Abstract
A prerequisite for the intracellular replication process of the Flavivirus West Nile virus (WNV) is the cyclization of the viral RNA genome, which enables the viral replicase to initiate RNA synthesis. Our earlier studies indicated that the p45 isoform of the cellular AU-rich element binding protein 1 (AUF1) has an RNA chaperone activity, which supports RNA cyclization and viral RNA synthesis by destabilizing a stem structure at the WNV RNA's 3′-end. Here we show that in mammalian cells, AUF1 p45 is consistently modified by arginine methylation of its C terminus. By a combination of different experimental approaches, we can demonstrate that the methyltransferase PRMT1 is necessary and sufficient for AUF1 p45 methylation and that PRMT1 is required for efficient WNV replication. Interestingly, in comparison to the nonmethylated AUF1 p45, the methylated AUF1 p45aDMA exhibits a significantly increased affinity to the WNV RNA termini. Further data also revealed that the RNA chaperone activity of AUF1 p45aDMA is improved and the methylated protein stimulates viral RNA synthesis considerably more efficiently than the nonmethylated AUF1 p45. In addition to its destabilizing RNA chaperone activity, we identified an RNA annealing activity of AUF1 p45, which is not affected by methylation. Arginine methylation of AUF1 p45 thus represents a specific determinant of its RNA chaperone activity while functioning as a WNV host factor. Our data suggest that the methylation modifies the conformation of AUF1 p45 and in this way affects its RNA binding and restructuring activities.
Keywords: AUF1, arginine methylation, West Nile virus, Flavivirus, host factor, RNA replication
INTRODUCTION
AU-rich element binding protein 1 (AUF1), also known as heterogeneous ribonucleoprotein D (hnRNPD), is a member of the RNA recognition motif (RRM) family of RNA binding proteins (Maris et al. 2005). In multiple processes, AUF1 acts during post-transcriptional regulation of gene expression by binding to AU-rich elements (AREs) in the 3′-untranslated regions (UTRs) of mRNAs and by supporting either the destabilization or stabilization of these RNAs (for review, see White et al. 2013). Besides this, AUF1 was demonstrated to be involved in telomere maintenance, transcriptional activation, and translation initiation (Tolnay et al. 2002; Liao et al. 2007; Pont et al. 2012). Four AUF1 isoforms are generated by alternative splicing from a common pre-mRNA (Wagner et al. 1998); according to their apparent molecular masses, they were named p37, p40, p42, and p45, respectively. The isoforms show differences in their phosphorylation pattern as well as in nuclear-cytoplasmic shuttling and ARE-binding; these differences are suspected to affect their activities in transcriptional and post-transcriptional regulation of gene expression (Wilson et al. 2001, 2003; Tolnay et al. 2002; Raineri et al. 2004; Gratacos and Brewer 2010; Zucconi et al. 2010). All isoforms share two tandemly arranged RRMs and an arginine-glycine-rich region, RGG/RG motif, within the C terminus (see Fig. 4B; Thandapani et al. 2013). The largest isoform, AUF1 p45, exhibits a characteristic composition with 19- and 49-amino acid inserts near the N and C termini that are encoded by the mRNA's exons 2 and 7, respectively (see Fig. 4B).
FIGURE 4.
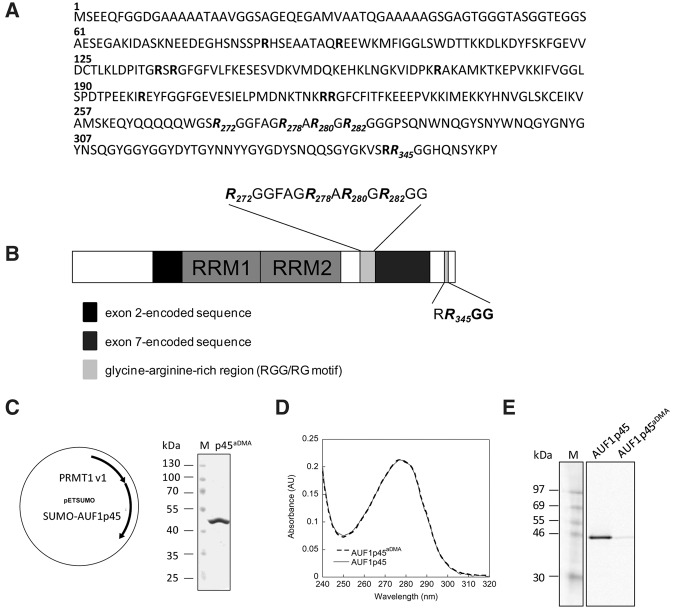
Identification of methylation sites; production and purification of methylated, recombinant AUF1 p45. (A) Amino acid sequence of AUF1 p45. All arginine residues in the sequence are highlighted in bold. The dimethylated arginines at positions 272, 278, 280, 282, and 345 that were identified by MALDI-TOF-MS after digestion with trypsin and chymotrypsin are highlighted in italics. (B) Schematic drawing of the domain organization of AUF1 p45. The RNA recognition motifs (RRM) are indicated as well as isoform-specific sequences encoded by exon 2 (black) and exon 7 (dark gray). The glycine-arginine-rich region (RGG/RG motif) within the C terminus of AUF1 p45 is indicated in light gray; the dimethylated arginines at positions 272, 278, 280, 282, and 345 that were identified by MALDI-TOF-MS are emphasized. (C) Schematic drawing of the bicistronic vector that was used to coexpress PRMT1 v1 and SUMO-AUF1 p45 in E. coli. The methylated AUF1 p45 (AUF1 p45aDMA) was purified as described in Materials and Methods. About 2 µg of purified AUF1 p45aDMA was analyzed on a Coomassie blue-stained SDS–PAGE in comparison to protein markers. (D) Absorption spectra of AUF1 p45 and AUF1 p45aDMA that were recombinantly expressed in and purified from E. coli. The ratio of the absorbance at 280 and 260 nm was about 1.8 for both protein preparations, indicating no contamination with nucleic acids. (E) In vitro methylation assay performed with 5 pmol purified PRMT1 v1 and 13 pmol of recombinant, nonmethylated AUF1 p45 and 13 pmol of recombinant, methylated AUF1 p45aDMA, respectively, using 400 pmol of [S-14C] adenosylmethionine as a substrate. The samples were taken after 2 h and analyzed by SDS–PAGE and the methylation state determined by phosphorimaging. One representative experiment is shown.
West Nile virus (WNV) is a widespread, causative agent of neuroinvasive and nonneuroinvasive diseases in humans. Viral infections occur in all continents except Antarctica (Ciota and Kramer 2013). WNV cycles between transmitting mosquitos and birds with humans being incidental hosts (Kramer et al. 2007). Together with other pathogens like Dengue virus (DENV), Japanese encephalitis virus (JEV), or tick-borne encephalitis virus (TBEV), WNV belongs to the genus Flavivirus of the Flaviviridae family. The viral genome is a single-stranded, type I capped, positive-strand RNA of ∼11 kilobases (kb), which consists of a single open reading frame (ORF) and flanking 5′ and 3′ UTRs. After entry and un-coating, the viral RNA acts as an mRNA and is translated in the cytoplasm. The ORF encodes a polyprotein that is co- and post-translationally processed by viral and cellular proteases to yield three structural proteins (core, membrane, and envelope) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Lindenbach et al. 2007). Replication of the WNV RNA genome, which also occurs in the cytoplasm of the infected cell, essentially requires the activity of the viral RNA-dependent RNA polymerase (RdRp or replicase) NS5. In the first step, NS5 transcribes the viral RNA genome synthesizing negative-strand RNA intermediates. The intermediates then serve as templates of the replicase to generate progeny positive-strand viral genomes (Lindenbach et al. 2007).
The Flavivirus genome is a dynamic RNA molecule that provides a set of cis-acting elements in the 5′ and 3′ UTRs, which are essential for the catalysis of viral RNA synthesis (Iglesias and Gamarnik 2011; Shi 2012). In vitro replicase assays, which applied the purified NS5 and viral RNA transcripts, or experiments with replicative viral RNAs in cultured cells demonstrated that the specific initiation of negative-strand RNA synthesis essentially involves both the 5′ UTR and the 3′ UTR (Khromykh et al. 2001; You et al. 2001; Corver et al. 2003; Nomaguchi et al. 2004; Alvarez et al. 2005). NS5 was shown to bind to a 5′-terminal stem–loop structure in the 5′ UTR, stem–loop A (SLA), which may act as a promoter for the initiation of viral RNA replication (Fig. 1; Brinton and Dispoto 1988; Filomatori et al. 2006). The 3′ UTR is composed of a 5′-variable region that contains an AU-rich element and a 3′-conserved region including the genomic 3′ terminus, which forms a stable stem–loop structure, 3′SL (Fig. 1). Accordingly, two requirements must be met that enable NS5 to initiate negative-strand RNA synthesis at the proper 3′ terminus. First, to relocate NS5 from the 5′- to the 3′-end of the viral genome, the 5′ UTR including the SLA promoter has to get into close proximity of the 3′ UTR. Second, the stem of the 3′ SL structure has to be opened such that the released 3′-terminal nucleotides may serve as the initiating template of the replicase (Fig. 1; Filomatori et al. 2006, 2011; Dong et al. 2008; Lodeiro et al. 2009). Mainly from the mosquito-borne DV and WNV, solid experimental evidence exists showing that these prerequisites are achieved by long-range RNA–RNA interactions of conserved complementary sequence elements, which support a 5′–3′ circularization of the viral genome. Considering their location in the viral genomes, the yet best characterized circularization elements were termed as CS, UAR, and DAR, respectively (see Fig. 1; for review, see Iglesias and Gamarnik 2011; Bidet and Garcia-Blanco 2014; Brinton and Basu 2015). Although circularization of the viral genome is essential for the initiation of RNA synthesis, the replication process nevertheless depends on a fine balance of the levels of the circular and linear forms of the flaviviral genome (Villordo et al. 2010). Importantly, in the context of the linear RNA form, several of the circularization sequences locate in stem–loop structures. This is particularly obvious with the 3′ UAR sequence, which is a major component of the 3′ SL stem (Fig. 1). Consequently, 5′–3′-hybridization of the UAR elements is supposed to be a main trigger for conformational changes that open the 3′ SL stem structure and thus release the genomic 3′-end to be recognized by NS5 (Dong et al. 2008; Filomatori et al. 2011).
FIGURE 1.
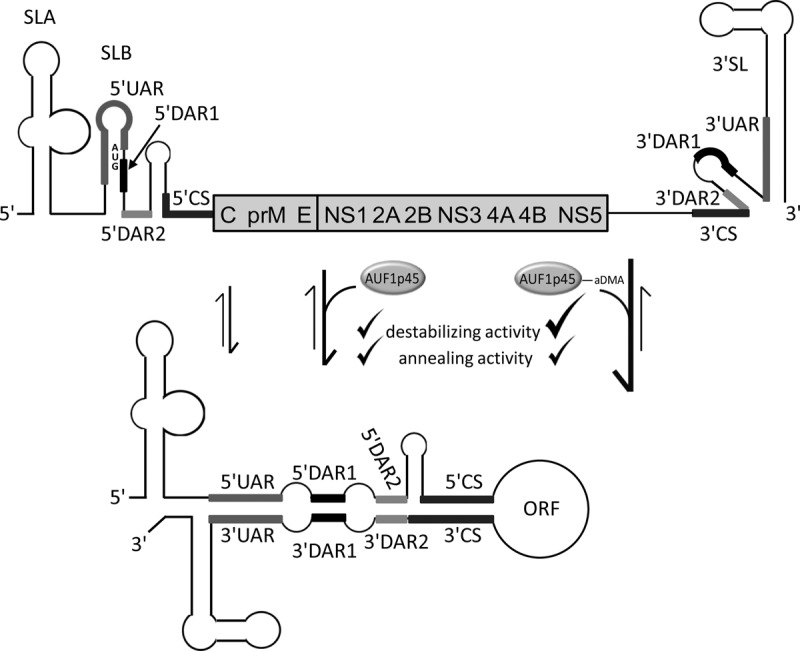
Model of AUF1 p45-mediated WNV genome cyclization. The genomic and structural organizations of the WNV RNA's 5′ and 3′ termini in the linear and circularized forms are shown. Conserved RNA elements that are required for WNV replication are highlighted by heavy lines: SLA (stem-loop A); SLB (stem–loop B); CS (conserved sequence); UAR (located upstream of the AUG translation initiation region); DAR (located downstream of the AUG translation initiation region); 3′ SL (3′ stem-loop). The RNA secondary structures of the UTRs and of the 5′-terminal region of the core (C) protein coding region are depicted according to the experimental data reported by Dong et al. (2008). The translational start codon AUG of the ORF is indicated; it is located within SLB. The residual part of the ORF encoding the viral structural (C, prM, and E) and nonstructural (NS) proteins is represented as a bar. (Upper panel) Linear form of the viral genome. (Lower panel) Circular conformation, mediated by complementary base pairing of sequences in the 5′- and 3′CS, UAR, DAR1, and DAR2 elements, respectively. Previously, we reported that AUF1 p45 stimulates the formation of the circular conformation of the WNV RNA (Friedrich et al. 2014). Here we show that the AUF1 p45-supported RNA cyclization and stimulation of viral RNA synthesis is most efficient when the protein is methylated (AUF1 p45aDMA). The cyclization of the WNV RNA is enhanced by the RNA chaperone activity of AUF1 p45 that comprises an RNA destabilizing as well as an RNA annealing activity. Although the RNA annealing activities of AUF1 p45 and AUF1 p45aDMA are comparable, the RNA destabilizing activity is more pronounced with the methylated AUF1 p45aDMA (indicated by a bold tick).
The limited coding capacity of positive-strand RNA viruses implies that viral propagation often depends on the auxiliary activity of cell-encoded proteins (“host factors”). Several host factors of WNV or DV were characterized (for review, see Bidet and Garcia-Blanco 2014; Brinton and Basu 2015), among these also AUF1. Thus, a certain intracellular level of AUF1 is crucial for the efficient initiation of WNV RNA replication. Most importantly, AUF1 isoform p45 was demonstrated having an RNA chaperone activity, which accelerates the structural rearrangement and cyclization of the WNV RNA that is required for the initiation of RNA replication by NS5 (Friedrich et al. 2014). In this study, we obtained experimental evidence that post-translational methylation of AUF1 p45 represents an important molecular feature that supports the function of the protein while acting as a WNV host factor.
RESULTS
AUF1 p45 interacts with the arginine-methyltransferase PRMT1 and is methylated in the cell
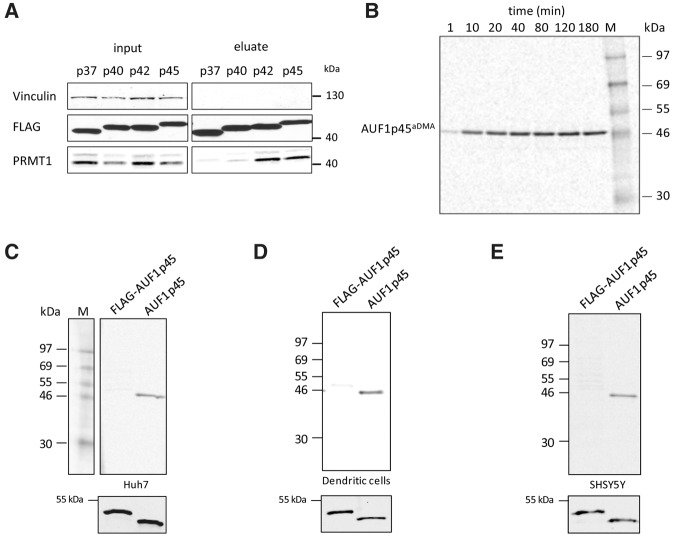
This study was originated by the question whether cellular factors affected the function of AUF1 p45 when acting as a WNV host factor. Accordingly, the initial experiment aimed at identifying binding partners of the protein. For this, we expressed FLAG-tagged versions of all AUF1 isoforms (p37, p40, p42, and p45) in Huh7 cells and purified the proteins via a standard immunoaffinity procedure (see Materials and Methods). Interestingly, when we analyzed such prepared FLAG–AUF1 p45 by mass spectrometry (MS), the protein was consistently found to copurify with the arginine-methyltransferase PRMT1 (see Supplemental Fig. S1; MS data not shown). Tests of the different, FLAG-purified AUF1 isoforms by Western blot with a PRMT1-specific antibody confirmed that PRMT1 reproducibly coprecipitated with the isoforms p42 and p45, respectively. In contrast, coprecipitation of PRMT1 with the isoforms p37 and p40 was only weakly detectable suggesting a favored interaction of the methyltransferase with the larger AUF1 isoforms (Fig. 2A; see Discussion). In the preparations containing the FLAG-purified AUF1 p42 and AUF1 p45, the anti-PRMT1 antibody stained at least two different proteins. Considering that four main isoforms of PRMT1 (variants v1, v2, v3, and v5; see also Supplemental Fig. S2A) were identified in hepatocytes (Goulet et al. 2007), we assumed that AUF1 p45 (and AUF1 p42) interacted with more than one of the PRMT1 isoforms (Fig. 2A).
FIGURE 2.
AUF1 p45 is methylated in vitro and in cells. (A) FLAG fusion proteins of the different AUF1 isoforms were expressed in Huh7 cells, immune-precipitated from cell extracts using anti-FLAG-antibody and eluted with FLAG peptide (see Materials and Methods; Friedrich et al. 2014). The presence of PRMT1 in the original extracts (input) and in the eluate was verified by Western blot with an anti-PRMT1-antibody. Vinculin was stained with a Vinculin-specific antibody as an internal control. (B) In vitro methylation assay of AUF1 p45. Thirteen picomoles of recombinant, Escherichia coli generated AUF1 p45 were methylated in vitro by 5 pmol PRMT1 in the presence of 400 pmol of [S-14C] adenosylmethionine as described in Materials and Methods. Samples were taken at the indicated time points and the methylation state analyzed by SDS–PAGE and phosphorimaging. (C) In vitro methylation assay with PRMT1 and different preparations of AUF1 p45. Applied was 5 pmol of AUF1 p45 that was expressed in and purified from E. coli and hence expected to be nonmethylated, and a comparable amount of FLAG–AUF1 p45 that was expressed in and purified from Huh7 cells. Both protein preparations were subjected to an in vitro methylation assay with 5 pmol of purified PRMT1 v1 and 400 pmol [S-14C] adenosylmethionine as a substrate. The samples were taken after 2 h and analyzed by SDS–PAGE and phosphorimaging. (Lower panel) Western blot with anti-AUF1 antibody demonstrating the use of equal amounts of protein in the in vitro methylation assay. One representative experiment is shown. (D) Same experiment as in C except that FLAG–AUF1 p45 was expressed in and purified from the dendritic cell line DC2.4. (E) Same experiment as in C except that FLAG–AUF1 p45 was expressed in and purified from SHSY5Y cells.
To next understand if AUF1 p45 is a PRMT1 substrate, AUF1 p45 as well as PRMT1 v1 were each expressed in and purified from E. coli (Supplemental Fig. S2B; see Friedrich et al. 2014). Both proteins were then applied to an in vitro methylation assay using [S-14C]adenosylmethionine as a methyl donor (see Materials and Methods). The results of this experiment demonstrated that AUF1 p45, in fact, was methylated by the purified PRMT1 v1 and that this process occurred in a time-dependent manner (Fig. 2B). To evaluate prospective isoform-specific differences, we also tested AUF1 p45 in the same assay for methylation by the other PRMT1 isoforms v2, v3, and v5; each was produced in and purified from E. coli. The resulting data showed that all tested PRMT1 isoforms catalyzed the methylation of AUF1 p45 at comparable efficiency (Supplemental Fig. S2B,C).
The fact that AUF1 p45 turned out to be accessible for methylation in vitro obviously raised the question on the methylation state of the protein in the cell. To address this, we applied FLAG-tagged AUF1 p45 that was affinity-purified from Huh7 cells side-by-side with the identical amount of nonmethylated AUF1 p45 that was purified from E. coli to the in vitro methylation assay with PRMT1 v1. Interestingly, efficient methylation was only detectable with the E. coli expressed nonmethylated AUF1 p45, suggesting that the hepatocyte-derived AUF1 p45 was partly or even fully methylated (Fig. 2C). The same experiment was repeated with FLAG-purified AUF1 p45 that was expressed in other cell lines such as the mouse-derived dendritic cell line DC2.4 and the human neuronal cell line SHSY5Y. In each case, we obtained identical results as earlier with the Huh7-derived protein, i.e., in comparison with the E. coli-expressed nonmethylated AUF1 p45, the cell-expressed FLAG-tagged AUF1 p45 proteins were only weak substrates of PRMT1 (Fig. 2D,E). These results suggested that in the investigated types of cells, most, if not all AUF1 p45 was considerably methylated.
PRMT1 is involved in the methylation of AUF1 p45 in cells, and it is required for efficient WNV replication
Having shown that PRMT1 methylates nonmethylated AUF1 p45 in vitro, it was next important to understand whether this methyltransferase definitely participates in the methylation of the protein in the cell. To address this question, we depleted PRMT1 in Huh7 cells by RNAi applying an siRNA, siPRMT1-1 (see Materials and Methods), which caused a highly effective knockdown of the protein in the cells (see below) without mediating an apparent cytopathic effect (Fig. 3A,C; Supplemental Fig. S3). Seventy-two hours post-transfection, we expressed the FLAG-tagged AUF1 p45 in these cells by transfection of the corresponding expression plasmid and purified the protein via its FLAG-tag 48 h later. The level of expression and recovery of FLAG–AUF1 p45 was not affected by the depletion of PRMT1. This became apparent when we performed the same experiment with mock-depleted (control siRNA-treated) cells (Fig. 3A). However, in these control cells, and in contrast to the PRMT1-depleted cells, PRMT1 co-eluted together with AUF1 p45. This was expected considering the earlier observed interaction of PRMT1 and AUF1 p45 (see above). The FLAG–AUF1 p45 proteins, which were obtained from the PRMT1- and mock-depleted cells, then were tested in the in vitro methylation assay, again side-by-side with the E. coli-derived nonmethylated protein (Fig. 3B). As earlier, we observed efficient methylation of the nonmethylated AUF1 p45 and only marginal methylation of the tagged AUF1 p45 that derived from the control cells. However, FLAG-tagged AUF1 p45, which was recovered from the PRMT1-depleted cells, was detectably methylated, although not as efficiently as the nonmethylated AUF1 p45. We interpreted these data as a strong hint that in human cells the methyltransferase PRMT1 is involved in the efficient methylation of AUF1 p45.
FIGURE 3.
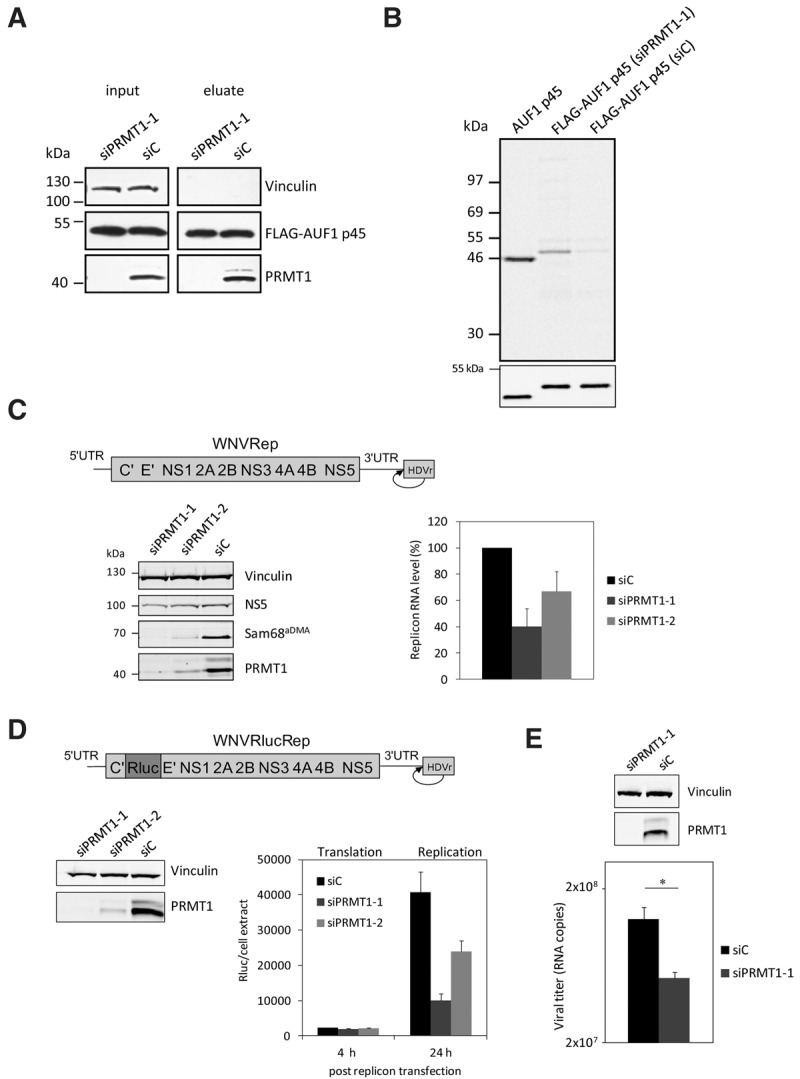
RNAi-mediated depletion of PRMT1 in Huh7 cells inhibits the methylation of FLAG–AUF1 p45 and WNV replication. (A) AUF1 p45 was expressed as a FLAG fusion protein in Huh7 cells and immune-affinity purified from extracts of cells that were earlier treated with a control siRNA (siC) or with an siRNA targeting the PRMT1 encoding mRNA (siPRMT1-1). The amounts of expressed FLAG–AUF1 p45 were determined by Western blot with anti-FLAG antibody comparing similar protein amounts of the original extracts (input) and of the eluate, respectively. The efficient depletion of PRMT1 by siPRMT1-1 was verified at 120 h post-transfection by Western blot with an anti-PRMT1 antibody. (B) In vitro methylation assay with PRMT1 and different preparations of AUF1 p45. Applied were 5 pmol of nonmethylated AUF1 p45 (expressed and purified from E. coli) and comparable amounts of FLAG–AUF1 p45 that was expressed in and purified from Huh7 cells (transfected with either siC or siPRMT1-1). The different AUF1 p45 preparations were methylated in vitro by 5 pmol of purified PRMT1 v1 using 400 pmol [S-14C] adenosylmethionine as a substrate. The samples were taken after 2 h and the methylation state analyzed by SDS–PAGE and phosphorimaging. (Lower panel) Western blot with anti-AUF1 antibody demonstrating the use of equal amounts of protein in the in vitro methylation assay. One representative experiment is shown. Note that the E. coli and Huh7-derived proteins show a slightly different migration behavior in the SDS–PAGE; this was explained by the FLAG-affinity tag that is present in the Huh7-expressed protein. (C) Scheme of the organization of the applied WNV replicon; the ORF is represented as a bar, the UTRs as single lines. The authentic 3′-end of the viral RNA is generated by the activity of a hepatitis delta ribozyme (HDVr) that was fused to the 3′ UTR (as indicated). Huh7 cells were transfected with control siRNA (siC) or siRNAs targeting the PRMT1 encoding mRNA (siPRMT1-1 and siPRMT1-2), respectively. Seventy-two hours later, all cells were transfected with the WNV replicon RNA. At 24 h post-transfection of the replicon, the protein levels of PRMT1, the viral NS5, Vinculin and of the methylated Sam68 were analyzed by Western blotting using appropriate antibodies (left panel; representative experiment shown). The RNA levels of the WNV replicon were quantified by real-time PCR at 24 h post-transfection (right panel). Results of three different experiments are shown; error bars reflect standard deviations. (D, top) Schematic representation of the applied WNVRluc replicon; the integrated Rluc ORF is indicated by different shading. Huh7 cells were treated with a control siRNA (siC) and siRNAs (siPRMT1-1 or siPRMT1-2) targeting PRMT1, respectively. The cytoplasmic depletion of PRMT1 120 h post siRNA transfection was confirmed by Western blotting (left), and the cells were transfected with the WNVRluc replicon RNA. At 4 h and 24 h post-transfection, the Renilla luciferase activity was measured (right). Results from three different experiments are shown; error bars reflect standard deviations. (E) Huh7 cells were treated with a control siRNA (siC) and an siRNA targeting PRMT1 (siPRMT1-1). After 120 h, the cytoplasmic depletion of PRMT1 was confirmed by Western blotting (top), and the cells were infected with WNV strain 3356. At 24 h post-infection, culture fluids were assayed for virus titers by qRT-PCR (see Materials and Methods). One representative experiment (performed in triplicate) is shown; (*) P ≤ 0.05.
The following experiments investigated the relevance of PRMT1 for the WNV RNA replication process. As a first approach, we reduced the intracellular level of PRMT1 with two different siRNAs and tested for the replication of a WNV replicon, which was transfected at 72 h post-transfection of the respective siRNAs. As shown in Figure 3C, with both RNAi approaches, which targeted different regions of the PRMT1 mRNA, we obtained a considerable drop in the quantity of intracellular PRMT1. Importantly, in each case, the siRNA-mediated depletion of PRMT1 also had an evident negative effect on the detectable amount of methylated Sam68 protein in the cell. Sam68 is a bona fide target of PRMT1 (Côté et al. 2003). The impact of the PRMT1 knockdown on viral RNA synthesis was tested at 24 h post-transfection of the WNV replicon. At this time point, viral RNA synthesis was earlier observed to be most significantly affected by an AUF1 knockdown (Friedrich et al. 2014). Most interestingly, siRNA-mediated depletion of PRMT1 had a clear effect on WNV RNA replication, i.e., in both types of knockdown experiments, we detected a considerable reduction of viral RNA replication corresponding to ∼40% and 60% of the control level, respectively (Fig. 3C). The idea that WNV replication depended on the presence of a certain amount of PRMT1 in the cell was supported by the fact that the level of viral replication correlated with the amount of residual PRMT1 that remained in the cells after RNAi. That is, the negative effect on replication was most apparent in cells that were depleted by siPRMT1-1 and that contained only marginal amounts of PRMT1. The negative effect was less pronounced in the cells where the protein was less effectively depleted by the second siRNA, siPRMT1-2 (Fig. 3C).
Next, we wanted to understand whether PRMT1 is required during protein or RNA synthesis of the WNV replication process. For this, we treated Huh7 cells with each of the PRMT1-specific siRNAs or with a control siRNA and, 120 h later, transfected the cells with a WNV reporter replicon that expressed Renilla luciferase (Rluc) in addition to the viral proteins (Fig. 3D). The long-term depletion was applied to reduce the endogenous amount of PRMT1 to a minimum. Again, we did not observe any cytopathic effect after siRNA treatment (Supplemental Fig. S3). When we measured the Rluc activity at 4 h post-transfection, a time point where translation of the viral RNA is detectable but no RNA replication, this revealed no significant differences in Rluc activity between the PRMT1- and mock-depleted control cells (Fig. 3D). In contrast, at 24 h post-transfection, where viral protein as well as viral RNA synthesis occurred, the Rluc activity was markedly reduced in the PRMT1-depleted cells (Fig. 3D).
In the last experiment of this series, we repeated the siRNA-mediated depletion of PRMT1 at identical conditions as above (Fig. 3E) but infected the cells this time with a WNV isolate (see Materials and Methods; Friedrich et al. 2014). Thus, in close accordance with the replicon experiments, at 24 h post-infection, we measured a significant drop of the virus titers in the supernatants of PRMT-depleted cells, although this was not the case in the mock-depleted cells (Fig. 3E).
In summary, these data revealed that PRMT1 is important for efficient replication of WNV RNA in human cells.
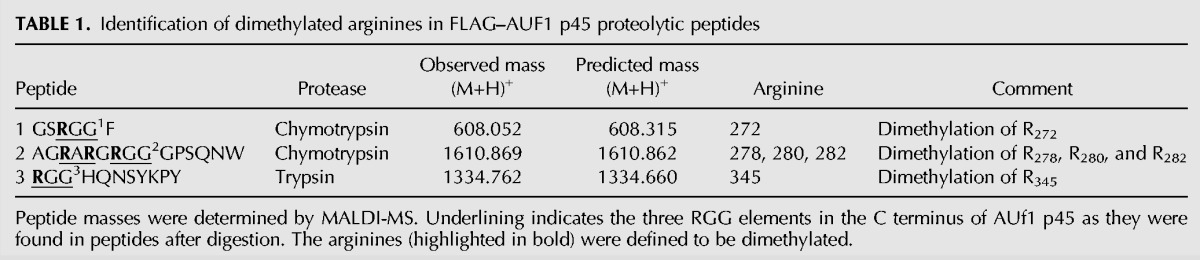
AUF1 p45 is dimethylated at five arginine residues in the C terminus
The next series of experiments addressed the questions whether and how the PRMT1-mediated methylation affected the activity of AUF1 p45 while acting as a WNV host factor. Along this line, we considered it first important to define the methylation sites of cell-derived AUF1 p45. Accordingly, AUF1 p45, which was FLAG-purified from Huh7 cells, was digested by trypsin or chymotrypsin and the resulting peptides analyzed by MALDI-TOF-MS. Thus, five methylated arginines were identified at positions 272, 278, 280, 282, and 345, i.e., within the RRG/RG motif in the C terminus of the protein, and all residues were indicated to be dimethylated (Table 1; Fig. 4A,B). In close accordance with the substrate preference of PRMT1 (Bedford and Clarke 2009), the methylated arginines are flanked by one or more glycine residues (Fig. 4A,B).
TABLE 1.
Identification of dimethylated arginines in FLAG–AUF1 p45 proteolytic peptides
For further studies on the biological significance of AUF1 p45 methylation, it was essential to obtain large-scale preparations of the methylated AUF1 p45 (hereafter termed as AUF1 p45aDMA) at a high and reproducible quality. For this, we expressed the PRMT1 methyltransferase and the target protein from the same plasmid in E. coli (Fig. 4C; Moritz et al. 2014) and purified AUF1 p45aDMA to homogeneity using the analogous purification protocol as earlier with the nonmethylated AUF1 p45 (Fig. 4D; Friedrich et al. 2014). Comparisons of the UV-absorption spectra of the methylated and nonmethylated AUF1 p45 variants showed that these were essentially congruent (Fig. 4D). Moreover, as an important prerequisite for protein–nucleic acids interaction studies, the 280/260 nm ratio of preparations of both types of proteins was determined to be in the range of 1.8 in each case, which demonstrated that these were mainly free of nucleic acids.
To confirm the methylation state of the E. coli-derived AUF1 p45aDMA, it was applied to the in vitro methylation assay, again side-by-side with the (also E. coli-derived) nonmethylated AUF1 p45. As shown in Figure 4E, efficient methylation was solely detectable with the nonmethylated version, indicating that AUF1 p45aDMA was highly methylated (Fig. 4E). This was confirmed by protease digestion of AUF1 p45aDMA and analysis of the resulting peptides by MS. Most importantly, the MS data revealed that the peptides that were obtained with the E. coli prepared AUF1 p45aDMA displayed essentially the same set of dimethylated arginines as the peptides that were earlier characterized from the Huh7-derived FLAG-tagged protein (Table 1; Fig. 4B). A further side-chain fragmentation analysis, which distinguished between symmetric and asymmetric dimethylation, revealed that the dimethylated peptide 2 (Table 1) containing arginines 278, 280, and 282 was asymmetrically dimethylated (Supplemental Fig. S4), which also is a characteristic of arginine methylation by type I PRMTs such as PRMT1 (Bedford and Clarke 2009). This set of experiments showed that we were able to generate authentically methylated, recombinant AUF1 p45aDMA. Importantly, the way of preparation and the quality (e.g., mainly nucleic acid free) of the prepared nonmethylated and methylated AUF1 p45 were essentially the same, which enabled functional studies aimed at directly comparing the characteristics of both protein variants. The fact that we obtained identical methylation data with the cell-derived and E. coli-derived AUF1 p45aDMA confirmed the notion that PRMT1 represents the main methyltransferase of AUF1 p45 in the cell (see above).
Methylation of AUF1 p45 induces structural changes in the protein
As an initial experiment to compare the methylated with the nonmethylated version of the protein, we tested the sedimentation behavior of AUF1 p45aDMA, applying analytical ultracentrifugation. This revealed an experimental molecular mass of 42.1 kDa, which was close to the theoretical weight of 38.3 kDa of the individual protein (Fig. 5A). With the nonmethylated AUF1 p45, it was already shown that at the purification conditions applied here (i.e., 150 mM KCl, pH 7.6; mainly free of nucleic acid contaminations), the protein exists as a monomer in solution (Friedrich et al. 2014). The ultracentrifugation data confirmed the monomeric nature of AUF1 p45 and revealed that it was not affected by the arginine methylation.
FIGURE 5.
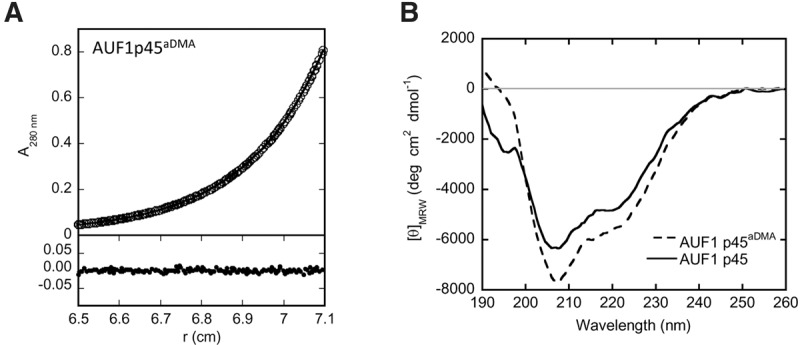
Features of AUF1 p45 and AUF1 p45aDMA. (A) In sedimentation equilibrium experiments (analytical ultracentrifugation), AUF1 p45aDMA (5 µM of protein) was analyzed at 14,000 rpm at 20°C using the software provided by Beckman Coulter. The upper panel shows the experimental data (circles) and the fit (line), and the lower panel shows the deviation of the experimental data and the fit. After reaching equilibrium, the molecular mass of AUF1 p45aDMA was determined to be 42.1 kDa (theoretical MW of the protein monomer = 38.3 kDa). No aggregation of AUF1 p45aDMA was observed in the course of the experiment. (B) Far-UV circular dichroism (CD) spectra of AUF1 p45 and AUF1 p45aDMA were recorded. The acquired data were normalized to mean residue weight (MRW) ellipticities.
Next, to compare the general structural features of the methylated and nonmethylated AUF1 p45, we measured both proteins by circular dichroism (CD) spectroscopy. The recorded spectrum of the nonmethylated AUF1 p45 revealed a maximum negative circular dichroism at 208 nm and a shoulder at ∼215 nm, indicating some helical content as well as the existence of some β-sheets. However, the overall CD signal (normalized to mean residue ellipticity ΘMRW) was rather low, signifying a high content of disordered regions (Fig. 5B; see also Discussion). Interestingly, with AUF1 p45aDMA, we observed evident conformational changes in comparison to the nonmethylated protein. In this case, the spectrum recorded a more pronounced CD at 208 nm and a less pronounced CD at <200 nm (Fig. 5B). Thus, arginine methylation appears to have a substantial effect on the secondary structure of AUF1 p45.
The methylation state of AUF1 p45 affects the RNA binding properties and the supportive effect of the protein on WNV RNA synthesis
The following experiments addressed the question whether the methylation of AUF1 p45 affected the RNA binding properties of the protein. For this, we used an earlier established RNA filter binding assay. As a first target RNA, we applied WNV sgRNA, which encodes the 5′-terminal 184 nucleotides (nt) and the 3′-terminal 611 nt of the viral genome and therefore contains all elements required to recapitulate the first step of the WNV RNA replication process in vitro (Friedrich et al. 2014; see also below). In addition, we tested RNA molecules, which encoded the WNV UTRs or individual segments of the UTRs, respectively (Fig. 6A). Increasing concentrations of the protein were applied to the radioactively labeled RNA substrates and the radioactive signal of the bound RNA quantified and plotted against the protein concentration (see example with the WNV sgRNA in Supplemental Fig. S5). As explained, the nonmethylated AUF1 p45 and AUF1 p45aDMA each were purified as monomeric proteins (Fig. 5A; Friedrich et al. 2014). Accordingly, during the initial tests, we assumed an equimolar RNA-to-protein ratio in the corresponding RNA–protein complexes, and the dissociation constants (KD) of these complexes were determined by fitting the data to a single-site binding model (Friedrich et al. 2014). However, in the course of this study, it became apparent that the interactions of AUF1 p45 to the viral RNAs applied here more likely display a sigmoidal binding behavior and that fitting of the curves to a cooperative binding mode (see Equation 1 in Materials and Methods) is more appropriate. This yielded the apparent binding constants (Kapp) of the reactions (see example in Supplemental Fig. S5), and binding of all tested viral RNAs revealed a cooperativity coefficient of n = ∼2 (data not shown), indicating that the binding of one protein molecule favors the binding of a second molecule.
FIGURE 6.
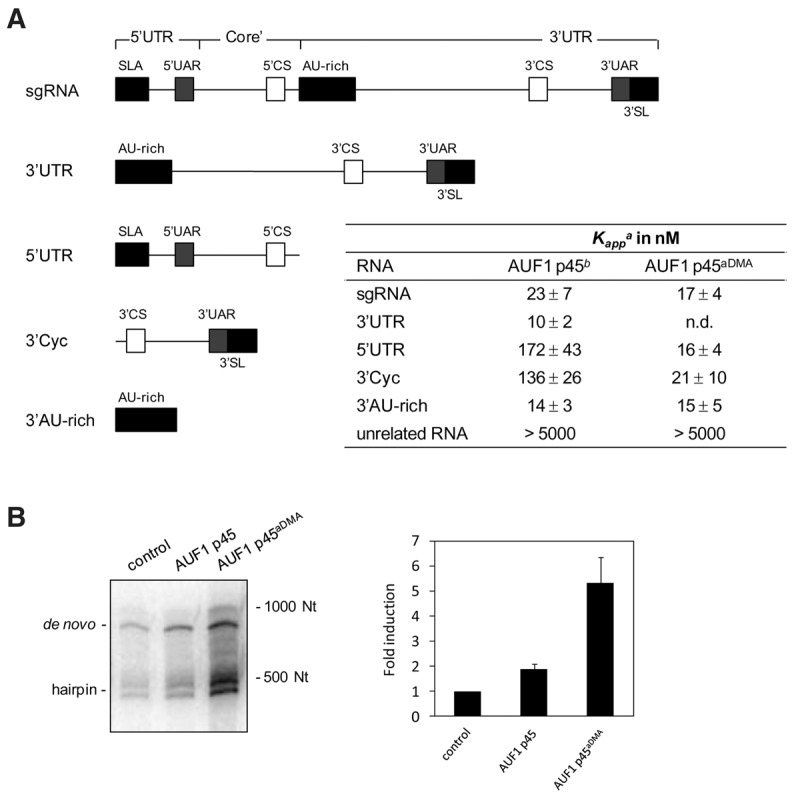
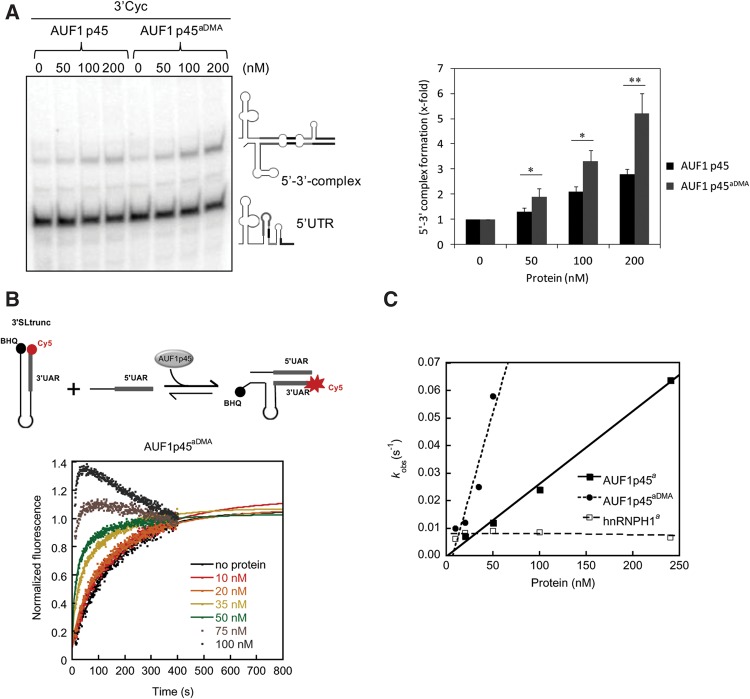
Methylation of AUF1 p45 increases the binding affinity of the protein to the WNV RNA termini and leads to an enhanced stimulation of WNV RNA synthesis. (A) Schematic drawing of the organization of the applied RNA transcripts. The longest transcript, the WNV sgRNA, consists of the 5′ and 3′ UTR and a part of the core coding sequence; accordingly, it contains the 5′ SLA, the 3′ SL, all cyclization elements, and the AU-rich element in the upstream portion of the 3′ UTR (indicated as differently shaded boxes). The organization of the other transcripts follows this scheme; the sequence of the unrelated RNA that was mostly used in these experiments is given in Supplemental Table S3. (Column 3) The binding affinities of AUF1 p45 and AUF1 p45aDMA with the different RNAs were determined by filter binding (see Materials and Methods). Fitting the curves according to a cooperative binding revealed the apparent binding constant Kapp. (Column 2) The Kapp values with AUF1 p45 were obtained by reevaluation of binding curves that were published earlier (Friedrich et al. 2014). The results and standard deviations from at least three different experiments are shown. (n.d.) Not determined. (B) 200 nM of nonmethylated AUF1 p45 and methylated AUF1 p45aDMA were tested by supplementation in the in vitro replicase assay with sgRNA template. Newly synthesized [32P]-labeled RNA products were analyzed on a denaturing Tris-Borate PAGE by phosphorimaging. The levels of induction of RNA synthesis are shown in comparison to the control (replicase assay in the absence of AUF1 p45; set to 1). Results of three independent experiments are shown; error bars reflect standard deviations.
Figure 6A shows the Kapp values that were measured with AUF1 p45aDMA in direct comparison to the Kapp values of the nonmethylated AUF1 p45; the latter were obtained by reevaluation of earlier data (Friedrich et al. 2014) with the cooperative binding model. Thus, both versions of the protein were determined to bind at high and comparable affinity (Kapp ∼20 nM) to the WNV sgRNA and to the AU-rich element that is located in the 5′-part of the 3′ UTR. As reported, the AU-rich stretch strongly resembles ARE target sequences of the AUF1 protein, and it is essential for efficient WNV RNA replication (Zhang et al. 1993; Friedrich et al. 2014). However, when we tested the binding of AUF1 p45aDMA to the WNV 5′ UTR and to the 3′-terminal 111 nt of the 3′ UTR (here called 3′ Cyc), we observed a significantly higher affinity of the methylated as compared to the nonmethylated version of the protein. This was indicated by considerably lower Kapp values (Kapp 5′ UTR = 16 nM and 3′ Cyc = 21 nM for AUF1 p45aDMA versus Kapp 5′ UTR = 172 nM and 3′ Cyc = 136 nM for the nonmethylated AUF1 p45; Fig. 6A). Importantly, methylation of AUF1 p45 did not generally increase the binding affinity of the protein to RNA. Different types of unrelated RNA substrates, such as a non-WNV stem–loop RNA that had approximately the same length and structure as 3′ Cyc, were bound at considerably lower affinity (Fig. 6A; data not shown).
The apparent differences in the affinity of AUF1 p45 and AUF1 p45aDMA to the termini of the WNV genome stimulated the next experiment to test whether the methylation of AUF1 p45 also affected the earlier observed supportive effect of the cellular protein on the initiation of viral negative-strand RNA synthesis. For this, we performed an in vitro replicase assay, which applied the purified NS5 and the WNV sgRNA as template (Friedrich et al. 2014), and complemented this assay with defined amounts of the purified nonmethylated and methylated AUF1 p45 variants. Note that this assay yields two types of NS5 synthesized RNA products that are detectable in the applied gel system. One product migrates at the same position as the original template and corresponds to a full-length negative-strand copy of the WNV sgRNA that is de novo synthesized by the NS5 RdRp. Several additional RNA species migrate significantly faster and likely represent hairpin by-products that are generated by priming of the replicase at the template's 3′ end, followed by a subsequent “copy-back” (Fig. 6B). Interestingly, when we performed the assay at essentially the same salt and buffer conditions as in the earlier RNA–protein interaction studies, complementation with the methylated AUF1 p45 protein stimulated viral negative-strand RNA synthesis at a significantly (two- to threefold) higher level than the nonmethylated protein. That is, in comparison with the noncomplemented control reaction, the NS5-catalyzed RNA synthesis was increased up to fivefold in the presence of AUF1 p45aDMA (Fig. 6B). The improved binding of the methylated AUF1 p45aDMA to each of the termini of the WNV RNA hence correlated with an enhanced stimulation of WNV negative-strand RNA synthesis.
Methylation of AUF1 p45 enhances the RNA chaperone activity that supports WNV 5′–3′ RNA–RNA interactions
During the earlier studies, we could exclude that the supportive effect of AUF1 p45 on the NS5-catalyzed negative-strand RNA synthesis was caused by a direct stimulation of the viral RdRp by the cellular protein. AUF1 p45 was rather shown to trigger the structural reorganization and cyclization of the WNV RNA, which is required for the effective initiation of RNA synthesis by NS5. This is achieved by an RNA chaperone activity of the protein, which is indicated to destabilize the 3′-terminal SL structure of the viral RNA (Fig. 1; Friedrich et al. 2014). Therefore, it was next interesting to prove whether the increased binding affinity of AUF1 p45aDMA to the WNV 5′ and 3′ UTRs (Fig. 6) affected this RNA chaperone activity.
Two types of previously established experimental assays were applied. First, to compare AUF1 p45 and AUF1 p45aDMA concerning their ability to stimulate cyclization of the 5′ and 3′ termini of the WNV RNA, we performed an RNA gel-shift assay. For this, the radioactively labeled RNA transcript of the WNV 5′ UTR was exposed to nonlabeled 3′ Cyc RNA, and this was done in the absence and presence of varying amounts of AUF1 p45 and AUF1 p45aDMA, respectively. After removal of the protein, 5′–3′ RNA–RNA duplex formation was detected as a mobility shift of the labeled 5′ UTR RNA on a native polyacrylamide gel (Fig. 7A). Although in the absence of protein, RNA–RNA interactions were only weakly detectable, the equilibrium of the individual 5′ and 3′ RNAs versus the 5′–3′ RNA–RNA duplex was significantly shifted toward the duplex when the RNAs were supplemented with AUF1 p45 or AUF1 p45aDMA. With both types of proteins, the WNV 5′–3′ RNA–RNA duplex formation was promoted in a dose-dependent manner, but it was most efficient with AUF1 p45aDMA (Fig. 7A). Notably, the measured levels of induction of RNA duplex formation (e.g., at 200 nM of protein concentration—threefold by AUF1 p45 and fivefold by AUF1 p45aDMA; Fig. 7A) properly correlated with the enhancing effects of the proteins on RNA synthesis in the in vitro replicase experiments (Fig. 6B). As already reported (Friedrich et al. 2014), an increase of the detectable level of RNA–RNA duplex formation was not observed if the experiment was performed with the RNA binding protein hnRNPH1, and a gel-shift was also undetectable when we performed the experiment in the absence of the 3′ Cyc RNA, i.e., solely with the 5′ UTR (see also Supplemental Fig. S6).
FIGURE 7.
The methylated AUF1 p45 shows an enhanced RNA chaperone activity. (A) Interactions of 3′ Cyc RNA and 5′ UTR RNA tested as a mobility shift on a native PAGE. The interactions were monitored in the absence and presence of AUF1 p45 and AUF1 p45aDMA, respectively. Two nanomoles of 3′ Cyc RNA was exposed to the indicated amounts of recombinant AUF1 p45 or AUF1 p45aDMA, followed by the addition of 2 nM [32P]-labeled 5′ UTR RNA; the added protein was removed before the analysis by PAGE. (Left) A representative gel is shown; the migration positions of the unbound 5′ UTR RNA and of the 5′–3′ RNA complex are indicated. (Right) The levels of 5′–3′ RNA complex formation were quantified and evaluated for statistical significance (RNA complex formation in the absence of protein set to 1); (*) P ≤ 0.05; (**) P ≤ 0.01. (B, top) Scheme of the fluorescence-based assay (3′SLtrunc–5′UAR interaction assay) to detect AUF1 p45-mediated conformational RNA rearrangement by dequenching of Cy5 (see text). (Bottom) The assay was performed with 10 nM of Cy5-labeled 3′ SLtrunc that had been incubated with the indicated concentrations of recombinant AUF1 p45aDMA. Following the addition of 100 nM 5′ UAR, the fluorescence emission was measured for 400 sec. The fluorescence signals were plotted as a function of time and fitted according to a first-order reaction (without protein; Equation 1) or second-order reaction (in the presence of protein; Equation 2). Note that no increase of fluorescence emission was detectable if the reaction was performed in the absence of 5′ UAR. (C) The observed rate constants kobs (sec−1) were plotted as a function of the protein concentration (see Supplemental Table S4). The kobs values for AUF1 p45 and hnRNPH1 were taken from Friedrich et al. (2014).
As a second approach, we performed an in vitro fluorescence-based assay that measures the kinetics of an RNA restructuring event, which, in consequence, enables the interaction of the 5′ UAR and 3′ UAR cyclization sequences (Friedrich et al. 2014) (in the following termed as 3′ SLtrunc–5′ UAR interaction assay). Essentially, this assay applies an unlabeled oligonucleotide consisting mainly of the 5′ UAR as well as a second RNA molecule, 3′ SLtrunc. 3′ SLtrunc mimics the lower part of the WNV 3′ SL stem structure by containing the 3′ UAR and the sequence of 3′ SL that is complementary to 3′ UAR. 3′ SLtrunc is 5′-labeled by Cy5 and 3′-labeled with a Black Hole Quencher (BHQ) and, at the given assay conditions and in the absence of any other reagent, the RNA forms a stem–loop structure with the 3′ UAR being a component of the stem. If the stem is formed, the BHQ causes a static quenching of the Cy5 fluorescence (Fig. 7B). Hybridization of the 3′ UAR in 3′ SLtrunc with the 5′ UAR–RNA accordingly requires a significant rearrangement of the stem structure, which is also a key event during the interaction of the intact termini of the WNV RNA genome (Fig. 1). In this case, dislocation from the BHQ and dequenching of the Cy5 fluorophore can be measured by fluorescence spectroscopy over time (Fig. 7B). The assay essentially involves two steps: (i) the opening of the stem of the 3′SLtrunc-RNA and (ii) the annealing of the complementary UAR sequences. It was designed in such a way that the data analysis focused exclusively on the destabilizing activity. For this, the 5′ UAR–RNA was applied in 10-fold excess to the 3′ SLtrunc-RNA in order to accelerate the second, annealing step of the reaction. During pilot studies we confirmed that under these conditions, the kinetics of the 3′ SLtrunc–5′ UAR interaction follows a first-order reaction. That is, the opening of the stem of the 3′ SLtrunc RNA was shown to be independent of its initial concentration and to represent the rate-limiting step (“kinetic bottleneck”) of the reaction. Accordingly, in an RNA-only experiment, hybridization of the UARs was found to occur rather slowly with a first-order reaction rate constant of 0.0062 sec−1 (determined with Equation 2; see Materials and Methods). This changed when we added the recombinant AUF1 p45 or AUF1 p45aDMA at increasing concentrations to the 3′SLtrunc RNA and then supplemented the reaction with the 5′ UAR RNA. Here, the kinetics followed a second-order reaction and was significantly accelerated in a protein concentration-dependent manner (determined with Equation 3). Interestingly, the reaction was most accelerated by the methylated AUF1 p45aDMA. For example, with 50 nM AUF1 p45, the rate constant was increased twofold , but in the presence of 50 nM AUF1 p45aDMA, we measured a ninefold increase (Fig. 7C; Supplemental Table S4). Note that at high protein concentrations, the fluorescence signal rapidly decreased after reaching the maximum of RNA complex formation. This suggested that the RNA complex served as a further substrate for AUF1 p45 and AUF1 p45aDMA when these were present in excess (Fig. 7B; Friedrich et al. 2014). An acceleration of the 3′SLtrunc–5′UAR interaction was undetectable when we applied the RNA binding protein hnRNPH1 to the reaction (Fig. 7C; Supplemental Table S4; Friedrich et al. 2014). These results demonstrated that the RNA chaperone activity of AUF1 p45, which leads to a destabilization of the 3′SL stem structure and in this way triggers 5′–3′ interactions of the WNV RNA, is considerably enhanced by arginine methylation.
AUF1 p45 is a true “RNA annealer,” which accelerates the hybridization of the viral cyclization sequences
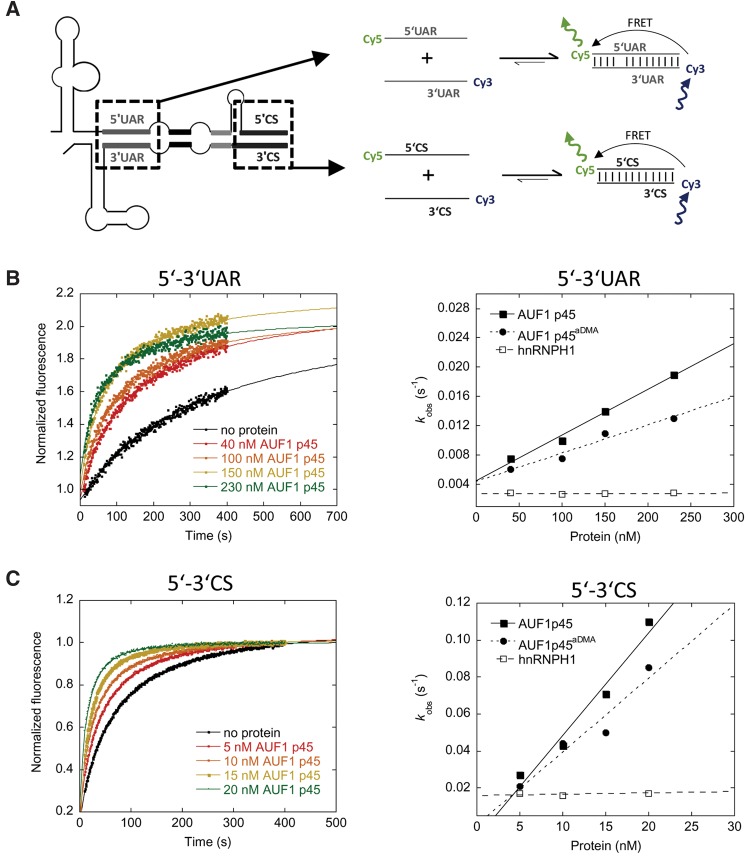
The earlier experiments indicated that AUF1 p45, and particularly the methylated AUF1 p45aDMA, support the “melting” of the stem structure that masks the 3′UAR cyclization sequence in the linear form of the viral genome. In this manner, the host factor increases the probability that the 3′UAR may hybridize with the 5′UAR in the other terminus of the viral genome (Fig. 1). In view of the scenario that several proteins with RNA chaperone activity show an RNA annealing activity (act as “RNA annealers”) (Rajkowitsch et al. 2007), we finally found it important to test whether AUF1 p45 and AUF1 p45aDMA also supported the hybridization of the UAR and/or the CS cyclization sequences. To address this, we took advantage of an RNA annealing assay of Rajkowitsch and Schroeder (2007) applying short, Cy5 and Cy3 fluorophore-labeled RNA molecules that corresponded to the 5′- and 3′UAR and 5′- and 3′CS elements, respectively. In this way, annealing could be measured by the development of a fluorescence resonance energy transfer (FRET) signal over time (Fig. 8A). Following this approach first with the RNAs only, we observed that hybridization of the 5′UAR and 3′UAR sequences occurred slowly, i.e., with an observed rate constant of 0.0029 sec−1 (Fig. 8B; Supplemental Table S5; determined with Equation 3). However, after the addition of AUF1 p45 to the reaction, the rate constant was significantly increased, and this occurred again in linear dependency of the protein concentration (Fig. 8B; Supplemental Table S5; determined with Equation 3). For example, in the presence of 150 nM AUF1 p45, we observed a fivefold increase of the rate constant in comparison to the RNA-only situation. As also shown in Figure 8B, supplementation of the reaction with AUF1 p45aDMA yielded comparable rate constants as with the nonmethylated AUF1 p45. Experiments that were performed with identical concentrations of hnRNPH1 showed no effect.
FIGURE 8.
AUF1 p45 and AUF1 p45aDMA support the annealing of the UAR and CS cyclization sequences of the WNV genome. (A) Scheme of the fluorescence-based assay to detect an RNA annealing activity of AUF1 p45 or AUF1 p45aDMA. Annealing of the complementary WNV 5′- and 3′UAR and WNV 5′- and 3′CS RNAs that are fluorescently labeled with Cy5 (5′UAR, 5′CS) and Cy3 (3′UAR, 3′CS), respectively, leads to a detectable FRET signal. (B, left panel) The assay (5′UAR–3′UAR RNA annealing assay) was performed with 10 nM of Cy5-labeled 5′UAR RNA that was incubated with the indicated concentrations of recombinant AUF1 p45 and AUF1 p45aDMA, respectively. Following the addition of 10 nM Cy3-labeled 3′UAR RNA, the fluorescence emission of the Cy5 fluorophore was measured for 400 sec. The fluorescence signals were plotted as a function of time and fitted according to a second-order reaction (Equation 3; see Materials and Methods). (Right panel) The observed rate constants kobs (sec−1) were plotted as a function of the concentration of AUF1 p45 and AUF1 p45aDMA; they are given in Supplemental Table S5. (C) Same as in B except that the complementary 5′- and 3′CS sequences of the WNV genome were used. The measured kobs (sec−1) values of the 5′CS–3′CS RNA annealing assay are given in Supplemental Table S6.
With the major WNV cyclization sequences, 5′CS and 3′CS, the RNA-only hybridization reaction revealed a rate constant of 0.017 sec−1. Thus, per se interactions of the CS sequences turned out to occur considerably more efficiently than interactions of the 5′- and 3′UARs (Fig. 8C). Nevertheless, also in this case, the measured rate constants were significantly increased as soon as we supplemented the reaction with AUF1 p45 or AUF1 p45aDMA. Again, we observed a linear dependency on the concentration of the proteins, and the activities of AUF1 p45 and AUF1 p45aDMA were very similar. In contrast, the addition of hnRNPH1 had no consequence. Notably, the annealing activities of AUF1 p45 or AUF1 p45aDMA were considerably higher with the 5′- and 3′CS elements than with the 5′ and 3′UARs. That is, already at 15 nM of AUF1 p45, the annealing rate constant was increased fourfold (Fig. 8C). These results demonstrate that AUF1 p45 displays a yet unknown, significant RNA annealing activity, which, however, is not affected by the methylation of the protein. Interestingly, the AUF1 p45 mediated RNA annealing turned out to be considerably more efficient with the WNV CS than with the UAR cyclization elements. This indicated an evident substrate preference of the RNA annealing activity of AUF1 p45 for the CS elements.
DISCUSSION
Arginine methylation has been demonstrated to affect protein translocation or to modulate protein activity in defined processes during signal transduction, transcriptional control or DNA repair (Bedford and Clarke 2009). Many RNA binding proteins are arginine methylated, signifying that this type of post-translational modification represents a critical element of their function, e.g., during post-transcriptional regulation of gene expression (Rajyaguru and Parker 2012). In mammalian cells, most hnRNP proteins were found to be methylated (Beyer et al. 1977; Wilk et al. 1985; Liu and Dreyfuss 1995), among them also hnRNPD/AUF1 (Liu and Dreyfuss 1995; Ong et al. 2004; Fellows et al. 2013). However, in contrast to other hnRNP proteins where methylation was shown to affect the cellular localization, protein–protein interactions or nucleic acid binding (Yu 2011), specific information on a physiological role of AUF1 methylation is sparse. Only very recently, methylation was determined to be required for the AUF1 p37-mediated repression of translation of the mRNA of vascular endothelial growth factor in macrophages (Fellows et al. 2013). The authors of this study performed MS analysis of p37 peptides and observed that in inactivated macrophages, the first and second RGG sequence elements of the protein were monomethylated while in activated cells these were dimethylated. In addition, the RAR motif, which is located between RGG1 and RGG2, was monomethylated in the activated macrophages. These findings fit well with the data presented in Figure 4 and Table 1. In AUF1 p45, the methylated R272 of peptide 1 is part of the first RGG, while R278, 280, 282 of peptide 2 locate in the RAR and second RGG sequences, respectively. We and Ong et al. (2004) also observed a methylation of the third RGG, corresponding to R345 of peptide 3 (Table 1), and we showed that with AUF1 p45 all five arginine residues are dimethylated in different cell lines. Recently, we also analyzed the methylation status of p37, p40, and p42 in Huh7 cells and observed all isoforms displaying the same methylation sites as AUF1 p45. However, with the smaller isoforms p37 and p40, several of the five arginines turned out to be only monomethylated (S Friedrich and A Schierhorn, unpubl.). These findings suggest that AUF1 p45 and p42 commonly exist fully methylated in the cell, although this appears to be not the case with p37 and p40, in which the methylation status may be affected by the cell type and/or external stimuli.
An explanation for the surprisingly constant methylation status of AUF1 p45 may be its tight interaction with PRMT1 (Fig. 2; Supplemental Fig. S1). PRMT1 is the most prevalent and conserved member of type I PRMT methyltransferases, which transfer either one or two methyl groups from an appropriate donor to a single guanidino nitrogen on a protein-internal arginine residue leading either to a monomethylarginine (MMA) or an asymmetric dimethylarginine (aDMA) (Bedford and Clarke 2009; Yu 2011). Although PRMT1 was already indicated to catalyze the modification of several hnRNPs (Nichols et al. 2000; Wada et al. 2002; Herrmann et al. 2004; Passos et al. 2006), we here present a complementary set of data demonstrating that PRMT1 is necessary and sufficient to catalyze the observed methylation pattern of AUF1 p45 (Fig. 3). AUF1 p42 and p45 bind PRMT1 considerably more efficiently than AUF1 p37 and p40 (Fig. 2). This supports the idea that the exon 7-encoded protein component represents an important determinant of the interaction with PRMT1. Because p37 and p40 are nevertheless methylated, though not as effectively as AUF1 p45, the exon 7-encoded protein component is suggested to enhance the binding of PRMT1 rather than to act as an exclusive binding site. Our data suggest that besides AUF1 p45 also PRMT1 is an important molecular determinant of WNV RNA replication. However, whether the host factor activity of PRMT1 involves solely the methylation of AUF1 p45 remains to be determined as the methyltransferase may modulate also other cellular or viral proteins that participate in WNV replication.
AUF1 is an RGG/RG motif protein (Kiledjian and Dreyfuss 1992; Thandapani et al. 2013). Members of this group of proteins are known to regulate various processes in RNA metabolism, and the RGG/RG motifs are indicated to essentially contribute to these functions, e.g., by interacting directly with protein partners or with the RNA substrates (Singh and Valcárcel 2005; Rajyaguru and Parker 2012). As outlined, all five identified methylation sites concentrate within the RGG/RG motif in the AUF1 p45 C terminus (Fig. 4). Methylation significantly enhanced the affinity of AUF1 p45 to the termini of the WNV genomic RNA (Fig. 6A) as well as the RNA chaperone activity of the protein (Fig. 7, see also below) implying that the RGG/RG motif plays a critical role in both functions. Currently, we can only speculate about the underlying mechanisms. The interactions of RGG/RG motifs with RNA molecules are believed to be based on the arginine's positive charge but also on hydrophobic interactions. Accordingly, the improved binding of AUF1 p45aDMA to the WNV RNA (Fig. 6A) may, to some extent, be related to a methylation-caused increment of the local basicity and hydrophobicity of the arginine residues (Bedford and Richard 2005; Pahlich et al. 2006). However, our data clearly show that the methylation of AUF1 p45 did not generally enhance the protein's RNA binding capacity because both the methylated and nonmethylated AUF1 p45 variants show a similar high affinity to the AU-rich element in the upstream portion of the WNV 3′ UTR and a comparable low affinity to unrelated RNA molecules (Fig. 6A). This supports the view that the interaction of AUF1 p45 with the WNV RNA termini involves specific features on the protein and/or RNA level. On the side of the protein, the CD data provide initial indications that the methylation of the RGG/RG motif correlates with a considerable change of the structure of AUF1 p45 (Fig. 5), which may explain the improved binding affinity of AUF1 p45aDMA to the WNV RNA termini. Thus, it is conceivable that methylation changes the positioning, and, with this, the functional interaction and RNA binding characteristics of the RGG/RG and RRM motifs, respectively. Our data are congruent with similar observations with the splicing factor proline- and glutamine-rich (SFPQ), which also shows an increased RNA binding affinity upon arginine methylation (Snijders et al. 2015).
How does the arginine methylation affect the RNA chaperone activity of AUF1 p45? RNA chaperones, per definition, increase the kinetics of intra- and intermolecular RNA–RNA interactions without consuming ATP. In this way, these proteins support the reorganization of misfolded toward kinetically favored and properly folded RNA molecules (Herschlag 1995; Schroeder et al. 2002; Rajkowitsch et al. 2007; Semrad 2011). As a general feature, RNA chaperones contain a high level of intrinsically disordered regions (Tompa and Csermely 2004; Ivanyi-Nagy et al. 2005), which, due to their high structural dynamics, enable these proteins to bind in an unspecific manner to a wide variety of RNA substrate molecules (Rajkowitsch et al. 2007). According to an entropy-transfer model, the activity of RNA chaperones is hypothesized to be mediated by disorder to order transitions of these regions (Tompa and Csermely 2004). As pointed out by a recent study, a central component of this activity appears to be the weakening of intramolecular interactions in the RNA substrate that involve guanosines (Grohman et al. 2013). The energy to unfold RNA structures is supposed to be obtained by the simultaneous folding of the chaperone and its substrate. In fact, for several chaperones a folding of disordered regions was observed after substrate binding (Dyson and Wright 2005; Kiefhaber et al. 2012). Our CD spectroscopy measurements (Fig. 5B) clearly indicate that AUF1 p45 comprises a high degree of disordered regions. According to secondary structure predictions (not shown) and in close analogy with the hnRNP proteins A1 and U, which also have an RNA chaperone activity (Pontius and Berg 1990; Kiledjian and Dreyfuss 1992; Munroe and Dong 1992; Portman and Dreyfuss 1994), a significant proportion of these regions locate in the AUF1 p45 C terminus, the part of the protein that encloses the RGG/RG motif and all methylation sites. The considerable increase in the RNA chaperone activity of AUF1 p45aDMA hence may be explained in two ways. As outlined, on the one hand, the methylated protein may be more active on the WNV 3′SL simply due to its enhanced binding affinity to the WNV 3′ terminus. On the other hand, the methylation-induced conformational change of AUF1 p45 (Fig. 5B) may have a direct impact on the protein's chaperone activity. Thus, methylation of the RGG/RG motif may cause a reorganization of the disordered regions in the AUF1 p45 C terminus, which, in turn, would facilitate disorder to order transitions in the restructuring of the WNV 3′SL. While this scenario is speculative, it is in agreement with earlier data of Raman et al. (2001) showing that the dimethylation of a protein's RGG motifs may alter the structure of the RNA to which these RGG motifs bind. Our findings also go well with studies indicating that the AUF1 isoforms may differently alter the local structures of model ARE substrates (Wilson et al. 2001; Zucconi et al. 2010) and that phosphorylation of AUF1 p40 may affect the conformation of a bound RNA (Wilson et al. 2003). Taken together, to our knowledge this is the first report showing that post-translational methylation of a protein enhances and possibly regulates its RNA chaperone activity.
Our study demonstrated that AUF1 p45 actually has two different RNA restructuring activities, namely the earlier identified RNA chaperone activity that supports the destabilization of the 3′SL stem structure, and a newly identified RNA annealing activity. In contrast to the destabilizing function of AUF1 p45, the annealing activity was not altered upon methylation (Fig. 8). One scenario that would explain this is that the annealing activity may be predominately mediated by the protein's RNA recognition motifs (RRM), which are not affected by arginine methylation (Fig. 4B). RRMs specifically bind to single-stranded nucleic acids (Maris et al. 2005) and thus could support the annealing of the single-stranded cyclization sequences. Remarkably, the annealing activity of AUF1 p45 had an evident preference for the 5′–3′ CS versus the 5′–3′UAR sequences (Fig. 8). This may relate to the sequence composition of the 5′–3′ CS elements, which are AU-rich and may facilitate the association of AUF1 p45. Overall, these data suggest that AUF1 p45 has two activities in the WNV replication process, each of which may support the transition of the linear form of the viral genome to the 5′–3′ cyclized form. On the one hand, the protein destabilizes 3′SL and in this way increases the accessibility of the 3′UAR sequence (Friedrich et al. 2014). On the other hand, AUF1 p45 may stimulate hybridization of the 5′- and 3′CS elements (Fig. 1). Whether AUF1 p45, besides destabilizing the 3′SL, may operate in a similar way on other RNA elements is currently uncertain. A candidate structure would be SLB within the 5′ UTR, where a destabilization could expose the 5′UAR element and thus support genome cyclization.
By binding to defined genomic elements and by modulating viral protein or RNA synthesis, AUF1/hnRNPD was found to accelerate or inhibit the life cycles of various RNA viruses (Paek et al. 2008; Lund et al. 2012; Cathcart et al. 2013; Hino et al. 2013; Lin et al. 2014). It is quite conceivable that the methylation-enhanced RNA restructuring activity of the AUF1 p45 isoform, which was demonstrated here to support WNV replication, may underlie several of the functions of AUF1 during the replication processes of other viruses as well. Moreover, the methylation of AUF1 may be a means of regulating the protein's RNA chaperone activity during various processes in RNA metabolism.
MATERIALS AND METHODS
In vitro transcription reactions, RNA extraction, and quantitative RT-PCR
These procedures were performed as previously described (Friedrich et al. 2014).
Cells, culturing, and transfection conditions
Human hepatoma cells (Huh7) were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal calf serum (FCS; PAN-Biotech), 1% penicillin/streptomycin (Gibco), 0.1% D-biotin and 0.1% hypoxanthine (Sigma). Duplex siRNAs targeting the mRNA of PRMT1 were purchased from Eurofins MWG Operon (see Supplemental Table S1). Transfection was performed with 70% confluent Huh7 cells and Lipofectamine RNAiMAX (Invitrogen) by using the manufacturer's instructions. To test for effects of an siRNA-mediated depletion of PRMT1 on WNV replication, approximately 107 cells were retransfected 72 h later with 300 ng WNV replicon RNA (99 fmol) or 120 h later with WNVRluc replicon RNA using the Bio-Rad Gene Pulser II (1 pulse without controller at 0.2 kV and 950 µF). A luciferase assay kit was used to quantify the activity of the replicon-encoded Renilla luciferase (Promega). To test for effects of PRMT1 depletion on propagation of infectious WNV, approximately 5 × 105 Huh7 cells were infected with WNV strain 3356 (1 h at a tissue culture infectious dose TCID50 of 1 × 103/mL). For the quantification of virus titers after infection with WNV, the viral RNA was extracted from culture fluids and quantified by a one-step qRT-PCR protocol using FRET probes targeting the 5′ UTR coding region (Eiden et al. 2010). The qRT-PCR was performed with the QuantiFast Multiplex RT-PCR Mix (Qiagen). The reactions contained the extracted RNA, RNase Inhibitor (20U, Thermo Scientific), primers WNV-INEID-f1-wob (8 pmol) and WNV-INEID-r1-mod (8 pmol), and the WNV-INEID-probe (4 pmol) (see Supplemental Table S2). The qRT-PCR conditions were 50°C for 20 min, 95°C for 5 min and 45 cycles at 95°C for 15 sec, 56°C for 10 sec, 60°C for 30 sec. To quantify the RNA copy number, the data were related to a standard curve that was assessed by plasmid dilution. Further details on RT and PCR conditions are given on request.
The dendritic cell line DC2.4 was kindly provided by Kenneth L. Rock (Dana Farber Cancer Institute, Boston, MA) and cultured in RPMI 1640 (Lonza) containing 10% fetal calf serum (FCS; PAN-Biotech), 2 mM l-glutamine (Lonza), 1% nonessential amino acids (Lonza), 1% penicillin/streptomycin (Gibco), and 0.054 µM 2-mercaptoethanol (Roth). The neuronal cell line SHSY5Y was cultured in DMEM F-12 (Gibco) supplemented with 10% fetal calf serum (FCS; PAN-Biotech), 1% penicillin/streptomycin (Gibco), and 2 mM l-glutamine (Lonza).
Plasmids encoding SUMO-fusion proteins
The pETSUMO expression system was used for the expression of recombinant proteins in E. coli. Cloning of the NS5 and AUF1 p45 coding sequences into pETSUMO has been previously described (Friedrich et al. 2014). The cDNAs coding for the different isoforms of PRMT1 (v1, v2, v3, and v5) were generated by reverse-transcription PCR of human total mRNA isolated from Huh7 cells using primers PRMT1v1v2BsaIFor, PRMT1v3BsaIFor, PRMT1v5BsaIFor, and PRMT1XhoI and inserted between the BsaI and XhoI restriction sites of the pETSUMO expression vector. To generate recombinant purified methylated AUF1 p45, the PRMT1 v1 gene was cloned upstream of the AUF1 p45 gene into pETSUMO to achieve a simultaneous expression of PRMT1 v1 and AUF1 p45. This was done by PCR amplification of the PRMT1 v1 gene using the primers PRMT1RBSFor and PRMT1RBSRev. In this way, also a ribosomal binding site was introduced via the forward primer. The PCR fragment was then blunt-end ligated upstream of the ribosomal binding site of the PCR amplified (primers pETSUMORBSFor and pETSUMORBSRev) pETSUMO-AUF1 p45. All primer sequences are provided in Supplemental Table S2.
Plasmids encoding FLAG–AUF1 fusion proteins and plasmids encoding WNV replicon and subgenomic RNAs
Construction of these plasmids has been described elsewhere (Friedrich et al. 2014).
Expression and purification of FLAG–AUF1 fusion proteins
Expression and purification of FLAG–AUF1 fusion proteins were done as previously described (Friedrich et al. 2014). Briefly, the plasmids encoding the FLAG fusion proteins were transfected into Huh7 cells using TurboFect in vitro transfection reagent (Fermentas). Forty-eight hours later, the cells were harvested and the cytoplasmic extracts were subjected to anti-FLAG M2 affinity gel (Sigma) in the presence of 0.1 mg/mL RNase A at 4°C overnight. Following precipitation and washing, according to the manufacturer's instructions, the FLAG fusion proteins were eluted with 3 × FLAG peptide (Sigma) at a final concentration of 500 ng/µL. The amount of the eluted FLAG fusion proteins were evaluated on silver-stained SDS–PAGE in comparison with mass standards of recombinant AUF1 p45 that was purified from E. coli (see below).
Expression and purification of PRMT1
The different PRMT1 isoforms were expressed as SUMO fusion proteins (see above) in E. coli BL21-CodonPlus (DE3)-RP after induction (0.4 mM IPTG). The proteins were purified from the soluble fraction using Nickel-agarose affinity chromatography (HisTrap HP, GE Healthcare), and, after cleavage with SUMO-protease, by anion exchange chromatography (RESOURCE Q, GE Healthcare). Active fractions were dialyzed and stored in 20 mM HEPES/NaOH, 100 mM KCl, 10% glycerol (pH 7.6) at −80°C. The protein concentration was determined by measuring the absorbance at 280 nm using ε = 56,435 M−1 cm−1.
Expression and purification of nonmethylated and methylated AUF1 p45
The nonmethylated AUF1 p45 was purified as previously described (Friedrich et al. 2014). Briefly, purification was performed from the soluble fraction of Escherichia coli BL21-CodonPlus (DE3)-RP cells using Nickel-agarose affinity chromatography (HisTrap HP, GE Healthcare) and, after cleavage with SUMO-protease, by Heparin sepharose affinity chromatography and gel-filtration (HiLoad 16/60 Superdex 200, GE Healthcare). To obtain recombinant purified methylated AUF1 p45, the PRMT1 v1 and AUF1 p45 genes were coexpressed in a bicistronic pETSUMO expression vector using the same E. coli cells. To allow for efficient methylation of AUF1 p45 by PRMT1, the bacteria were incubated for an additional 2 h with the translation inhibitor erythromycin (10 µg/mL). The methylated AUF1 p45aDMA was purified using the analogous purification protocol as with the nonmethylated AUF1 p45. UV/Vis absorption spectra were measured using a JASCO V-550 spectrometer. The protein concentration was determined by measuring the absorbance at 280 nm using ε = 58,915 M−1 cm−1. The nonmethylated and methylated AUF1 p45 were stored at −80°C in 20 mM Tris/HCl (pH 7.6), 150 mM KCl, 1 mM Tris(2-carboxyethyl)phosphine (TCEP).
Expression and purification of WNV NS5 and hnRNPH1
Expression and purification of WNV NS5 and hnRNPH1 was performed as previously described (Friedrich et al. 2014).
In vitro methylation assay
Thirteen picomoles of AUF1 p45 and 5 pmol of PRMT1 v1 were incubated in a reaction mixture (10 µL total volume) that contained 50 mM HEPES/NaOH (pH 8.0), 10 mM KCl, 5 mM MgCl2, 0.2 mg/mL bovine serum albumin, 0.5 mM dithiothreitol, and 40 µM [S-14C]adenosylmethionine (60 mCi/mmol, Amersham Biosciences). The reaction was performed at 30°C for the time indicated. The degree of methylation was analyzed by SDS–PAGE and phosphorimaging. To test for the methylation status of transiently expressed FLAG–AUF1 p45 in human cells, 5 pmol of the FLAG–AUF1 p45 that was purified from the indicated cell line and 5 pmol of recombinant nonmethylated AUF1 p45 that was prepared from E. coli were methylated in vitro by 5 pmol PRMT1 v1. The samples were taken after 2 h and analyzed as described.
Western blotting and antibodies
To detect NS5 in Western blots, a rabbit polyclonal antiserum was applied that was raised against the recombinant, purified full-length protein (Eurogentec). Anti-Vinculin antibody and anti-FLAG antibody were purchased from Sigma. Anti-PRMT1 antibody and anti-dimethyl-arginine antibody were purchased from Millipore. Secondary antibodies were purchased from GE Healthcare and LICOR. Western blots were performed using standard protocols and by following the instructions of the manufacturers.
Mass spectrometry
The protein bands were excised from the gels and the slices incubated for 45 min at 50°C with 10 mM dithiothreitol (Sigma) in 100 mM ammonium bicarbonate. After removal of the solution, the slices were treated for 45 min with 55 mM iodoacetamide (Sigma) in 100 mM ammonium bicarbonate in the dark to modify cysteine residues. The gel pieces were then washed three times with water, twice with 50 mM ammonium bicarbonate and finally with 50 mM ammonium bicarbonate in 50% acetonitrile. After drying under a gentle stream of nitrogen, the pieces were re-swollen in 20 µL 50 mM ammonium bicarbonate (pH 8.0) and digested with trypsin or chymotrypsin (Promega) overnight at 37°C. The peptide mass fingerprint spectra were recorded on an Ultraflex-II TOF/TOF mass spectrometer (Bruker Daltonic) equipped with MALDI source, nitrogen laser, LIFT cell for fragment ion post-acceleration, and a gridless ion reflector. The software Flex Control 2.4, Flex Analysis 2.4, and Biotools 3.0 were used to operate the instrument and to analyze the data. For external calibration, a peptide calibration mixture (Bruker Daltonics) was used. The methylated peptides were fragmented using the LIFT method (Suckau et al. 2003). MALDI samples were prepared as follows. One microliter of a DHB-matrix (7 mg 2.5-dihydroxybenzoic acid in 100 µL methanol) was mixed with 1 µL digest and deposited on the target.
Filter binding assay
The assay has been previously described in detail (Friedrich et al. 2014). Briefly, the binding reactions contained 50 mM Tris/HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.1 nM 32P-labeled target RNA, and increasing concentrations of AUF1 p45 or AUF1 p45aDMA (total volume 20 µL). After incubation for 30 min at room temperature, the reaction was applied to a nitrocellulose membrane (Whatman) that was presoaked with 50 mM Tris-HCl, 100 mM KCl and 5 mM MgCl2 (pH 8.0). The membrane was air dried and visualized by autoradiography. The radioactive signals were quantified using ImageQuant software (GE Healthcare), plotted against the protein concentration, and fitted by KaleidaGraph (Synergy) to a single-site binding model (Friedrich et al. 2014) or with the following equation to determine the apparent binding constant Kapp:
| (1) |
where Sb is the radioactive signal bound, c is the total concentration of the protein, n is the cooperativity coefficient, Kapp is the apparent binding constant, Smax is the maximum signal amplitude, and Soffset is the offset (see also Supplemental Fig. S4).
RNA gel-shift assay
RNA–RNA interactions were analyzed by electrophoretic mobility shift assays as performed earlier (Friedrich et al. 2014). The binding reaction contained 5 mM HEPES/NaOH (pH 7.9), 100 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, uniformly 32P-labeled 5′ UTR (2 nM), and nonlabeled 3′Cyc-RNA (2 nM) in a final volume of 20 µL. The purified, recombinant AUF1 p45 or AUF1 p45aDMA was added to the RNAs at final concentrations of 0, 50, 100, and 200 nM, respectively. The binding reaction was performed at room temperature for 30 min and quenched by the addition of stop solution (0.5% SDS, 20 mM EDTA). After removal of the proteins by phenol–chloroform extraction, the RNA–RNA complexes were resolved by native 5% polyacrylamide gel electrophoresis in 0.5× TBE at 4°C, analyzed by phosphorimaging and quantified by ImageQuant Software (GE Healthcare).
Fluorescence-based RNA–RNA interaction assay
Briefly, the purified, recombinant AUF1 p45aDMA was added to 10 nM of 5′-Cy5-labeled 3′SLtrunc-RNA (purchased from IBA; Fig. 7; Supplemental Table S3) at different concentrations in assay buffer (50 mM HEPES/NaOH, pH 8.0, 100 mM KCl, 5 mM MgCl2). Then, 100 nM of nonlabeled 5′UAR RNA was added and readings were taken for another 400 sec. Changes in the fluorescence signals were monitored in a Fluoromax-4 Spectrofluorometer (Jobin Yvon) at 22°C with the following parameters: excitation at 643 nm, emission at 667 nm, slit widths 5 and 5 nm, respectively. Fluorescence intensities relative to the starting fluorescence were plotted against the time and fitted by KaleidaGraph (Synergy) to first-order reaction when AUF1 p45aDMA was omitted (Equation 2) and second-order reaction when AUF1 p45aDMA was included (Equation 3), yielding the corresponding rate constants kobs. For further detail, see Friedrich et al. (2014).
| (2) |
| (3) |
where ΔF is the total change of relative fluorescence amplitude, Foffset is the fluorescence intensity at the start point of the reaction, Fmax is the maximum signal amplitude, kobs is the observed rate constant, and t is the time.
Replicase assay
The replicase assay was done as previously described (Friedrich et al. 2014) except for a different buffer system to adjust the ionic strength of the assay to the conditions used in the filter-binding assay. Hence, the assay was performed in buffer containing 50 mM HEPES–NaOH (pH 8.0), 100 mM KCl, 5 mM MgCl2, 0.5 mM MnCl2, 1 mM dithiothreitol.
FRET-based RNA annealing of complementary cyclization sequences
The fluorescent oligonucleotides (Supplemental Table S3) were purchased from IBA. The purified, recombinant AUF1 p45 or AUF1 p45aDMA were added at different concentrations (as indicated) to 10 nM of 5′-Cy3-labeled 3′UAR-RNA or 5′-Cy3-labeled 3′CS-RNA in assay buffer (50 mM HEPES/NaOH (pH 8.0), 100 mM KCl, 5 mM MgCl2). The reaction was allowed to proceed for 100 sec. Then, 10 nM of 5′-Cy5-labeled 5′UAR-RNA or 5′-Cy5-labeled 5′CS-RNA was added and readings were taken for another 400 sec. Changes in the fluorescence signals were monitored in a Fluoromax-4 Spectrofluorometer (Jobin Yvon) at 22°C. The Cy3 fluorophore was excited at 535 nm wavelength and readings were taken at the Cy5 emission wavelength 680 nm. RNA–RNA annealing of the two fluorophore-labeled RNAs results in a fluorescence resonance energy transfer (FRET). Fluorescence intensities relative to the starting fluorescence were plotted against the time and fitted by KaleidaGraph (Synergy software) to a second-order reaction (Equation 3, see above) yielding the corresponding rate constants kobs.
Analytical ultracentrifugation
Sedimentation equilibrium measurements were performed in an analytical ultracentrifuge Optima XL-A (Beckman Instruments, Inc.). Double sector cells were used at 14,000 rpm and 20°C in an An50Ti rotor. Analysis was performed at a protein concentration of 5 µM AUF1 p45aDMA in 20 mM Tris/HCl, pH 7.6, 150 mM KCl, 1 mM TCEP. The data obtained were analyzed using the program Sedfit (Schuck 2000). For calculations, a partial specific volume of 0.73 mL/mg was assumed.
Circular dichroism
Measurements of dichroic properties of AUF1 p45 and AUF1 p45aDMA were performed using a JASCO J-810 spectropolarimeter. Far-UV circular dichroism spectra were recorded at a protein concentration of 10 µM in 20 mM Tris/HCl, pH 7.6, 150 mM KCl, 1 mM TCEP at 22°C using cuvettes with optical path lengths of 0.1 mm. Acquired protein spectra were corrected for buffer contribution and smoothed using the Spectra Manager I software (JASCO). The data were converted to mean residue ellipticity ΘMRW.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
Pei-Yong Shi is acknowledged for supplying the cDNA clone of the WNV replicon. We thank Christine Hamann and Nancy Funk for technical assistance.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.055269.115.
REFERENCES
- Alvarez DE, Lodeiro MF, Ludueña SJ, Pietrasanta LI, Gamarnik AV. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol 79: 6631–6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. 2009. Protein arginine methylation in mammals: who, what, and why. Mol Cell 33: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Richard S. 2005. Arginine methylation an emerging regulator of protein function. Mol Cell 18: 263–272. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Christensen ME, Walker BW, LeStourgeon WM. 1977. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell 11: 127–138. [DOI] [PubMed] [Google Scholar]
- Bidet K, Garcia-Blanco MA. 2014. Flaviviral RNAs: weapons and targets in the war between virus and host. Biochem J 462: 215–230. [DOI] [PubMed] [Google Scholar]
- Brinton MA, Basu M. 2015. Functions of the 3′ and 5′ genome RNA regions of members of the genus Flavivirus. Virus Res 206: 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton MA, Dispoto JH. 1988. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162: 290–299. [DOI] [PubMed] [Google Scholar]
- Cathcart AL, Rozovics JM, Semler BL. 2013. Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. J Virol 87: 10423–10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota AT, Kramer LD. 2013. Vector-virus interactions and transmission dynamics of West Nile virus. Viruses 5: 3021–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corver J, Lenches E, Smith K, Robison RA, Sando T, Strauss EG, Strauss JH. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J Virol 77: 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J, Boisvert FM, Boulanger MC, Bedford MT, Richard S. 2003. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol Biol Cell 14: 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhang B, Shi PY. 2008. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology 381: 123–135. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. 2005. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208. [DOI] [PubMed] [Google Scholar]
- Eiden M, Vina-Rodriguez A, Hoffmann B, Ziegler U, Groschup MH. 2010. Two new real-time quantitative reverse transcription polymerase chain reaction assays with unique target sites for the specific and sensitive detection of lineages 1 and 2 West Nile virus strains. J Vet Diagn Invest 22: 748–753. [DOI] [PubMed] [Google Scholar]
- Fellows A, Deng B, Mierke D, Robey RB, Nichols RC. 2013. Peptides modeled on the RGG domain of AUF1/hnRNP-D regulate 3′ UTR-dependent gene expression. Int Immunopharmacol 17: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev 20: 2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. 2011. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J Biol Chem 286: 6929–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich S, Schmidt T, Geissler R, Lilie H, Chabierski S, Ulbert S, Liebert UG, Golbik RP, Behrens SE. 2014. AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J Virol 88: 11586–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet I, Gauvin G, Boisvenue S, Côté J. 2007. Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J Biol Chem 282: 33009–33021. [DOI] [PubMed] [Google Scholar]
- Gratacos FM, Brewer G. 2010. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA 1: 457–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohman JK, Gorelick RJ, Lickwar CR, Lieb JD, Bower BD, Znosko BM, Weeks KM. 2013. A guanosine-centric mechanism for RNA chaperone function. Science 340: 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann F, Bossert M, Schwander A, Akgün E, Fackelmayer FO. 2004. Arginine methylation of scaffold attachment factor A by heterogeneous nuclear ribonucleoprotein particle-associated PRMT1. J Biol Chem 279: 48774–48779. [DOI] [PubMed] [Google Scholar]
- Herschlag D. 1995. RNA chaperones and the RNA folding problem. J Biol Chem 270: 20871–20874. [DOI] [PubMed] [Google Scholar]
- Hino K, Sato H, Sugai A, Kato M, Yoneda M, Kai C. 2013. Downregulation of Nipah virus N mRNA occurs through interaction between its 3′ untranslated region and hnRNP D. J Virol 87: 6582–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias NG, Gamarnik AV. 2011. Dynamic RNA structures in the dengue virus genome. RNA Biol 8: 249–257. [DOI] [PubMed] [Google Scholar]
- Ivanyi-Nagy R, Davidovic L, Khandjian EW, Darlix JL. 2005. Disordered RNA chaperone proteins: from functions to disease. Cell Mol Life Sci 62: 1409–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J Virol 75: 6719–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefhaber T, Bachmann A, Jensen KS. 2012. Dynamics and mechanisms of coupled protein folding and binding reactions. Curr Opin Struct Biol 22: 21–29. [DOI] [PubMed] [Google Scholar]
- Kiledjian M, Dreyfuss G. 1992. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J 11: 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer LD, Li J, Shi PY. 2007. West Nile virus. Lancet Neurol 6: 171–181. [DOI] [PubMed] [Google Scholar]
- Liao B, Hu Y, Brewer G. 2007. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol 14: 511–518. [DOI] [PubMed] [Google Scholar]
- Lin JY, Li ML, Brewer G. 2014. mRNA decay factor AUF1 binds the internal ribosomal entry site of enterovirus 71 and inhibits virus replication. PLoS One 9: e103827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel HJ, Rice CM. 2007. Flaviviridae: the viruses and their replication. In Fields virology, 5th ed. (ed. Knipe DM, Howley PM), pp. 1101–1151. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- Liu Q, Dreyfuss G. 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol 15: 2800–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodeiro MF, Filomatori CV, Gamarnik AV. 2009. Structural and functional studies of the promoter element for dengue virus RNA replication. J Virol 83: 993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund N, Milev MP, Wong R, Sanmuganantham T, Woolaway K, Chabot B, Abou Elela S, Mouland AJ, Cochrane A. 2012. Differential effects of hnRNP D/AUF1 isoforms on HIV-1 gene expression. Nucleic Acids Res 40: 3663–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272: 2118–2131. [DOI] [PubMed] [Google Scholar]
- Moritz B, Lilie H, Naarmann-de Vries IS, Urlaub H, Wahle E, Ostareck-Lederer A, Ostareck DH. 2014. Biophysical and biochemical analysis of hnRNP K: arginine methylation, reversible aggregation and combinatorial binding to nucleic acids. Biol Chem 395: 837–853. [DOI] [PubMed] [Google Scholar]
- Munroe SH, Dong XF. 1992. Heterogeneous nuclear ribonucleoprotein A1 catalyzes RNA.RNA annealing. Proc Natl Acad Sci 89: 895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RC, Wang XW, Tang J, Hamilton BJ, High FA, Herschman HR, Rigby WF. 2000. The RGG domain in hnRNP A2 affects subcellular localization. Exp Cell Res 256: 522–532. [DOI] [PubMed] [Google Scholar]
- Nomaguchi M, Teramoto T, Yu L, Markoff L, Padmanabhan R. 2004. Requirements for West Nile virus (−)- and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J Biol Chem 279: 12141–12151. [DOI] [PubMed] [Google Scholar]
- Ong SE, Mittler G, Mann M. 2004. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods 1: 119–126. [DOI] [PubMed] [Google Scholar]
- Paek KY, Kim CS, Park SM, Kim JH, Jang SK. 2008. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J Virol 82: 12082–12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlich S, Zakaryan RP, Gehring H. 2006. Protein arginine methylation: cellular functions and methods of analysis. Biochim Biophys Acta 1764: 1890–1903. [DOI] [PubMed] [Google Scholar]
- Passos DO, Quaresma AJ, Kobarg J. 2006. The methylation of the C-terminal region of hnRNPQ (NSAP1) is important for its nuclear localization. Biochem Biophys Res Commun 346: 517–525. [DOI] [PubMed] [Google Scholar]
- Pont AR, Sadri N, Hsiao SJ, Smith S, Schneider RJ. 2012. mRNA decay factor AUF1 maintains normal aging, telomere maintenance, and suppression of senescence by activation of telomerase transcription. Mol Cell 47: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontius BW, Berg P. 1990. Renaturation of complementary DNA strands mediated by purified mammalian heterogeneous nuclear ribonucleoprotein A1 protein: implications for a mechanism for rapid molecular assembly. Proc Natl Acad Sci 87: 8403–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman DS, Dreyfuss G. 1994. RNA annealing activities in HeLa nuclei. EMBO J 13: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res 32: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Schroeder R. 2007. Dissecting RNA chaperone activity. RNA 13: 2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. 2007. RNA chaperones, RNA annealers and RNA helicases. RNA Biol 4: 118–130. [DOI] [PubMed] [Google Scholar]
- Rajyaguru P, Parker R. 2012. RGG motif proteins: modulators of mRNA functional states. Cell Cycle 11: 2594–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman B, Guarnaccia C, Nadassy K, Zakhariev S, Pintar A, Zanuttin F, Frigyes D, Acatrinei C, Vindigni A, Pongor G, et al. 2001. Nω-arginine dimethylation modulates the interaction between a Gly/Arg-rich peptide from human nucleolin and nucleic acids. Nucleic Acids Res 29: 3377–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R, Grossberger R, Pichler A, Waldsich C. 2002. RNA folding in vivo. Curr Opin Struct Biol 12: 296–300. [DOI] [PubMed] [Google Scholar]
- Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78: 1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrad K. 2011. Proteins with RNA chaperone activity: a world of diverse proteins with a common task-impediment of RNA misfolding. Biochem Res Int 2011: 532908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY, ed. 2012. Molecular virology and control of flaviviruses. Caister Academic Press, Norfolk, UK. [Google Scholar]
- Singh R, Valcárcel J. 2005. Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol 12: 645–653. [DOI] [PubMed] [Google Scholar]
- Snijders AP, Hautbergue GM, Bloom A, Williamson JC, Minshull TC, Phillips HL, Mihaylov SR, Gjerde DT, Hornby DP, Wilson SA, et al. 2015. Arginine methylation and citrullination of splicing factor proline- and glutamine-rich (SFPQ/PSF) regulates its association with mRNA. RNA 21: 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, Holle A. 2003. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal Bioanal Chem 376: 952–965. [DOI] [PubMed] [Google Scholar]
- Thandapani P, O'Connor TR, Bailey TL, Richard S. 2013. Defining the RGG/RG motif. Mol Cell 50: 613–623. [DOI] [PubMed] [Google Scholar]
- Tolnay M, Juang YT, Tsokos GC. 2002. Protein kinase A enhances, whereas glycogen synthase kinase-3 β inhibits, the activity of the exon 2-encoded transactivator domain of heterogeneous nuclear ribonucleoprotein D in a hierarchical fashion. Biochem J 363: 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Csermely P. 2004. The role of structural disorder in the function of RNA and protein chaperones. FASEB J 18: 1169–1175. [DOI] [PubMed] [Google Scholar]
- Villordo SM, Alvarez DE, Gamarnik AV. 2010. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 16: 2325–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Inoue K, Hagiwara M. 2002. Identification of methylated proteins by protein arginine N-methyltransferase 1, PRMT1, with a new expression cloning strategy. Biochim Biophys Acta 1591: 1–10. [DOI] [PubMed] [Google Scholar]
- Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. 1998. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics 48: 195–202. [DOI] [PubMed] [Google Scholar]
- White EJ, Brewer G, Wilson GM. 2013. Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation. Biochim Biophys Acta 1829: 680–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk HE, Werr H, Friedrich D, Kiltz HH, Schäfer KP. 1985. The core proteins of 35S hnRNP complexes. Characterization of nine different species. Eur J Biochem 146: 71–81. [DOI] [PubMed] [Google Scholar]
- Wilson GM, Sutphen K, Moutafis M, Sinha S, Brewer G. 2001. Structural remodeling of an A + U-rich RNA element by cation or AUF1 binding. J Biol Chem 276: 38400–38409. [DOI] [PubMed] [Google Scholar]
- Wilson GM, Lu J, Sutphen K, Suarez Y, Sinha S, Brewer B, Villanueva-Feliciano EC, Ysla RM, Charles S, Brewer G. 2003. Phosphorylation of p40AUF1 regulates binding to A + U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J Biol Chem 278: 33039–33048. [DOI] [PubMed] [Google Scholar]
- You S, Falgout B, Markoff L, Padmanabhan R. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J Biol Chem 276: 15581–15591. [DOI] [PubMed] [Google Scholar]
- Yu MC. 2011. The role of protein arginine methylation in mRNP dynamics. Mol Biol Int 2011: 163827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol 13: 7652–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucconi BE, Ballin JD, Brewer BY, Ross CR, Huang J, Toth EA, Wilson GM. 2010. Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands. J Biol Chem 285: 39127–39139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.