Abstract
Endoplasmic reticulum (ER) stress results from changes in ER homeostasis and folding of proteins. ER stress initiates cellular adaptive mechanisms to rescue cell homeostasis or, if that does not work, to elicit apoptosis. We have previously shown that mouse SDF2 is sublocalized in the ER, is ubiquitously expressed, and shows strong similarities with stromal cell-derived factor (SDF) 2L1 and SDF2-like from Arabidopsis, ER proteins involved in chaperone network and protein folding. Thus, we hypothesized that SDF2 plays a role in the ER stress and unfolded protein response. In this study, we investigated the possible role of SDF2 in the human placenta. Expression of SDF2 was present throughout gestation and was expressed by several cell types. Second-trimester cytotrophoblast cells (CTBs) in the differentiation process, monitored through chorionic gonadotropin production, showed upregulation of SDF2 protein. SDF2 expression, however, was significantly diminished in placentas from neonates small for gestational age and in hypoxic in vitro conditions (P ≤ 0.001, 2% O2), suggesting a link with cellular stress. ER stress-induced cells—CTB and BeWo—also showed SDF2 downregulation in different time points, emphasizing this relationship. SDF2 downregulation was also followed by an increase in binding immunoglobulin protein (BiP) expression, an ER protein-associated chaperone acting as a sensor for misfolded proteins and an ER stress cell survival marker. In line with this, SDF2 siRNA resulted in significant anticipation of BiP expression. Downregulation of SDF2 also interfered with C/EBP homologous protein expression, one of the highest inducible genes during ER stress. These findings suggest that SDF2 may be an important regulatory factor by which trophoblast cells can control cell survival under ER stress. In conclusion, this study identifies a novel factor with the ability to interfere with ER stress proteins, which may contribute to the understanding of ER stress associated with placental-related diseases of pregnancy.
Keywords: cell survival/apoptosis, ER Stress, human placenta, SDF2, UPR
INTRODUCTION
The stromal cell-derived factor (SDF) 2 gene was first described by Hamada et al. [1]. It is well conserved in mammals, but its underlying function is still open to conjectures. Few studies have shown expression of SDF2 in human and mouse tissues. Human SDF2 mRNA and other SDFs, such as SDF1, SDF2L1, SDF4, and SDF5, are all ubiquitously expressed in breast cancer tissues and cell lines [2]. Gene expression of SDF2 is reduced along with a poor prognosis (metastasis and death) in breast and colorectal cancer [2, 3]. In human endothelial cells, SDF2 was identified as a component of Hsp90-eNOS complex, required for eNOS phosphorylation and activation [4]. Analyzing mouse placental tissues during postimplantation steps, Hoshida et al. [5, 6] have also shown an overexpression of Sdf2 mRNA. Furthermore, knockout mice for Tie2, an angiogenic factor receptor essential for embryonic vascular development, showed reduced Sdf2 mRNA in the yolk sac at gestation by Day 8.5 [7].
Our previous studies [8] have reported the predicted mouse and human Sdf2 amino acid sequence being similar to the human and mouse SDF2L1 sequence (an endoplasmic reticulum [ER] stress-inducible gene); the predicted mouse Sdf2 structure is also similar to Arabidopsis thaliana SDF2-like protein, a target of unfolded protein response (UPR) in the ER stress pathway [8]. We have also shown that the protein is sublocalized in the ER, being widely expressed in mouse tissues and organs [8]. Based on the close similarity of SDF2, SDF2L1, and SDF2-like from Arabidopsis, we hypothesized that SDF2 may also play a role in the UPR and ER stress.
Secreted and membrane proteins are folded and released by the ER. In adverse conditions, such as hypoxia and glucose deprivation, ER stops folding proteins appropriately and begins accumulating misfolded proteins in the lumen, which activates the UPR [9]. Three transmembrane proteins act as an ER sensor for misfolded proteins, which then activate the UPR: inositol-requiring enzyme (IRE1) 1, PKR-like eukaryotic initiation factor (eIF) 2α kinase (PERK), and activating transcription factor (ATF) 6. They are activated (IRE1 and PERK are phosphorylated, and ATF6 is translocated and cleaved in Golgi) through the luminal release of the chaperone binding immunoglobulin protein (BiP)/glucose-related protein (GRP) 78, normally coupled to these sensor proteins [9]. In adverse or stressful conditions, BiP/GRP78 detaches from IRE1, PERK, and ATF6 and binds to the misfolded/unfolded proteins in the lumen, which activates several signaling cascades to improve chaperone production, protein degradation, and downregulation of de novo protein synthesis (the UPR). These responses increase the cell's capacity to survive even under stress. If the cell fails to recover its homeostasis, the UPR activates the apoptotic pathway mainly through the proapoptotic transcription factor C/EBP homologous protein (CHOP). CHOP is primarily induced through PERK and ATF6 sensors [9], but it has also been supposed to be activated by an IRE1 sensor [10].
When BiP/GRP78 is released from IRE1 under ER stress, this factor activates an endonuclease that promotes the splicing of X-box binding factor (XBP) 1 to its smaller form, spliced XBP1 (sXBP1) [9]. Spliced XBP1 is considered to be a protective transcriptional factor for several cell types [11]. XBP1 is the principal transcriptional factor involved in ER stress adaptive response, and is also crucial for growth and survival of solid tumors under hypoxic stress [11]. Increase in sXBP1 expression is seen in several cancer types, such as breast cancer and hepatocellular carcinoma [11].
The chaperone BiP/GRP78 seems to be involved in the prosurvival and cytoprotective response in cancer, acting as a biomarker predictor of treatment outcomes and as a possible target for anticancer therapy [11]. After PERK is activated upon releasing BiP/GRP78, this transmembrane protein phosphorylates eIF2α, which inhibits protein synthesis. However, some factors escape this regulation, such as ATF4, which upregulates the transcription of CHOP, the main apoptotic mediator in UPR [9]. When ATF6 is translocated to the Golgi complex, this protein is cleaved (ATF6f) and activates transcription of several genes, including BiP and XBP1 [9].
Organs that have intense endocrine/exocrine activity, such as the liver and placenta, show basal levels of ER stress due to high protein transduction and, therefore, increased probability of inaccuracies during the synthesis process [12, 13]. In placenta, approximately 30% of its oxygen is used in protein synthesis [14], the dysfunction of which is associated with severe gestational problems, such as intrauterine growth restriction and preeclampsia [15].
ER stress and UPR activation have been reported in human gestational diseases that can affect placental development. ER stress and UPR markers are upregulated in placentas from intrauterine growth-restricted newborns [16] and patients with severe preeclampsia [17]. In vitro assays using JEG-3 cells also showed an increase of the proapoptotic factor, CHOP, in an ischemia/reperfusion model [18]. In pregnant mice, systemic exposure to tunicamycin, a classical ER stress inducer, increased CHOP expression in the placenta, which seems to contribute to the development of growth-restricted fetuses [19]. In addition, IRE1α knockout mice were embryonically and placentally impaired, succumbing at Gestation Day 12.5 [13]. Normal fetal development and live progeny were produced only when Ire1−/− embryos developed with Ire1+/+ placentas (conditioned knockout). These data reinforce the relevant putative role played by ER stress and UPR in gestation and successful embryo development. Furthermore, disruption in ER homeostasis and activation of UPR during gestation are also of biological relevance, as they can affect the production of key factors (hormones, growth factors, and regulatory proteins) associated with the development of gestational diseases.
In this study, we have mapped human SDF2 expression through all gestation phases, placental compartments, and cell types, and carried out functional assays of differentiation, hypoxia, and ER stress using primary cytotrophoblast cells (CTBs) and the BeWo trophoblast cell line. The data suggest a role for SDF2 in UPR in trophoblast cell survival/apoptosis, a crucial balance closely associated with placental fate. The cellular decision in eliminating cells that are producing nonfunctional proteins during pregnancy may be the turning point that determines the health of a pregnancy without fetal consequences or placental changes that lead to changes in fetal development, as occurs, for example, in intrauterine growth restriction and preeclampsia.
MATERIALS AND METHODS
Human Tissue Collection and Reagents
This study was approved by University of California San Francisco (UCSF) Human Research Protection Program/Committee on Human Research. Written informed consent was obtained from the donors. Biopsies of normal placentas from elective termination for psychosocial reasons were obtained in two clinics in San Francisco city (6–24 wk of gestation, n = 12); placental samples from term delivery were collected at the UCSF Medical Center (37–39 wk, n = 11). Term neonates were classified as: appropriate-for-gestational-age (AGA; birthweight between the 10th and 90th percentile [n = 5]) and small-for-gestational-age (SGA; birthweight below the 10th percentile [n = 6]). Placental samples were dissected to: 1) collect protein lysates, 2) prepare fresh-frozen and paraformaldehyde-fixed samples for immunofluorescence, and 3) immediately process for CTB isolation. Reagents were purchased from Sigma-Aldrich unless otherwise specified.
Western Blot
Placental tissues were obtained as villous, decidua, and membrane samples (first trimester), and as villous, basal plate, amnion, smooth chorion, and chorionic plate samples (second trimester and term). A placental diagram identifying these regions is available as Supplemental Figure S1 (Supplemental Data are available online at www.biolreprod.org). Samples were homogenized using lysis buffer (150 mM NaCl, 50 mM Tris, 2 mM EDTA, 1% NP-40, 1× protease mix) with Western blotting carried out as previously described [20]. Briefly, samples were resolved by SDS-PAGE in 15% acrylamide gels (Bio-Rad) and transferred to nitrocellulose membranes (0.45 μm; Bio-Rad). Membranes were blocked in 3% nonfat milk (Bio-Rad) for 1 h followed by primary antibody incubation (anti-SDF2 [Sigma] polyclonal antibody was used at 2.5 ng/ml, polyclonal anti-GRP78 [Santa Cruz Biotechnology] at 0.15 μg/ml, polyclonal caspase 3 [Santa Cruz Biotechnology] at 0.6 μg/ml, and monoclonal anti-β-actin (clone AC-15; Sigma) were used as loading control at 0.15 μg/ml). Secondary antibodies (Jackson ImmunoResearch Laboratories) were used at 1.25 μg/ml. Immunoreactive bands were detected using the ECL method, and were captured with high-sensitivity film (GE Healthcare).
Immunofluorescence
Sample tissues were washed in PBS, fixed in 4% paraformaldehyde, infiltrated with 5%–15% sucrose, followed by O.C.T. compound, frozen in liquid nitrogen [21] and cryosectioned at 5 μm. CTBs were plated in matrigel on coverslips and fixed in 4% paraformaldehyde in PBS for 5 min before being treated as described previously [22]. Primary antibodies are mentioned in each figure legend, and a list—with antibodies and their concentrations—is given in Supplemental Table S1. Donkey anti-rabbit/mouse/rat IgG (H+L) rhodamine (red) or fluorescein (green) (Jackson ImmunoResearch) were used as secondary antibodies, as specified in the figure legends. Primary antibodies were omitted for all negative controls. Images were acquired using a Leica DM5000B Microscope System (Leica Microsystems) and analyzed using Leica Application Suite v3.5 Software.
Cell Isolation—Differentiation, Hypoxia, and ER Stress Assays
CTBs were isolated as described by Hunkapiller and Fisher [23] and cultured up to 48 h (n = 5). Tissues were digested in collagenase and trypsin, followed by using a Percoll gradient. Cells (purity higher than 85%; data not shown) were plated on matrigel (BD) in 24-well dishes (0.1 × 106 cells per well) and maintained under culture conditions for 30 min or 24 or 48 h at 37°C, air plus 5% CO2, for differentiation assays. An additional group was maintained in culture for 24 h before being made hypoxic for 1 h or overnight in 2% O2. CTBs under ER stress were treated with tunicamycin—an inhibitor of protein glycosylation leading to accumulation of misfolded proteins in ER lumen, commonly used as an ER stress inducer—or dimethyl sulfoxide (DMSO; tunicamycin vehicle, control) 0, 1.0, 2.5, or 5.0 μg/ml on the following day after isolation and maintained for 30 min, 3 h, 24 h, and 30 h in culture. After analysis of SDF2 expression, the following experiments involved using 2.5 and 5.0 μg/ml tunicamycin/DMSO for 3 or 24 h. Cells were lysed as described above and Western blotted for analysis of GRP78, caspase 3, SDF2, and β-actin. Human umbilical vein endothelial cells (HUVECs; Clonetics, Biowhittaker, Inc.) were maintained in EBM-2 medium supplemented with EGM-2 and EGM-2-MV Singlequots (Clonetics). Mesenchymal stromal cells from chorionic villi were isolated using the same protocol used for CTBs, but without percoll gradient. Purity of mesenchymal cells was assessed by the presence/absence of cell markers: cytokeratin clone 7D3 (−), cytokeratin 7 (−), vimentin (+), CD45 (−), CD34 (−), and CD3 (−) (data not shown). They were maintained in gelatin-coated coverslips for immunofluorescence.
ELISA—Human Chorionic Gonadotropin Quantification
Supernatants from 30 min, 24 h, and 48 h CTB (n = 5) cultures were used in an ELISA assay detecting human chorionic gonadotropin (hCG). Samples were immediately centrifuged at 800 rpm and frozen assayed (Abazyme, LLC). Results were collected using Bio-Rad Spectrophotometer Model 680.
BeWo—Differentiation, Hypoxia, and ER Stress Assays
BeWo cell line was kindly provided by Eloisa A. Ferro, Ph.D. (Federal University of Uberlandia, Brazil). Cells were maintained in RPMI media supplemented with 10% FBS and antibiotics (gentamicin and amphoterecin) at 37°C and air with 5% CO2. Cells were plated in 24-well dishes (0.1 × 106 cells/well) and maintained overnight before being treated with tunicamycin 2.5 or 5.0 μg/ml for 3, 6, or 24 h. In hypoxia assays, 1 mM N-[methoxyoxoacetyl]-glycine methyl ester (DMOG; Cayman Chemical) was used for 24–48 h, or DMSO as control. Cells were lysed for protein analysis (Western blots for GRP78, caspase 3, SDF2, and β-actin for ER stress assays, and SDF2, HIF1α, and β-actin for hypoxia assays) and to extract total mRNA using a Rneasy Mini Kit (QIAGEN). Quality and quantification of mRNA was assessed by NanoVue (GE Healthcare). These experiments were performed in triplicate.
SDF2 Gene Silencing—siRNA
BeWo cells were treated with human SDF2 siRNA (Santa Cruz Biotechnology). Briefly, cells were plated in 24-well dishes in an antibiotic-free medium (RPMI plus 10% FBS only) for 18 h. Silenced RNA and transfection reagent were diluted in transfection medium and incubated for 30 min. Cells were incubated with siRNA solution for 7 h, then the medium was replaced by RPMI plus FBS and antibiotics; the cells were maintained for 24 h before receiving 5 μg/ml tunicamycin/DMSO for 3, 6, and 24 h, when their mRNA was isolated using the Rneasy Mini Kit and protein lysate for Western blotting. Apoptosis and cell viability were assessed using Caspase-Glo 3/7 and CellTiter-Glo Luminescent Cell Viability assays (Promega) and analyzed by the Glomax Multi Detection System (Promega). These experiments were performed in triplicate.
Real-Time Quantitative PCR
Complementary DNA was produced after treatment with Dnase I (Invitrogen, Life Technologies) using a High-Capacity cDNA kit (Invitrogen, Life Technologies). Briefly, 0.2 μg of total RNA was treated with Dnase I and incubated with kit buffers for 10 min at 25°C, 120 min at 37°C, and 5 sec at 85°C in a thermocycler (Eppendorf). Primers for sXBP1, CHOP, SDF2, and YWHAZ (endogenous control) and amplification conditions are listed in Supplemental Table S2. Real-time quantitative PCR was performed with SYBR Green with StepOnePlus equipment standard conditions and analyzed using StepOnePlus Software 2.0 (Life Technologies).
Statistical Analysis
Statistical analysis was performed comparing different doses (1.0, 2.5, and 5.0 μg/ml) in the same period—30 min or 3, 6, 24, or 30 h—and comparing the same dose over the time in ER stress assays using CTB and BeWo cells. Data are presented as mean ± SD and were analyzed by one-way ANOVA followed by Tukey multiple comparison test. SGA and AGA placentas were analyzed by two-tailed Student t-test followed by nonparametric Mann-Whitney U post test. P ≤ 0.05 was considered statistically significant.
RESULTS
SDF2 Protein Expression Mapping
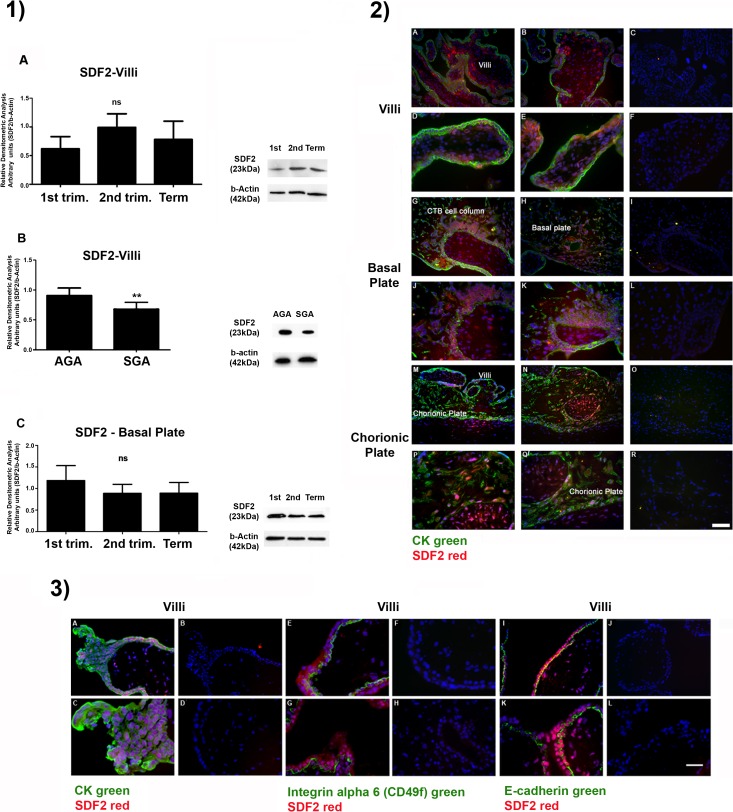
Expression of SDF2 in placental tissues was first assessed using Western blot and immunofluorescence in all placental compartments (Fig. 1 and Supplemental Fig. S1). SDF2 was ubiquitously present in the different cell types and compartments. No differences were seen for SDF2 expression along the gestational trimesters in the fetal area of the placenta, villi, and basal plate (Fig. 1, Part 1, A and C). SDF2 expression in placental villi from AGA neonates was higher than those seen in villi from SGA neonates (Fig. 1, Part 1, B, clinical data in Supplemental Table S3). There were also differences among placental compartments that were statistically significant—increased SDF2 protein expression—in decidua during the first trimester in comparison with villi (P < 0.05) and chorionic membranes (P < 0.01; Supplemental Fig. S1, Part 1, A), at term in smooth chorionic membrane (P < 0.05) compared to chorionic plate (Supplemental Fig. S1, Part 1, C) and in smooth chorion membrane (P < 0.01) compared to chorion membranes and chorionic plate (Supplemental Fig. S1, Part 1, D). Immunofluorescence also followed expression mapping of SDF2 (Fig. 1, Part 2). Placental compartments showed a broad distribution for SDF2, a pattern that did not change throughout the gestational phases. Trophoblast (colocalized with cytokeratin) and nontrophoblast cells were SDF2 reactive at the villi stroma (Fig. 1, Part 2, A–F) and basal plate (the maternal portion of the placenta with reactive CTB columns; Fig. 1, Part 2, G–L).
FIG. 1.
SDF2 protein expression mapping. Part 1) Western blotting analysis. A–C) No statistical difference is seen in SDF2 expression in villous (A) and decidua/basal (C) plate compartments throughout gestation. Values are plotted as mean ± SD. One-way ANOVA followed by Tukey multiple comparison test. B) Term placentas (villi) from AGA and SGA neonates. SDF2 expression is downregulated in SGA placentas. Values are plotted as mean ± SD. Two-tailed Student t-test and nonparametric Mann-Whitney U post test; **P ≤ 0.01. Beta-actin was used as loading control. Part 2) Immunofluorescence (15- to 16-wk placenta). SDF2 (red), cytokeratin (CK; green), and nuclei (blue; DAPI). A–F) Villi: SDF2 is stained in trophoblastic (cyto- and syncytiotrophoblast) and nontrophoblastic cells (villous core). G–L) Basal plate: SDF2 is also expressed in invasive CTB (cell columns) and nontrophoblastic cells. M–R) Smooth chorionic membrane: SDF2 is expressed in chorionic and amniotic epithelium and stroma. S–X) The 23.6-wk placenta. Chorion plate: expression of SDF2 is concentrated near the villi. Part 3) Immunofluorescence for CTB and syncytiotrophoblast. A–D) First-trimester villi stained for CK (green), SDF2 (red), and nuclei (blue; DAPI). E–H) First-trimester villi stained for integrin alpha 6 (green; CTBs) and SDF2 (red). I–L) First-trimester villi stained for E-cadherin (green; CTB), SDF2 (red), and nuclei (blue; DAPI). Negative controls (no primary antibody incubation) are on the right of each column. Bar in Part 2 = 100 μm for A–C, G–I, M–O, S–U, and V–X and 50 μm for D–F, J–L, and P–R. Bar in Part 3 = 100 μm for A, B, E, F, I, and J and 50 μm for C, D, G, H, K, and L.
CTBs in anchoring villi (cell column CTBs, first trimester) also expressed SDF2 (Fig. 1, Part 3, A–D). First-trimester SDF2 staining in CTB and syncytiotrophoblast was confirmed by colocalization with integrin alpha-6 (Fig. 1, Part 3, E–H) and E-cadherin (Fig. 1, Part 3, I–L) [24]. Remaining structures, such as smooth chorionic and amniotic membranes, also expressed SDF2 in trophoblast and nontrophoblast cells (Supplemental Fig. S1, Part 2, A–F), as in the chorionic plate (Fig. 1, Part 2, M–R).
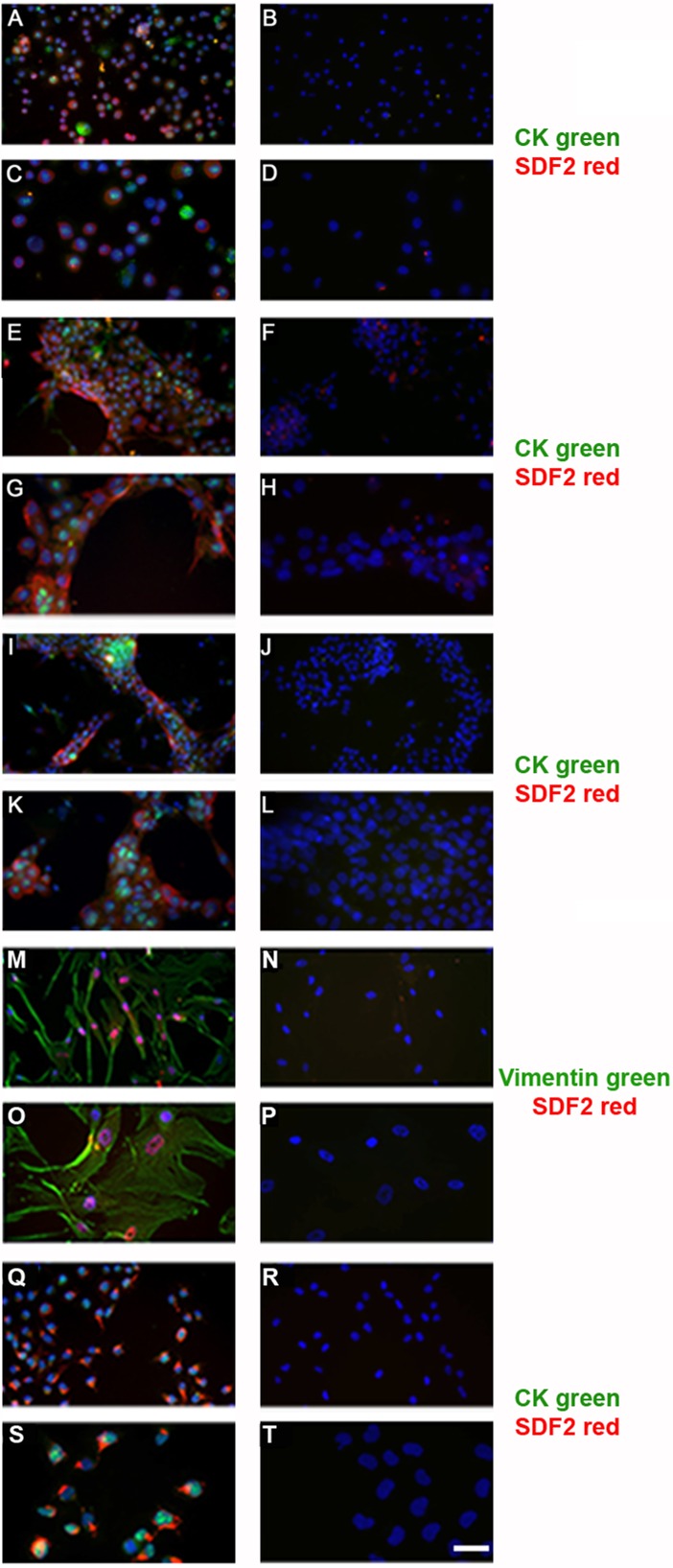
Since functional assays have to be done in culture systems, SDF2 expression was also investigated in second-trimester placentas in CTB villous cells cultured on matrigel (Fig. 2) for 30 min (time to allow viable cells to attach in matrigel; Fig. 2, A–D), 24 h (Fig. 2, E–H), or 48 h (Fig. 2, I–L) by using double-staining immunofluorescence to identify cell types. SDF2 expression was evident after 24 h in differentiated CTBs (Fig. 2, E–H), stromal cells (Fig. 2, M–P), and, in placental macrophages, the Hofbauer cells (Supplemental Fig. S2, A–D). The breast cancer cell line, MDA-MB231, used as a positive SDF2 immune control, also expressed SDF2 (Fig. 2, Q–T), as did HUVECs (Supplemental Fig. S2, E–H).
FIG. 2.
Double-staining of SDF2 in cell culture types using immunofluorescence. Primary villous CTBs in culture for 30 min (A and C), 24 h (E and G), and 48 h (I and K); SDF2 (green), CK (red). M,O) Primary villous stromal cells from term placenta; SDF2 (red), vimentin (green). Q and S) MDA-MB-231 breast cancer cell lineage; SDF2 (green) and CK (red). Negative controls on the right of each column were not incubated with primary antibodies (B, D, F, H, J, L, N, P, R, and T). Bar = 100 μm in A, B, E, F, I, J, M, N, Q, and R and 50 μm in C, D, G, H, K, L, O, P, S, and T.
Differentiation and Hypoxia
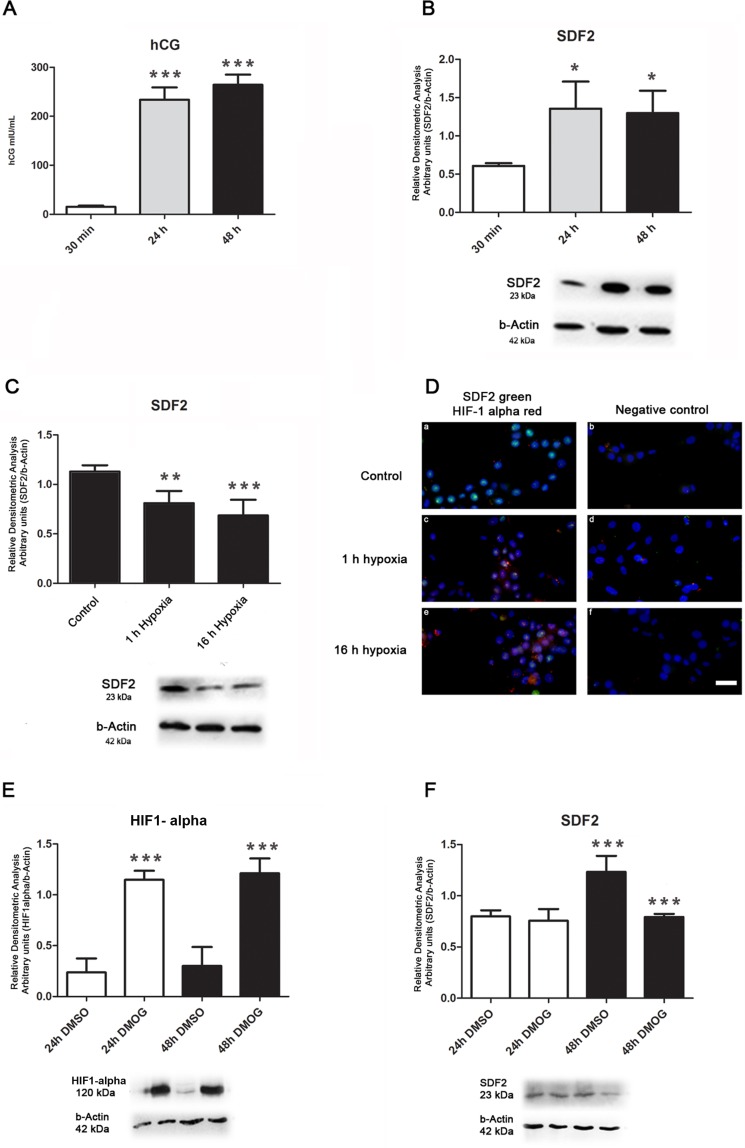
In vitro, CTBs differentiate into syncytiotrophoblast within 24–48 h after plating, as shown by morphological studies [25]. This process can also be followed by increased secretion of hCG [25]. In our model, CTBs were cultured for 30 min, 24 h, or 48 h, and hCG levels monitored in the culture supernatant. Human CG was elevated after 24 and 48 h (P ≤ 0.001, Fig. 3A), confirming trophoblast cell differentiation. SDF2 expression also increased after 24 and 48 h compared to 30 min (Fig. 3B, P ≤ 0.05), in parallel with the differentiation process.
FIG. 3.
Differentiation and hypoxia assays. A) The increased production of β-hCG confirms trophoblast differentiation after 24 and 48 h of cultured CTBs when compared to control (30 min). B) In CTBs, SDF2 protein concentration is upregulated after 24 and 48 h when compared to control (30 min). C) After 24 h in normoxia (Control), CTBs were subjected to 1 or 16 h at 2% O2. Protein concentration of SDF2 is downregulated in hypoxia, at both 1 and 16 h when compared to control. D) Immunofluorescence of HIF-1α assay confirms hypoxia (1 and 16 h, 2% O2); control: CTBs in normoxia for 24 h. SDF2 (green) and HIF-1α (red). Primary antibodies were omitted in negative controls. Bar = 50 μm (a–f). E) Upregulation of HIF-1α in DMOG-treated BeWo cells confirms hypoxia when compared to the control (DMSO only) in the same period (24 or 48 h). F) BeWo cells show upregulation of SDF2 after 48 h when compared to 24-h cells in the control group (DMSO). In DMOG-treated cells, SDF2 is downregulated after 48 h when compared to control in the same period; β-actin was used as the loading control. Values are plotted as mean ± SD. One-way ANOVA followed by Tukey multiple comparison test, *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
Placental physiology is highly susceptible to oxygen conditions [26]. Hypoxic insult to the placenta has been associated with several gestational disorders, such as preeclampsia [26]. In this study, we tested exposure to hypoxia to disturb placental homeostasis in villous explants for 1 and 16 h. When CTBs were maintained in culture conditions for 24 h (5% CO2 and 21% O2 in air—standard conditions), and then placed in conditions of hypoxia (5% CO2 and 2% O2 in air) for 1 or 16 h, SDF2 expression decreased (P ≤ 0.01 and P ≤ 0.001, respectively; Fig. 3C). Hypoxia was confirmed with HIF-1α staining (Fig. 3D).
The trophoblast-derived choriocarcinoma cell line BeWo was also tested in hypoxic conditions, using 1 mM DMOG for 24 and 48 h (Fig. 3, E–F). Increased HIF-1α expression in DMOG-treated cells confirmed hypoxic conditions at 24 and 48 h (Fig. 3E, P ≤ 0.001). SDF2 expression decreased, but only after 48 h in culture compared to untreated cells (Fig. 3E, P ≤ 0.001). Expression was higher at 48 than 24 h (Fig. 3E, P ≤ 0.001).
ER Stress
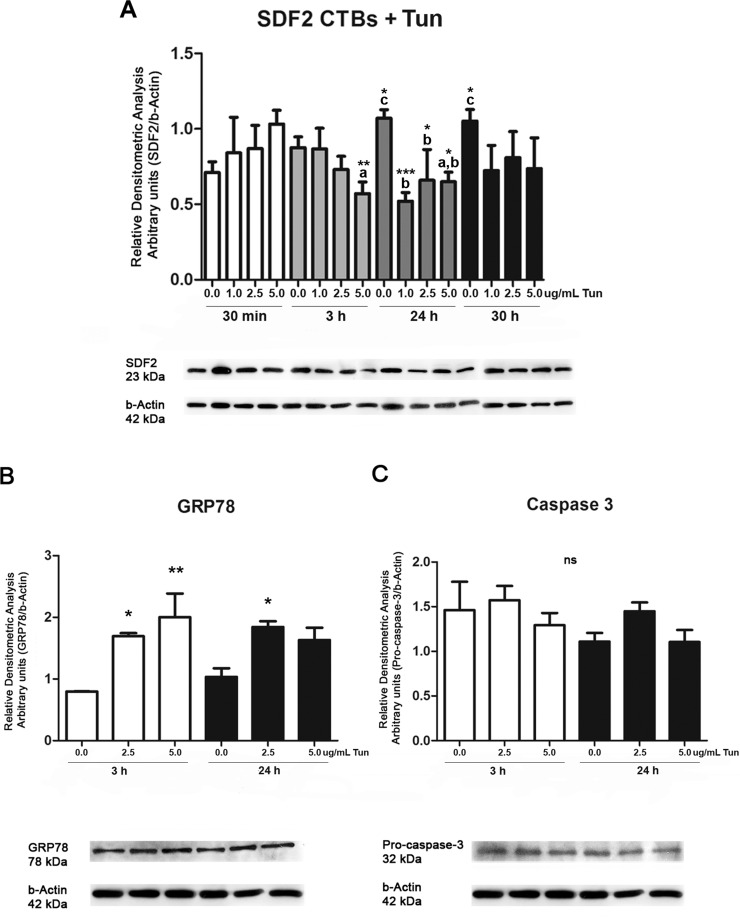
Several pharmacological agents can induce ER stress [27]. Here, ER stress was induced during the second-trimester CTBs cells through tunicamycin exposure. Figure 4A shows SDF2 expression is dose responsive to tunicamycin treatment. SDF2 protein expression decreased after 3 h (P ≤ 0.01) and 24 h (P ≤ 0.05) at 5.0 μg/ml compared to 30 min of exposure. After 24 h, all tunicamycin concentrations (1.0, 2.5, and 5.0 μg/ml) decreased SDF2 expression compared to 24-h samples that had received only DMSO, which contrasts with the increased SDF2 expression in CTBs cells that were untreated after 24 h (P ≤ 0.05) and 30 h (P ≤ 0.05) during the differentiation process.
FIG. 4.
Protein expression of SDF2, GRP78/BiP, and caspase-3 after treatment with tunicamycin (Tun) in CTBs. A) SDF2 expression is downregulated at 3 and 24 h of tunicamycin treatment (5 μg/ml) when compared to the same concentration at 30 min of treatment (a). At 24 h, all concentrations showed downregulation of SDF2 (1–2.5 and 5 μg/ml [b]). Without tunicamycin, expression of SDF2 is upregulated at 24 and 30 h when compared to 30 min (c). B) GRP78/BiP is upregulated at 3 h (2.5 and 5 μg/ml tunicamycin) and 24 h (2.5 μg/ml) when compared to cells that received only DMSO during the same period of treatment. No statistical differences were seen among the same concentrations at 3 and 24 h. C) No statistical difference is seen in procaspase-3 expression. Values are plotted as mean ± SD. One-way ANOVA followed by Tukey multiple comparison test, *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
We chose 2.5 and 5.0 μg/ml tunicamycin and 3- and 24-h treatment to analyze the expression of ER stress markers, GRP78/BiP and caspase-3. Increased expression of GRP78/BiP was seen at both tunicamycin dosages after 3 h of treatment (P ≤ 0.05 and P ≤ 0.01), and after 24 h of treatment at 2.5 μg/ml (Fig. 4B, P ≤ 0.05). No statistical difference was seen in procaspase-3 expression (Fig. 4C). There was no evidence of cleaved caspase, although the antibody was able to recognize both procaspase and caspase-3-cleavage subunits (Supplemental Fig. S3).
Based on the CTBs results, BeWo cells were also tested for ER stress after being treated with 2.5 or 5.0 μg/ml tunicamycin for 3, 6, and 24 h (Fig. 5A).
FIG. 5.
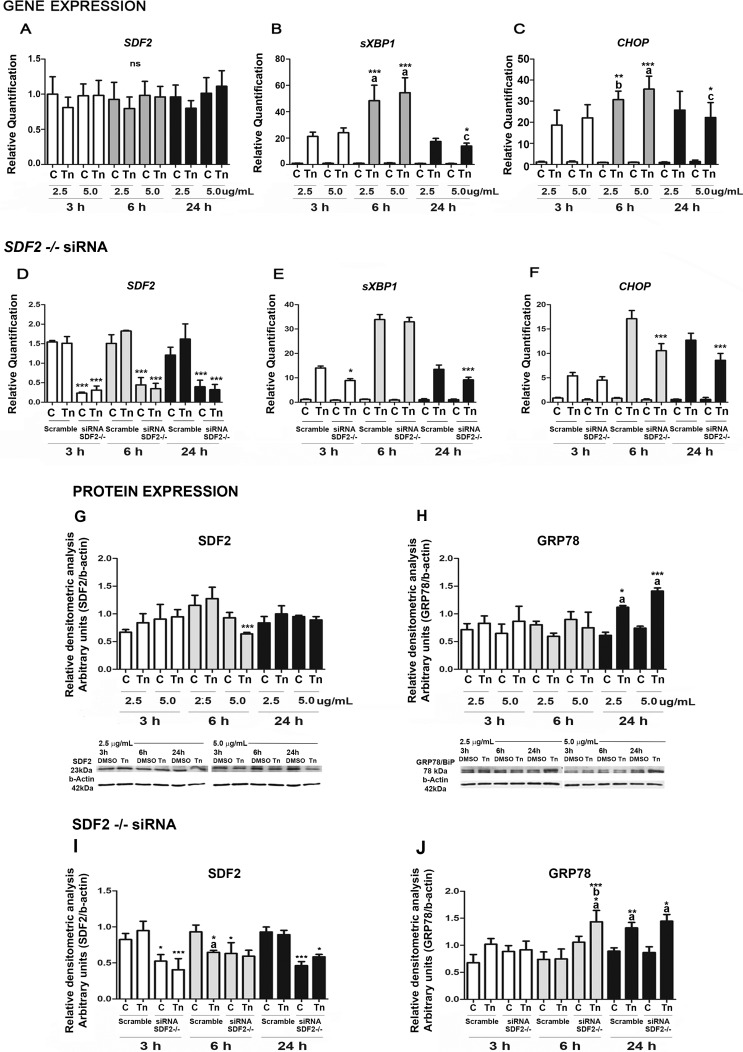
ER stress assays in BeWo cells. A–C) Real-time PCR for sXBP1, CHOP, and SDF2 factors after treatment with 2.5 or 5 μg/ml tunicamycin (Tun). A) Cells unsilenced for SDF2 show no statistical difference. B and C) Spliced XBP1 and CHOP show high levels with 5 μg/ml tunicamycin at 6 h (when compared to 3 and 24 h [a]; when compared to 3 h [b]; when compared to 24 h [c]). At 24 h, both factors showed similar levels to the 3-h treatment. D–F) BeWo cells silenced for SDF2 with siRNA, treated, or not, with 5 μg/ml tunicamycin. D) Silenced RNA treatment downregulated SDF2 mRNA expression in cells treated or not with tunicamycin. Asterisks represent the difference between cells silenced and not silenced within the same time and tunicamycin concentration. E) Spliced XBP1 shows lower expression at 3 and 24 h of treatment in silenced cells when compared to cells not silenced in the same conditions. F) CHOP has reduced expression at 6 and 24 h of treatment in silenced cells when compared to cells that were not silenced under the same conditions. G–H) Western blot for SDF2 and GRP78/BiP in the presence of 2.5 or 5 μg/ml tunicamycin. G) SDF2 expression is downregulated at 6 h of tunicamycin (5 μg/ml) treatment when compared to cells treated with 2.5 μg/ml of tunicamycin. H) GRP78/BiP is upregulated after 24 h at both tunicamycin concentrations (when compared to 3 and 6 h [a]). I and J) Western blot in BeWo silenced cells. I) SDF2 downregulation confirms silenced cells. Asterisks represent the difference between cells silenced and not silenced within the same time and tunicamycin concentration. Asterisks followed by a represents the difference between cells treated with tunicamycin compared to cells treated with DMSO (control [C]). J) GRP78/BiP is upregulated at 6 h of tunicamycin treatment (5 μg/ml) in siRNA-treated cells when compared to control (a) and unsilenced cells also treated with tunicamycin (asterisks followed by b), and at 24 h in both silenced and unsilenced cells when compared to controls (asterisks followed by a). Values are plotted as mean ± SD. One-way ANOVA followed by Tukey multiple comparison test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
CHOP, a transcription factor the expression of which is induced to high levels by ER stress [28], and spliced XBP1, a potent UPR transcriptional activator [9], were also assessed to confirm ER stress condition. Spliced XBP1 (Fig. 5B) and CHOP (Fig. 5C) mRNA levels at both tunicamycin concentrations were detected at all culture times. There was increased XBP1 and CHOP expression after 6-h treatment at both concentrations (Fig. 5, B and C).
Gene expression of SDF2 (Fig. 5A) did not change throughout the treatment. However, SDF2 silencing changed the expression of the ER stress markers. Using siRNA for SDF2, ∼70% SDF2 mRNA silencing was achieved (Fig. 5D). In silenced cells, expression of mRNA sXBP1 decreased after 3 h (P ≤ 0.05) and 24 h (P ≤ 0.001) compared to nonsilenced cells (Fig. 5E). A decrease in mRNA levels was seen after 6 h (P ≤ 0.001) and 24 h (P ≤ 0.001) when CHOP expression was analyzed (Fig. 5F) following tunicamycin treatment (5.0 μg/ml) of SDF2 silenced cells.
Protein expression of tunicamycin-treated BeWo cells was also analyzed by Western blot for SDF2 (Fig. 5G) and GRP78/BiP (Fig. 5H). SDF2 expression decreased (P ≤ 0.001) after 6 h at 5.0 μg/ml. ER stress marker GRP78/BiP increased after 24 h at both concentrations (P ≤ 0.05 and P ≤ 0.001). Decreased SDF2 protein expression confirmed siRNA silencing treatment (Fig. 5I). Expression of GRP78/BiP (Fig. 5J) increased earlier, after 6 h (P ≤ 0.05, 5.0 μg/ml), and was maintained after 24 h at both concentrations (P ≤ 0.001).
Cell Viability and Apoptosis in SDF2−/− BeWo Cells
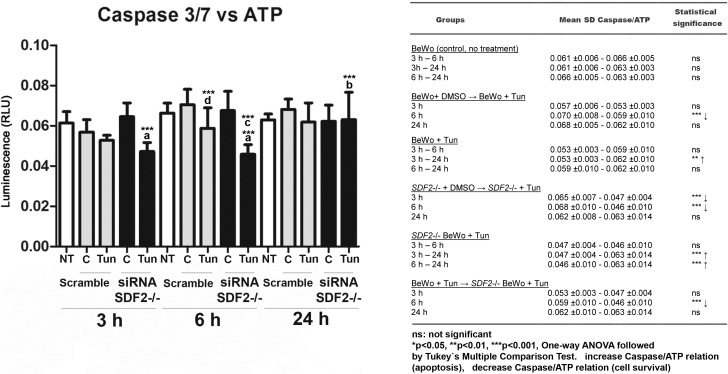
As the expression of GRP78/BiP and CHOP changed in SDF2−/− cells (Fig. 5), which are classical markers for cell survival/apoptosis in UPR, we performed an assay to show whether caspase-3 and -7 cleavage activity (apoptosis) versus ATP production (cell viability) changed in our treated groups (Fig. 6). The analyses of cell viability and apoptosis in BeWo cells silenced for SDF2, with or without tunicamycin treatment, were assessed by caspase-Glo 3/7 and CellTiter-Glo Luminescent Cell Viability assays. The relationship between caspase-3/7 activity and ATP production (caspase-3/7 vs. ATP) is shown for each group in Figure 6, A and B; cells treated with staurosporine (apoptosis-positive control) are shown in Supplemental Figure S4. After 3 and 6 h in SDF2−/− cells, this relationship decreased (P ≤ 0.001) compared to the same group treated with DMSO, suggesting an increase in survival. After 24 h, this relationship was normalized in SDF2−/− tunicamycin-treated cells (P ≤ 0.001). After 6 h of tunicamycin treatment, nonsilenced cells also showed a decrease in the relative caspase-3/7 and ATP production levels (P ≤ 0.001). Importantly, the same group showed decreased SDF2 protein expression (Fig. 5G). Furthermore, cells SDF2−/− treated with tunicamycin for 6 h had a lower caspase-3/7 and ATP relationship compared to BeWo cells (P ≤ 0.001) that were treated for 6 h. Data from caspase-3/7 and ATP production for each group and positive controls are shown in Supplemental Figures S5–S7.
FIG. 6.
Ratio of caspase-3/7 activity:ATP production in BeWo cells treated with tunicamycin (3, 6, and 24 h) with or without SDF2 silencing. Cells untreated, white bars; cells treated with tunicamycin or DMSO, gray bars; cells silenced and treated with tunicamycin or DMSO, black bars. Caspase-3/7:ATP ratio decreases in SDF2-silenced cells and in those treated with tunicamycin compared to cells treated only with DMSO at 3 and 6 h (asterisks followed by a). At 24 h, this ratio becomes normalized (asterisks followed by b). At 6 h, note also the decrease in caspase/ATP ratio in silenced cells treated with tunicamycin compared to unsilenced cells treated with tunicamycin (asterisks followed by c). Unsilenced cells showed a decrease in caspase/ATP ratio in those treated with tunicamycin when compared to control in the 6-h group (asterisks followed by d). Right panel shows a table containing all groups and statistical analyses. C = controls treated with DMSO; NT = nontreated cells; Tun = tunicamycin. Values are plotted as mean ± SD. One-way ANOVA followed by Tukey multiple comparison test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
DISCUSSION
We suggested earlier that mouse Sdf2 is involved in ER stress and UPR due to protein localization and high-protein structural similarities with mouse and human Sdf2l1 and SDF2-like from Arabidopsis thaliana, two proteins known to participate in these cellular responses [8]. This study confirms our hypothesis using the human placenta model.
Gene expression of SDF2 in human placenta was previously demonstrated in profile studies in villi and basal plate from different gestational stages [29, 30]. These studies showed that SDF2 mRNA is highly expressed and did not change during the gestational phases. Consistent with this pattern in our protein expression mapping, SDF2 was expressed during all gestational phases and placental compartments, and also in several cell types. These findings indicate a relationship with gene expression, and suggest that this protein may be involved in fundamental cellular processes.
Higher expression of SDF2 in some placental compartments, as found in decidua during the first trimester, in the smooth chorionic membrane in term placenta, and in placentas from AGA when compared to SGA neonates, might be related with the intensity of tissue activity. As a site for angiogenesis, immune regulation, extravillous CTB invasion, and vessel remodeling, the decidua exhibits intense activity during the first trimester [31]. In term placenta, the smooth chorionic membrane is a key compartment for labor preparation [32]. Its early rupture is one of the major causes of preterm labor and a leading cause of infant mortality and morbidity [33]. Notably, at term, smooth chorionic and amniotic membranes are much more susceptible to infections [34]. In addition, increase of inflammatory cytokines, such as TNF-α, IL-1B, and IL-6, and of leukocytes, macrophages, and neutrophils, is also reported in the fetal membranes [35]. The increase in SDF2 expression in smooth chorionic membrane may be thus related to the greater physiological stress due to susceptibility to infectious agents, to an inflammatory environment, and/or to uterine contractions.
Interestingly, term placentas from SGA neonates showed lower SDF2 expression when compared to placentas from AGA. SGA or fetal growth-restricted neonates are usually classified as birthweight below the 10th percentile [36]. Studies investigating the possible cause for the lack of achievement of fetal potential growth point to inadequate nutritional transfer through the placenta [37]. This low physiological efficiency in placental nutrient transport seems to be consistent with fetal growth restriction [37]. Intrauterine growth restriction pathophysiology has been associated with increase in ER stress markers, mainly reducing placental protein translation [16]. In this study, downregulation of SDF2 in placentas from SGA neonates parallels the data showing that SDF2 is also reduced in ER stress conditions, and emphasizes the putative role of this protein in placental adaptation failures.
In our differentiation assays, SDF2 expression also increased in the villous CTB, but decreased in cells upon hypoxia, a differentiation disruptor [26]. Similar results were found using BeWo cells, but with a delayed response. Primary cells come from a complex environment and, as such, their sensitivity in responding to surrounding changes might be higher than established cell lines. Differentiation of the villous CTB involves intense metabolic processes and cell fusion, a process dependent of caspase-8 activation, which can initiate the apoptosis cascade [38]. Pharmacological and biological ER stress inducers in several cell types have been shown to activate caspase-8 [39]. Using caspase-8 siRNA, caspase-3/7 activation and apoptosis decreased [40]. Increase of SDF2 in differentiation assays on human CTBs might therefore be related to the augmentation of caspase-8 activity in this process and to ER stress.
Our experiments also detected decreased SDF2 levels caused by hypoxic conditions. In placenta, approximately 30% of its oxygen is used in protein synthesis [14]. A possible mechanism may result from protein synthesis inhibition, also a factor associated with ER stress. Hypoxia might therefore restrict the synthetic activity, further activating ER stress and UPR in placental cells.
It is widely known that when UPR is activated, the first goal is to reverse the stress stage and attain cellular homeostasis [9]; however, to reach this state, misfolded proteins in the ER lumen must be repaired or eliminated. Increase in the chaperone GRP78/BiP is pivotal in this process as the main response leading to repair of misfolded proteins and possible cell survival [9]. If UPR fails to reestablish homeostasis, it will activate apoptosis mainly through the transcription factor, CHOP [40]. XBP1 is the principal transcriptional factor involved in the ER stress-adaptive response, being crucial for growth and survival of solid tumors under hypoxic stress [11]. Increase in XBP1 expression and splicing occurs in breast cancer and hepatocellular carcinoma, which may result from the regulatory function of XBP1 toward BiP [11].
While analyzing some markers of ER stress in cultured placental cells, we detected a reduction in SDF2 and an increase in GRP78/BiP after ER stress-inducing treatment with tunicamycin. Moreover, caspase-3 cleavage remained unaffected, supporting the idea that SDF2 is negatively related to ER stress activation and cell survival.
Given these observations, we assessed the effects of the tunicamycin treatment in BeWo cells. SDF2 gene expression did not change with tunicamycin treatment, although high expression of sXBP1 and CHOP confirmed the ER stress condition. Both factors showed maximum mRNA expression after 6-h tunicamycin treatment, similar to other cell types [41]. SDF2 protein expression decreased upon tunicamycin treatment, but only 6 h after treatment, and recovered after 24 h. This delay of expression compared to CTBs was also observed in GRP78/BiP, which increased only 24 h after tunicamycin treatment. As we have discussed above, cells from primary culture and from cell lines appear to have different temporal UPR responses, but still show SDF2 switching in the presence of ER stress/UPR markers.
To determine how SDF2 interferes in the ER stress and UPR pathways, BeWo cells were SDF2 silenced using siRNA and tested under the same tunicamycin treatment. We observed two main differences in gene expression: a decrease in sXBP1 expression after 3 and 6 h of tunicamycin treatment and a decrease in CHOP expression after 6 and 24 h of treatment. Clearly, silenced SDF2 changed the pattern of the UPR biomarker expression. The changes seen in the UPR marker expression were also seen in protein expression. Levels of GRP78/BiP were increased after 6 h of tunicamycin treatment in silenced cells, which occurred only after 24-h treatment when SDF2 had not been silenced. Since the increase in BiP and reduction in CHOP has been associated with ER stress attenuation of apoptotic ER signals, these data seem to indicate that SDF2 may be related to survival/apoptosis via UPR.
To test this hypothesis, the relationship between caspase-3/7 activity and ATP production was assessed in BeWo SDF2-silenced cells. As observed, the caspase/ATP ratio was reduced in silenced cells in which ER stress was induced after 3 and 6 h, which suggests an increase in cell survival response. Lack of SDF2 during UPR facilitated the cell survival profile, which corroborates our hypothesis.
ER stress and UPR mechanisms should be carefully considered during placental development. The placenta faces several changes in environmental conditions during development, which could cause UPR onset. During gestation, hormones, growth factors, and regulatory proteins are extremely important for proper development of the placenta. A lack in production in any of these factors or release of nonfunctional peptides can contribute to the development of a gestational disease, such as preeclampsia or intrauterine growth restriction [16]. In this context, our data suggest the inclusion of SDF2 as a novel factor in ER stress and UPR, which may shed new light on key targets in this response and help investigations into gestational development and several diseases.
We have shown that SDF2 is widely expressed in several cell types in human placenta and its function is probably related to ER stress. Downregulation of CHOP, anticipation of GRP78/BiP, and lower caspase-3/ATP production in SDF2-silenced cells indicate a role of this protein in the control of survival/apoptosis in the trophoblast ER stress response. Thus, SDF2 may be a valuable target in UPR research, not only in gestation, but in models such as cancer, in which cells can escape from survival/apoptosis regulation [38].
Supplementary Material
ACKNOWLEDGMENT
The authors thank Rosangela Augusto de Oliveira and Luiz Gustavo Sparvoli for their excellent technical assistance and Jusciele Brogin Moreli, Mara Sandra Hoshida, and Rossana Pulcineli Vieira Francisco for scientific support.
Footnotes
This work was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) grant 2009/03510-4 and 2009/11869-2, Conselho Nacional de Desenvolvimento Científico e Tecnológico, Coordernação de Aperfeiçoamento de Pessoal de Nível Superior, and National Institutes of Health grant R37 HD076253. A.R.L.-O. received a Ph.D. scholarship from Fundação de Amparo a Pesquisa do Estado de São Paulo-FAPESP grant 2009/03510-4.
REFERENCES
- Hamada T, Tashiro K, Tada H, Inazawa J, Shirozu M, Shibahara K, Nakamura T, Martina N, Nakano T, Honjo T. Isolation and characterization of a novel secretory protein, stromal cell-derived factor-2 (SDF-2) using the signal sequence trap method. Gene. 1996;176:211–214. doi: 10.1016/0378-1119(96)00251-x. [DOI] [PubMed] [Google Scholar]
- Kang H, Escudero-Esparza A, Douglas-Jones A, Mansel RE, Jiang WG. Transcript analyses of stromal cell derived factors (SDFs): SDF-2, SDF-4 and SDF-5 reveal a different pattern of expression and prognostic association in human breast cancer. Int J Oncol. 2009;35:205–211. doi: 10.3892/ijo_00000330. [DOI] [PubMed] [Google Scholar]
- Vendrell E, Ribas M, Valls J, Solé X, Grau M, Moreno V, Capellà G, Peinado MA. Genomic and transcriptomic prognostic factors in R0 Dukes B and C colorectal cancer patients. Int J Oncol. 2007;30:1099–1107. [PubMed] [Google Scholar]
- Siragusa M, Fröhlich F, Park EJ, Schleicher M, Walther TC, Sessa WC. Stromal cell-derived factor 2 is critical for Hsp90-dependent eNOS activation Sci Signal 2015. 8:ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida MS, Gorjão R, Lima C, Daher S, Curi R, Bevilacqua E. Regulation of gene expression in mouse trophoblast cells by interferon-gamma. Placenta. 2007;28:1059–1072. doi: 10.1016/j.placenta.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Hoshida MS, Gorjão R, Bevilacqua E. Profiling expression patterns of interferon-gamma treated trophoblast cells by cDNA macroarray Placenta 2008. 29 103. [DOI] [PubMed] [Google Scholar]
- Chen SH, Babichev Y, Rodrigues N, Voskas D, Ling L, Nguyen VP, Dumont DJ. Gene expression analysis of Tek/Tie2 signaling. Physiol Genomics. 2005;22:257–267. doi: 10.1152/physiolgenomics.00063.2005. [DOI] [PubMed] [Google Scholar]
- Lorenzon-Ojea AR, Caldeira W, Ribeiro AF, Fisher SJ, Guzzo CR, Bevilacqua E. Stromal cell derived factor-2 (Sdf2): a novel protein expressed in mouse. Int J Biochem Cell Biol. 2014;53:262–270. doi: 10.1016/j.biocel.2014.05.024. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Liu AX, He WH, Yin LJ, Lv PP, Zhang Y, Sheng JZ, Leung PC, Huang HF. Sustained endoplasmic reticulum stress as a cofactor of oxidative stress in decidual cells from patients with early pregnancy loss. J Clin Endocrinol Metab. 2011;96:E493–497. doi: 10.1210/jc.2010-2192. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang K, Li Z. Unfolded protein response in cancer: the physician's perspective. J Hematol Oncol. 2011;4:8. doi: 10.1186/1756-8722-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci U S A. 2009;106:16657–16662. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AM. Placental oxygen consumption. Part I: in vivo studies—a review Placenta 2000. 21 (suppl A): S31 S37 [DOI] [PubMed] [Google Scholar]
- Fisher SJ. The placental problem: linking abnormal cytotrophoblast differentiation to the maternal symptoms of preeclampsia. Reprod Biol Endocrinol. 2004;2:53. doi: 10.1186/1477-7827-2-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung HW, Calabrese S, Hynx D, Hemmings BA, Cetin I, Charnock-Jones DS, Burton GJ. Evidence of placental translation inhibition and endoplasmic reticulum stress in the etiology of human intrauterine growth restriction. Am J Pathol. 2008;173:451–462. doi: 10.2353/ajpath.2008.071193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton GJ, Yung HW. Endoplasmic reticulum stress in the pathogenesis of early-onset pre-eclampsia. Pregnancy Hypertens. 2011;1:72–78. doi: 10.1016/j.preghy.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung HW, Korolchuk S, Tolkovsky AM, Charnock-Jones DS, Burton GJ. Endoplasmic reticulum stress exacerbates ischemia-reperfusion-induced apoptosis through attenuation of Akt protein synthesis in human choriocarcinoma cells. FASEB J. 2007;21:872–884. doi: 10.1096/fj.06-6054com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Yoshimi M, Kadota Y, Inoue M, Sato M, Suzuki S. Prolonged endoplasmic reticulum stress alters placental morphology and causes low birth weight. Toxicol Appl Pharmacol. 2014;275:134–144. doi: 10.1016/j.taap.2013.12.008. [DOI] [PubMed] [Google Scholar]
- Faria MR, Hoshida MS, Ferro EA, Ietta F, Paulesu L, Bevilacqua E. Spatiotemporal patterns of macrophage migration inhibitory factor (Mif) expression in the mouse placenta. Reprod Biol Endocrinol. 2010;8:95. doi: 10.1186/1477-7827-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Fisher SJ, Janatpour M, Genbacev O, Dejana E, Wheelock M, Damsky CH. Human cytotrophoblasts adopt a vascular phenotype as they differentiate: a strategy for successful endovascular invasion? J Clin Invest. 1997;99:2139–2151. doi: 10.1172/JCI119387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hromatka BS, Ngeleza S, Adibi JJ, Niles RK, Tshefu AK, Fisher SJ. Histopathologies, immunolocalization, and a glycan binding screen provide insights into Plasmodium falciparum interactions with the human placenta. Biol Reprod. 2013;88:154. doi: 10.1095/biolreprod.112.106195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller NM, Fisher SJ. Chapter 12: placental remodeling of the uterine vasculature. Methods Enzymol. 2008;445:281–302. doi: 10.1016/S0076-6879(08)03012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky CH, Librach C, Lim KH, Fitzgerald ML, McMaster MT, Janatpour M, Zhou Y, Logan SK, Fisher SJ. Integrin switching regulates normal trophoblast invasion Development 1994. 120 12: 3657 3666 [DOI] [PubMed] [Google Scholar]
- Handschuh K, Guibourdenche J, Cocquebert M, Tsatsaris V, Vidaud M, Evain-Brion D, Fournier T. Expression and regulation by PPARgamma of hCG alpha- and beta-subunits: comparison between villous and invasive extravillous trophoblastic cells Placenta 2009. 30 12: 1016 1022 [DOI] [PubMed] [Google Scholar]
- Genbacev O, Zhou Y, Ludlow JW, Fisher SJ. Regulation of human placental development by oxygen tension. Science. 1997;277:1669–1672. doi: 10.1126/science.277.5332.1669. [DOI] [PubMed] [Google Scholar]
- Minamino T, Komuro I, Kitakaze M. Endoplasmic reticulum stress as a therapeutic target in cardiovascular disease Circ Res 2010. 107 9: 1071 1082 [DOI] [PubMed] [Google Scholar]
- Oyadomari S1, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress Cell Death Differ 2004. 11 4: 381 389 [DOI] [PubMed] [Google Scholar]
- Mikheev AM, Nabekura T, Kaddoumi A, Bammler TK, Govindarajan R, Hebert MF, Unadkat JD. Profiling gene expression in human placentae of different gestational ages: an OPRU Network and UW SCOR Study. Reprod Sci. 2008;15:866–877. doi: 10.1177/1933719108322425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn VD, Haimov-Kochman R, Paquet AC, Yang YJ, Madhusudhan MS, Gormley M, Feng KT, Bernlohr DA, McDonagh S, Pereira L, Sali A, Fisher SJ. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology. 2007;148:1059–1079. doi: 10.1210/en.2006-0683. [DOI] [PubMed] [Google Scholar]
- Harris LK. Review: trophoblast-vascular cell interactions in early pregnancy: how to remodel a vessel Placenta 2010. 31 (suppl): S93 S98 [DOI] [PubMed] [Google Scholar]
- Lavery JP, Miller CE, Knight RD. The effect of labor on the rheologic response of chorioamniotic membranes. Obstet Gynecol. 1982;60:87–92. [PubMed] [Google Scholar]
- Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science. 2014;345:760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher N. Adams Waldorf K, Hein M, Uldbjerg N. The cervical mucus plug: structured review of the literature. Acta Obstet Gynecol Scand. 2009;88:502–513. doi: 10.1080/00016340902852898. [DOI] [PubMed] [Google Scholar]
- Rinaldi SF, Catalano RD, Wade J, Rossi AG, Norman JE. Decidual neutrophil infiltration is not required for preterm birth in a mouse model of infection-induced preterm labor. J Immunol. 2014;192:2315–2325. doi: 10.4049/jimmunol.1302891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Epstein LH, Eiden RD, Shenassa ED, Li X, Liao Y, Wen X. Stunting at 5 years among SGA newborns. Pediatrics. 2016;137:1–10. doi: 10.1542/peds.2015-2636. [DOI] [PubMed] [Google Scholar]
- Hayward CE, Lean S, Sibley CP, Jones RL, Wareing M, Greenwood SL, Dilworth MR. Placental adaptation: what can we learn from birthweight: placental weight ratio? Front Physiol. 2016;7:28. doi: 10.3389/fphys.2016.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black S, Kadyrov M, Kaufmann P, Ugele B, Emans N, Huppertz B. Syncytial fusion of human trophoblast depends on caspase 8. Cell Death Differ. 2004;11:90–98. doi: 10.1038/sj.cdd.4401307. [DOI] [PubMed] [Google Scholar]
- Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A. Cell death: opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WA, Groenendyk J, Michalak M. Endoplasmic reticulum stress associated responses in cancer. Biochim Biophys Acta. 2014;1843:2143–2149. doi: 10.1016/j.bbamcr.2014.01.012. [DOI] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.