FIGURE 2.
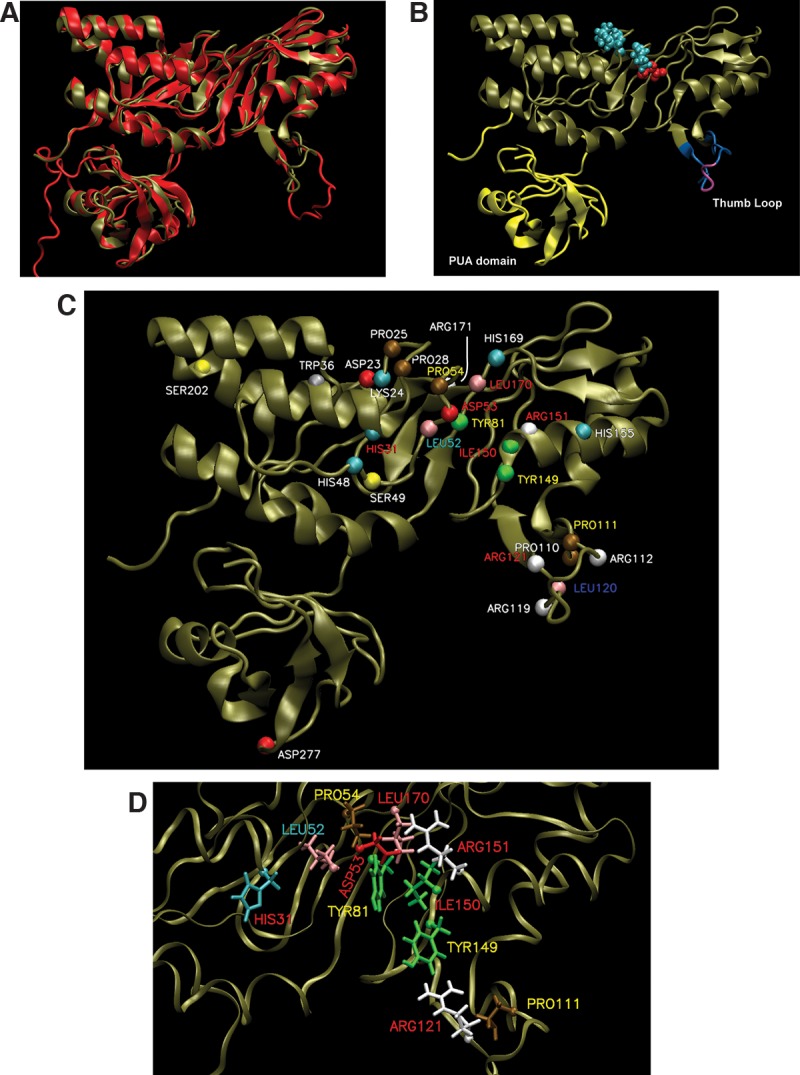
Homology model of HvCbf5. (A) The structure of PfuCbf5 (red) has been extracted from the Cbf5-Nop10-Gar1 crystal structure (PDB 2RFK) (Liang et al. 2007). The homology model of HvCbf5 (tan) based on I-TASSER predicted structure is overlaid. (B) The HvCbf5 model structure from A is shown with thumb loop in blue and purple and the PUA domain in yellow. The purple part of the thumb loop denotes the C loop (Ala115, Val116, and Ser117). The catalytic Asp53 (red) and Cbf5 residues (Pro25, Pro28, and Pro54 in cyan) of the “proline spine” of the RNP complex are shown as van der Waal's spheres. (C) Model of HvCbf5 showing positions of the residues that are individually substituted in this study. The α-carbons of these residues are shown as colored spheres. The colors of these spheres are by residue type as defined in VMD. Effects of Ala substitution of residues are indicated by colors of the labels: absence of (red) or partial (yellow) modification at the three positions (1940, 1942, and 2605) of 23S rRNA; partial modification at positions 1940 and 2605, but no modification at position 1942 (cyan); partial modification at position 1940, and normal modification at positions 1942 and 2605 (blue); and no effect (white), i.e., normal modification at all three positions. (See Table 1 for the effects of mutations of single and multiple residues of HvCbf5.) (D) Structural details near the active site of HvCbf5. Most residues that affected activity of HvCbf5 after mutation are shown. Colors of residues and labels are as in C.
