Abstract
In Eukarya and Archaea, in addition to protein-only pseudouridine (Ψ) synthases, complexes containing one guide RNA and four proteins can also produce Ψ. Cbf5 protein is the Ψ synthase in the complex. Previously, we showed that Ψ’s at positions 1940, 1942, and 2605 of Haloferax volcanii 23S rRNA are absent in a cbf5-deleted strain, and a plasmid-borne copy of cbf5 can rescue the synthesis of these Ψ’s. Based on published reports of the structure of archaeal Cbf5 complexed with other proteins and RNAs, we identified several potential residues and structures in H. volcanii Cbf5, which were expected to play important roles in pseudouridylation. We mutated these structures and determined their effects on Ψ production at the three rRNA positions under in vivo conditions. Mutations of several residues in the catalytic domain and certain residues in the thumb loop either abolished Ψ’s or produced partial modification; the latter indicates a slower rate of Ψ formation. The universal catalytic aspartate of Ψ synthases could be replaced by glutamate in Cbf5. A conserved histidine, which is common to Cbf5 and TruB is not needed, but another conserved histidine of Cbf5 is required for the in vivo RNA-guided Ψ formation. We also identified a previously unreported novelty in the pseudouridylation activity of Cbf5 where a single stem–loop of a guide H/ACA RNA is used to produce two closely placed Ψ’s and mutations of certain residues of Cbf5 abolished one of these two Ψ’s. In summary, this first in vivo study identifies several structures of an archaeal Cbf5 protein that are important for its RNA-guided pseudouridylation activity.
Keywords: RNA modification, ribonucleoprotein, protein structure, mutagenesis, H/ACA RNA, Haloferax volcanii
INTRODUCTION
Pseudouridine (Ψ) is the most common modified nucleoside found in different RNAs in all organisms (Charette and Gray 2000; Hamma and Ferré-D'Amaré 2006; Grosjean 2009; Mueller and Ferré-D'Amaré 2009; Cantara et al. 2011). It is a C5-ribosyl isomer of uridine (U) produced post-transcriptionally. In comparison to U, Ψ can provide an additional hydrogen bond and increased base stacking (Davis 1995; Charette and Gray 2000; Hamma and Ferré-D'Amaré 2006). Ψ residues are common in functionally important regions of RNAs, suggesting their possible roles in structure stabilization and function of RNAs (Charette and Gray 2000; Ofengand et al. 2001b; Decatur and Fournier 2002; Lecointe et al. 2002; Namy et al. 2005; Baudin-Baillieu et al. 2009; Karijolich and Yu 2010; Wu et al. 2011; Spenkuch et al. 2014). New techniques have determined that Ψ is also abundant in eukaryal mRNAs (Carlile et al. 2014; Lovejoy et al. 2014; Schwartz et al. 2014; Li et al. 2015). Single- or multi-site-specific single-protein Ψ synthases have been recognized in all three domains of life. Based on sequence similarities, these Ψ synthases have been classified into six families: TruA, TruB, TruD, RluA, RsuA, and Pus10 (Mueller and Ferré-D'Amaré 2009; Spenkuch et al. 2014).
In addition to single-protein Ψ synthases, Eukarya and Archaea have specific ribonucleoprotein (RNP) complexes that can also produce Ψ at many sites in different cellular RNAs (Kiss 2002; Decatur and Fournier 2003; Rozhdestvensky et al. 2003; Henras et al. 2004; Wang and Meier 2004; Dennis and Omer 2005; Meier 2005; Yu et al. 2005; Matera et al. 2007; Hamma and Ferré-D'Amaré 2010; Kiss et al. 2010; Watkins and Bohnsack 2012; Ge and Yu 2013). Each complex contains a distinct box H/ACA guide RNA and four core proteins. Cbf5 (Dyskerin in human and NAP57 in rodents) is the Ψ synthase in these complexes. The three accessory proteins are Gar1, Nop10, and L7Ae (Nhp2 in Eukarya). Several crystal structures of RNA-free and RNA-bound forms of these complexes are now available (Hamma et al. 2005; Li and Ye 2006; Manival et al. 2006; Rashid et al. 2006; Ye 2007; Duan et al. 2009; Liang et al. 2009; Li et al. 2011). The RNA component of the RNP contains an internal loop, called the Ψ pocket, between two double-stranded stem regions. Two strands of the Ψ pocket pair with the target RNA on both sides of the U, which is converted to Ψ. Guide RNAs can contain one to three of these stem–loops. In box H/ACA RNAs, a highly conserved sequence, known as box ACA or box H (ANANNA), is situated at the 3′-end of each stem–loop. These conserved sequences are important for Cbf5 binding and formation of a stable RNA–protein complex (Baker et al. 2005). Archaeal Cbf5 protein interacts with box H/ACA guide RNA, independently of the other three core proteins (Baker et al. 2005; Charpentier et al. 2005; Li and Ye 2006). Archaeal L7Ae protein interacts with a kink turn motif in the guide RNA to assist with RNA folding (Hamma and Ferré-D'Amaré 2004). L7Ae does not interact with the other three proteins in the absence of the guide RNA (Hamma and Ferré-D'Amaré 2004).
Cbf5 belongs to the TruB family (Koonin 1996) of Ψ synthases. Bacterial TruB (and Pus4, its eukaryal ortholog) produces Ψ55 in tRNAs in a guide RNA-independent manner. Cbf5 has also been shown in vitro to produce Ψ55 in tRNAs and some other Ψ in rRNAs in a guide RNA-independent manner and this activity is enhanced by Gar1 and Nop10 (Roovers et al. 2006; Gurha et al. 2007; Muller et al. 2007, 2008; Kamalampeta and Kothe 2012). Based on the structural and functional similarities of Cbf5 and TruB, certain common features about their mechanisms of RNA recognition and action have been proposed. Cbf5 and TruB share a highly conserved RNA-binding PUA (PseudoUridine synthase and Archaeosine transglycosylase) domain, which is larger in Cbf5 than in TruB (Li and Ye 2006; Manival et al. 2006; Rashid et al. 2006; Ye 2007; Li 2008; Duan et al. 2009; Hamma and Ferré-D'Amaré 2010). The PUA domain of Cbf5 also has sequence and structural similarities with two other RNA processing enzymes, Nip7p and archaeosine transglycosylase (ArcTGT) (Rashid et al. 2006). The ArcTGT PUA domain specifically contacts the 3′-terminal CCA trinucleotides of the tRNA (Ishitani et al. 2003; Rashid et al. 2006). This similarity and other structural studies have suggested that the PUA domain of Cbf5 binds to the lower stem and the 3′ ACA trinucleotides of box H/ACA RNA (Li and Ye 2006; Rashid et al. 2006; Duan et al. 2009; Hamma and Ferré-D'Amaré 2010). The accessory proteins Nop10 and Gar1 also help Cbf5 to bind to the specific structures of the guide and target RNAs.
Both TruB and Cbf5 contain a thumb loop, which plays a crucial role in substrate turnover (Hoang et al. 2005; Hamma and Ferré-D'Amaré 2010), but their interactions are somewhat different. The thumb loop of Cbf5, also called the β7_10 loop, is located between its β7 and β10 strands (Li and Ye 2006; Liang et al. 2008, 2009; Duan et al. 2009; Hamma and Ferré-D'Amaré 2010; Li et al. 2011). It interacts with the Gar1 protein and the substrate RNA but not with the guide RNA. It changes conformation during the activity of the RNP. Its Gar1-bound conformation in the substrate-free RNP is referred to as the open state and its substrate RNA-bound state is referred to as the closed state.
Bacterial TruB positions U55 of tRNA to convert it to Ψ55 by flipping the base out of the RNA helix (Hoang and Ferré-D'Amaré 2001). The imidazole ring of a histidine in TruB (His43 in Escherichia coli) occupies the space vacated by the flipped-out uridine. This histidine is also conserved in Cbf5. There is another conserved histidine in Cbf5, which is not conserved in TruB. Both of these histidines have been proposed to have certain roles in the activity of Cbf5 (Muller et al. 2007; Duan et al. 2009; Liang et al. 2009; Hamma and Ferré-D'Amaré 2010; Tillault et al. 2015b).
In the guide RNP complex, Nop10 interacts with Cbf5 along its entire length. Nop10 stabilizes the active-site structure of Cbf5 and the interface between the proteins contains many highly conserved residues (Hamma et al. 2005; Manival et al. 2006; Rashid et al. 2006; Hamma and Ferré-D'Amaré 2010). The N-terminal region of Cbf5 contains a highly conserved region called Motif I (Hamma et al. 2005). It is well conserved in other Ψ synthases and it is important for the stability and function of Cbf5 (Koonin 1996; Spedaliere et al. 2000). Motif I has certain conserved proline residues, which form a structure called a “proline spine” with other conserved prolines in Cbf5, Nop10, and L7Ae in the RNP complex (Hamma and Ferré-D'Amaré 2010).
The gene encoding Cbf5 is essential in Eukarya (Jiang et al. 1993; Meier and Blobel 1994; Heiss et al. 1998; Phillips et al. 1998; Giordano et al. 1999; He et al. 2002), which may be due to different roles of H/ACA RNPs in ribosome biogenesis, mRNA splicing, and telomere maintenance (Meier 2005; Karijolich and Yu 2008). The human homolog of Cbf5 is called dyskerin. Several mutations and deletions in DKC1 (encoding dyskerin) cause the X-linked recessive disorder dyskeratosis congenita (DC) (Ruggero et al. 2003; Mason et al. 2005; Kirwan and Dokal 2008). The molecular basis of DC is not yet clear. Both short telomeric repeats and dysfunctional rRNAs have been noticed in DC patients (Mitchell et al. 1999).
Previously, by deleting the cbf5 gene in Haloferax volcanii, we showed that the Cbf5 protein and Cbf5-mediated Ψ residues are not essential in this archaeon (Blaby et al. 2011). In the present study, we used this deletion strain to express mutants of H. volcanii Cbf5 (HvCbf5) for several residues and structures that are predicted to interact with guide and target RNAs, and H. volcanii Gar1 and Nop10 (HvGar1 and HvNop10) proteins. The selection of residues and structures for mutations was primarily based on the previous structural studies done with Pyrococcus furiosus Cbf5, Gar1, and Nop10 (PfuCbf5, PfuGar1, and PfuNop10) proteins. We show here that several structures of archaeal Cbf5 are essential, whereas certain other structures are not. In addition, we also report for the first time that archaeal Cbf5 can produce two closely placed Ψ's that are guided by the same single stem–loop of H/ACA RNA and that certain residues of Cbf5 are essential for the production of one of these two Ψ's.
RESULTS
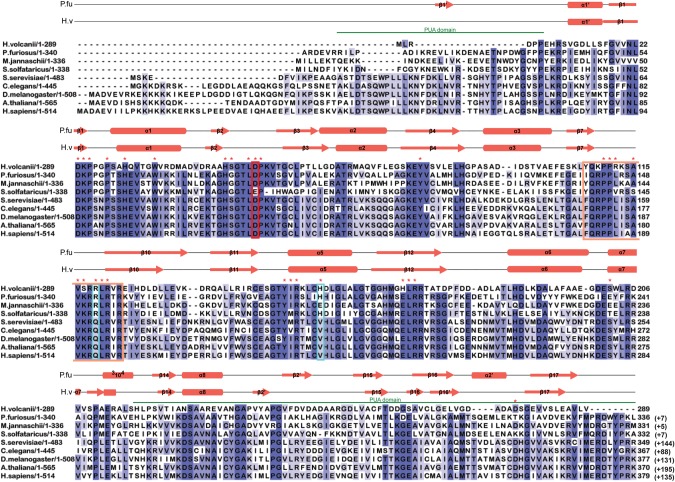
Several structural and in vitro studies of Cbf5 protein have either shown or predicted the importance of certain amino acid residues and structures for the Ψ formation. Our objective was to determine the relevance of different conserved residues and segments of archaeal Cbf5 in RNA-guided Ψ formation under in vivo conditions where, in addition to interactions within the RNP, other factors may also have some role. Previously, we showed that Cbf5-mediated modifications were restored when HvCbf5 was expressed from a plasmid-encoded H. volcanii cbf5 gene in the Δcbf5 strain of H. volcanii (Blaby et al. 2011). Here we used this system for our objective. We expressed various mutants of HvCbf5 in the Δcbf5 strain of H. volcanii and determined the presence or absence of Cbf5-mediated Ψ in 23S rRNA of those cells. Specific residues and structures of HvCbf5 selected for mutations (mostly Ala substitutions) were based on the comparative sequence alignment (Fig. 1) of Cbf5 proteins of several organisms and a homology model of HvCbf5 based on the available structure of PfuCbf5 (Fig. 2) as well as various reports in the literature. Positions of the residues of HvCbf5 selected for substitution mutations in this study are shown in Figures 1, 2C and D. Results of the reactions are shown in Figures 3–7 and Supplemental Figure S1 and are summarized in Table 1.
FIGURE 1.
Multiple sequence alignment of archaeal and eukaryal Cbf5 proteins. Sequences represented here are from four Archaea: Haloferax volcanii (H. volcanii), Pyrococcus furiosus (P. furiosus), Methanocaldococcus jannaschii (M. jannaschii), and Sulfolobus solfataricus (S. solfataricus); and five Eukarya: Saccharomyces cerevisiae (S. cerevisiae), Caenorhabditis elegans (C. elegans), Drosophila melanogaster (D. melanogaster), Arabidopsis thaliana (A. thaliana), and Homo sapiens (H. sapiens). Secondary structural elements are marked above the sequences, with α-helices depicted as cylinders and β-strands as arrows. Secondary structure of P. furiosus protein is based on the crystal structure (PDB 2EY4) (Rashid et al. 2006). Secondary structure of H. volcanii protein is the resulting modeled structure from I-TASSER. One new secondary structural element for HvCbf5 was predicted by I-TASSER, which is not shown in the cited reference (Rashid et al. 2006). The PUA domain regions comprising the structures at the two termini are marked by green lines above the sequence. Thumb loop sequences are boxed in orange. The conserved catalytic aspartate (D) residue is enclosed within a red box. Certain residues that differ between Archaea and Eukarya are enclosed in blue boxes. Residues used for mutagenesis in this study are indicated with red asterisks above the sequence. Highly conserved residues (>80%) among all proteins are shaded in dark blue, and at least 60% conserved residues are shown in medium blue. Light blue represents at least 40% conservation. Numbers after each sequence denote ending residues of each block. Numbering of P. furiosus Cbf5 has been adjusted to match a previous report (Rashid et al. 2006), which is commonly used in the literature. Numbers in parentheses indicate the length of the sequence not shown here.
FIGURE 2.
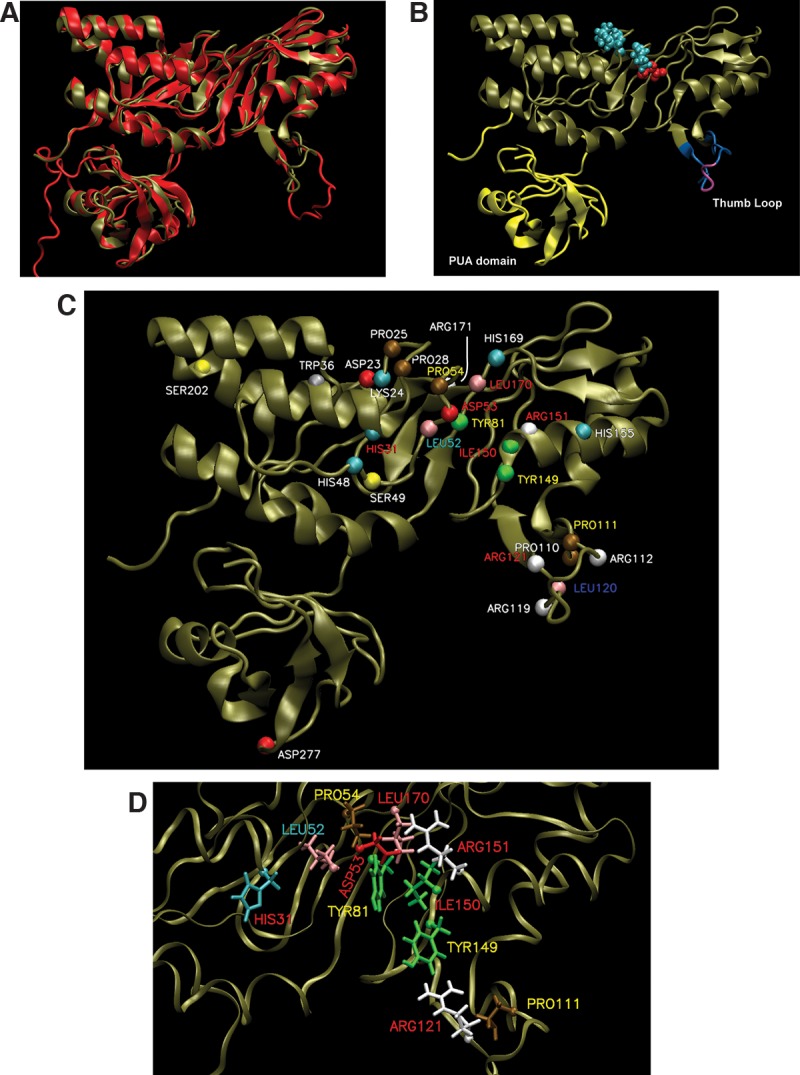
Homology model of HvCbf5. (A) The structure of PfuCbf5 (red) has been extracted from the Cbf5-Nop10-Gar1 crystal structure (PDB 2RFK) (Liang et al. 2007). The homology model of HvCbf5 (tan) based on I-TASSER predicted structure is overlaid. (B) The HvCbf5 model structure from A is shown with thumb loop in blue and purple and the PUA domain in yellow. The purple part of the thumb loop denotes the C loop (Ala115, Val116, and Ser117). The catalytic Asp53 (red) and Cbf5 residues (Pro25, Pro28, and Pro54 in cyan) of the “proline spine” of the RNP complex are shown as van der Waal's spheres. (C) Model of HvCbf5 showing positions of the residues that are individually substituted in this study. The α-carbons of these residues are shown as colored spheres. The colors of these spheres are by residue type as defined in VMD. Effects of Ala substitution of residues are indicated by colors of the labels: absence of (red) or partial (yellow) modification at the three positions (1940, 1942, and 2605) of 23S rRNA; partial modification at positions 1940 and 2605, but no modification at position 1942 (cyan); partial modification at position 1940, and normal modification at positions 1942 and 2605 (blue); and no effect (white), i.e., normal modification at all three positions. (See Table 1 for the effects of mutations of single and multiple residues of HvCbf5.) (D) Structural details near the active site of HvCbf5. Most residues that affected activity of HvCbf5 after mutation are shown. Colors of residues and labels are as in C.
FIGURE 3.
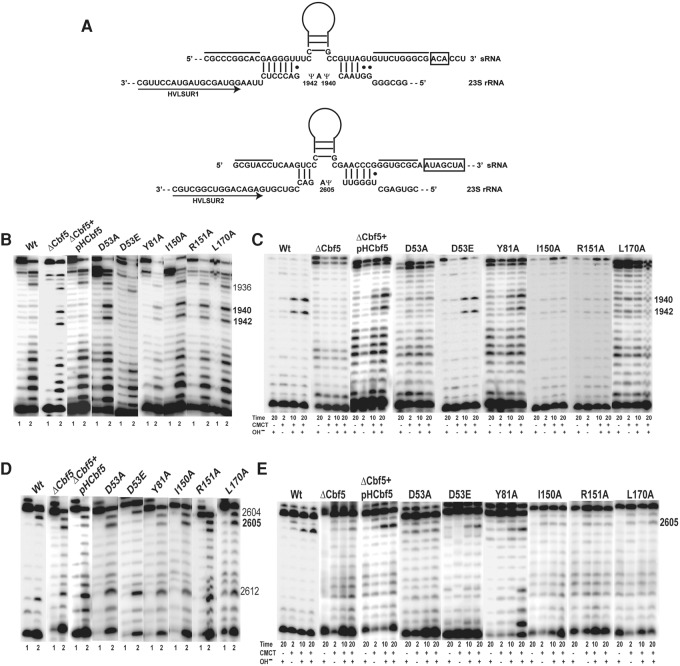
Ala substitutions of HvCbf5 residues conserved across all Ψ synthases abolish or reduce Cbf5-mediated Ψ formation at the three positions of 23S rRNA. (A) Interactions between the two H/ACA motifs of the sRNA and their 23S rRNA target sequences are shown. The two lines above the sRNA indicate the sequences that pair to form the lower stem of the H/ACA RNA. H and ACA sequences are boxed. Regions of sRNAs above the Ψ pocket are illustrated as stem–loops. Positions of the primers (arrows) HVLSUR1 and HVLSUR2 relative to rRNA sequences are shown. Positions of Ψ’s (1940, 1942, and 2605) in the rRNA sequence are indicated. (B) U-specific analyses to determine the modification status of U1940 and U1942 of 23S rRNA were done using primer HVLSUR1 (see panel A) and total RNA of Δcbf5 strain transformed with different mutant pHCbf5 plasmids (marked above each panel). Lanes 1 and 2: Primer extensions on untreated RNA and on RNA following U-specific reactions, respectively. A dark band at a position in lane 2, but not in lane 1 indicates an unmodified U. Positions of certain U's in 23S rRNA are indicated on the side. In WT cells, U1940 and U1942 are converted to Ψ, and U1936 (used as indicator for the positions) remains unmodified. (C) The primer and total RNA used in B are also used for CMCT-primer extension analyses. Total RNAs were either untreated (−) or treated with CMCT (+) for the indicated time (in minutes), followed by alkali (OH−) treatment (+) or no treatment (−). Positions of Cbf5-mediated modifications are indicated on the side. A dark band in CMCT followed by alkali, in 10 min and 20 min lanes, indicates Ψ. (D,E) Analyses similar to those in B and C, respectively, using primer HVLSUR2 (see panel A), were done to determine the modification status of U2605 of 23S rRNA. Unmodified U2604 and U2612 served as indicators for positions in D.
FIGURE 7.
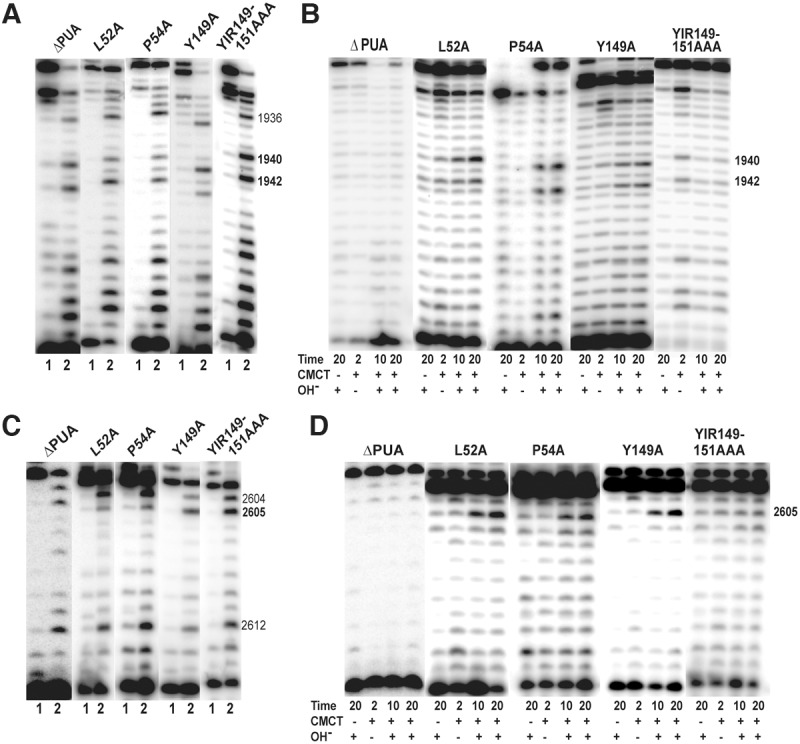
Relevance of PUA domain and certain catalytic core residues of Cbf5 in Ψ formation in vivo. Effects of deletion of PUA domain and Ala substitution of certain conserved residues (marked above each panel) of HvCbf5 for Ψ formation at positions 1940, 1942, and 2605 were determined by primer extensions following U-specific (A,C) and CMCT reactions (B,D). The reactions were done as in Figure 3.
TABLE 1.
Summary of in vivo Ψ production at positions 1940, 1942, and 2605 of H. volcanii 23S rRNA using different mutants of HvCbf5
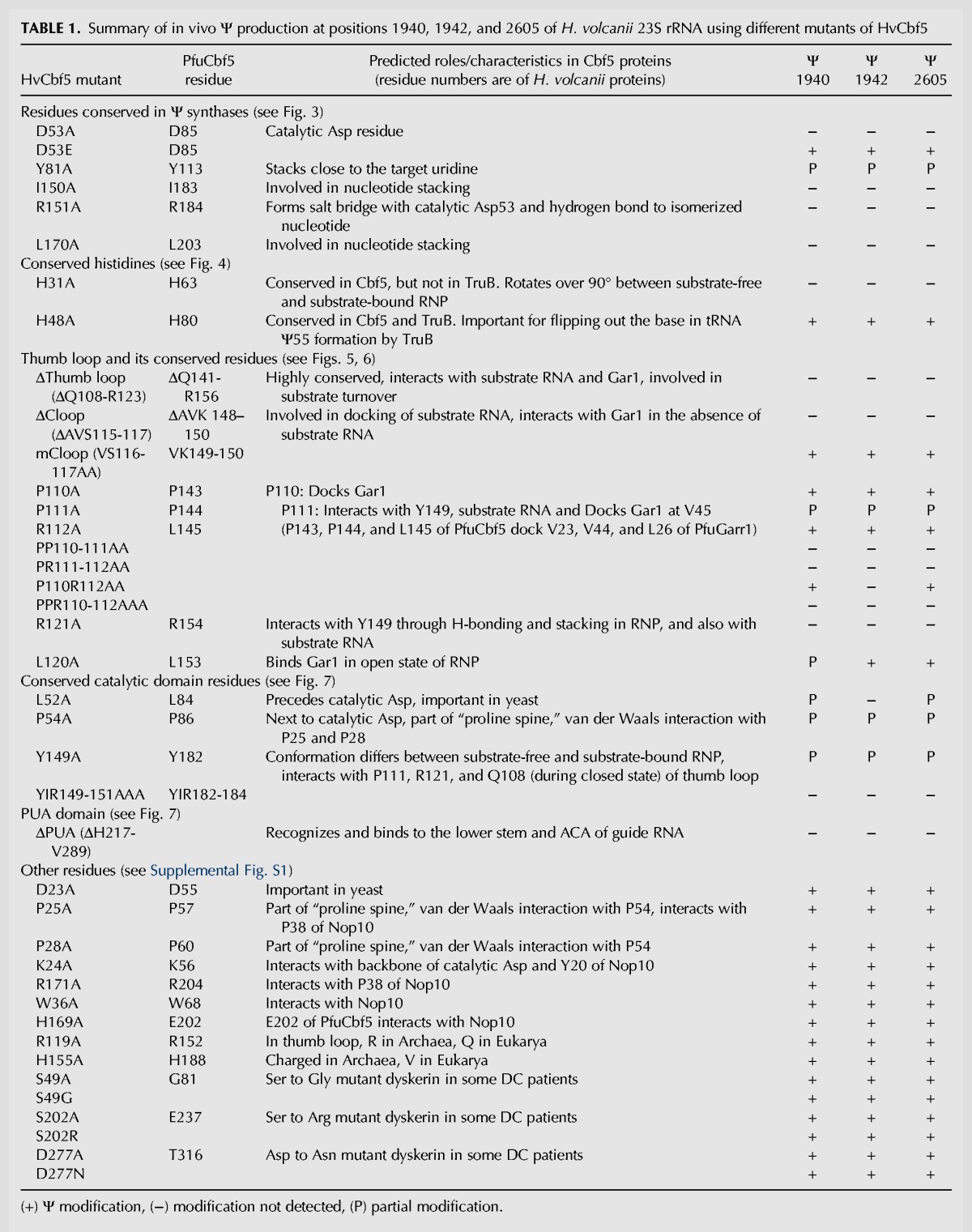
The Ψ formation at positions 1940, 1942, and 2605 of the 23S rRNA of H. volcanii (E. coli positions 1915, 1917, and 2572) is guided by a box H/ACA sRNA (Blaby et al. 2011). Two stem–loops of this sRNA have been proposed to guide all three pseudouridylations, one for Ψ2605 and the other one for both Ψ1940 and Ψ1942 (Grosjean et al. 2008). Deletion of the gene for this box H/ACA sRNA in H. volcanii abolishes all three Ψ's (M Majumder and R Gupta, unpubl.). Interactions between the sRNA and the target sites are shown in Figure 3A. In this in vivo study, we checked the effects of Cbf5 mutants on Ψ formation at all three positions, i.e., at position 2605 for single modification and at positions 1940 and 1942 for double modification, to determine if the mutants show different effects in the two cases.
Sequence alignment and homology model of HvCbf5
A multiple sequence alignment using sequences of HvCbf5 and the Cbf5 proteins of several other archaeal and eukaryal organisms was created (Fig. 1). These proteins contain the five conserved residues observed in most Ψ synthases. These residues are (i) a catalytic Asp, (ii) a basic (Arg/Lys) residue, which forms a salt bridge with the catalytic Asp, (iii) an aromatic (Tyr/Phe) residue, which stacks close to the target uridine, orienting the base for catalysis, (iv) a hydrophobic residue, and (v) a Leu (McCleverty et al. 2007; Mueller and Ferré-D'Amaré 2009; Hamma and Ferré-D'Amaré 2010). The last two residues are involved in nucleotide stacking and are not observed in some Ψ synthases. Corresponding HvCbf5 residues are D53, R151, Y81, I150, and L170, respectively. The primary sequence, GTLDPK (D is the catalytic Asp) of the catalytic core is extremely conserved, with the exception of S. solfataricus, where catalytic Asp is replaced by Glu. The thumb loop is highly conserved in all proteins. HvCbf5 is shorter than PfuCbf5 at both termini. Even then, the two structures are nearly superimposable (Fig. 2A). HvCbf5 has a smaller PUA domain, especially at the N terminus, when compared to its homologs. Certain residues at homologous positions are different between archaeal and eukaryal Cbf5, but are similar among members of each of the two domains of life. HvCbf5 residues at several positions differ from those of other Archaea. This may be a reflection of the internal environment of H. volcanii. Halophilic Archaea have a very high concentration of K+ in their cytoplasm, approaching saturation (Madern et al. 2000). HvCbf5 residues at certain positions (S29, S49, L157, and S202) differ from other archaeal Cbf5, but are the same as in eukaryal Cbf5. Interestingly, serines of dyskerin (human Cbf5) at positions homologous to HvCbf5 S49 and S202 are mutated in DC patients (Heiss et al. 1998; Knight et al. 1999).
Catalytic core residues of HvCbf5 that are conserved in most Ψ synthases are responsible for Ψ formation at positions 1940, 1942, and 2605 of H. volcanii 23S rRNA
All five residues of HvCbf5 that are conserved in most Ψ synthases were independently changed to Ala, and Ψ formation at positions 1940, 1942, and 2605 of 23S rRNA of these transformed strains was determined. Strains expressing D53A, I150A, R151A, and L170A did not show Ψ at any of the three positions (Fig. 3C,E). Instead, these strains showed the presence of unmodified U at these positions (Table 1; Fig. 3B,D). RNA of the strain containing the Y81A mutation showed the presence of both Ψ and unmodified U at all three positions, suggesting partial modification (Table 1; Fig. 3). Replacing catalytic Asp (D53E) of HvCbf5 with Glu did not affect its activity. It behaved like the WT protein under in vivo conditions, producing Ψ at all three positions (Table 1; Fig. 3).
The histidine conserved in both Cbf5 and TruB is not required for in vivo RNA-guided Ψ formation, but another conserved histidine of Cbf5 is required
We mutated H31 and H48, two conserved His of HvCbf5. Bacterial TruB contains a conserved His, homologous to H48, but none corresponding to H31. The H31A mutation abolished all three Ψ’s while the H48A mutation had no effect on the Cbf5 activity (Table 1; Fig. 4).
FIGURE 4.
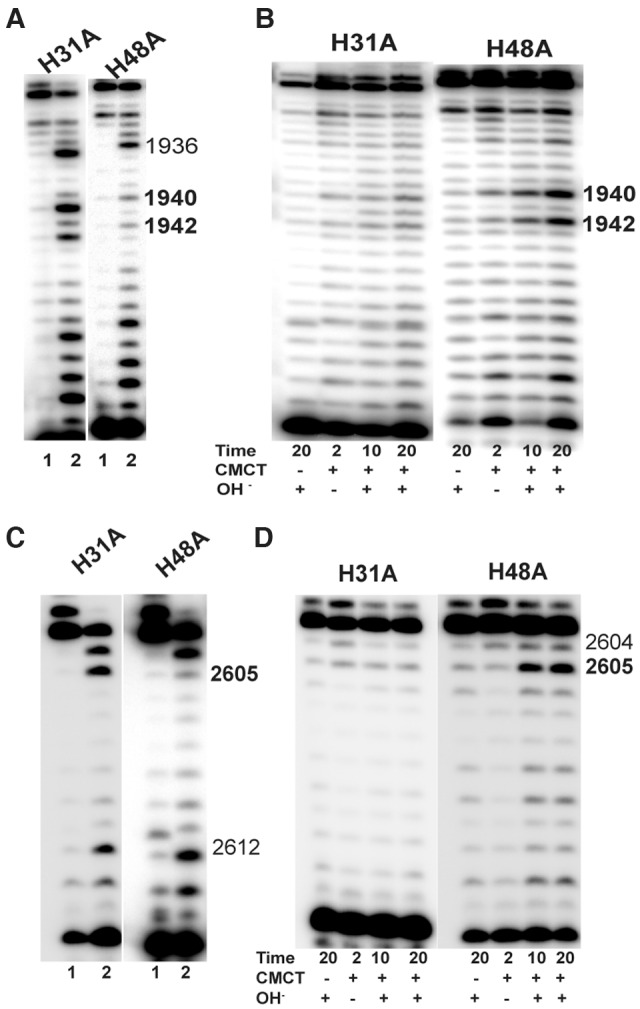
Of the two conserved histidines of HvCbf5, H31 is required but H48 is not for rRNA Ψ formation in vivo. Effects of Ala substitution of conserved histidines (marked above each panel) of HvCbf5 for Ψ formation at positions 1940, 1942, and 2605 were determined by primer extensions following U-specific (A,C) and CMCT reactions (B,D). The reactions were done as in Figure 3.
The thumb loop of Cbf5 plays a major role in RNA-guided Ψ formation
We deleted 16 residues (Q108-R123) of HvCbf5 to create a ΔThumb loop mutant. This mutant did not produce Ψ at any of the three positions (Table 1; Fig. 5). A structure at the end of the thumb loop of PfuCbf5 is referred to as the C loop (Liang et al. 2009). We created a mutant, ΔCloop, where the corresponding region (A115, V116, and S117) of HvCbf5 was deleted. This mutant also did not produce Ψ at any of the three positions (Table 1; Fig. 5). However, when we changed both V116 and S117 of the C loop to Ala (retaining A115), the mutation (mCloop) had no effect at Ψ production at any of the three positions (Table 1; Fig. 5).
FIGURE 5.
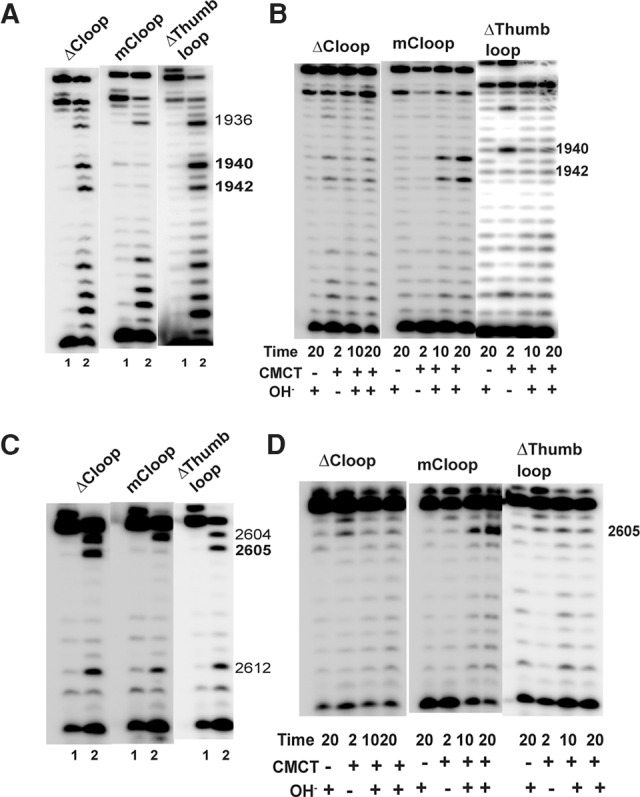
Proper structure of the thumb loop is important for the activity of Cbf5. Effects of the mutations (marked above each panel) in the thumb loop of HvCbf5 on Ψ formation at positions 1940, 1942, and 2605 were determined by primer extension following U-specific (A,C) and CMCT (B,D) reactions. The reactions were done as in Figure 3.
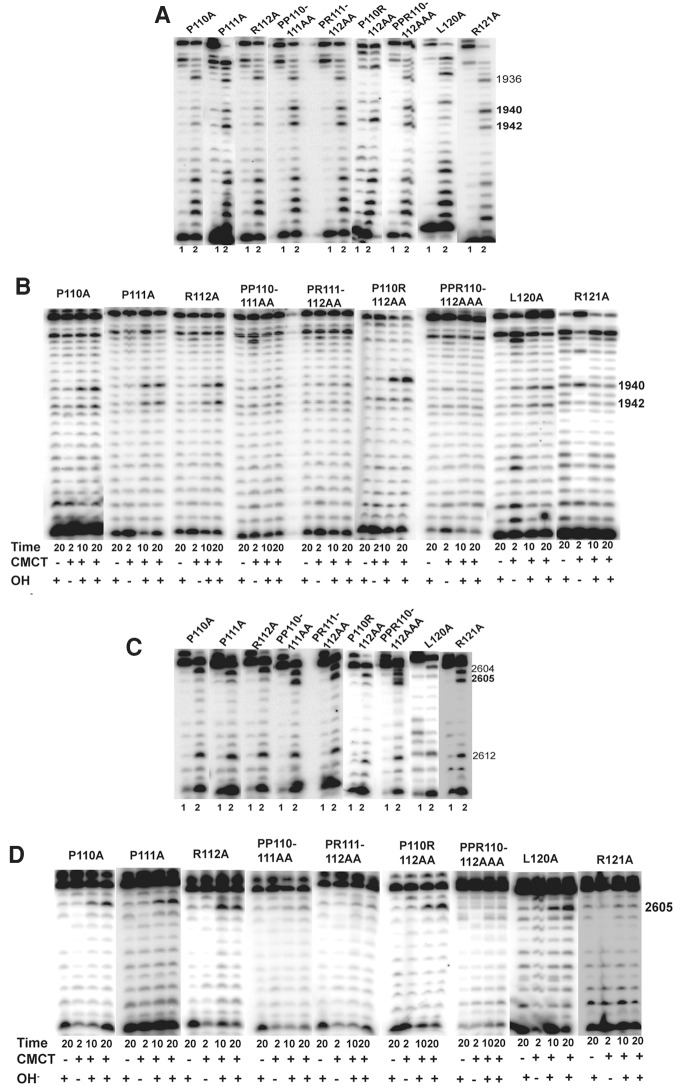
We substituted the three consecutive residues (P110, P111, and R112) of HvCbf5 that precede the C loop with Ala independently and in different combinations. The results are shown in Figure 6 and Table 1. Individually, P110A and R112A did not show any defect in modification and P111A only partially modified the three U's. Mutating all three residues to Ala (PPR110-112AAA) abolished all three modifications. Double mutants PP110-111AA and PR111-112AA, where two consecutive residues were changed, abolished all modifications. However, the P110R112AA mutant (where P111 was not changed) abolished modification of U1942 but did not affect modifications of U1940 and U2605. Ala substitution of P111, either alone or in combination with other mutations, in addition to affecting the activity of HvCbf5 (Table 1; Fig. 6), appears to reduce the amount of the protein in the cell (cf. panels F and G with panel B in Supplemental Fig. S2).
FIGURE 6.
Certain residues of the thumb loop are important for the guide RNA-dependent activity of Cbf5. Effects of alanine substitution of some conserved residues (marked above each panel) of the thumb loop of HvCbf5 for Ψ formation at positions 1940, 1942, and 2605 were determined by primer extensions following U-specific (A,C) and CMCT reactions (B,D). The reactions were done as in Figure 3.
We also substituted the three consecutive residues (R119, L120, and R121) of HvCbf5 that are after the C loop, independently with Ala. The results are shown in Figure 6 and Supplemental Figure S1 (panels E–H), and Table 1. Residue R119 is Archaea specific. R119A did not affect the modifications. L120A showed partial modification of U1940 but retained normal modification of U1942 and U2605. R121A lost all modifications.
Certain other residues of the catalytic domain of Cbf5 are relevant for Ψ formation
Mutations of certain residues of HvCbf5 that are adjacent to conserved residues present in most Ψ synthases affected its activity. Mutation of the Leu (L52A) that precedes catalytic Asp showed partial loss of modification at positions 1940 and 2605, and absence of modification at position 1942 (Table 1; Fig. 7). Mutation of Pro (P54A) that follows catalytic Asp showed partial modification at all three positions (Table 1; Fig. 7). There is a conserved Tyr (Y149) that precedes I150 and R151 of HvCbf5, the conserved hydrophobic and basic residues of Ψ synthases. Mutation of this Tyr (Y149A) also showed partial modification at all three positions (Table 1; Fig. 7). As expected when all three residues Y149, I150, and R151 were replaced with alanines, the modifications were lost (Table 1; Fig. 7), because as mentioned, I150A and R151A mutants did not show any modification.
The C-terminal PUA domain is needed for Cbf5-mediated Ψ formation
Both N- and C-terminal sequences of Cbf5 share parts of the PUA domain, which holds the lower stem of the guide RNA. Relative positions of the PUA domain and the Thumb loop of HvCbf5 are shown in Figure 2. Deletion of the PUA domain drastically reduces the in vitro activity of Cbf5 (Baker et al. 2005; Manival et al. 2006). We deleted the C-terminal PUA domain (positions 217–289) of HvCbf5. As expected, this abolished Ψ production at all three positions (Table 1; Fig. 7).
Individual mutations of several conserved Cbf5 residues involved in interaction with Nop10 do not completely abolish Ψ formation
Three prolines of HvCbf5, P25, P28, and P54 (P57, P60, and P86, respectively, of PfuCbf5) form a part of the “proline spine” (see Fig. 2B), which as mentioned before also includes prolines of Nop10 and L7Ae in the RNP complex. P25 is also expected to interact with certain conserved Pro residues like P38 of HvNop10 (P32 of PfuNop10) (Hamma and Ferré-D'Amaré 2010). Ala substitution of HvCbf5 P25 and P28 did not have any effect on modification (Table 1; Supplemental Fig. S1). This is similar to a yeast study where a mutant (P67A of S. cerevisiae Cbf5) similar to P25A of HvCbf5 also did not show any reduction of total Ψ content of rRNAs (Zebarjadian et al. 1999). However, as mentioned, the third Pro mutant (P54A) of HvCbf5 did show reduction in activity (Table 1; Fig. 7).
Based on crystal structures, it is expected that K24 and R171 of HvCbf5 (K56 and R204, respectively, of PfuCbf5) would interact with Y20 and P38, respectively, of HvNop10 (Y14 and P32, respectively, of PfuNop10) (Hamma et al. 2005; Rashid et al. 2006). In vitro studies showed that Ala substitutions of the corresponding residues in P. abyssi Cbf5 (K53 and R202, respectively) significantly impaired RNA-guided Ψ synthesis (Muller et al. 2007). However, K24A and R171A mutants of HvCbf5 did not show any defect in Ψ synthesis (Table 1; Supplemental Fig. S1). A conserved Asp (D23) precedes K24 of HvCbf5. Yeast strains containing Ala substitutions of corresponding Asp (D65, of S. cerevisiae Cbf5, Fig. 1) showed a sharp reduction of total Ψ content of rRNAs of the cells (Zebarjadian et al. 1999). The corresponding D23A mutant did not affect Ψ formation at any of the three positions (Table 1; Supplemental Fig. S1).
We also changed W36 and H169 of HvCbf5 to Ala. PfuCbf5 contains conserved Trp (W69) and Glu (E202) at corresponding positions (Fig. 1). These residues interact with Nop10 (Li and Ye 2006). As shown in Figure 1, Trp is conserved in all Cbf5, but His replaces the conserved Glu in HvCbf5. W36A and H169A mutants of HvCbf5 did not affect modification (Table 1; Supplemental Fig. S1).
Ala substitution of certain Archaea-specific charged residues of HvCbf5 did not affect Ψ formation
Archaeal Cbf5 proteins share most of the domains and motifs with their eukaryal homologs. However, eukaryal Cbf5 is larger than archaeal Cbf5 (Fig. 1). The eukaryal extensions are beyond the N and C termini of the archaeal proteins. These extensions may be reflections of multiple functions of eukaryal Cbf5. There are certain residues at the homologous positions of archaeal and eukaryal proteins that are conserved but the conservations between the two groups are different, i.e., these are either Archaea specific or Eukarya specific. These differences could be either due to multiple functions of eukaryal Cbf5 or to accommodate interactions with other accessory proteins, which may require some structural differences between Archaea and Eukarya. We selected two residues, R119 (described before with thumb loop mutants) and H155 of HvCbf5, which are charged residues in Archaea but not charged (Gln and Val, respectively) in Eukarya and mutated these to Ala. Neither of these mutations affected Ψ synthesis at any of the three positions (Table 1; Supplemental Fig. S1). We also changed R119 of HvCbf5 to Gln, its eukaryal counterpart, but we could not successfully transform this R119Q mutant-containing plasmid into a Δcbf5 strain in spite of multiple attempts.
Mutations of HvCbf5 corresponding to DKC1 mutations causing dyskeratosis congenita do not abolish Ψ modification
Mutations of DKC1 that cause DC are well dispersed on the dyskerin structure (Mason et al. 2005). Most of the residues involved in DC cluster in the N- and C-terminal extensions of dyskerin that are not present in archaeal Cbf5 (Li et al. 2011). However, there are three residues (S121, S280, and D369) within the central part of dyskerin that are mutated in multiple families of DC patients (Heiss et al. 1998; Knight et al. 1999). HvCbf5 has corresponding residues (S49, S202, and D277, respectively). Surprisingly, the HvCbf5 and dyskerin residues are identical at these three positions, whereas in PfuCbf5 (and other archaeal Cbf5) residues at these positions are different. Dyskerin mutations at these positions in DC patients are S121G, S280R, and D369N. We changed the corresponding positions of HvCbf5 to Ala (S49A, S202A, and D277A) as well as to those observed in DC patients (S49G, S202R, and D277N). None of these modifications affected Ψ formation at any of the three positions (Table 1; Supplemental Fig. S1). This suggests that these mutations in DC patients affect an activity or interaction of dyskerin that might not be involved in Ψ synthesis.
DISCUSSION
Partial Ψ modifications in vivo may indicate an overall slower rate of Ψ formation
Analyses of Ψ formation using primer extensions following U-specific and CMCT reactions allow us to determine partial modification at a specific position of rRNA. The cells showing partial modification at a particular site would have ribosomes containing two populations of 23S rRNA; one with Ψ and the other having unmodified U at that site. We believe that partial modifications indicate an overall slower rate of Ψ formation. Our reasoning is that any particular rRNA modification occurs in a short window of time between pre-rRNA transcription and ribosome biogenesis. Therefore, an optimum amount of specific modifying RNPs (or enzymes) has to be available to modify all rRNA molecules of the cell. We believe that a slower rate of Ψ formation would effectively reduce functioning RNPs that are available to modify all of the rRNA molecules of the cell. This delay can occur at any stage of Ψ formation, e.g., substrate recognition and its placement in catalytic site, catalysis, or product release. We observed partial modifications for L52A, P54A, Y81A, P111A, L120A, and Y149A mutants of HvCbf5. L52 and P54 residues precede and follow the catalytic D53 and probably provide optimum conditions for Ψ formation. Ala substitution of Leu of yeast Cbf5 (L94 of S. cerevisiae Cbf5, Fig. 1) corresponding to L52 of HvCbf5 also showed a sharp reduction of total Ψ in rRNAs (Zebarjadian et al. 1999). P54 of HvCbf5 is the first residue of the “proline spine” nearest to the catalytic Asp and is in van der Waals interaction with P25 and P28 of Motif I (Spedaliere et al. 2000; Hamma and Ferré-D'Amaré 2010), the other two Pro of the “proline spine” of Cbf5. The “proline spine” probably provides a communication path between the L7Ae protein and the active site of the Cbf5 (Hamma and Ferré-D'Amaré 2010). Correct positioning of this Pro may enhance Ψ formation. Y81 is conserved in all Ψ synthases and is predicted to stack close to the target uridine, orienting the base for catalysis (Mueller and Ferré-D'Amaré 2009; Hamma and Ferré-D'Amaré 2010). P111 of the thumb loop and Y149 (which precedes conserved I150 and R151, present in other Ψ synthases also) interact with each other as well as with other residues of Cbf5 and Gar1 and with the substrate RNA (Table 1). Conformations of Y149 are also different under substrate-bound and substrate-free states (see later). Partial Ψ modification by mutations of these two residues suggests that although not necessary, these interactions help in proper placement of the substrate and release of the product. L120 of HvCbf5 (L153 of PfuCbf5) is conserved in the thumb loop of Cbf5 proteins. It is at the surface that binds Gar1 in the open state of the RNP (Duan et al. 2009). The mutant L120A is somewhat unique. It showed normal modification of U1942 and U2605, but partial modification of U1940 (Table 1; Fig. 6). When the corresponding L153 of PfuCbf5 was changed to Gly, it retained substantial in vitro activity under single-turnover conditions, but showed only ∼25% activity under multiple-turnover conditions (Duan et al. 2009). This suggests that the same structures of Cbf5 may be involved both in multiple turnover (in vitro) and in multiple site (in vivo) activity.
Certain Cbf5 residues are specifically needed for one of the two modifications guided by a single stem–loop of H/ACA RNA
As mentioned previously, the 5′ stem–loop of the box H/ACA sRNA guides the formation of Ψ2605 and the 3′ stem–loop guides both Ψ1940 and Ψ1942, the last two modifications being the most conserved from bacteria to human. This case of the same stem–loop converting two uridines of an RNA that are close to each other is unique. A search of different s(no)RNA databases indicates that there are several instances where one stem–loop of a box H/ACA RNA can guide more than one Ψ modification. However, in these cases, the sites are either in two different RNAs or at two places far apart in the same RNA. The two conserved modifications of human and yeast large subunit rRNAs that correspond to Ψ1940 and Ψ1942 of H. volcanii 23S rRNA (Ψ3741 and Ψ3743 in human, Ψ2258 and Ψ2260 in yeast) are guided by one box H/ACA RNA each (U19 in human, snR191 in yeast) but by two separate stem–loops of these RNAs (Ganot et al. 1997; Bortolin and Kiss 1998; Badis et al. 2003), not by a single stem–loop as in H. volcanii.
Two mutants of HvCbf5 (L52A and P110R112AA) showed partial or nearly complete modification at position 1940 (and position 2605) but no modification at position 1942 (Table 1). This suggested that these two modifications may occur sequentially, i.e., Ψ1942 modification occurs only after the production of Ψ1940. The target RNA, in this case, may be recruited once and is released after formation of both Ψ’s, or recruitment of the two target uridines may occur independently of each other. In the latter case, U1942 is recruited when the RNA contains Ψ1940. In both cases, U1942 is recognized in an unconventional manner and certain mutations of HvCbf5 affect only this recognition. Residue L120 appears to play some role in this recognition, because the above-mentioned L120A mutant shows partial modification at position 1940 but near normal modification at position 1942 (and position 2605). Specific features of the guide-target RNA interactions (cf. two interactions shown in Fig. 3A) would also be important in production of these double modifications. This specific double modification activity of Cbf5 (or box H/ACA RNPs) and the role of structural elements of Cbf5 in this activity were not detected before our present study, because no modification of two closely placed sites by a single stem–loop was previously known.
Most predicted catalytic core residues are important for Cbf5 activity
Four (D53, R151, I150, and L170) of the five conserved residues of the catalytic core of the Ψ synthases are essential for HvCbf5 activity because Ψ formation is abolished when any one of these is individually substituted by Ala. Mutation of the fifth residue (Y81A) showed partial modification. Different mutants of Tyr of Pyrococcus proteins (Y113A, Y113H, and Y113L of PfuCbf5, and Y110A and Y110F of P. abyssi Cbf5) that correspond to Y81 of HvCbf5 also retained some in vitro activity (Zhou et al. 2010; Tillault et al. 2015a). Therefore, this nonessential but enhancing role of Y81 in Ψ formation is somewhat surprising, because it (or another aromatic residue) is conserved in all Ψ synthases.
All Cbf5 proteins contain a conserved Leu and Pro (L52 and P54 of HvCbf5) on two sides of the catalytic Asp (D53) and a conserved Tyr (Y149 of HvCbf5) before the adjacent hydrophobic and basic residues (I150 and R151) present in most Ψ synthases (Fig. 1). The Pro is also part of the “proline spine” (see Fig. 2B) and the Tyr also interacts with certain residues of the thumb loop (Hamma and Ferré-D'Amaré 2010). Individual Ala substitutions of all three residues show partial modification (Table 1). Furthermore, as mentioned above, the L52A mutant, although it partially modified U1940 and U2605, did not show modification of U1942. The Y182 of PfuCbf5, which corresponds to Y149 of HvCbf5, acquires two different conformations under substrate-free and substrate-bound conditions and interacts with conserved P144 and R154 (P111and R121 of HvCbf5) of the thumb loop (Hamma and Ferré-D'Amaré 2010) and with Q141 (Q108 of HvCbf5) during the closed state of the thumb loop (Duan et al. 2009). Arg, His, and Ser mutation of Tyr182 of PfuCbf5 abolished enzymatic activity under in vitro conditions, but an aromatic replacement (Y182F) only reduced the activity (Zhou et al. 2010). Similarly, the Y179F mutant of P. abyssi Cbf5 retained some in vitro activity and the Y179A mutant almost lost this activity (Tillault et al. 2015a). Therefore, it appears that the residues of Cbf5 that are adjacent to the conserved residues of Ψ synthases are also important for the optimum activity of Cbf5.
A Cbf5 segment of nearly conserved residues GTYIR (positions 180–184 in PfuCbf5, 147–151 in HvCbf5, see Fig. 1), which includes I150 and R151 is conserved in most Ψ synthases, interacts with newly formed Ψ, and is suggested to communicate the isomerization of U to the thumb loop (and Gar1) for product release (Hamma and Ferré-D'Amaré 2010). R151 forms a salt bridge with catalytic D53, hydrogen bonds to Ψ, and signals the chemical change in the catalytic site for product release. Furthermore, the conformation of Y149 in this segment shows a difference between substrate-free and bound RNP. Loss or reduction in Ψ formation in Y149A, I150A, and R151A mutants of HvCbf5 supports the communication role of this segment of Cbf5.
The conserved catalytic aspartate of Ψ synthases can be replaced by glutamate in Cbf5
As stated before, the conserved catalytic Asp observed in all six families of Ψ synthases is replaced by Glu in S. solfataricus (Fig. 1) and S. acidocaldarius (not shown). This Glu in Cbf5 is not characteristic of the genus Sulfolobus or the kingdom Crenarchaeota (to which Sulfolobus belongs) of the domain Archaea. Cbf5 of Sulfolobus shibatae, Sulfolobus metallicus, Sulfolobus tokodaii, and other members of the Crenarchaeota contain Asp as the catalytic residue (data not shown). Furthermore, there was no previous report that Glu-containing Cbf5 of S. Solfataricus or S. acidocaldarius do produce Ψ. Conversion of catalytic Asp of PfuCbf5 to Glu also abolished its activity under in vitro conditions (Zhou et al. 2010). Therefore, the D53E mutant of HvCbf5 behaving like the WT protein under in vivo conditions was surprising. Although the physical structures of these two amino acids are somewhat similar, nature has consistently selected Asp as the catalytic residue in (nearly) all known Ψ synthases.
Roles of conserved histidines during in vivo RNA-guided activity of archaeal Cbf5
Archaeal Cbf5 in vitro has been shown to have both RNA-guided and guide RNA-independent pseudouridylation activities (Baker et al. 2005; Charpentier et al. 2005; Roovers et al. 2006; Gurha et al. 2007; Muller et al. 2007, 2008). Histidines equivalent to H31 and H48 of HvCbf5 play important but distinct roles in these two activities (Muller et al. 2007; Duan et al. 2009; Liang et al. 2009; Hamma and Ferré-D'Amaré 2010; Tillault et al. 2015b). Histidines comparable to H48 of HvCbf5 (H80 in PfuCbf5, H77 in Pyrococcus abyssi Cbf5) are conserved both in Cbf5 and in TruB (H43 of E. coli TruB). This His of TruB interacts with the U54•A58 reverse Hoogsteen pair of tRNA and is critical for flipping out the target U55 (Hoang and Ferré-D'Amaré 2001; Duan et al. 2009; Hamma and Ferré-D'Amaré 2010). Though this His of Cbf5 has been suggested to have an interaction with the substrate U in guide RNA-dependent activity, its confirmation does not change between the substrate-free and substrate-bound H/ACA RNP (Hamma and Ferré-D'Amaré 2010). The other His (H31 in HvCbf5, H63 in PfuCbf5, H60 in P. abyssi Cbf5) that is conserved only in Cbf5 interacts with a Watson-Crick pair formed between guide and target RNA in RNA-mediated activity of Cbf5 (Duan et al. 2009; Liang et al. 2009). This second His may also be involved in the RNA-independent conversion of U55 to Ψ55 when the tRNA does not contain the U54•A58 reverse Hoogsteen pair (Gurha et al. 2007; Duan et al. 2009). Unlike the His common to both Cbf5 and TruB, this particular His rotates over 90° from substrate-free to substrate-bound state of the RNP during RNA-dependent activity of Cbf5 (Hamma and Ferré-D'Amaré 2010). Our results agree with the importance of the Cbf5-specific histidine in the RNA-dependent activity of Cbf5; the H31A mutation abolishes all three Ψ’s in the rRNA (Table 1; Fig. 4). The other His (H48), which is conserved in both Cbf5 and TruB, is not important for in vivo RNA-dependent activity of Cbf5; all three Ψ’s are present in the H48A mutant (Table 1; Fig. 4). In agreement with our present in vivo study, previous in vitro studies (Muller et al. 2007; Tillault et al. 2015b) showed that H77A mutants of P. abyssi Cbf5 (similar to the H48A mutant of HvCbf5) did not affect its guide RNA-dependent activity. One of these studies also showed that H60, the other His of P. abyssi Cbf5 (comparable to H31 of HvCbf5), is not essential for either the RNA-independent or guide RNA-dependent activity of Cbf5 (Tillault et al. 2015b). Our in vivo results do not agree with their in vitro results for guide RNA-dependent activity of Cbf5. We show here that this activity is abolished in the comparable H31A mutant (Table 1; Fig. 4). It is possible that one or the other His of Cbf5 may be important for the guide RNA-independent activity of Cbf5 depending on the structure of the substrate RNA (e.g., presence or absence of U54•A58 reverse Hoogsteen pair in a tRNA). Furthermore, the question may be raised whether under certain conditions (or for some substrates) the observed results of in vitro activities of H/ACA RNPs may be a combination of guide RNA-dependent and independent activities of Cbf5, i.e., whether a particular substrate in vitro can be modified by both activities of Cbf5. An in vivo study of RNA-independent activity of Cbf5, if it occurs, may help in resolving some of these issues.
Several residues of the thumb loop involved in interactions with Gar1 and substrate RNA are important for Cbf5 activity
The Thumb loop locks the target nucleotide at the active site cleft. Residues near the closed end (tip) of this loop interact with Gar1 in the substrate-free open state and dock the substrate in the closed state (Duan et al. 2009; Liang et al. 2009; Hamma and Ferré-D'Amaré 2010). In one publication, this segment is referred to as the C loop, consisting of A148, V149, and K150 of PfuCbf5 (Liang et al. 2009), corresponding to HvCbf5 A115, V116, and S117 (Figs. 1, 2B). In another publication, it is referred to as the tip of the thumb loop, consisting of S147, A148, and V149 of PfuCbf5 (Duan et al. 2009), corresponding to HvCbf5 S114, A115, and V116. Our results of ΔCloop and mCloop mutants (Table 1) indicate that the exact size of the tip rather than its specific sequence is essential for HvCbf5 activity. Interestingly, the ΔCloop mutant of PfuCbf5, comparable to the ΔCloop of HvCbf5, did not completely abolish in vitro Ψ formation under single turnover conditions (Liang et al. 2009).
Residues preceding the tip (108–112 of HvCbf5) mainly interact with Gar1 in the open state and flip over in the closed state (Duan et al. 2009). Three consecutive residues P143, P144, and L145 of PfuCbf5 dock at a hydrophobic patch (V23, V44, and L26) of PfuGar1 (Li and Ye 2006). Only two of the three residues are conserved in HvCbf5 (P110 and P111). The third HvCbf5 residue is a charged R112 instead of the hydrophobic residue observed in other Cbf5 proteins (Fig. 1). (HvGar1 residues that correspond to the hydrophobic patch of PfuGar1 are T24, V45 and I27.) P144 of PfuCbf5 (P111 of HvCbf5) also interacts with Y182 of PfuCbf5 (Y149 of HvCbf5) of the catalytic core region through hydrogen bonding and stacking in the fully assembled RNP and with substrate RNA through water-mediated hydrogen bonding (Hamma and Ferré-D'Amaré 2010). Although this Pro (P111) appears to be very important, P110 and R112 do play some accessory roles. By itself, P111A showed partial modifications, while P110A and R112A did not affect modifications. However, P111A in any combination with mutations of the other two residues (PP110-111AA, PR111-112AA, and PPR110-112AAA) abolished all modifications. Furthermore, mutant P110R112AA (retaining P111) showed modification of U1940 (and U2605) but not of U1942. This mutant is somewhat like the above-mentioned L52A mutant that showed partial modification of U1940 but no modification of U1942.
Several basic residues after the tip (e.g., R119 and R121 in HvCbf5) interact with phosphate groups of the substrate RNA on the 5′ side of the target uridine (Duan et al. 2009; Hamma and Ferré-D'Amaré 2010). R154 of PfuCbf5 (R121 of HvCbf5) also interacts with Y182 (Y149 of HvCbf5) of the catalytic core region through hydrogen bonding and stacking in the fully assembled RNP (Hamma and Ferré-D'Amaré 2010). Although the R119A mutation did not affect modification, residue at this particular position must have some special role in Eukarya (in other than H/ACA RNP activity) because, as mentioned before, we did not succeed in preparing H. volcanii strains that contained R119Q. Eukaryal Cbf5 contains Gln instead of Archaea-specific Arg at this position (see Fig. 1). Interactions of R121 (R154 of PfuCbf5) must be essential because the R121A mutant lost all modifications. Mutant R154Q of PfuCbf5 also lost all activity under in vitro conditions (Duan et al. 2009). A conserved Leu (L153 in PfuCbf5, L120 in HvCbf5) between the two Arg (R119 and R121 of HvCbf5) is observed in Cbf5 proteins. It is at the surface that binds to Gar1 in the open state of the RNP (Duan et al. 2009). This Leu appears to have some role during modification of U1940 and U1942, but not during that of U2605. Mutant L120A showed normal modification of U1942 and U2605, but no modification of U1940.
Most mutations of individual residues that form part of an interacting surface of Cbf5 do not affect its activity
Single mutations of individual residues of certain structures or surfaces that are involved in intra- or intermolecular interactions in general do not affect modifications, as we observed for the mutants of the residues of the proline spine and of those on the Cbf5 surface that interacts with Nop10 (Table 1). The Pro (P54 of HvCbf5, next to catalytic D53) at one end of the proline spine stacks next to the flipped out base of the residue 3′ to the target uridine of the substrate RNA, and the Pro (of L7Ae) at the other end contacts the flipped out uridine of the K-loop of the H/ACA guide RNA (Hamma and Ferré-D'Amaré 2010). Mutation of P54 showed partial modification, but mutations of P25 and P28 did not affect modification. Probably, inside the cell, individual mutation of residues forming part of a structure either slow down Ψ formation or are compensated by accessory factors or other interactions. On the other hand, the identity of the residues, whose chemical status or orientation changes extensively during Ψ formation, appears to be essential.
This is the first report of an in vivo study of an archaeal box H/ACA sRNP. By changing several residues and regions of the Ψ synthase Cbf5, we identified several conserved residues that are crucial for and others that facilitate the RNA-guided activity of archaeal Cbf5. Furthermore, we also showed for the first time that activities of archaeal Cbf5 are not identical when it uses one stem–loop of guide RNA to modify only one uridine and when it uses one stem–loop to modify two closely placed uridines in a sequential manner. These differences could be seen because mutations of certain residues abolished only one of the two modifications.
MATERIALS AND METHODS
Standard molecular biology procedures (Sambrook and Russell 2001) were used unless specifically described.
Strains, media, and transformation procedures
H. volcanii H26 and VDC2364 (Blaby et al. 2011) were used as wild type (WT) and cbf5-deleted (Δcbf5) strains. E. coli was routinely grown in LB (Fisher) or LB agar (Fisher) at 37°C, supplemented when required with ampicillin (100 µg/mL), chloramphenicol (34 µg/mL), IPTG (0.2 mM), and X-Gal (40 µg/mL). H. volcanii cells were routinely grown at 42°C–44°C in Hv-YPC medium, as described in the HaloHandbook (http://www.haloarchaea.com/resources/halohandbook/Halohandbook_2008_v7.pdf) or in a medium described previously (Gupta 1984), supplemented with novobiocin (0.8 µg/mL) when required. Transformation of H. volcanii was performed as described before (Blaby et al. 2011).
Cloning and expression of wild type and mutants of H. volcanii Cbf5 for in vivo analysis
Plasmid pHCbf5 (Blaby et al. 2011) was used to express WT HvCbf5 in H. volcanii. An ∼1000 bp HindIII-EcoRI fragment of pHCbf5 containing cbf5 was excised and recloned in pBluescript KS+ (Stratagene) to prepare the mutants of HvCbf5 in E. coli. Mutants were created by site-directed mutagenesis. Oligonucleotides used in this study are listed in Supplemental Table S1. After confirmation of mutation by sequencing, the mutant HindIII-EcoRI fragment was cut out from the plasmid and used to replace the corresponding WT fragment of pHCbf5 for expression in H. volcanii. The sequence of the mutant gene was again checked to ensure proper replacement of the WT fragment. A commonly used system, e.g., D53A (Asp53 changed to Ala) and YIR149-151AAA (Tyr, Ile and Arg at positions 149–151 simultaneously changed to three Ala), was used to name the substitution mutants.
The presence of mutant Cbf5 protein in every H. volcanii strain that did not show Ψ at all three positions or had Ψ at positions 1940 and 2605 but not at position 1942 was checked by Western blot analyses (see Supplemental Fig. S2). Antibody preparation and these analyses are described in the Methods section of the Supplemental Material.
Determination of the presence or absence of Ψ at specific positions in 23S rRNA
All strains of H. volcanii were grown to A600 of approximately 1.0 and total RNA was isolated using Tri Reagent (Molecular Research Center) according to manufacturer's protocol. The presence of Ψ and its absence (the presence of unmodified U) in the RNA was determined by primer extension following 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluenesulfonate (CMCT) treatment (Ofengand et al. 2001a; Motorin et al. 2007) and U-specific sequencing reaction (Peattie 1979; Gupta 1984), respectively, as described previously (Blaby et al. 2011). Essentially, CMCT reaction involves treatment of RNA with CMCT, ethanol precipitation, alkali treatment, and ethanol precipitation again, followed by primer extension. CMCT forms adducts with Ψ, U, and G. Alkali removes all CMCT groups except those attached to N3 of Ψ (Bakin and Ofengand 1993). This method is not suitable for quantitation of Ψ at a particular position in the RNA, because partial reaction conditions are used here for the CMCT reactions. Furthermore, reverse transcriptase reactions used for primer extensions show varying amounts of sensitivity to secondary structures and different modified residues present in the RNA. Similarly, U-specific reactions are also partial. Here uridines in RNA are hydrazinolysed using aqueous hydrazine followed by strand scission at the position of damaged bases using acidified aniline. Both CMCT and U-specific reactions were done on the same batch of RNA. Reactions for the effects of each mutant protein were done at least twice on the RNAs prepared from independent cell cultures harvested at different times. Results of repeat reactions were similar. We consider Ψ present at a position in the RNA if a dark band is observed at that position in the gel after CMCT but not after U-specific reaction and absent when a dark band is observed after U-specific but not after CMCT reaction. A band is considered “dark” when its intensity is considerably more in the lane containing CMCT- or hydrazine-treated RNA than the untreated RNA. Since these primer extension reactions are not quantitative, we scored them as partial Ψ production when dark bands were observed at a particular position after both CMCT and U-specific reactions.
Multiple sequence alignment, homology modeling, and bioinformatic analyses
Archaeal Cbf5 protein sequences were obtained from the UCSC Archaeal Genome Browser (http://archaea.ucsc.edu/) and all other (from yeast to human) sequences were obtained from the NCBI protein database. Multiple sequence alignments of the Cbf5 protein homologs were done in Clustal W (Larkin et al. 2007). The color-coding in Figure 1 represents conservation of residues done by Jalview software (Waterhouse et al. 2009). Secondary structure and homology models of H. volcanii Cbf5 were prepared using the I-Tasser protein structure prediction suite (Zhang 2008; Roy et al. 2010; Yang et al. 2015; Yang and Zhang 2015). In this pipeline, 10 template protein structures were identified by sequence and initially modeled using the LOMETS threading algorithm. These structures included Pyrococcous furiosus Cbf5 (PDB accession code 2EY4) and H/ACA ribonucleoprotein complex (2RFK; 2HVY), the Cbf5/NOP complex in Methanococcus jannaschii (2APO), and the Cbf5-Nop10-Gar1 complex of Saccharomyces cerevisiae (3U28) as threading templates. These templates had between 35% and 45% primary sequence identities compared to H. volcanii Cbf5. After threading the primary sequence of H. volcanii Cbf5 around these template structures, a consensus centroid position for α-carbons and the side-chain center of mass was calculated from the resulting lowest energy structures. The model was then pipelined into the I-TASSER algorithm for model refinement to determine lowest energy guided by RMSD and steric clashes. The output-refined model was then assigned a C-score based on RMSD in a consensus scoring of the multiple threading templates, and a TM-score of structural similarity of the final structure with the reference template structures. The refined structure for our homology model of H. volcanii Cbf5 had a C-score of 1.13, indicating a high confidence model (typical reported range is −5 to 2); a TM-score of 0.87 ± 0.07, indicating a model with the correct topology (>0.5 is a typical cutoff); and an estimated RMSD of 3.8 ± 2.6Å compared to reference Cbf5 models. Examination of local accuracy estimates using ResQ (Yang et al. 2016) indicated an accuracy of better than 2 Å for most residues except for a small region (residues 111–119) where estimated accuracy was ∼10 Å. Dihedral angles of the refined model were examined on a Ramachandran plot and compared to template structure (PDB 2RFK). Secondary structures were within the most favored regions of the plot or in similar locations as the template structure for most residues with the exception of one β strand (residues 137–144). This final refined structure for H. volcanii Cbf5 was stored as a PDB file, visualized in the Visual Molecular Dynamics (VMD) suite (Humphrey et al. 1996), structurally aligned with P. furiosus Cbf5 using the MultiSeq extension (Roberts et al. 2006), and rendered using Tachyon (www.photonlimited.com/~johns/tachyon).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matt Geisler for help and advice in preparing molecular models and David Clark for critical reading of the manuscript. This work was supported by National Institutes of Health, National Institute of General Medical Sciences (grant number GM055945 to R.G.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.057547.116.
REFERENCES
- Badis G, Fromont-Racine M, Jacquier A. 2003. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA 9: 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP. 2005. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev 19: 1238–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin A, Ofengand J. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762. [DOI] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Fabret C, Liang XH, Piekna-Przybylska D, Fournier MJ, Rousset JP. 2009. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res 37: 7665–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby IK, Majumder M, Chatterjee K, Jana S, Grosjean H, de Crecy-Lagard V, Gupta R. 2011. Pseudouridine formation in archaeal RNAs: the case of Haloferax volcanii. RNA 17: 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin ML, Kiss T. 1998. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA 4: 445–454. [PMC free article] [PubMed] [Google Scholar]
- Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. 2011. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res 39: D195–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. 2014. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette M, Gray MW. 2000. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49: 341–351. [DOI] [PubMed] [Google Scholar]
- Charpentier B, Muller S, Branlant C. 2005. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res 33: 3133–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DR. 1995. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23: 5020–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. 2002. rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351. [DOI] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J Biol Chem 278: 695–698. [DOI] [PubMed] [Google Scholar]
- Dennis PP, Omer A. 2005. Small non-coding RNAs in Archaea. Curr Opin Microbiol 8: 685–694. [DOI] [PubMed] [Google Scholar]
- Duan J, Li L, Lu J, Wang W, Ye K. 2009. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell 34: 427–439. [DOI] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T. 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809. [DOI] [PubMed] [Google Scholar]
- Ge J, Yu YT. 2013. RNA pseudouridylation: new insights into an old modification. Trends Biochem Sci 38: 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano E, Peluso I, Senger S, Furia M. 1999. minifly, a Drosophila gene required for ribosome biogenesis. J Cell Biol 144: 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H. 2009. Nucleic acids are not boring long polymers of only four types of nucleotides: a guided tour. In DNA and RNA modifying enzymes: structure, mechanism, function and evolution (ed. Grosjean H), pp. 1–18. Landes Bioscience, Austin, TX. [Google Scholar]
- Grosjean H, Gaspin C, Marck C, Decatur WA, de Crecy-Lagard V. 2008. RNomics and Modomics in the halophilic archaea Haloferax volcanii: identification of RNA modification genes. BMC Genomics 9: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. 1984. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem 259: 9461–9471. [PubMed] [Google Scholar]
- Gurha P, Joardar A, Chaurasia P, Gupta R. 2007. Differential roles of archaeal box H/ACA proteins in guide RNA-dependent and independent pseudouridine formation. RNA Biol 4: 101–109. [DOI] [PubMed] [Google Scholar]
- Hamma T, Ferré-D'Amaré AR. 2004. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure 12: 893–903. [DOI] [PubMed] [Google Scholar]
- Hamma T, Ferré-D'Amaré AR. 2006. Pseudouridine synthases. Chem Biol 13: 1125–1135. [DOI] [PubMed] [Google Scholar]
- Hamma T, Ferré-D'Amaré AR. 2010. The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. J Biol Chem 285: 805–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamma T, Reichow SL, Varani G, Ferré-D'Amaré AR. 2005. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat Struct Mol Biol 12: 1101–1107. [DOI] [PubMed] [Google Scholar]
- He J, Navarrete S, Jasinski M, Vulliamy T, Dokal I, Bessler M, Mason PJ. 2002. Targeted disruption of Dkc1, the gene mutated in X-linked dyskeratosis congenita, causes embryonic lethality in mice. Oncogene 21: 7740–7744. [DOI] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. 1998. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 19: 32–38. [DOI] [PubMed] [Google Scholar]
- Henras AK, Dez C, Henry Y. 2004. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr Opin Struct Biol 14: 335–343. [DOI] [PubMed] [Google Scholar]
- Hoang C, Ferré-D'Amaré AR. 2001. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107: 929–939. [DOI] [PubMed] [Google Scholar]
- Hoang C, Hamilton CS, Mueller EG, Ferré-D'Amaré AR. 2005. Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain. Protein Sci 14: 2201–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14: 33–38. [DOI] [PubMed] [Google Scholar]
- Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. 2003. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell 113: 383–394. [DOI] [PubMed] [Google Scholar]
- Jiang W, Middleton K, Yoon HJ, Fouquet C, Carbon J. 1993. An essential yeast protein, CBF5p, binds in vitro to centromeres and microtubules. Mol Cell Biol 13: 4884–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalampeta R, Kothe U. 2012. Archaeal proteins Nop10 and Gar1 increase the catalytic activity of Cbf5 in pseudouridylating tRNA. Sci Rep 2: 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Yu YT. 2008. Insight into the protein components of the box H/ACA RNP. Curr Proteomics 5: 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Yu YT. 2010. Spliceosomal snRNA modifications and their function. RNA Biol 7: 192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan MD, Dokal I. 2008. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet 73: 103–112. [DOI] [PubMed] [Google Scholar]
- Kiss T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Kiss T, Fayet-Lebaron E, Jady BE. 2010. Box H/ACA small ribonucleoproteins. Mol Cell 37: 597–606. [DOI] [PubMed] [Google Scholar]
- Knight SW, Heiss NS, Vulliamy TJ, Greschner S, Stavrides G, Pai GS, Lestringant G, Varma N, Mason PJ, Dokal I, et al. 1999. X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am J Hum Genet 65: 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 1996. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res 24: 2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lecointe F, Namy O, Hatin I, Simos G, Rousset JP, Grosjean H. 2002. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J Biol Chem 277: 30445–30453. [DOI] [PubMed] [Google Scholar]
- Li H. 2008. Unveiling substrate RNA binding to H/ACA RNPs: one side fits all. Curr Opin Struct Biol 18: 78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye K. 2006. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443: 302–307. [DOI] [PubMed] [Google Scholar]
- Li S, Duan J, Li D, Yang B, Dong M, Ye K. 2011. Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev 25: 2409–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. 2015. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11: 592–597. [DOI] [PubMed] [Google Scholar]
- Liang B, Xue S, Terns RM, Terns MP, Li H. 2007. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat Struct Mol Biol 14: 1189–1195. [DOI] [PubMed] [Google Scholar]
- Liang B, Kahen EJ, Calvin K, Zhou J, Blanco M, Li H. 2008. Long-distance placement of substrate RNA by H/ACA proteins. RNA 14: 2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H. 2009. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat Struct Mol Biol 16: 740–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy AF, Riordan DP, Brown PO. 2014. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9: e110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madern D, Ebel C, Zaccai G. 2000. Halophilic adaptation of enzymes. Extremophiles 4: 91–98. [DOI] [PubMed] [Google Scholar]
- Manival X, Charron C, Fourmann J, Godard F, Charpentier B, Branlant C. 2006. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res 34: 826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PJ, Wilson DB, Bessler M. 2005. Dyskeratosis congenita—a disease of dysfunctional telomere maintenance. Curr Mol Med 5: 159–170. [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209–220. [DOI] [PubMed] [Google Scholar]
- McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. 2007. Crystal structure of human Pus10, a novel pseudouridine synthase. J Mol Biol 373: 1243–1254. [DOI] [PubMed] [Google Scholar]
- Meier UT. 2005. The many facets of H/ACA ribonucleoproteins. Chromosoma 114: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT, Blobel G. 1994. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol 127: 1505–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555. [DOI] [PubMed] [Google Scholar]
- Motorin Y, Muller S, Behm-Ansmant I, Branlant C. 2007. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol 425: 21–53. [DOI] [PubMed] [Google Scholar]
- Mueller EG, Ferré-D'Amaré AR. 2009. Pseudouridine formation, the most common transglycosylation in RNA. In DNA and RNA modification enzymes: structure, mechanism, function and evolution (ed. Grosjean H), pp. 363–376. Landes Bioscience, Austin, TX. [Google Scholar]
- Muller S, Fourmann JB, Loegler C, Charpentier B, Branlant C. 2007. Identification of determinants in the protein partners aCBF5 and aNOP10 necessary for the tRNA:Ψ55-synthase and RNA-guided RNA:Ψ-synthase activities. Nucleic Acids Res 35: 5610–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Leclerc F, Behm-Ansmant I, Fourmann JB, Charpentier B, Branlant C. 2008. Combined in silico and experimental identification of the Pyrococcus abyssi H/ACA sRNAs and their target sites in ribosomal RNAs. Nucleic Acids Res 36: 2459–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Lecointe F, Grosjean H, Rousset J-P. 2005. Translational recoding and RNA modifications. In Fine-tuning of RNA function by modification and editing. Topics in Current Genetics, Vol. 12 (ed. Grosjean H), pp. 309–340. Springer-Verlag, Berlin-Heidelberg, Germany. [Google Scholar]
- Ofengand J, Del Campo M, Kaya Y. 2001a. Mapping pseudouridines in RNA molecules. Methods 25: 365–373. [DOI] [PubMed] [Google Scholar]
- Ofengand J, Malhotra A, Remme J, Gutgsell NS, Del Campo M, Jean-Charles S, Peil L, Kaya Y. 2001b. Pseudouridines and pseudouridine synthases of the riobosome. Cold Spring Harb Symp Quant Biol 66: 147–159. [DOI] [PubMed] [Google Scholar]
- Peattie DA. 1979. Direct chemical method for sequencing RNA. Proc Natl Acad Sci 76: 1760–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips B, Billin AN, Cadwell C, Buchholz R, Erickson C, Merriam JR, Carbon J, Poole SJ. 1998. The Nop60B gene of Drosophila encodes an essential nucleolar protein that functions in yeast. Mol Gen Genet 260: 20–29. [DOI] [PubMed] [Google Scholar]
- Rashid R, Liang B, Baker DL, Youssef OA, He Y, Phipps K, Terns RM, Terns MP, Li H. 2006. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol Cell 21: 249–260. [DOI] [PubMed] [Google Scholar]
- Roberts E, Eargle J, Wright D, Luthey-Schulten Z. 2006. MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics 7: 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L. 2006. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res 34: 4293–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A. 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res 31: 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. 2003. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299: 259–262. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. 2014. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedaliere CJ, Hamilton CS, Mueller EG. 2000. Functional importance of motif I of pseudouridine synthases: mutagenesis of aligned lysine and proline residues. Biochemistry 39: 9459–9465. [DOI] [PubMed] [Google Scholar]
- Spenkuch F, Motorin Y, Helm M. 2014. Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol 11: 1540–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillault AS, Fourmann JB, Loegler C, Blaud M, Branlant C, Charpentier B. 2015a. RNA size is a critical factor for U-containing substrate selectivity and permanent pseudouridylated product release during the RNA:Ψ-synthase reaction catalyzed by box H/ACA sRNP enzyme at high temperature. Biochimie 113: 134–142. [DOI] [PubMed] [Google Scholar]
- Tillault AS, Fourmann JB, Loegler C, Wieden HJ, Kothe U, Charpentier B. 2015b. Contribution of two conserved histidines to the dual activity of archaeal RNA guide-dependent and -independent pseudouridine synthase Cbf5. RNA 21: 1233–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Meier UT. 2004. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J 23: 1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT. 2012. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414. [DOI] [PubMed] [Google Scholar]
- Wu G, Xiao M, Yang C, Yu YT. 2011. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J 30: 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang Y. 2015. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43: W174–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods 12: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang Y, Zhang Y. 2016. ResQ: an approach to unified estimation of B-factor and residue-specific error in protein structure prediction. J Mol Biol 428: 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K. 2007. H/ACA guide RNAs, proteins and complexes. Curr Opin Struct Biol 17: 287–292. [DOI] [PubMed] [Google Scholar]
- Yu YT, Terns RM, Terns MP. 2005. Mechanisms and functions of RNA-guided RNA modification. In Fine-tuning of RNA functions by modification and editing (ed. Grosjean H), pp. 223–262. Springer-Verlag, New York. [Google Scholar]
- Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. 1999. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol 19: 7461–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liang B, Li H. 2010. Functional and structural impact of target uridine substitutions on the H/ACA ribonucleoprotein particle pseudouridine synthase. Biochemistry 49: 6276–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.