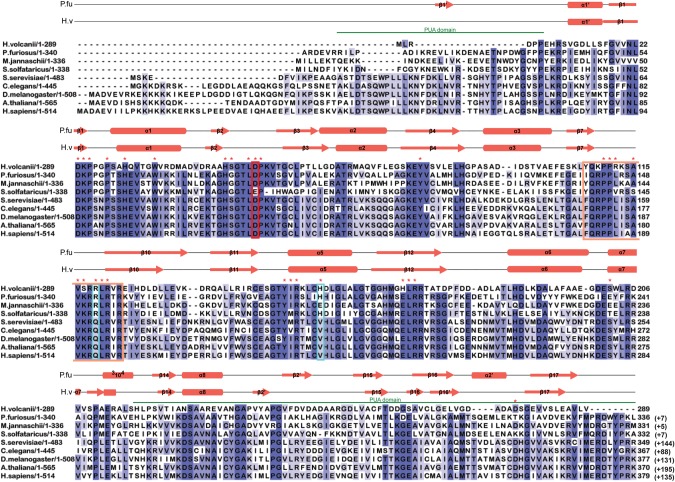
FIGURE 1.
Multiple sequence alignment of archaeal and eukaryal Cbf5 proteins. Sequences represented here are from four Archaea: Haloferax volcanii (H. volcanii), Pyrococcus furiosus (P. furiosus), Methanocaldococcus jannaschii (M. jannaschii), and Sulfolobus solfataricus (S. solfataricus); and five Eukarya: Saccharomyces cerevisiae (S. cerevisiae), Caenorhabditis elegans (C. elegans), Drosophila melanogaster (D. melanogaster), Arabidopsis thaliana (A. thaliana), and Homo sapiens (H. sapiens). Secondary structural elements are marked above the sequences, with α-helices depicted as cylinders and β-strands as arrows. Secondary structure of P. furiosus protein is based on the crystal structure (PDB 2EY4) (Rashid et al. 2006). Secondary structure of H. volcanii protein is the resulting modeled structure from I-TASSER. One new secondary structural element for HvCbf5 was predicted by I-TASSER, which is not shown in the cited reference (Rashid et al. 2006). The PUA domain regions comprising the structures at the two termini are marked by green lines above the sequence. Thumb loop sequences are boxed in orange. The conserved catalytic aspartate (D) residue is enclosed within a red box. Certain residues that differ between Archaea and Eukarya are enclosed in blue boxes. Residues used for mutagenesis in this study are indicated with red asterisks above the sequence. Highly conserved residues (>80%) among all proteins are shaded in dark blue, and at least 60% conserved residues are shown in medium blue. Light blue represents at least 40% conservation. Numbers after each sequence denote ending residues of each block. Numbering of P. furiosus Cbf5 has been adjusted to match a previous report (Rashid et al. 2006), which is commonly used in the literature. Numbers in parentheses indicate the length of the sequence not shown here.