006-3e | Isoform Glt 1B of Excitatory amino acid transporter 2 | Slc1a2 | 926 | 6 | 1.24 | ** |
| E9Q579 | Mitochondrial glutamate carrier 1 | Slc25a22 | 47 | 4 | 1.23 | ** |
| G3UYZ1 | Immunoglobulin superfamily member 8 | Igsf8 | 26 | 3 | 0.86 | ** |
| F6QPR1 | Prohibitin-2 (Fragment) | Phb2 | 34 | 3 | 1.10 | * |
| Q9Z1S5-2e | Isoform 2 of Neuronal-specific septin-3 | SEPT3 | 41 | 3 | 1.12 | * |
| P31786 | Acyl-CoA-binding protein | Dbi | 219 | 3 | 0.85 | * |
| Q9D172 | ES1 protein homolog, mitochondrial | D10Jhu81e | 101 | 3 | 1.11 | * |
| Q8VDN2 | Sodium/potassium-transporting ATPase subunit alpha-1 | Atp1a1 | 219 | 29 | 1.09 | * |
| P55258 | Ras-related protein Rab-8A | Rab8a | 72 | 7 | 1.79 | * |
| Q922F4 | Tubulin beta-6 chain | Tubb6 | 35 | 13 | 0.55 | * |
| Q9QYG0-2e | Isoform 2 of Protein NDRG2 | Ndrg2 | 120 | 6 | 0.92 | * |
| P05064 | Fructose-bisphosphate aldolase A | Aldoa | 1200 | 16 | 1.11 | * |
| P46660 | Alpha-internexin | Ina | 132 | 18 | 0.74 | * |
| E9Q453 | Tropomyosin alpha-1 chain | Tpm1 | 66 | 4 | 1.07 | * |
| P11499 | Heat shock protein HSP 90-beta | Hsp90ab1 | 211 | 19 | 1.10 | * |
| P35802 | Neuronal membrane glycoprotein M6-a | Gpm6a | 625 | 3 | 1.14 | * |
| P18872 | Guanine nucleotide-binding protein G(o) subunit alpha | Gnao1 | 611 | 10 | 1.11 | * |
| Q3UHL1 | CaM kinase-like vesicle associated protein | Camkv | 82 | 9 | 1.25 | * |
| P51881 | ADP/ATP translocase 2 | Slc25a5 | 227 | 11 | 1.16 | * |
| P08752 | Guanine nucleotide binding protein G(i) subunit alpha-2 | Gnai2 | 38 | 6 | 0.88 | * |
| G3UX26 | Voltage-dependent anion-selective channel protein 2 (Fragment) | Vdac2 | 187 | 7 | 1.06 | * |
| P00405 | Cytochrome c oxidase subunit 2 | Mtco2 | 445 | 4 | 1.16 | * |
| P59999 | Actin-related protein 2/3 complex subunit 4 | Arpc4 | 44 | 4 | 1.21 | * |
| P40124 | Adenylyl cyclase-associated protein 1 | Cap1 | 75 | 6 | 1.15 | * |
| D3YVA2 | Neurocalcin-delta (Fragment) | Ncald | 114 | 5 | 1.17 | * |
| Q6P1J1 | Crmp1 protein | Crmp1 | 132 | 11 | 1.22 | * |
| Q8BFR5-2e | Isoform 2 of Elongation factor Tu, mitochondrial | Tufm | 42 | 4 | 1.08 | * |
Abundance level estimate based on the use of 100 fmol yeast enolase digest as internal standard spiked in prior to LC-IMS MSE analysis
average number of peptides identified for each protein
fold change was calculated based on the intensity of same protein in irradiated group versus the sham group
* p-value<0.05, ** p-value<0.01, *** p-value<0.001
putative identification; isoforms produced by alternative splicing. Note of caution, non-detection of peptides does not allow assignment of distinct isoforms per se.
3.3. Protein pathways and networks perturbed by irradiation
The molecular function analysis revealed that, catalytic activity represented the largest group in both up-regulated and down-regulated proteins, while translation regulator activity and transporter activity were two groups exclusively found in up-regulated proteins. The proteins involved in these two groups were mostly carrier proteins and channel proteins. The myelin-associated protein Mag was exclusively defined as the “function of receptor activity” among the down-regulated proteins (Figure S2A). The biological process analysis revealed that metabolic processes and cellular processes were two dominant groups in both up-regulated proteins and down-regulated proteins (Figure S2B). Cellular compartment analysis showed that most up-regulated proteins were associated with the mitochondrial and plasma membranes, while the down-regulated proteins were associated with the cytosol and cytoskeleton (data not shown). The proteins classified by molecular function and biological processes are compiled in Table S3.
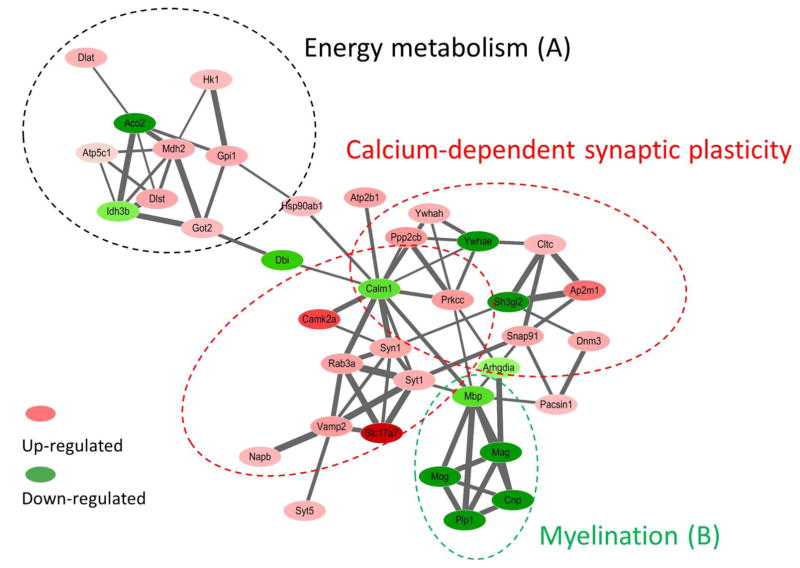
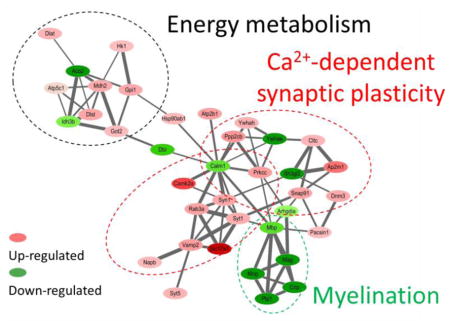
In order to better understand the functional consequences of radiation exposure on the hippocampal biology, we evaluated to which protein networks the proteins that displayed deregulation were associated with. The analysis of the protein interactions showed three distinct but interconnected clusters of proteins, that were named based on the annotation of the proteins and prior citations in the literature (Figure 5): (1) energy metabolism (mainly tricarboxylic acid cycle (TCA) cycle), (2) calcium-dependent synaptic plasticity, and (3) myelination. The accession numbers, gene names and corresponding fold changes are listed in Table 1.
Figure 5.
The STRING protein network shows three distinct but interconnected clusters of proteins that are associated with (a) energy metabolism, (B) calcium-dependent synaptic plasticity, and (C) myelination. Red/pink nodes represent proteins that increased their amounts as a consequence of radiation exposure, dark/light green represents those that decreased. The intensity of a node is proportional to the fold change between conditions (irradiated versus sham). The width of a line connecting proteins represents the strength of the proteins interaction, as extracted from STRING software.
To gain a better understanding of the signaling pathways and networks affected by irradiation, we used the IPA software to analyze the deregulated proteins. The analysis revealed that the networks “molecular transport,” “cell-to-cell signaling and interaction,” and “cellular assembly and organization” were the most significantly affected networks after irradiation (Figure S3, Table S6). The IPA analysis also showed that the canonical pathways “TCA cycle”, “nNOS signaling in neurons”, and “endocytic pathways” were significantly affected after irradiation. Compilation of the top 20 canonical pathways of the significantly regulated proteins provided several overlapping and interconnected pathways that were mainly involved in metabolic activity and cellular signaling (Table S7). In addition, IPA also suggested that PPARα/RXRα activation, DNA checkpoint regulation, and mitochondrial dysfunction were the main targets of radiation-induced cellular changes (Table S8). Furthermore, the IPA predicted that the transcriptional factors MYRF and mTOR were inhibited by irradiation.
3.4. Western blotting analysis
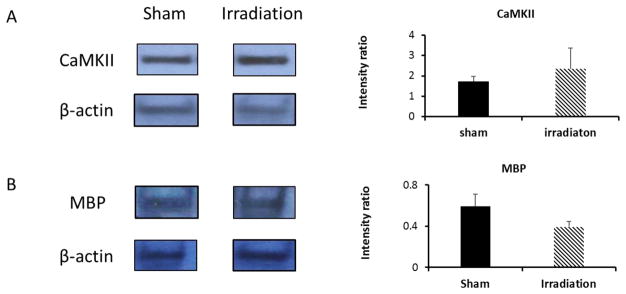
Western blotting analysis was used to complement the mass spectrometry-derived protein abundance estimates. Calcium/calmodulin-dependent protein kinase II (CaMKII) and myelin basic protein (MBP) were selected as representatives for the subgroup of proteins reporting on Calcium-dependent synaptic plasticity and myelination (Table 1), respectively. Figure 6A shows a representative Western-blot of CaMKII and reference protein actin. The elevated levels of CaMKII in the irradiated group (FC 1.4, n=2) is consistent with the findings from the proteomic dataset (Table 1, FC 1.4, p < 0.01). Figure 6B shows a representative Western-blot of MBP using actin as reference protein. The decreased levels of MBP in the irradiated group (FC 0.7, n=3) indicates down-regulation of MBP and this result is close to the findings obtained from the proteomic dataset (Table 1, FC 0.88, p < 0.05).
Figure 6.
Western-blotting analysis of representative proteins identified by IMS-enhanced MSE.
Left panel: Western blotting analysis of CaMKII (A) and MBP (B) expression levels in the hippocampal tissues. Hippocampal protein lysate was loaded onto a SDS-PAGE gel and probed with antibody against CaMKII and MBP. β-actin was used as loading control and for data normalization; Right panel: bar chart of relative band intensity ratio of CaMKII versus reference protein (β-actin) and MBP versus reference. Average and standard deviation calculated from biological replicates (n=2 for CaMKII, n=3 for MBP).
4. DISCUSSION
4.1. Overall performance of the ion-mobility enhanced MSE-based label-free proteomics approach
The aim of this study was to develop, apply a label-free proteomics approach for determining hippocampal proteome changes as the consequences of low-level whole body irradiation (X-rays, 1 Gy) of mice following a hippocampus-dependent memory test. We used the hippocampal tissue of individual mice as biological replicate rather than pooling tissue of hippocampi to avoid the “dilution effect” whereby less abundant proteins are pushed under the detection limit [41]. We chose an ion-mobility enhanced DIA method, MSE, in conjunction with accurate mass high resolution mass spectrometry to take advantage of the quantification capability of the MSE method, which is particularly beneficial for determining abundance estimates for low abundant proteins [42]. In addition, because in the MSE acquisition mode precursor ions are subjected to fragmentation without selection bias, MSE provides more identification for less abundant proteins [43].
In a label-free quantitative proteomic study design the inclusion of technical and biological replicates is essential for the downstream data analysis. Technical replicates address the instrumental noise and error in the measurement with multiple measurements reducing uncertainty. Biological replicates address the biological noise from random sampling [41]. Multiple biological replicates reduce the biological variability thereby indicating the true effect of the treatment. High natural variation among biological individuals interfere with analyzing protein expression levels and evaluating the effects of treatment [44]. In this study, five biological replicates were available within each group and each sample was injected three times. The low median CV values (0.99%) in both the sham and irradiated groups of the technical replicates ensured the robustness of the mass spectrometric measurements and the data analysis workflow. The biological variance was as low as 1.61% for the sham group and 1.31% for the irradiated group. The biological replicates encompass the technical noise in this system. The low biological variance ensured a high accuracy (ability to detect changes in expression) in assessing differences between the two groups.
4.2. Hippocampal proteome alterations
In this dataset, the proteins that showed fold changes in abundance with p value <0.05 were considered as significantly different without consideration of large fold change cut-offs (i.e. larger than 1.5). Stringent fold change cut-offs might alter the data interpretation [45] especially for this dataset with small fold changes (ranging from 0.56 (down-regulated) to 1.7 (up-regulated)). Given the small biological variances in both groups, 1.61% and 1.31% for the sham and irradiated group, respectively, the fold changes were accurately evaluated even though the biological replicate size was moderate with 5 biological replicates per group. On contrary, the p values can be highly affected by the sample size, and we chose the cut-off at 0.05 as commonly used in the literature [46; 47].
Seventy-three proteins were identified as being significantly deregulated following 1 Gy of total body irradiation. The most significantly impacted protein pathway (i.e. with the lowest p value) was involved in energy metabolism, i.e. the tricarboxylic acid cycle (TCA cycle) with p=2.58E-07. This was consistent with previous reports that the TCA cycle and mitochondrial structure and function were affected after exposure to X-ray radiation at 1 Gy [9; 48]. Up to 20% of them were associated with calcium-dependent synaptic plasticity. More specifically, they were involved in synaptic vesicle recycling, long-term potentiation (LTP) and long-term depression (LTD). Multiple significant deregulated proteins indicated impairment of myelination. In addition, IPA predicted that PPARα/RXRα activation, DNA checkpoint regulation, and mitochondrial dysfunction were main targets of radiation-induced cellular changes (Table S7). Impairment of PPARα activity has been described previously although discussed in the context of pathological changes of the heart after high dose X-ray irradiation at 8 Gy [49].
4.2.1. Alteration of proteins associated with energy metabolism pathways
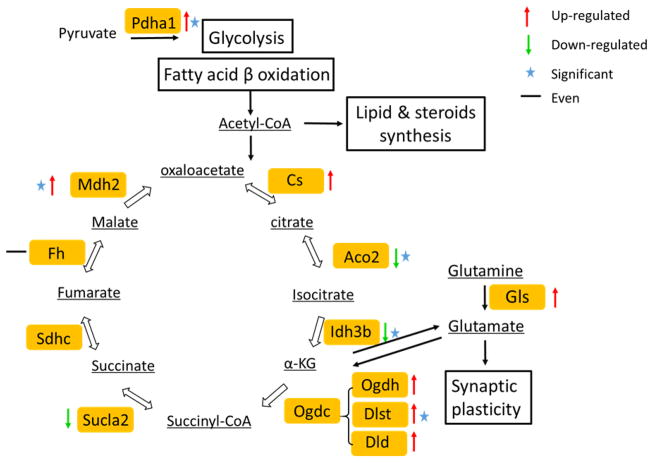
The two bioinformatics analysis tools (IPA and STRING) predicted that the TCA cycle was significantly affected following radiation exposure. The TCA cycle is composed of a set of eight enzymes involved in oxidation of acetyl-CoA, which can be generated through glycolysis and fatty acid β-oxidation. Figure 7 illustrates the TCA cycle and other interactive pathways. Four enzymes in this cycle, isocitrate dehydrogenase (Idh), aconitase (Aco2), dihydrolipoyllysine-residue succinyltransferase (Dlst) from the ketoglutarate dehydrogenase (Ogdc) complex, and malate dehydrogenase (Mdh1), showed significant protein level changes. The ratio of proteins with significant changes was up to 0.25 of all enzymes involved in TCA cycle. Idh generates the 5-carbon ketoglutarate (KG) from isocitrate by decarboxylation. This is the rate-limiting step in the TCA cycle, and as such, Idh is considered as playing an important role in the regulation of the TCA cycle and maintaining the 2-oxoglutarate level. The cycle is completed by reversible oxidation of malate to produce oxaloacetate (OAA) by Mdh. In vivo, the OAA is continuously removed by citrate synthase, coupled with the consumption of NADH by the respiratory chain, and this event pushes the equilibrium towards favoring malate oxidation [50]. Our data suggest that whole body irradiation (1 Gy) diminishes the TCA cycle efficiency, which in turn impacts energy supply to support cellular metabolism [51]. Consequently, limited energy supply may impose constraints on synaptic transmission and information processing within the hippocampus [52].
Figure 7.
Radiation-induced modulation of proteins associated with energy metabolism.
Enzymes associated with energy metabolism are highlighted in yellow boxes while metabolites are underlined. Proteins that showed level changes as a consequence of radiation exposure are annotated as follows: up-regulated (red arrow up), down-regulated (green arrow down), and even (black equal). Proteins that showed a statistically significant fold change are marked with an asterisk. Table S4 compiles the proteins (uniprot ID, gene name) that showed altered expression levels (fold change, p values) upon irradiation.
Interestingly, we observed increased abundances of glutaminase (Gls) which suggests that oxidation of glutamate is used as an alternative way of providing energy to satisfy intracellular ATP requirements [53]. Glutamate is an important substrate in hippocampus to support energy metabolism [53]. Glutamate is also critically involved as neurotransmitter in synaptic plasticity [54] and elevated levels of glutamate would be consistent with the proteome changes observed for synaptic plasticity as discussed below.
4.2.2. Alteration of Calcium-dependent synaptic plasticity
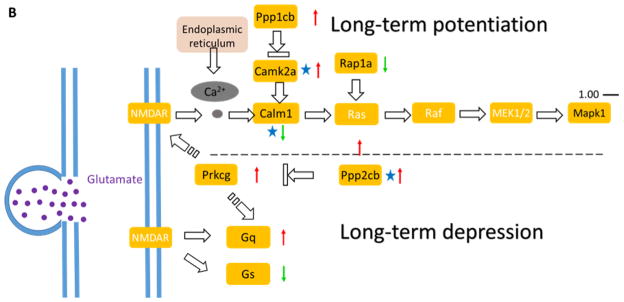
In this dataset, up to 20% of the proteins with significant changes identified here were involved in calcium (Ca2+)-dependent synaptic plasticity. Synaptic plasticity is important for storing and retrieving information in many brain regions, including the hippocampus, striatum and cerebellum [55]. Synaptic plasticity modulates the robustness and flexibility of neuronal networks [56]. Synaptic plasticity involves synaptic vesicle (SV) recycling (Figure 8A), LTP and LTD (Figure 8B).
Figure 8.
Radiation-induced modulation of proteins associated with synaptic plasticity regulated by calcium (Ca2+).
Proteins higlighted showed changes in abundance as a consequence of radiation exposure. The pathway consists of synaptic vesicle (SV) cycle (A) and as well as LTP and LTD (B). The majority of proteins involved in SV cycle were up-regulated. Glutamate release mediates down-stream effects of LTP and LTD triggered by calcium. Protein level changes are shown as up-regulated (red arrow up), down-regulated (green arrow down), and even (black equal). Proteins that showed a statistically significant fold change are marked with an asterisk. Table S5 compiles the proteins (uniprot ID, gene name) that showed altered expression levels (fold change) upon irradiation.
The SV cycle consists of neurotransmitter loading (exocytosis), recovery (endocytosis), refilling, and release from the presynaptic bouton. Both exocytosis and endocytosis are strictly regulated, and fluctuation of the number of vesicles and re-filling of SV might affect the amount of neurotransmitter released [57]. Following radiation exposure, most of the proteins involved in synaptic vesicle (SV) recycling were up-regulated and up to three quarters of the proteins identified in this process were significantly changed (Figure 8A). The up-regulated proteins included the core SNARE complex proteins VAMP2, Syntaxin-1, and SNAP-25 as well as other proteins necessary for vesicle fusion such as NSF and the Rab family of GTPase [58]. VAMP2 facilitates pore formation and stabilizes fusion intermediates. An absence of VAPM2 leads to failure of vesicle fusion and a decrease in Ca2+-triggered fusion [59]. Syntaxin is a transmembrane protein that has the ability to form a stable complex between VAMP2 and SNAP-25 for physiological α-SNAP binding [60]. SNAP-25 is necessary for Ca2+-triggered exocytosis in neuronal cells by interacting with other regulatory proteins [61]. All three proteins were up-regulated indicating increased SV recycle dynamics.
Furthermore, several of the proteins necessary for the endocytosis of SV from the membrane were identified (Figure 8A). The clathrin-mediated endocytosis mechanism carries proteins into the cell in the form of vesicles coated on the outside with a clathrin [62]. Key proteins, besides clathrin, involved in this event, are the adaptor proteins AP2 and Dynamin-1. They are both necessary for SV invagination and fusion with the plasma membrane [63]. Our proteomic screen also identified proteins involved in vesicle fission/fusion such as the ARFs (ADP-ribosylation factors) and Rab proteins, including Rab3, which regulates a late step in synaptic vesicle fusion. ARFs are small GTPases regulating the assembly of clathrin coat complexes [64]. The multi-subunit vacuolar adenosine triphosphatase (V-ATPase) is critical for neurotransmitter release and acidification of the SV lumen [65]. The increased abundance of the proteins discussed above indicates an increase in the amount of neurotransmitter release from endocytosis resulting in LTP (Figure 8B) and this, in turn, might contribute to the comparable memory retention of the groups despite other profound effects of irradiation on the hippocampal proteome.
LTP refers to the long-lasting increase in synaptic strength in response to short periods of synapse’s elevated activity (Figure 8B). During this event, glutamate activates AMPA-type glutamate receptors (AMPARs) triggers a Ca2+-mediated signaling cascade. From our quantitative dataset it can be seen that the Ca2+/calmodulin-dependent protein kinase II (CaMKII) alpha was significantly upregulated in the irradiated group (up to 1.41 fold, Table 1) compared to the sham group. The increased abundance of CaMKII alpha was also confirmed by Western-blotting (Figure 6A). CaMKII has been shown to be essential for synaptic plasticity, synaptic organization, LTP as well as LTD [66]. Ca2+ triggers the autophosphorylation site of CaMKIIα at Thr286 and increases the affinity of Ca2+/CaM dramatically [67], which, in turn, results in autonomous CaMKIIα activity. The binding of CaMKII to the NMDAR increases binding of other proteins that produce the synapse enlargement in late LTP. Increased CAMKII activity is essential for the consolidation of long-term object recognition memory [68]. Genetic deletion of CaMKII leads to a significantly reduced LTP induction. In our recent work we showed that 28Si radiation induced cognitive changes and increased the magnitude of LTP in CA1 region of the hippocampus at low doses [28]. The increased levels of LTP improved synaptic transmission and corresponding hippocampal compensation. Our quantitative dataset proved enhanced efficacy of neurotransmitter release by up-regulation of all the proteins in the claritin-mediated endocytosis process as we discussed above. These findings are consistent and supportive of that enhanced neurotransmitter release may function as a compensatory mechanism for other radiation effects in concordance to that no memory impairment was observed in the current study.
The opposing process of LTP is LTD, which refers to a long-lasting decrease in the strength of synaptic transmission (Figure 8B). Inhibition of CaMKII has been shown to prevent the mGluR-mediated induction of protein synthesis [69]. Besides CaMKII, other phosphatases such as protein phosphatase 1 (PP1) and protein phosphatase 2B (PP2B, a.k.a. calcineurin) are also required for LTP and LTD. LTP induction is blocked in the presence of PP1/2A inhibitors [70]. In addition, a calcineurin inhibitor led to a complete rescue of LTD calcineurin inhibition. This may be investigated further as a neuroprotective treatment to stop or slow down synaptic alterations. The increased level of PP2B suggests elevated LTP and decreased LTD, indicating enhancement of memory acquisition and consolation. Calcium input activates both calcineurin and CaMKII at all frequencies, which is in good accordance to the dataset generated in this study [71].
The protein alterations observed for SV recycling, LTP and LTD may point to low radiation-induced enhancement of synaptic plasticity which would be supportive of our experimental finding that irradiation (X-rays, 1 Gy) had little impact on the cognitive performance of the adult mice despite indications of deregulation of other processes at the protein level.
4.2.3. Impairment of Myelination
Myelination dysfunction was indicated by another group of proteins that was significantly affected following radiation exposure (Table 1). Myelin is a biologically active membrane sheath formed by oligodendrocyte. It provides trophic and metabolic support to axons, assists energy-efficient salutatory conduction and maintains proper neuronal function [72]. Deposition or loss of the myelin sheath results in irreversible axonal degeneration and affects neuronal connectivity [73]. Defective myelination, due to either genetic or acquired factors, can lead to neurological diseases including multiple sclerosis [74], leukodystrophies [75], and peripheral neuropathies [76]. Exposure to high dose of X-ray radiation reduced the numbers of migratory neuroblasts, and significantly less brain myelination was observed as well [77].
In this study, the abundance estimates of four proteins related to myelination, namely myelin oligodendrocyte glycoprotein (MOG), myelin proteolipid protein (PLP), myelin associated glycoprotein (MAG), and myelin basic protein (MBP) were reduced. Ingenuity Pathway Analysis predicted that the myelin regulatory factor (MYRF) was inhibited after radiation exposure (Figure S4). MYRF has been recently identified as a membrane-associated transcription factor that specifically activates expression of four myelin genes (e.g. Mog, Plp1, Mag and Mbp) during oligodendrocyte (OL) maturation.[77]
MYRF is highly induced during oligodendrocyte differentiation and absent in other central nervous system cell types. Conditional ablation of MYRF in mature oligodendrocytes led to a rapid down-regulation of myelin gene expression and delayed demyelination.[78] The transcripts for the myelin genes, Mog, Plp1, Mag and Mbp, were also rapidly down-regulated after ablation, indicating the importance of MYRF in myelination.[79]. This is in good accordance with our proteomics findings. Down-regulation of MBP was described previously but at high doses of irradiation (≥10 Gy) [14; 80]. Also, a much higher radiation dose (22 Gy) was used to mimic delayed demyelination of the spinal cord following irradiation injury [14]. In our study, the down regulation of myelination was observed at a dose of as low as 1 Gy, providing proof for irradiation damage on myelination and these effects were seen at a relative early time point (24 hr post radiation) following exposure to a much lower dose of irradiation.
Other signaling pathways, including ERK1/2 MAPK [81] and Akt/mTOR [82], are associated with myelination regulation as well. Interesting, Akt/mTOR activated by oxidative stress [83] has the ability to disrupt the TCA cycle, which is consistent with our dataset as discussed above.
5. CONCLUSION
This study was designed to investigate the effects of post-training low dose irradiation on the hippocampal proteome. We utilized an IMS-enhanced MSE acquisition method for the generation of a label-free quantitative proteomic dataset to determine the protein abundance level changes in response to the low dose irradiation (1 Gy). The robust IMS-enhanced MSE acquisition method in conjunction with IsoQuant software resulted in an accurate quantitative proteomic dataset with low median CV values (< 1%) for the technical and for biological replicates ( < 2%), which ensured the accuracy of the proteomic findings even if subtle fold changes were observed. Overall, the study revealed deregulation of 73 proteins (out of 399 proteins).
The deregulated proteins indicated adverse effects of irradiation on myelination. Other observed protein level changes indicate perturbation of energy metabolism pathways possibly involving a shift from TCA cycle to glutamate utilization to cover ATP requirements. Notably, level estimates of proteins associated with synaptic activity including vesicle recycling and neurotransmission were altered in the irradiated mice. We confirmed these conclusions by validating the expression level changes of MBP from myelination pathway and CaMKII from synaptic network by western-blotting. Our data suggest that the elevated LTP and decreased LTD improved synaptic transmission and enhanced efficiency of neurotransmitter release which would be consistent with the observed comparable contextual fear memory performance of the mice following post-training whole body or sham-irradiation.
To conclude, our results underscore the importance of conducting low dose radiation experiments for illuminating the sensitivity of biochemical pathways to radiation, and the modulation of potential repair and compensatory response mechanisms. Our findings may also ultimately lead to the design of attenuating strategies for ameliorating hippocampal and CNS injury following radiation exposure as part of medical therapies or as a consequence of occupational circumstances.
The current study established a robust mass-spectrometry based quantitative proteomics workflow which is adaptable to other studies designed for determining the effects of various radiation exposures on hippocampus and possible other regions of the brain. This may include future studies to address the potential biological effects of low dose radiation exposure during early life stages in which brain maturation takes place and/or studies that may attempt to assess the therapeutic strategies to mitigate brain injuries caused by radiation exposure.
Supplementary Material
Significance.
This study is significant because the biological consequences of low dose radiation on learning and memory are complex and not yet well understood. We conducted a IMS-enhanced MSE-based label-free quantitative proteomic analysis of hippocampal tissue with the goal of determining protein alteration associated with low-dose whole body ionizing radiation (X-ray, 1 Gy) of 5.5-month-old male C57BL/6J mice post contextual fear conditioning training. The IMS-enhanced MSE approach in conjunction with IsoQuant software was robust and accurate with low median CV values of 0.99% for the technical replicates for samples from both the sham and irradiated groups. The biological variance was as low as 1.61% for the sham group and 1.31% for the irradiated group. The applied data genereation and processing workflow allowed the quantitative evaluation of 399 proteins. The current proteomic analysis indicates that myelination is sensitive to low dose radiation. The observed protein level changes imply modulation of energy metabolism pathways in the radiation exposed group, specifically changes in protein abundance levels suggest a shift from TCA cycle to glutamate oxidation to statisfy energy demands. Most significantly, our study reveals deregulation of proteins involved in processes that govern synaptic activity including enhanced synaptic vesicle cycling, and altered long-term potentiation (LTP) and depression (LTD). An elevated LTP and decreased LTD suggest improved synaptic transmission and enhanced efficiency of neurotransmitter release which is consistent with the observed comparable contextual fear memory performance of the mice following post-training whole body or sham-irradiation. Overall, our results underscore the importance of low dose radiation experiments for illuminating the sensitivity of biochemical pathways to radiation, and the modulation of potential repair and compensatory response mechanisms. This kind of studies and associated findings may ultimately lead to the design of strategies for ameliorating hippocampal and CNS injury following radiation exposure as part of medical therapies or as a consequence of occupational hazards.
HIGHLIGHTS.
IMS-MSE-based proteomics reveals deregulation of 73 hippocampal proteins
Impairment of myelination is discovered even at low dose ionizing radiation (IR);
Deregulation of energy metabolism pathways is observed;
Pathways related to synaptic activity and vesicle recycling are enhanced;
Changes are consistent with comparable memory performance of irradiated mice;
Acknowledgments
This work was supported in part by NIH grants P30ES000210, S10RR025628, and T32AA007468, NASA Grants #NNX10AD59G and #NNJ12ZSA001N and the development account of Dr. Raber.
ABBREVIATIONS
- Aco2
aconitase
- AMPAR
AMPA-type glutamate receptor
- ARFs
ADP-ribosylation factors
- CaMKII
calcium calmodulin dependent protein kinase type II
- CV
coefficient of variation
- DIA
data-independent acquisition
- Dlst
dihydrolipoyllysine-residue succinyltransferase
- EMRT
exact mass retention
- IPA
Ingenuity Pathway Analysis
- ISI
inter-shock-intervals
- Idh
isocitrate dehydrogenase
- KG
ketoglutarate
- Mdh1
malate dehydrogenase
- MAPK
mitogen-activated protein kinase (MAPK)
- NMDA
N-methyl-D-aspartate
- PLP
myelin proteolipid protein
- MAG
myelin associated glycoprotein
- MBP
myelin basic protein
- MOG
myelin oligodendrocyte glycoprotein
- LC-MS
liquid chromatography mass spectrometry
- LTP
long-term potentiation
- LTD
long-term depression
- MYRF
myelin regulatory factor
- OAA
oxaloacetate
- PPARα
peroxisome proliferator-activated receptor alpha
- PP1
protein phosphatase 1
- PP2B
protein phosphatase 2B
- ROS
reactive oxygen species
- TCA
tricarboxylic acid
- SV
synaptic vesicle
- TWIMS
traveling wave ion mobility spectrometry
Footnotes
NOTES
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Azimzadeh O, Atkinson M, Tapio S. Proteomics in radiation research: present status and future perspectives. Radiat Environ Biophys. 2014;53:31–38. doi: 10.1007/s00411-013-0495-4. [DOI] [PubMed] [Google Scholar]
- 2.Leszczynski D. Radiation proteomics: A brief overview. Proteomics. 2014;14:481–488. doi: 10.1002/pmic.201300390. [DOI] [PubMed] [Google Scholar]
- 3.Tapio S. Ionizing Radiation Effects on Cells, Organelles and Tissues on Proteome Level. In: Leszczynski D, editor. Radiation Proteomics. Springer; Netherlands: 2013. pp. 37–48. [DOI] [PubMed] [Google Scholar]
- 4.Johannesen TB, Lien HH, Hole KH, Lote K. Radiological and clinical assessment of long-term brain tumour survivors after radiotherapy. Radiother Oncol. 2003;69:169–76. doi: 10.1016/s0167-8140(03)00192-0. [DOI] [PubMed] [Google Scholar]
- 5.Mulhern RK, Merchant TE, Gajjar A, Reddick WE, Kun LE. Late neurocognitive sequelae in survivors of brain tumours in childhood. Lancet Oncol. 2004;5:399–408. doi: 10.1016/S1470-2045(04)01507-4. [DOI] [PubMed] [Google Scholar]
- 6.Lafay–Cousin L, Bouffet E, Hawkins C, Amid A, Huang A, Mabbott DJ. Impact of radiation avoidance on survival and neurocognitive outcome in infant medulloblastoma. Curr Oncol. 2009;16:21–28. doi: 10.3747/co.v16i6.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffman KE, Yock TI. Radiation therapy for pediatric central nervous system tumors. J Child Neurol. 2009;24:1387–96. doi: 10.1177/0883073809342275. [DOI] [PubMed] [Google Scholar]
- 8.Kirsch DG, Tarbell NJ. Conformal radiation therapy for childhood CNS tumors. Oncologist. 2004;9:442–50. doi: 10.1634/theoncologist.9-4-442. [DOI] [PubMed] [Google Scholar]
- 9.Kempf SJ, Moertl S, Sepe S, von Toerne C, Hauck SM, Atkinson MJ, Mastroberardino PG, Tapio S. Low-dose ionizing radiation rapidly affects mitochondrial and synaptic signaling pathways in murine hippocampus and cortex. J Proteome Res. 2015;14:2055–64. doi: 10.1021/acs.jproteome.5b00114. [DOI] [PubMed] [Google Scholar]
- 10.Sokolova IV, Schneider CJ, Bezaire M, Soltesz I, Vlkolinsky R, Nelson GA. Proton radiation alters intrinsic and synaptic properties of CA1 pyramidal neurons of the mouse hippocampus. Radiation research. 2015;183:208–18. doi: 10.1667/RR13785.1. [DOI] [PubMed] [Google Scholar]
- 11.Chakraborti A, Allen A, Allen B, Rosi S, Fike JR. Cranial irradiation alters dendritic spine density and morphology in the hippocampus. PLoS One. 2012;7:e40844. doi: 10.1371/journal.pone.0040844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeung Maggie SY, Zdunek S, Bergmann O, Bernard S, Salehpour M, Alkass K, Perl S, Tisdale J, Possnert G, Brundin L, Druid H, Frisén J. Dynamics of Oligodendrocyte Generation and Myelination in the Human Brain. Cell. 2014;159:766–774. doi: 10.1016/j.cell.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Wong CS, Van der Kogel AJ. Mechanisms of radiation injury to the central nervous system: implications for neuroprotection. Mol Interv. 2004;4:273–84. doi: 10.1124/mi.4.5.7. [DOI] [PubMed] [Google Scholar]
- 14.Sun Y, Xu CC, Li J, Guan XY, Gao L, Ma LX, Li RX, Peng YW, Zhu GP. Transplantation of Oligodendrocyte Precursor Cells Improves Locomotion Deficits in Rats with Spinal Cord Irradiation Injury. PLoS ONE. 2013;8:e57534. doi: 10.1371/journal.pone.0057534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SE, Turkington K, Kushmerick C, Kim JH. Central dysmyelination reduces the temporal fidelity of synaptic transmission and the reliability of postsynaptic firing during high-frequency stimulation. J Neurophysiol. 2013;110:1621–30. doi: 10.1152/jn.00117.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grishchuk Y, Sri S, Rudinskiy N, Ma W, Stember KG, Cottle MW, Sapp E, Difiglia M, Muzikansky A, Betensky RA, Wong AM, Bacskai BJ, Hyman BT, Kelleher RJ, 3rd, Cooper JD, Slaugenhaupt SA. Behavioral deficits, early gliosis, dysmyelination and synaptic dysfunction in a mouse model of mucolipidosis IV. Acta Neuropathol Commun. 2014;2:133. doi: 10.1186/s40478-014-0133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parihar VK, Limoli CL. Cranial irradiation compromises neuronal architecture in the hippocampus. Proc Natl Acad Sci USA. 2013;110:12822–12827. doi: 10.1073/pnas.1307301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kempf SJ, Casciati A, Buratovic S, Janik D, von Toerne C, Ueffing M, Neff F, Moertl S, Stenerlow B, Saran A, Atkinson MJ, Eriksson P, Pazzaglia S, Tapio S. The cognitive defects of neonatally irradiated mice are accompanied by changed synaptic plasticity, adult neurogenesis and neuroinflammation. Mol Neurodegener. 2014;9:57. doi: 10.1186/1750-1326-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fazel R, Krumholz HM, Wang Y, Ross JS, Chen J, Ting HH, Shah ND, Nasir K, Einstein AJ, Nallamothu BK. Exposure to Low-Dose Ionizing Radiation from Medical Imaging Procedures. N Engl J Med. 2009;361:849–857. doi: 10.1056/NEJMoa0901249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doss M. Low Dose Radiation Adaptive Protection to Control Neurodegenerative Diseases. Dose-Response. 2014;12:277–287. doi: 10.2203/dose-response.13-030.Doss. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luckey TD. Radiation Hormesis: The Good, the Bad, and the Ugly. Dose-Response. 2006;4:169–190. doi: 10.2203/dose-response.06-102.Luckey. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stranahan AM, Mattson MP. Recruiting adaptive cellular stress responses for successful brain ageing. Nat Rev Neurosci. 2012;13:209–216. doi: 10.1038/nrn3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei LC, Ding YX, Liu YH, Duan L, Bai Y, Shi M, Chen LW. Low-dose radiation stimulates Wnt/beta-catenin signaling, neural stem cell proliferation and neurogenesis of the mouse hippocampus in vitro and in vivo. Current Alzheimer research. 2012;9:278–89. doi: 10.2174/156720512800107627. [DOI] [PubMed] [Google Scholar]
- 24.Rola R, Raber J, Rizk A, Otsuka S, VandenBerg SR, Morhardt DR, Fike JR. Radiation-induced impairment of hippocampal neurogenesis is associated with cognitive deficits in young mice. Experimental neurology. 2004;188:316–30. doi: 10.1016/j.expneurol.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Raber J, Rosi S, Chakraborti A, Fishman K, Dayger C, Davis MJ, Villasana L, Fike JR. Effects of 56Fe-particle cranial radiation on hippocampus-dependent cognition depend on the salience of the environmental stimuli. Radiation research. 2011;176:521–6. doi: 10.1667/rr2635.1. [DOI] [PubMed] [Google Scholar]
- 26.Anagnostaras SG, Gale GD, Fanselow MS. Hippocampus and contextual fear conditioning: recent controversies and advances. Hippocampus. 2001;11:8–17. doi: 10.1002/1098-1063(2001)11:1<8::AID-HIPO1015>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Milad M, Igoe S, Orr S. Fear Conditioning in Rodents and Humans. In: Raber J, editor. Animal Models of Behavioral Analysis. Humana Press; 2011. pp. 111–132. [Google Scholar]
- 28.Raber J, Rudobeck E, Campbell-Beachler M, Allen AR, Allen B, Rosi S, Nelson GA, Ramachandran S, Turner J, Fike JR, Vlkolinsky R. 28Silicon radiation-induced enhancement of synaptic plasticity in the hippocampus of naive and cognitively tested mice. Radiation research. 2014;181:362–8. doi: 10.1667/RR13347.1. [DOI] [PubMed] [Google Scholar]
- 29.Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Analytical chemistry. 2005;77:2187–200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 30.Distler U, Kuharev J, Tenzer S. Biomedical applications of ion mobility-enhanced data-independent acquisition-based label-free quantitative proteomics. Expert review of proteomics. 2014;11:675–84. doi: 10.1586/14789450.2014.971114. [DOI] [PubMed] [Google Scholar]
- 31.Shliaha PV, Bond NJ, Gatto L, Lilley KS. Effects of traveling wave ion mobility separation on data independent acquisition in proteomics studies. J Proteome Res. 2013;12:2323–39. doi: 10.1021/pr300775k. [DOI] [PubMed] [Google Scholar]
- 32.Distler U, Kuharev J, Navarro P, Levin Y, Schild H, Tenzer S. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat Methods. 2014;11:167–70. doi: 10.1038/nmeth.2767. [DOI] [PubMed] [Google Scholar]
- 33.Olsen RH, Marzulla T, Raber J. Impairment in extinction of contextual and cued fear following post-training whole-body irradiation. Front Behav Neurosci. 2014;8:231. doi: 10.3389/fnbeh.2014.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 35.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–56. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Distler U, Schmeisser MJ, Pelosi A, Reim D, Kuharev J, Weiczner R, Baumgart J, Boeckers TM, Nitsch R, Vogt J, Tenzer S. In-depth protein profiling of the postsynaptic density from mouse hippocampus using data-independent acquisition proteomics. Proteomics. 2014;14:2607–13. doi: 10.1002/pmic.201300520. [DOI] [PubMed] [Google Scholar]
- 37.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic acids research. 2015;43:D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with cytoscape 3. In: Baxevanis Andreas D, et al., editors. Current protocols in bioinformatics/editoral board. Vol. 47. 2014. pp. 8.13.1–8.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding C, Jiang J, Wei J, Liu W, Zhang W, Liu M, Fu T, Lu T, Song L, Ying W, Chang C, Zhang Y, Ma J, Wei L, Malovannaya A, Jia L, Zhen B, Wang Y, He F, Qian X, Qin J. A Fast Workflow for Identification and Quantification of Proteomes. Mol Cell Proteomics. 2013;12:2370–2380. doi: 10.1074/mcp.O112.025023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krämer A, Green J, Pollard J, Tugendreich S. Causal Analysis Approaches in Ingenuity Pathway Analysis (IPA) Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karp NA, Spencer M, Lindsay H, O’Dell K, Lilley KS. Impact of replicate types on proteomic expression analysis. J Proteome Res. 2005;4:1867–71. doi: 10.1021/pr050084g. [DOI] [PubMed] [Google Scholar]
- 42.Muntel J, Fromion V, Goelzer A, Maabeta S, Mader U, Buttner K, Hecker M, Becher D. Comprehensive absolute quantification of the cytosolic proteome of Bacillus subtilis by data independent, parallel fragmentation in liquid chromatography/mass spectrometry (LC/MS(E)) Mol Cell Proteomics. 2014;13:1008–19. doi: 10.1074/mcp.M113.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bilbao A, Varesio E, Luban J, Strambio-De-Castillia C, Hopfgartner G, Muller M, Lisacek F. Processing strategies and software solutions for data-independent acquisition in mass spectrometry. Proteomics. 2015;15:964–80. doi: 10.1002/pmic.201400323. [DOI] [PubMed] [Google Scholar]
- 44.Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci USA. 2005;102:4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalman MR, Deeter A, Nimishakavi G, Duan ZH. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinformatics. 2012;13(Suppl 2):S11. doi: 10.1186/1471-2105-13-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao Z, Chang JG, Hendriks IA, Sigurdsson JO, Olsen JV, Vertegaal AC. System-wide analysis of SUMOylation dynamics in response to replication stress reveals novel SUMO target proteins and acceptor lysines relevant for genome stability. Mol Cell Proteomics. 2015 doi: 10.1074/mcp.O114.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang CY, Sabido E, Aebersold R, Vitek O. Targeted protein quantification using sparse reference labeling. Nat Methods. 2014;11:301–4. doi: 10.1038/nmeth.2806. [DOI] [PubMed] [Google Scholar]
- 48.Bakshi MV, Azimzadeh O, Barjaktarovic Z, Kempf SJ, Merl-Pham J, Hauck SM, Buratovic S, Eriksson P, Atkinson MJ, Tapio S. Total body exposure to low-dose ionizing radiation induces long-term alterations to the liver proteome of neonatally exposed mice. J Proteome Res. 2015;14:366–73. doi: 10.1021/pr500890n. [DOI] [PubMed] [Google Scholar]
- 49.Azimzadeh O, Sievert W, Sarioglu H, Yentrapalli R, Barjaktarovic Z, Sriharshan A, Ueffing M, Janik D, Aichler M, Atkinson MJ, Multhoff G, Tapio S. PPAR alpha: a novel radiation target in locally exposed Mus musculus heart revealed by quantitative proteomics. J Proteome Res. 2013;12:2700–14. doi: 10.1021/pr400071g. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Li C, Chuo W, Liu Z, Ouyang Y, Li D, Han J, Wu Y, Guo S, Wang W. Integrated proteomic and metabolomic analysis reveals the NADH-mediated TCA cycle and energy metabolism disorders based on a new model of chronic progressive heart failure. Mol Biosyst. 2013;9:3135–45. doi: 10.1039/c3mb70263d. [DOI] [PubMed] [Google Scholar]
- 51.Liesa M, Shirihai Orian S. Mitochondrial Dynamics in the Regulation of Nutrient Utilization and Energy Expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris Julia J, Jolivet R, Attwell D. Synaptic Energy Use and Supply. Neuron. 2012;75:762–777. doi: 10.1016/j.neuron.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 53.Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 54.Van Huijstee AN, Mansvelder HD. Glutamatergic synaptic plasticity in the mesocorticolimbic system in addiction. Frontiers in cellular neuroscience. 2014;8:466. doi: 10.3389/fncel.2014.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hubener M, Bonhoeffer T. Neuronal Plasticity: Beyond the Critical Period. Cell. 2014;159:727–737. doi: 10.1016/j.cell.2014.10.035. [DOI] [PubMed] [Google Scholar]
- 56.Stone DB, Tesche CD. Topological dynamics in spike-timing dependent plastic model neural networks. Front Neural Circuits. 2013;7:70. doi: 10.3389/fncir.2013.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rizzoli SO. Synaptic vesicle recycling: steps and principles. The EMBO Journal. 2014;33:788–822. doi: 10.1002/embj.201386357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramakrishnan NA, Drescher MJ, Drescher DG. The SNARE complex in neuronal and sensory cells. Mol Cell Neurosci. 2012;50:58–69. doi: 10.1016/j.mcn.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schoch S, Deak F, Konigstorfer A, Mozhayeva M, Sara Y, Sudhof TC, Kavalali ET. SNARE function analyzed in synaptobrevin/VAMP knockout mice. Science. 2001;294:1117–22. doi: 10.1126/science.1064335. [DOI] [PubMed] [Google Scholar]
- 60.McMahon HT, Sudhof TC. Synaptic core complex of synaptobrevin, syntaxin, and SNAP25 forms high affinity alpha-SNAP binding site. J Biol Chem. 1995;270:2213–7. doi: 10.1074/jbc.270.5.2213. [DOI] [PubMed] [Google Scholar]
- 61.Sorensen JB, Matti U, Wei SH, Nehring RB, Voets T, Ashery U, Binz T, Neher E, Rettig J. The SNARE protein SNAP-25 is linked to fast calcium triggering of exocytosis. Proc Natl Acad Sci U S A. 2002;99:1627–32. doi: 10.1073/pnas.251673298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young A. Structural insights into the clathrin coat. Seminars in cell & developmental biology. 2007;18:448–58. doi: 10.1016/j.semcdb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 63.Popova NV, Deyev IE, Petrenko AG. Clathrin-mediated endocytosis and adaptor proteins. Acta naturae. 2013;5:62–73. [PMC free article] [PubMed] [Google Scholar]
- 64.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 65.El Far O, Seagar M. A role for V-ATPase subunits in synaptic vesicle fusion? J Neurochem. 2011;117:603–12. doi: 10.1111/j.1471-4159.2011.07234.x. [DOI] [PubMed] [Google Scholar]
- 66.de Jong AP, Fioravante D. Translating neuronal activity at the synapse: presynaptic calcium sensors in short-term plasticity. Frontiers in cellular neuroscience. 2014;8:356. doi: 10.3389/fncel.2014.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lisman J, Raghavachari S. Biochemical principles underlying the stable maintenance of LTP by the CaMKII/NMDAR complex. Brain Res. 2014 doi: 10.1016/j.brainres.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tinsley CJ, Narduzzo KE, Ho JW, Barker GR, Brown MW, Warburton EC. A role for calcium-calmodulin-dependent protein kinase II in the consolidation of visual object recognition memory. Eur J Neurosci. 2009;30:1128–39. doi: 10.1111/j.1460-9568.2009.06917.x. [DOI] [PubMed] [Google Scholar]
- 69.Mockett BG, Guevremont D, Wutte M, Hulme SR, Williams JM, Abraham WC. Calcium/calmodulin-dependent protein kinase II mediates group I metabotropic glutamate receptor-dependent protein synthesis and long-term depression in rat hippocampus. J Neurosci. 2011;31:7380–91. doi: 10.1523/JNEUROSCI.6656-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Belmeguenai A, Hansel C. A role for protein phosphatases 1, 2A, and 2B in cerebellar long-term potentiation. J Neurosci. 2005;25:10768–72. doi: 10.1523/JNEUROSCI.2876-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L, Stefan MI, Le Novere N. Calcium input frequency, duration and amplitude differentially modulate the relative activation of calcineurin and CaMKII. PLoS One. 2012;7:e43810. doi: 10.1371/journal.pone.0043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aggarwal S, Snaidero N, Pahler G, Frey S, Sanchez P, Zweckstetter M, Janshoff A, Schneider A, Weil MT, Schaap IA, Gorlich D, Simons M. Myelin membrane assembly is driven by a phase transition of myelin basic proteins into a cohesive protein meshwork. PLoS Biol. 2013;11:e1001577. doi: 10.1371/journal.pbio.1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mighdoll MI, Tao R, Kleinman JE, Hyde TM. Myelin, myelin-related disorders, and psychosis. Schizophr Res. 2015;161:85–93. doi: 10.1016/j.schres.2014.09.040. [DOI] [PubMed] [Google Scholar]
- 74.Wang P, Xie K, Wang C, Bi J. Oxidative stress induced by lipid peroxidation is related with inflammation of demyelination and neurodegeneration in multiple sclerosis. Eur Neurol. 2014;72:249–54. doi: 10.1159/000363515. [DOI] [PubMed] [Google Scholar]
- 75.Prukop T, Epplen DB, Nientiedt T, Wichert SP, Fledrich R, Stassart RM, Rossner MJ, Edgar JM, Werner HB, Nave KA, Sereda MW. Progesterone antagonist therapy in a Pelizaeus-Merzbacher mouse model. Am J Hum Genet. 2014;94:533–46. doi: 10.1016/j.ajhg.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tapinos N, Ohnishi M, Rambukkana A. ErbB2 receptor tyrosine kinase signaling mediates early demyelination induced by leprosy bacilli. Nat Med. 2006;12:961–6. doi: 10.1038/nm1433. [DOI] [PubMed] [Google Scholar]
- 77.Bujalka H, Koenning M, Jackson S, Perreau VM, Pope B, Hay CM, Mitew S, Hill AF, Lu QR, Wegner M, Srinivasan R, Svaren J, Willingham M, Barres BA, Emery B. MYRF is a membrane-associated transcription factor that autoproteolytically cleaves to directly activate myelin genes. PLoS Biol. 2013;11:e1001625. doi: 10.1371/journal.pbio.1001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koenning M, Jackson S, Hay CM, Faux C, Kilpatrick TJ, Willingham M, Emery B. Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. J Neurosci. 2012;32:12528–42. doi: 10.1523/JNEUROSCI.1069-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koenning M, Jackson S, Hay CM, Faux C, Kilpatrick TJ, Willingham M, Emery B. Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. J Neurosci. 2012;32:12528–42. doi: 10.1523/JNEUROSCI.1069-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tian Y, Shi Z, Yang S, Chen Y, Bao S. Changes in myelin basic protein and demyelination in the rat brain within 3 months of single 2-, 10-, or 30-Gy whole-brain radiation treatments. J Neurosurg. 2008;109:881–8. doi: 10.3171/JNS/2008/109/11/0881. [DOI] [PubMed] [Google Scholar]
- 81.Ishii A, Fyffe-Maricich SL, Furusho M, Miller RH, Bansal R. ERK1/ERK2 MAPK signaling is required to increase myelin thickness independent of oligodendrocyte differentiation and initiation of myelination. J Neurosci. 2012;32:8855–64. doi: 10.1523/JNEUROSCI.0137-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Norrmen C, Suter U. Akt/mTOR signalling in myelination. Biochem Soc Trans. 2013;41:944–50. doi: 10.1042/BST20130046. [DOI] [PubMed] [Google Scholar]
- 83.Morita M, Gravel SP, Hulea L, Larsson O, Pollak M, St-Pierre J, Topisirovic I. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle. 2015;14:473–80. doi: 10.4161/15384101.2014.991572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.