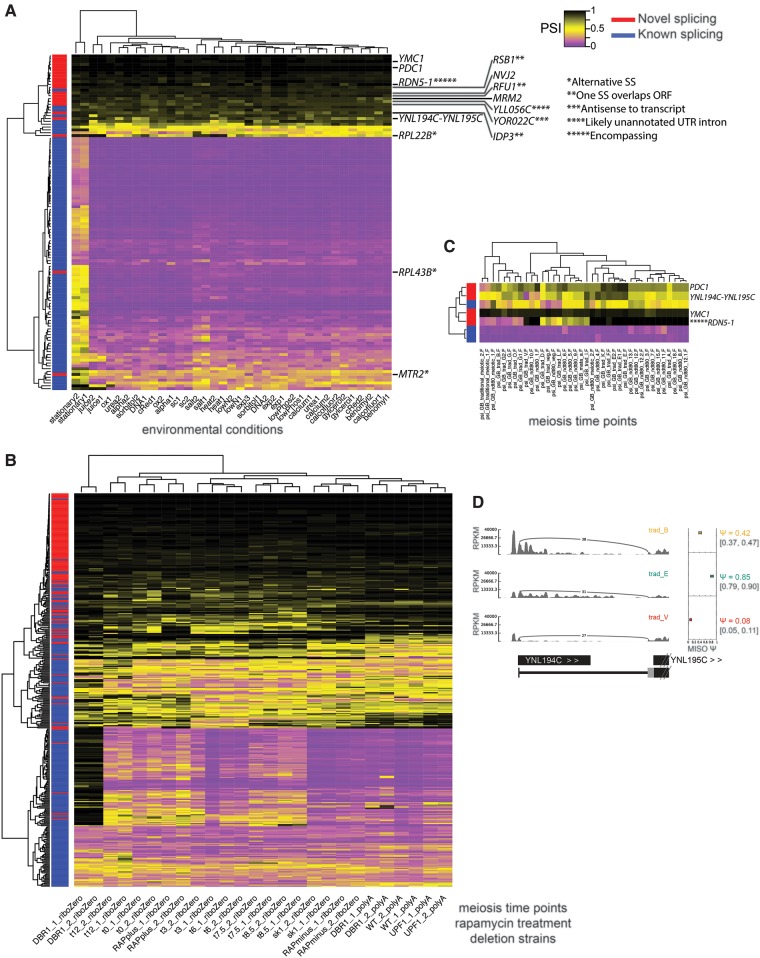
FIGURE 6.
Rare splicing of novel retained introns mirrors splicing patterns of known introns. (A) Clustering of PSI values calculated by MISO for retained introns in RNA-seq data from 18 environmental conditions (Waern and Snyder 2013), including 136 novel introns. PSI ranges from 0 (complete splicing, purple) to 1 (complete retention, black). (*) Alternative splice site. (**) One splice site overlaps gene ORF listed. (***) Antisense to an annotated transcript. (****) Intron likely in unannotated UTR. (*****) Intron encompasses gene. (YLL056C) 5′UTR supported by RNA-seq. (IDP3) 5′SS inside ORF. (RFU1 and RSB1) 3′SS inside ORF. Conditions are listed in Supplemental Methods. (B) Clustering of PSI of retained introns and alternative splice sites from RNA-seq of a meiosis time course, rapamycin treatment, and deletion strains, including additional novel introns. (C) Clustering of retained intron PSI from ribosome footprint profiling data from a meiosis time course (Brar et al. 2012). (D) Sashimi plot (Katz et al. 2010) depiction of ribosome footprint profiling splice junction reads from B joining YNL194 and YNL195 transcripts at a few stages of meiosis.