Abstract
Polyhydroxyalkanoate (PHA) biopolymers are widely recognised as outstanding candidates to replace conventional petroleum-derived polymers. Their mechanical properties are good and can be tailored through copolymer composition, they are biodegradable, and unlike many alternatives, they do not rely on oil-based feedstocks. Further, they are the only commodity polymer that can be synthesised intracellularly, ensuring stereoregularity and high molecular weight. However, despite offering enormous potential for many years, they are still not making a significant impact. This is broadly because commercial uptake has been limited by variable performance (inconsistent polymer properties) and high production costs of the raw polymer. Additionally, the main type of PHA produced naturally is poly-3-hydroxybutyrate (PHB), which has limited scope due to its brittle nature and low thermal stability, as well as its tendency to embrittle over time. Production cost is strongly impacted by the type of the feedstock used. In this article we consider: the production of PHAs from methanotrophs using methane as a cost-effective substrate; the use of mixed cultures, as opposed to pure strains; and strategies to generate a poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer (PHBV), which has more desirable qualities such as toughness and elasticity.
Keywords: PHA, PHB, PHBV, methane, syngas methanotroph, gas fermentation, biopolymer
1. Introduction
Polyhydroxyalkanoate (PHA) biopolymers are high molecular weight, stereoregular, linear thermoplastic polymers that are naturally produced by bacteria in an aqueous environment. Their mechanical properties are good, they are biodegradable, and, unlike many alternatives, they do not rely on oil-based feedstocks [1,2]. PHAs are intra-cellular storage granules that serve as a source of carbon (C), energy, or reducing-power and may comprise up to 90% of a microbe’s dry weight [3,4]. The industrial production of poly-3-hydroxybutyrate (PHB), the simplest and most widely produced form of PHA, involves accumulation of polymer in pure strains using plant-derived carbon sources, typically sugars, as the feedstock. PHA accumulation is generally enhanced by culturing the microbes under a nutrient limitation in the presence of excess carbon. The availability of alternative carbon sources, pH, temperature, oxygen, methane, carbon dioxide, macronutrients (nitrogen, phosphorus, sulphur, potassium, magnesium sodium), and trace metals (copper, iron, zinc, manganese, and/or cobalt) can all affect PHA yield and quality [5,6,7].
However, the production of PHA by conventional means also uses expensive refined substrates and requires sterilisation, limiting widespread commercialisation [8]. Techno-economic studies have shown that a major cost of pure culture production is the carbon feedstock, estimated to be up to 40% of the product cost [9,10,11]. Consequently, many research groups are investigating the potential of using waste streams for PHA production, such as dairy whey waste, waste lipids, sugar industry waste streams, agricultural crop residues, petrochemical waste, syngas and glycerol [12,13]. The problems with waste streams, however, are their limited abundance and distributed nature. In contrast, methane is a cheap, abundant and widely available carbon source. Additionally, the robust, self-regulating nature of mixed methanotrophic cultures [14] offers the opportunity to operate under non-sterile conditions, thereby reducing operating costs on an industrial scale. Over 300 bacterial strains, including methanotrophs, have shown potential to synthesise and store PHB [15].
Methane is the principal component of natural gas, is abundantly available in many oilfields and coal deposits, and is widely produced during the biological degradation of organic matter, either in engineered processes or natural environments. Methane is considered to be the second most abundant greenhouse gas (GHG) after carbon dioxide, contributing to 18% of the total atmospheric radiative forcing [16,17]. Globally, over 60% of total methane emissions are anthropogenic emissions, the majority of which are the result of microbial metabolism [16,18]. Microbial methane production occurs during the anaerobic biodegradation of complex organic matter. Complex molecules and solids are solubilised (hydrolysis), degraded to various organic acids, such as acetic, propionic, butyric acids (acidogenesis), converted to acetic acid (acetogenesis), and finally transformed into methane during methanogenesis [19].
Microbial methane production and capture is an established and sustainable strategy for the treatment of organic waste streams in a number of industries (e.g., municipal solid waste, wastewater treatment, brewing, food processing, agriculture, crop residues). When captured, microbially-produced methane is a potentially valuable and renewable resource [20]. Significant sources of renewable methane that are either captured, or present the opportunity for capture and reuse, include landfills (38 Mt/year), wastewater treatment (21 Mt/year), agriculture (11–30 Mt/year), and biomass (10 Mt/year). However, with natural gas alone, the World Bank estimates wastage of 92 Mt/year that is flared or vented [21] which represents an enormous opportunity for converting methane to higher value products [18].
The rapid expansion of global methane production and capture, both in the form of natural gas and generation of methane-rich biogas, has improved the accessibility of methane on the global market and consequently reduced the commodity price. This has generated increased interest in methane as a carbon source for novel, value-added processes. The thermochemical processing of methane can produce syngas and methanol [22,23], which are robust processes, but require large capital investments that can exceed a billion USD and prohibit many applications. There has been growing interest in the biological conversion of C1 compounds to fine and commodity chemicals due to recent economic changes and the mind shift towards sustainably-produced chemicals.
Pure methane is an energy-rich feedstock with an energy density of 55.7 MJ/kg at 1.013 bar, 15 °C [24]. Figure 1 illustrates the degree of reduction of various organic feedstocks, metabolites, and products. It shows that the oxidation potential of methane (the energy content) is greater than that of the product PHB, greater than the widely-used alternative feedstocks, e.g., sugar (glucose) and organic acids (acetic, propionic and butyric acids), and much greater than CO2. Using CO2 as a feedstock to generate PHB would, thus, be akin to “pushing up hill”, requiring significant electron donor supplementation. This translates to more favourable energy balances when using methane. For example, recent life cycle assessments of biogas generated at wastewater treatment plants and landfills and used as a feedstock for PHA production indicate a lower energy requirement for PHB production from methane (37 MJ/kg PHB from biogas) compared to PHB production from alternative renewable substrates (42 MJ/kg PHB from corn-derived sugar) [25]. There is also significant potential for enhanced carbon sequestration (at ~2 kg CO2 equivalents fixed/kg methane compared to 0.1 kg CO2 equivalents fixed/kg of corn-derived sugar). What is more, the cost of methane is around $520/tonne carbon, based on a commercial price for natural gas in the United States of US $7.74/thousand cubic feet [26]. If the fraction of carbon converted to PHA is considered, at, for example, a PHB yield of around 0.55 g/g methane as reported by Wendlandt et al. [27], methane has potentially strong economic benefits [28].
Figure 1.
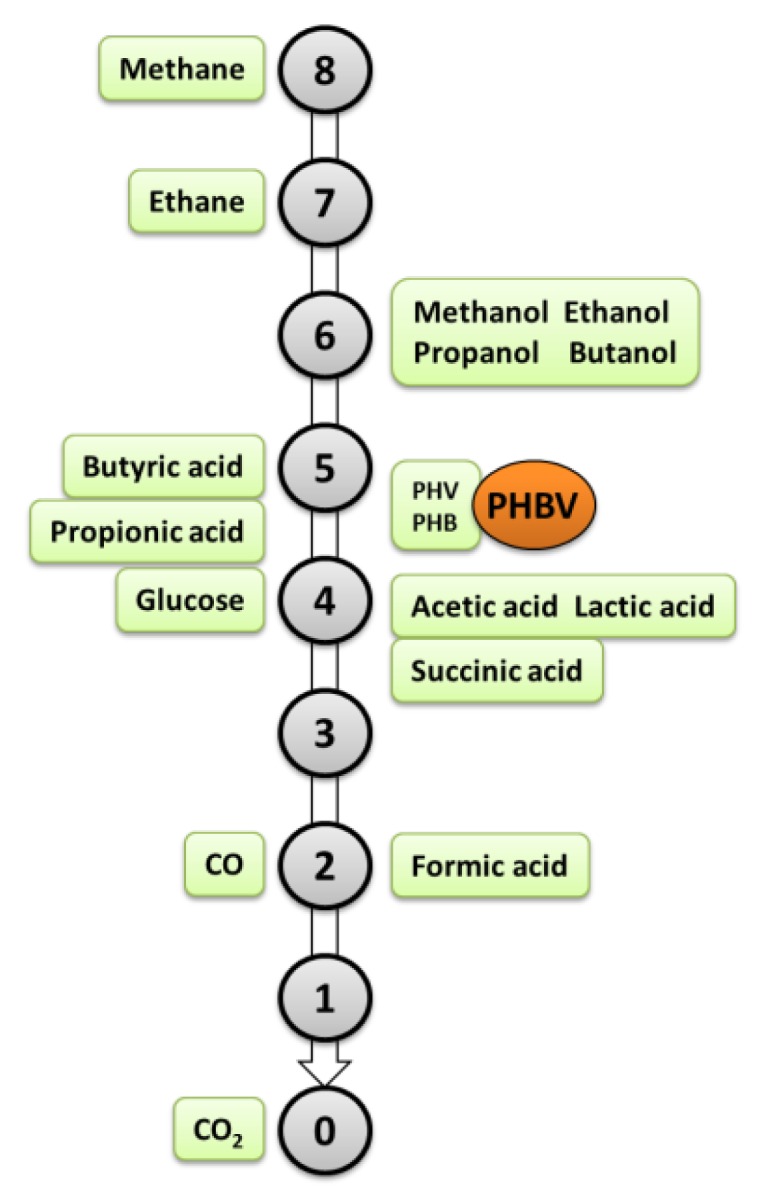
Degree of reduction of methane relative to other feedstocks, metabolites, PHB (CH1.5O0.5: 4.5), and PHV (CH1.6O0.4: 4.8). The degree of reduction is of a measure of the oxidation potential, with high oxidation potential indicating higher potential for energy release upon oxidation. The figure is modified from Kracke and Kromer [29] and Pratt [30].
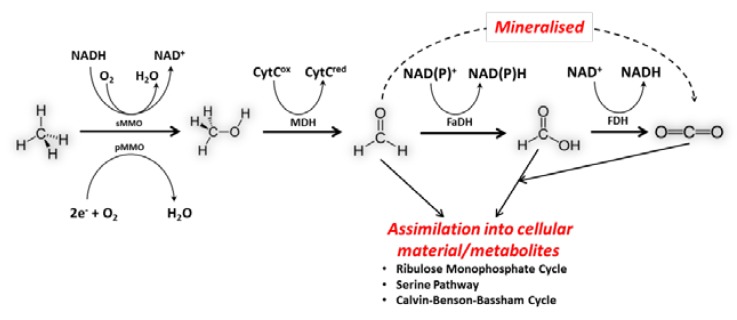
The methanotrophic conversion of methane into biomass and CO2 plays a large part in regulating the global methane cycle by serving as a biological CH4 sink [31]. Methane monooxygenase is the key enzyme responsible for CH4 oxidation to methanol, which is further oxidised to CO2 to regenerate reducing equivalents, or assimilated into cellular components (Figure 2). Traditionally, these bacteria were classified as Type I (γ-proteobacteria) or Type II (α-proteobacteria) methanotrophs, primarily according to their use of the ribulose monophosphate pathway (Type I) or serine (Type II) pathways for formaldehyde assimilation). They were further subdivided into a Type X group, which contained biochemical capabilities of Type I and II. Methanotrophs are now typically grouped as γ-proteobacteria or α-proteobacteria (where Type X is a subdivision of γ-proteobacteria). A recently-discovered thermophilic subset, Verrucomicrobia (Methylacidiphilum and Methylomirabilis spp.), was added to the group (see Kalyuzhnaya et al. [32] and Strong et al. [33] and references therein).
Figure 2.
Generalised reaction scheme for methane oxidation via methanotrophs. Adapted from Kalyuzhnaya et al. [32] and Hanson and Hanson [31]; sMMO: soluble methane mono-oxygenase, pMMO: particulate methane mono-oxygenase, MDH: methanol dehydrogenase, FaDH: formaldehyde dehydrogenase, FDH: formate dehydrogenase and CytC: cytochrome C.
Certain methanotrophs can synthesise the PHB homopolymer from methane under nutrient limited conditions [6]. The general ability of methanotrophic and methylotrophic bacteria to produce PHB was described for several strains as early as 1970, in the seminal work by Whittenbury, et al. [34], and has been of consistent interest for decades [6,35,36,37,38,39]. The use of methanotrophic bacteria is viewed as a powerful route to the microbial production of PHB from methane, as it provides a collective solution for three major environmental problems:
-
(1)
A potential approach for carbon sequestration and GHGs emission reduction;
-
(2)
Production of biodegradable polymers for replacing conventional fossil fuel-derived plastics;
-
(3)
Reducing the use of organic carbon sources such as sugars for PHB production [6].
Overall, therefore, this review seeks to provide an overview of the current state of research into the production of PHAs from methanotrophs using methane as a cost-effective substrate as well as the use of mixed cultures as opposed to pure strains and strategies to generate a poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer (PHBV), which has more desirable qualities, such as toughness and elasticity.
2. Metabolism of Methanotrophs and Biosynthesis of PHB from Methane
It is important to consider the biochemical capabilities of methanotrophs and their microbial consortia with regard to biopolymer synthesis. This enables better selection of culture conditions that favour PHB-producing strains, or can establish whether the mixed culture is capable of PHB synthesis at all. The first step in aerobic methane catalysis consumes methane, oxygen, and reducing equivalents. Methane mono-oxygenase breaks the O-O bond in the oxygen molecule by using two reducing equivalents [31]. One of the oxygen atoms is incorporated into methane to form methanol, while the second is converted to water. Methane mono-oxygenase can occur as a particulate, membrane-bound form (pMMO) or in a soluble form (sMMO). The sMMO requires NAD(P)H and O2 to convert methane to methanol in the following manner: CH4 + NAD(P)H + H+ + O2 = CH3OH + NAD(P)+ + H2O, while the pMMO requires cytochromes b559/569 or c553 or artificial reductants such as duroquinol and NADH to complete the reaction [40]. However, despite a great deal of effort in this area, the physiological source of the electron donor to the pMMO remains unresolved [32]. The pMMO and the ammonia mono-oxygenase share many similarities and an endogenenous quinol may serve as a reductant for both enzymes, and may potentially abstract electrons from the subsequent methanol oxidation via the periplasmic PQQ-linked methanol dehydrogenase, which is coupled to a cytochrome c [41]. Direct coupling of pMMO and methanol dehydrogenase catalysis is supported by their close proximity and reports of a super-complex of the two enzymes [42,43,44,45]. The ability to convert methane into organic acids and H2 under oxygen-limiting conditions suggests that pMMO can use electron donors other than NADH [46]. Considering the range of metabolic capabilities of the methane-oxidising bacteria, it is likely that there are multiple systems for electron abstraction, and that these may vary between Gammaproteobacteria, Alphaproteobacteria, and Verrucomicrobia.
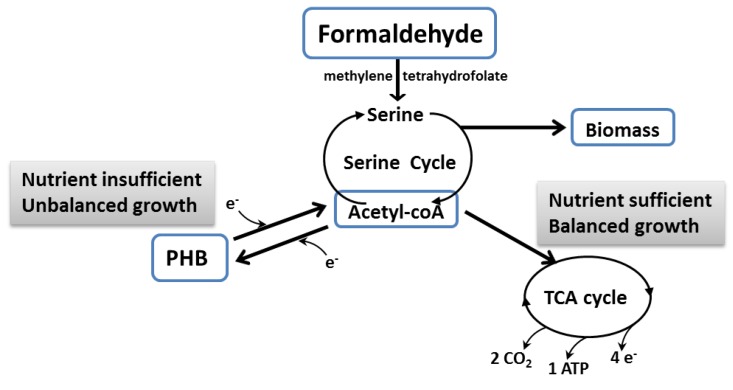
After methane oxidation, methanol is oxidised to formaldehyde (by methanol dehydrogenase), which is the central metabolite in the anabolic and catabolic pathways (Figure 2). In the catabolic pathway, formaldehyde is further converted to formate and then to CO2, regenerating two NADH. Formaldehyde is, in turn, assimilated using different pathways in α-proteobacteria or γ-proteobacteria. Typically, γ-proteobacteria use the ribulose monophosphate (RuMP) pathway, in which the 3-hexulose-phosphate-synthase (HPS) catalyses formaldehyde conversion into (D-arabino)-3-hexulose-6-phosphate. D-arabino-3-hexulose-6-phosphate is then converted into different intermediates and assimilated as cell biomass or oxidised to CO2 via an incomplete TCA cycle. The α-proteobacteria typically assimilate formaldehyde using the serine pathway [47]. Formaldehyde oxidation is activated by the pterin co-factor catalysed by methylene tetrahydrofolate, an enzyme that combines formaldehyde with glycine to generate serine [48,49]. Serine then either enters the TCA cycle under nutrient-sufficient balanced growth, or the PHB cycle under nutrient-deficient conditions (Figure 3).
Figure 3.
Schematic indicating carbon flow from the serine cycle with regard to balanced or unbalanced growth (adapted by combining Karthikeyan et al. [6] and Pieja et al. [54]).
PHB formation in methanotrophs starts from acetyl-CoA molecules and proceeds via the enzyme-mediated reactions. The genes coding for the primary enzymes involved in the biosynthesis of PHA are the phaA (β-ketothiolase), phaB (acetoacetyl-CoA reductase), and phaC (PHA synthase) genes [50], which commonly serve as a screen for microbial PHB production capacity as they are relatively well conserved [51,52]. Typically, proteins known as phasins accumulate during PHA synthesis, which bind PHA granules and promote further PHA production [53]. PHA synthases use coenzyme A (CoA) thioesters of hydroxyalkanoic acids (HAs) as substrates and catalyse the polymerisation of HAs into PHA with the concomitant release of CoA. Although PHB can function as a sole growth substrate in aerobic cultures enriched on acetate during periods of carbon deficiency, in an elegant set of experiments using a methanotrophs (Methylocystis parvus OBBP), PHB was used as a source of reducing power to aid methane consumption, as opposed to the supply of C2 units for synthesis [54].
PHB synthesis in methylotrophs, such as Methylobacter extorquens [55,56], and methanotrophs [35,36,37,38] is well documented for C1 substrates, such as methane or methanol. In the seminal paper on PHB production by methanotrophs, Asenjo and Suk [38] analysed the biochemical pathways in type II methanotrophs to establish the preliminary kinetic analysis and stoichiometry of PHB synthesis. Their overall equation for PHB accumulation for methanotrophs using the serine pathway was presented as:
| 8 CH4 + 12 O2 + FP → C4H6O2 (PHB monomer) + 4 CO2 + 12 ATP + FPH2 |
where FP = oxidised succinate dehydrogenase, and FPH2 = reduced succinate dehydrogenase.
Although the theoretical mass yield for this bioconversion based on this equation is 67% (86 g PHB/128 g methane), this excludes the fraction of the methane and oxygen that has to be converted to CO2 to regenerate the NADH required for biosynthesis. Wendlandt et al. [27,37,57] obtained yields of 0.55 g PHB/g methane in an enriched (>90%) culture of Methylocystis sp. GB 25 DSMZ 7674, close to the theoretical maximum, with PHB contents in the biomass of 51 wt%. In this series of studies, the methane-utilizing mixed culture was maintained under non-sterile found to have the potential of self-regulation resulting in a stable composition even under such non-aseptic conditions [58]. However, Zúňiga et al. [59] reported higher yields of 0.81 g PHB/g methane (reported as 0.60 g carbon from PHB with respect to g carbon in methane consumed) for a mixed methanotrophic community including a Methylobacterium organophilum strain, although with only 34% PHB content overall. The isolated strain by itself, though, had up to 58% PHB content after accumulation at a mass yield of 0.59 g PHB/g methane.
Most quantitative PHB production studies are reported from Type II α-proteobacter genera, such as Methylocystis and Methylosinus, Methylococcus, and Methylomonas spp. [38], with molecular weights (Mw) up to 3.1 MDa being recorded [37]. For example, Asenjo and Suk [38] reported PHB production of up to 67% of cell dry weight using a pure culture of Methylosinus trichosporium OB3b—the highest PHB content reported to date from methane. Overall, total PHB contents obtained for accumulations from pure and mixed methanotrophic cultures are summarised in Table 1; less data is available for mass or carbon yields in these systems. While these concentrations of PHB are lower than those produced in bacteria that feed on sugars, they are still significant and show promise for industrial applications.
Table 1.
Summary of the methylotrophic and methanotrophic production of PHB. Adapted from Khosravi-Darani et al. [66] and Karthikeyan et al. [6].
| Microorganisms (% in Mixed Culture) | Carbon Source | PHB Content (% of Total Biomass) | Yield (g PHB/g Carbon Source) | References |
|---|---|---|---|---|
| Methlocystis sp. GB25 DSMZ 7674 (>90%) | Methane | 28.3–51.3 | 0.55 | [27,57,58] |
| Methlocystis sp. GB25 DSMZ 7674 (>86%) | Methane | 10.4–33.6 | 0.45 | [37] |
| Methylosinus/methylocystis (percent unknown) + Type I genera | Methane | 7–46 | n/a | [63] |
| Mixed consortium including Methylobacterium organophilum (percent unknown) | Methane | 34 | 0.80* | [59] |
| Methylosinus and Methylocystis spp. dominant in mixed consortium (percent unknown) | Methane | 17–26 | n/a | [67] |
| Methylocystis (~77%) | Methane | 39 | 0.64 * | [68] |
| Methylocystis (> 76%) | Methane | 40 | 0.49 * | [69] |
| Methylomicrobium (96%) | Methane | 23 | n/a | [70] |
| Methylocystis sp. (71.6%) | Methane | 2.5–8.5 | n/a | [71] |
| Methylosinus trichosporium | Methane | 20–25 | n/a | [72] |
| Methylocystis parvus OBBP | Methane | 68 | n/a | [38] |
| Methylosinus trichosporium OB3b | Methane | 30–50 | n/a | [73] |
| Type II methanotrophic strain MTS | Methane | 3 | n/a | [35] |
| Methylosinus trichosporium OB3b | Methane | 30 | n/a | [74] |
| Microorganisms (Pure Culture) | Carbon Source | PHB Content (% of Total Biomass) | Yield (g PHB/g Carbon Source) | References |
|
Methylosinus trichosporium OB3b Methylobacterium organophilum strains |
Methane Methane |
25 38–57 |
n/a 0.48–0.59 * |
[59] |
|
Methylocystis spp. Methylosinus spp. |
Methane Methane |
7–36 9–38 |
n/a n/a |
[63] |
| Methylocystis hirsute | Methane | 51.6 | n/a | [75] |
|
Methylocystis parvus OBBP Methylosinus trichosporium OB3b |
Methane | 60 29 |
0.88 1.13 |
[76] |
| Methylocystis parvus OBBP | Methane | 49.4 | n/a | [77] |
| Methylosinus trichosporium IMV3011 | Methane + methanol | 40 | n/a | [36] |
| Methylosinus trichosporium IMV 3011 | Methane + Methanol | 46 | n/a | [78] |
| Pseudomonas sp. K | Methanol | 66 | 0.18 | [79] |
| Methylobacterium rhodesianum | Methanol | 45–55 | n/a | [65] |
|
Methylobacterium extorquens K Paracoccus denitrificans |
Methanol + n-amyl alcohol | 44 57 |
0.11 (0.97 on n-amyl alcohol) |
[80] |
| Pseudomonas 135 | Methanol | 55 | n/a | [81] |
| Methylobacterium extorquens NCIMB 9133 | Methanol | 7–21 | [82] | |
| Methylobacterium extorquens ATCC 55366 | Methanol | 40–46 | 0.09–0.12 | [55] |
| Methylobacterium organophilum | Methanol | 52–56 | 0.19 | [83] |
| Methylobacterium sp V49 | Methanol | 11 | n/a | [84] |
| Methylobacterium extorquens AM1 | Methanol | 34–42 | n/a | [85] |
| Methylobacterium extorquens AM1 | Methanol | 22–25 | n/a | [56] |
| Methylobacterium sp. GW2 | Methanol | 40 | n/a | [86] |
| Methylobacterium extorquens DSMZ 1340 | Methanol | 35 | 0.3 | [87] |
| Methylosinus trichosporium IMV 3011 | Methanol | 32 | n/a | [88] |
| Methylobacterium extorquens AM1 | Methanol | 27 | n/a | [89] |
|
Methylobacteria extorquens G10 Methyloligella halotolerans C2 |
Methanol | 40 17 |
n/a | [90] |
* Based on reported yields of g carbon from PHB with respect to g carbon from methane or mole carbon from PHB with respect to mole carbon from methane
Although qualitative data regarding PHB production has been reported in RuMP-pathway γ-proteobacteria [60,61,62], production is still uncertain [63]. Previous methylotroph studies suggest that PHB synthesis may be linked to the serine cycle, and that RuMP-pathway methanotrophs may be incapable of PHB synthesis. An early study detected no 3-ketothiolase, acetoacetyl-coenzyme A reductase or PHA synthase in RuMP-pathway methylotrophs, and none of the bacteria produced a measurable amount of PHB [64]. Pieja et al. [63] evaluated twelve strains from six different genera of methanotrophs for encoding of PHB synthase (phaC), as well as PHB production under nitrogen-limited conditions in γ-proteobacter and α-proteobacter methanotrophs. The γ-proteobacter strains tested negative for phaC gene, as well as PHB production, while all α-proteobacter strains tested positive for phaC and PHB synthesis. Babel [65] hypothesised that serine-pathway methylotrophs produce PHB as a carbon storage polymer, whereas RuMP-pathway methylotrophs produce exopolysaccharides under unbalanced growth conditions. More recent research suggests that PHB accumulation is associated with the supply of reducing equivalents, rather than providing cell components during growth [54].
3. Process Conditions for PHB Production from Methane
The effects of operating conditions on PHB production using methanotrophs have been reviewed in some detail [6,66]. In summary, PHB production is typically achieved using a two stage process, with an initial period of cell growth under nutrient-sufficient conditions, then PHB accumulation is triggered by the absence of one or more major nutrients needed for cell division (unbalanced growth), with this being exaggerated under conditions of transient nitrogen and methane availability [69,91]. In mixed cultures, this two-stage process can be repeated multiple times to select for PHA accumulators.
There are a great many studies that have explored the relative merits of mixed cultures versus pure strains for PHA accumulation in general, including some that are specific to the use of methane as a carbon source [14,58,70,91,92,93,94]. Overall, these studies suggest that such mixed cultures may provide specific benefits such as: co-culture bacteria removing potentially toxic byproducts (e.g., methanol or formaldehyde) from the medium; beneficial nutrients being supplied by co-culture bacteria; operation under non-sterile conditions; and even possibly higher yields and production rates. However, many questions still remain with respect to optimizing the conditions for PHA production and culture selection.
It is known, for example, that the use of relatively high dissolved oxygen (DO, 9 mg/L) and nitrate as the nitrogen source will favour the growth of the Type I γ-proteobacteria [67], while the use of dissolved nitrogen as the nitrogen source, with a low influent DO (2 mg/L) was found to shift the culture to Type II α-proteobacteria [67], although the Methylocystis species may be an exception to this, tolerating a wide range of CH4/O2 ratios [95]. It is also known that nutrient concentration affects γ-proteobacteria and that they prefer a neutral pH, while α-proteobacteria are typically more acidophilic and, thus, acid-tolerant [63,67,96,97,98]. In pure strains, methanotrophic acitivity can be inhibited at DO levels >1 mg/L [99]. The addition of carbon dioxide as a gas or as bicarbonate may increase productivity [100], while iron deficiency can have a negative impact [37]. Metal ions such as copper, nickel, and zinc are important in the regulation of the activity of the pMMO and sMMO enzymes [101,102], with inhibitory effects on sMMO under some concentrations, while potassium deficiency has been shown to cause increased molecular weight in a Methylocystis dominated mixed culture [37]. Overall, there are many critical factors that affect PHA accumulation in methanotrophic bacteria, and these need to be carefully controlled.
According to the literature to date, the gases that make up biogas and natural gas do not inhibit the bacteria [75,92]. Natural gas is a mixture of several hydrocarbon gases containing predominantly methane (80%–95%), and may contain other heavier alkanes such as ethane, propane, butane and pentane [39]. Typically, biogas contains primarily methane (60%–70%) and carbon dioxide (30%–40%), with traces of nitrogen and hydrogen sulfide [103]. The CO2 in the biogas may even assist the growth of α-proteobacterial (PHB producing) methanotrophs, as described above, as they use CO2 in the serine cycle. For the most part the α-proteobacteria methanotrophs are also more resilient to gas fluctuation and variation than the γ-proteobacterial methanotrophs, since they are most robust to higher methane concentrations [104].
There have been a wide range of reactor types used in the laboratory scale production of PHB from methane. In the majority of cases, simple batch reactors (such as bottles, shake flasks, tanks, or fermenters) have been used, with either a static head or flow through of gas and under static, shaken or stirred conditions. On a micro scale, Sundstrom and Criddle used a high-throughput microbioreactor with aerated microtiter plates for rapid screening and isolation [77]. On a larger lab scale, pressurised bioreactors were used by Wendlandt et al. [27] while Zúñiga et al. [105] used a two-phase partitioning bioreactor, Rahnama et al. explored the use of a bubble column and vertical loop reactor [75], and Pfluger et al. adapted a laboratory scale fluidized bed reactor for growth of methanotrophs [67]. All of these processes were found to be viable, although none at this stage appear to show very significant improvements in productivity relative to the others.
The scale-up of biological processes is never a simple process. In the case of gas-liquid transfer reactions, there are a variety of reactor types available, ranging from continuous stirred tank reactors (CSTRs) to bubble lift and airlift reactors. CSTRs do not scale up well for gas fermentation, and airlift reactors are generally employed at scale, frequently using static mixers to ensure gas distribution [106,107]. Companies such as Calysta (Menlo Park, CA, USA) that use methane, or Lanzatech (Skokie, IL, USA) that use syngas, have spent considerable effort enhancing fluid dynamics and mass transfer of gases into the liquid phase. Detailed process and plant options from Unibio A/S website indicate the use of a U-tube reactor with a significant headspace (essentially an airlift reactor where the downcomer is equal in proportion to the riser) that has been implemented in a pilot-scale plant in Trinidad and Tobago—which uses natural gas as a methane source for single-cell protein production. The economic feasibility of PHB production from methane was first reported by Listewnik et al. [92]. They studied a relatively small-scale process (500 t/a) and found that the cost of production of PHB from methane at this scale was of the order of $8.5/kg. Since that time, Newlight Technologies have also commercialised a proprietary greenhouse gas-to-plastic technology, ramping up from pilot to commercial scale production in 2013. Newlight Technologies has added 22.7 thousand t/a production capacity (and signed an 8.6 million tonne offtake agreement with Vinmar), although process details for this technology are unavailable [108]. Likewise Mango Materials uses proprietary technologies to produce PHB from waste biogas (methane) in a process that is claimed to be economically competitive with conventional oil-based plastics [109].
In a recent paper, a techno-economic assessment of the potential for the production of 100,000 t/a of PHB from methane with at least 98% purity was undertaken [28]. Capital and operating costs were estimated, with air-lift bioreactors with a concentric internal draft tube being selected to reduce mixing costs at this large scale. The surface area to volume ratio is restricted at this scale, limiting the potential for heat removal so that heat removal from a two-stage bioreactor process running at 38 °C contributed 28% of the overall operating cost. Energy consumption for air compression and biomass drying were also identified as significant capital and operating costs, and the effects of bioreactor height and pressure and biomass moisture content need to be understood.
In designing such processes, one also needs to consider that while the use of air is frequently employed at lab scale, at an industrial scale this is problematic because 78% of the gas volume is wasted (unnecessary gas holdup volume) and the effectively dilute oxygen (21%) is much less efficient for mass transfer. Pure oxygen is expensive to provide at an industrial scale. Oxygen-enriched air can provide a higher concentration gradient between the gas and liquid phase (and occupy less gas volume in the reactor), but has greater capital costs than an air compressor. Thus, it is also necessary to consider the cost of supplying oxygen (up to half the total production cost), the hazardous nature of combining explosive/flammability gases, the risk of culture contamination affecting the culture stability and biopolymer yield/characteristics, as well as culture instability at low oxygen supply [24]. The downstream isolation technologies also need to be optimised, with many options having been proposed in the literature, including bead milling, high-pressure homogenisation, flotation, supercritical fluid extraction, chemical and enzymatic digestion, and solvent extraction [110].
4. High-Performance Biomaterials from Methane: PHA Co-Polymers
As discussed, many methanotrophic bacteria are known to synthesise PHB under N-limited conditions, with methane as the sole carbon source. PHB is an attractive polymer for many reasons, including its inherent biodegradability in aerobic and anaerobic environments, particularly marine, its water resistance, biocompatibility, optical purity, and piezoelectric properties [1]. However, the range of applications for PHB is currently limited due to its stiffness and brittleness, caused by its high crystallinity and also large spherulite size (if not processed to minimise this). This is reflected particularly in its elongation to break, which is around 100 times less than that of low density polyethylene [1]. In addition, the high melting temperature for PHB is matched by a low degradation temperature for the untreated polymer without stabiliser. This means that it is difficult to process while retaining material properties. Despite these limitations, a number of companies have commercialised the production of PHB, with applications in many areas including: biomedical devices, controlled-release and drug delivery applications, and injection molded articles, such as bottles, food containers, and cutlery, and many others [111].
Co-polymerisation of 3-hydroxybutyrate (3HB) with alternative hydroxyalkanoate (HA) monomers during biological synthesis is one alternative to generate biopolymers with more desirable properties. Through this approach, PHAs with different and tailored mechanical properties can be made, ranging from hard and crystalline to tough and flexible to elastic and rubbery. Again this is an approach that has been adopted by commercial manufacturers, using pure strains and sugar-based feedstocks. An example of such a copolymer is poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV), a polymer potentially tougher and an order of magnitude more elastic than PHB (Table 2). The lower melting temperature of PHBV, which is matched with the same degradation temperature as for PHB, also means that these polymers are much more readily processed without loss of properties.
Table 2.
Properties of PHB compared to PHBV and other PHAs [112]
| Polymer | Melting Temperature Tm (◦C) | Glass-Transition Temperature Tg (◦C) | Young’s Modulus (GPa) | Tensile Strength (MPa) | Elongation to Break (%) |
|---|---|---|---|---|---|
| PHB | 180 | 4 | 3.5 | 40 | 5 |
| P(3HB-co-20 mol%3HV) | 145 | −1 | 0.8 | 20 | 50 |
| P(3HB-co-6 mol%3HA) * | 133 | −8 | 0.2 | 17 | 680 |
| Polypropylene | 176 | −10 | 1.7 | 38 | 400 |
| Low-density polyethylene | 130 | −30 | 0.2 | 10 | 620 |
* HA, mixed hydroxyalkanoate co-monomer units including 3-hydroxydecanoate (3 mol%), 3-hydroxydodecanoate (3 mol%), 3-hydroxyoctanoate (1 mol%), 3-hydroxy-cis-5-dodecenoate (1 mol%).
5. Strategies for Producing PHA Co-Polymers from Methane
There are various potential strategies for directly converting methane into more versatile PHAs (such as PHBV copolymers) using biological means. The growth media can be supplemented with a co-substrate that is directly metabolised to form a fatty acid-CoA, such as propionyl-CoA, which is then condensed with acetyl-CoA to form starting materials for copolymer synthesis. In the case of propionyl-CoA, 3-ketovaleryl-CoA is formed which, in turn, is condensed with 3-hydroxybuturyl-CoA to form the biopolymer PHBV. Another potential route involves a co-culture/mixed culture where the non-methanotrophic strain is capable of generating a PHA, while surviving off a carbon source that is essentially a byproduct of methanotroph metabolism. Alternatively, there are various combinations of biological and chemical means that could be integrated to yield more attractive PHA co-polymers and their derivatives. These strategies are not without their challenges and are briefly summarised in Table 3 and discussed further in this section.
Table 3.
Strategies for PHA and PHA derivative production using methane as the primary substrate.
| Process | Pros | Cons | Unknowns |
|---|---|---|---|
| Direct production of PHB from CH4 | Has been achieved | Average to poor mechanical properties | Processes for maximising yields and rates |
| Methanotroph PHBV production through the co-addition of a fatty acid with odd carbon numbers | Has been achieved | The cost of the Codd feedstock offsets the savings of CH4; may produce blends of polymer product if there is variation in uptake rates amongst the different members of the community | Yields, homogeneity and maximum co-monomer content |
| Methanotrophs facilitating growth of a co-culture capable of PHBV production (direct or co-substrate addition) | Potentially achievable using Ralstonia sp. | The cost of the Codd feedstock offsets the savings of CH4; may produce blends of polymer product if there is variation in uptake rates amongst the different members of the community | Yields, homogeneity and maximum co-monomer content |
| Generate alternative copolymers through supply of appropriate feed. | Delivers broader range of mechanical properties | Cost of adding the monomers | Yields, homogeneity and maximum co-monomer content |
| Use a co-feeding strategy of timed pulses of methane and alternative feeds to tailor copolymer compositional distribution | Tailored, e.g., block copolymers already produced in the literature using alternating feeding strategy | Not proven in methanotrophs. May produce blends | Pulses of gas feed alternating with soluble carbon feed may prove difficult for cells to adapt to |
| Generate monomers biologically and polymerise ex situ | Can achieve desired copolymer composition | Monomer concentration and purification | Currently being commercialised |
| Downstream polymer modification/functionalisation/ depolymerisation into oligomers and use as building block | Processes are well established | Costly, intensive additional processes and can be time-consuming | Potential for development of unique material properties for niche applications. |
5.1. Direct Methanotrophic PHA Synthesis by Adding External Precursors
A copolymerisation strategy relying on the addition of precursor compounds that are structurally related to the desired co-monomer units is possible and well-established. Ueda et al. [80] supplemented a methanol feedstock with a 3HV precursor, n-amyl alcohol (a C5 alcohol), to synthesise a PHBV copolymer using methylotrophs (Paracoccus denitrificans and Methylobacterium extorquens). Babel used fructose as the co-substrate in another early study using methanol for PHA production from Methylobacterium rhodesianum and M. extorquens producing, in some cases, very high proportions of HV in the copolymers produced [65]. Yezza et al. [86] likewise produced PHBV at 67 mol% HV content using valeric acid as a co-substrate with methanol in Methylobacterium sp. GW2. Orita et al. [89] were able to use cobalt deficient conditions to synthesis PHBV from methanol alone.
The successful production of PHBV copolymers from methane was also demonstrated by Zúñiga et al. [105] in Methylobacterium organophilum CZ-2 using citrate or propionate as a co-substrate. The largest PHA yield to date (at 82% w/w) was obtained with citrate as a co-substrate, and had a 3HB:3HV:3HO ratio of 86:14:0, where 3HO is 3-hydroxyoctanoate. An 80% w/w yield was also obtained using propionate as a co-substrate, with the copolymer having a 3HB:3HV:3HO ratio of 70:30:0. NMR analyses revealed six different monomers with citrate in the media (3HB, 3HV, 4-hydroxyvalerate, 4-hydroxyhexanoate, 3HO, and 4-hydroxyoctanoate), indicating a remarkable diversity in the PHA produced by this methanotrophic strain. Myung et al. [69] recently demonstrated the potential to tailor poly(3-hydroxybutyrate-co-3-hydroxyvalerate) from methane using an enriched culture dominated by a Methylocystis species under non-aseptic conditions. When fed CH4 plus valerate, PHBV was synthesized. As expected, the mol% of 3HV increased with additional valerate. Adding different HAs also allowed the tailoring of PHA copolymer composition in an enriched culture of methanotrophs and in two pure strains (Methylocystis parvus OBBP and Methylosinus trichosporium OB3b) [113]. Only PHB was synthesised when using methane as the carbon source. Adding 3-hydroxybutyric acid to the media increased the PHB yield; adding propionate led to PHBV synthesis; and including valerate further increased PHBV content, with higher levels of valerate being associated with higher 3HV contents in the copolymer produced [113]. While this strategy has proven successful and would seem compatible with generating PHBV using a mixed consortium, the addition of these precursors is an additional cost.
The use of alternating carbon feed sources to produce block copolymers has also been demonstrated in pure and mixed cultures [114] and represents a route to novel materials with unique microstructure and material properties. This could be an interesting process to assess, using methane as the primary substrate, alternating it with other C1 compounds such as methanol or formate, or C3 compounds, such as propionate or propanol. Again, the addition of other carbon sources represents an additional cost that would have to be balanced against the value of the copolymer production yields and rates.
5.2. Indirect Biological PHA Production: Synthesis in Consortia
Aerobic methanotrophs naturally exist in microbial consortia, where carbon derived from methane is distributed to non-methanotrophic heterotrophs [32]. In natural ecosystems and laboratory enrichment cultures, methane assimilation and distribution has been observed to support a diversity of microbes that may comprise a complex, multi-tiered microbial food web [115]. Kalyuzhnaya et al. [46] demonstrated that methane assimilation using a highly efficient pyrophosphate-mediated glycolytic pathway under low oxygen tension. Here, a methanotroph (Methylomicrobium alcaliphilum 20Z) secreted fermentation products such as lactate, acetate, and H2 under low oxygen tension, all potential metabolites that could be used by other bacteria within the consortia. A striking example of this symbiotic mechanism occurs within the consortia in reactors used to produce single cell protein. The stability of continuous cultures of a methanotroph (Methylococcus capsulatus str. Bath) is dependent on the consortium to maintain stability [116].
Various bioreactors of single-cell protein-producing Methylococcus capsulatus Bath were consistently invaded by three bacteria: an Aneurinibacillus species, a Brevibacillus agri strain, and a Ralstonia species [116]. The Ralstonia species is of particular significance as this acetate-oxidising bacterium is a known PHA accumulator. Ralstonia eutropha produces both the homopolymer PHB [117]. When provided with the appropriate substrate, it can produce PHBV. When levulinic acid was added to a Ralstonia eutropha culture, the 3HV content in PHBV was up to 41% [118].
A low-substrate-specificity PHA synthase PhaC2Ps was expressed in a PHB negative Ralstonia eutropha strain, allowing it to accumulate short and medium chain length PHA copolymers when grown on mixed carbon sources [119]. This suggests that mixed culture used for single cell production may also serve as a feasible example for co-cultures that generate PHBV from methane-derived carbon. The methanotrophs would provide the carbon for Ralstonia, and imbalanced growth with HA supplementation could facilitate copolymer production. Although feasible, the yields may be low as the PHBV content would be a fraction of Ralstonia sp. and other biomass in the mixed population.
5.3. Downstream Processing of PHA—Including Precursor Production from Methane
There is also significant potential for value-adding by broadening the range of products derived from PHAs through further downstream product development. Polyhydroxyalkanoates are readily modified, and processes, such as grafting other functional groups onto the polymer chain, are well-established [120,121,122]. Blends, composites and highly-elongated fibres of unique strength and toughness have all been well studied [123,124]. The production of oligomeric blocks of PHA also provides a valuable route to alternative materials, such as synthetic block copolymers and other novel structures [125,126].
An alternative strategy for biopolymer synthesis is to produce other types of biologically-derived monomers or oligomers from methane, which may be subsequently polymerised or synthetically-coupled to PHA or its derivatives into a material of choice. A current example is the production of lactic acid as a precursor for chemical synthesis. Calysta have genetically modified a methanotroph with an exogenous nucleic acid that expresses lactate dehydrogenase. This delivers significantly enhanced lactate production from the reverse reaction, where pyruvate is converted to lactate [127], while presumably regenerating reducing equivalents (NAD+ is reduced to NADH) in the catalysis. This process is currently being commercialised by Calysta in conjunction with Natureworks in the US.
5.4. Synopsis
The options discussed in this article are summarised in Table 3. Each process has inherent restrictions, some of which are unknown and will require further research to elucidate. While the production of PHBV using methanotrophs is established, the decreased substrate cost still needs to be balanced against provision of other carbon sources/supplements, yield, production rate, extraction and processing, and capital expenditure for bioreactors and gas provision. Establishing the range of these values will provide clarity as to where improvements are required to progress towards financial viability for this versatile biopolymer.
6. PHA Production from Other Gas Streams
It should be noted that methane is not the only gaseous feedstock that can be considered for PHA production using microbes. As mentioned in the introduction, syngas (a gas mixture consisting primarily of hydrogen, carbon monoxide, with potentially some carbon dioxide) is well known as a feedstock for PHA production, with a techno-economic analysis having been recently conducted on a gasification-based hybrid biorefinery producing both hydrogen gas and PHA [128]. Hydrogen-oxidising bacteria (or Knallgas bacteria) have the ability to utilize hydrogen as an electron donor and oxygen as an electron acceptor to fix CO2 via the ribulose biphosphate or reverse tricarboxylic cycle [129]. Cupriavidus necator is one example of such an organism that is well known for producing PHA [130]. In addition, many cyanobacteria have the capacity to produce PHA from CO2, in the presence of light, although additional nutrients accelerate the production rates and at this stage the process is not economic [131].
7. Conclusions
Over the past decades, the production of PHA from methane by methanotrophic bacteria and the study of their PHA biosynthesis pathways were generally limited to pure cultures. The most widely produced PHA is the homopolymer PHB, but this is a low performance biopolymer that is highly crystalline and brittle, with poor elastic properties that limit the process and end-use applications. The copolymer PHBV can be tougher and more elastic with the correct composition and microstructure. Potentially, PHBV copolymers could be produced from mixed cultures of methanotrophic bacteria using methane as a feedstock, reducing the production costs associated with substrate and reactor sterility. However, the production of a consistent PHBV copolymer from methane is not a straightforward process. We present production scenarios that include (1) enriching a consortia, or using a stable mixed culture, and supplementing the feed with C3 or C5 co-substrates that could be used directly by the methanotrophs or (2) enriching their consortia with known PHBV accumulators such as Ralstonia spp. and (3) alternating the feed regime between methane and an alternative C source to tailor the desired copolymer, or (4) use methane to generate copolymer precursors that are synthetically assembled into the desired co-polymer. A fundamental understanding of polymer properties from methanotrophic mixed cultures is necessary, as it is important to characterise the relative purity of the product within the polymer blend, as this significantly affects polymer properties. Additional research is still required to establish the feasibility and shortcomings of ideas presented in this article.
Acknowledgments
Bronwyn Laycock, Paul Jensen, Paul Lant, Gene Tyson and Steven Pratt gratefully acknowledge financial support from the Australian Research Commission (DP 150103060). James Strong gratefully acknowledges financial support from Remondis and the Australian Research Commission (DP 140104572). Nuraqmar Mahamud gratefully acknowledges financial support from the Ministry of Higher Education Malaysia and Universiti Malaysia Perlis.
Author Contributions
Peter James Strong and Bronwyn Laycock wrote the bulk of the manuscript. Steven Pratt, Syarifah Nuraqmar Syed Mahamud, Paul Douglas Jensen oversaw specific portions of the manuscript. Paul Andrew Lant, Gene Tyson and Steven Pratt contributed to the intellectual content, proofread the manuscript drafts, adding necessary adjustments where required.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Laycock B., Halley P., Pratt S., Werker A., Lant P. The chemomechanical properties of microbial polyhydroxyalkanoates. Prog. Polym. Sci. 2013;38:536–583. doi: 10.1016/j.progpolymsci.2012.06.003. [DOI] [Google Scholar]
- 2.Chen G. Plastics completely synthesized by bacteria: Polyhydroxyalkanoates. In: Chen G., editor. Plastics from Bacteria: Natural Functions and Applications. Volume 14. Springer-Verlag; Berlin Heidelberg: 2010. pp. 17–37. [Google Scholar]
- 3.Anderson A.J., Dawes E.A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S.Y. Bacterial polyhydroxyalkanoates. Biotechnol. Bioeng. 1996;49:1–14. doi: 10.1002/(SICI)1097-0290(19960105)49:1<1::AID-BIT1>3.3.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 5.Jendrossek D., Knoke I., Habibian R., Steinbüchel A., Schlegel H. Degradation of poly(3-hydroxybutyrate), PHB, by bacteria and purification of a novel PHB depolymerase from Comamonas sp. J. Environ. Polym. Degr. 1993;1:53–63. doi: 10.1007/BF01457653. [DOI] [Google Scholar]
- 6.Karthikeyan O., Karthigeyan C., Cirés S., Heimann K. Review of sustainable methane mitigation and biopolymer production. Crit. Rev. Environ. Sci. Technol. 2015;45:1579–1610. doi: 10.1080/10643389.2014.966422. [DOI] [Google Scholar]
- 7.Tempest D.W., Wouters J.T.M. Properties and performance of microorganisms in chemostat culture. Enzyme Microb. Technol. 1981;3:283–290. doi: 10.1016/0141-0229(81)90001-6. [DOI] [Google Scholar]
- 8.Gurieff N., Lant P. Comparative life cycle assessment and financial analysis of mixed culture polyhydroxyalkanoate production. Bioresour. Technol. 2007;98:3393–3403. doi: 10.1016/j.biortech.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 9.Halami P.M. Production of polyhydroxyalkanoate from starch by the native isolate Bacillus cereus CFR06. World J. Microb. Biotechnol. 2008;24:805–812. doi: 10.1007/s11274-007-9543-z. [DOI] [Google Scholar]
- 10.Choi J.I., Lee S.Y. High-level production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by fed-batch culture of recombinant Escherichia coli. Appl. Environ. Microbiol. 1999;65:4363–4368. doi: 10.1128/aem.65.10.4363-4368.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamane T. Yield of poly-d(−)-3-hydroxybutyrate from various carbon-sources—A theoretical study. Biotechnol. Bioeng. 1993;41:165–170. doi: 10.1002/bit.260410122. [DOI] [PubMed] [Google Scholar]
- 12.Nikodinovic-Runic J., Guzik M., Kenny S.T., Babu R., Werker A., O Connor K.E. Carbon-rich wastes as feedstocks for biodegradable polymer (polyhydroxyalkanoate) production using bacteria. Adv. Appl. Microbiol. 2013;84:139–200. doi: 10.1016/B978-0-12-407673-0.00004-7. [DOI] [PubMed] [Google Scholar]
- 13.Koller M., Salerno A., Muhr A., Reiterer A., Braunegg G. Polyhydroxyalkanoates: Biodegradable polymers and plastics from renewable resources. Mater. Tehnol. 2013;47:5–12. [Google Scholar]
- 14.Wendlandt K.D., Stottmeister U., Helm J., Soltmann B., Jechorek M., Beck M. The potential of methane-oxidizing bacteria for applications in environmental biotechnology. Eng. Life Sci. 2010;10:87–102. doi: 10.1002/elsc.200900093. [DOI] [Google Scholar]
- 15.Rehm B.H.A. Polyester synthases: Natural catalysts for plastics. Biochem. J. 2003;376:15–33. doi: 10.1042/bj20031254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.US Environmental Protection Agency Methane and Nitrous Oxide Emissions from Natural Sources. EPA 430-R-10–001. [(accessed on 20 November 2015)]; Available online: http://nepis.epa.gov.
- 17.Intergovernmental Panel on Climate Change (IPCC) IPCC Fourth Assessment Report (AR4). Climate Change 2007: The Physical Science Basis. [(accessed on 20 November 2015)]. Available online: http://www.ipcc.ch/publications_and_data/publications_ipcc_fourth_assessment_report_wg1_report_the_physical_science_basis.htm.
- 18.Abbasi T., Tauseef S.M., Abbasi S.A. Anaerobic digestion for global warming control and energy generation—An overview. Renew Sust. Energ. Rev. 2012;16:3228–3242. doi: 10.1016/j.rser.2012.02.046. [DOI] [Google Scholar]
- 19.Batstone D.J., Jensen P.D. 4.17—Anaerobic processes. In: Wilderer P., editor. Treatise on Water Science. Elsevier; Oxford, UK: 2011. pp. 615–639. [Google Scholar]
- 20.Rittmann B.E. Opportunities for renewable bioenergy using microorganisms. Biotechnol. Bioeng. 2008;100:203–212. doi: 10.1002/bit.21875. [DOI] [PubMed] [Google Scholar]
- 21.WorldBank Zero Routine Flaring by 2030. [(accessed on 20 November 2015)]. Available online: http://www.worldbank.org/en/programs/zero-routine-flaring-by-2030.
- 22.Tijm P.J.A., Waller F.J., Brown D.M. Methanol technology developments for the new millennium. Appl. Catal. Gen. 2001;221:275–282. doi: 10.1016/S0926-860X(01)00805-5. [DOI] [Google Scholar]
- 23.Kuo J.W. Engineering evaluation of direct methane conversion processes. In: Wolf E.E., editor. Methane Conversion by Oxidative Processes. Springer Netherlands; Dordrecht, The Netherlands: 1992. pp. 483–526. [Google Scholar]
- 24.Dorin H., Demmin P., Gabriel D.L. Chemistry—The Study of Matter. 4th Ed. Prentice Hall; Needham, MA, USA: 1987. [Google Scholar]
- 25.Rostkowski K.H. Ph.D. Thesis. Stanford University; Stanford, CA, USA: 2012. Understanding Methanotrophic Polyhydroxybutyrate (PHB) Production Across Scale: Life Cycle Assessment, Pure Culture Experimentation, and Pathway/Genome Database Development. [Google Scholar]
- 26.U.S. Energy Information Administration, U.S. Price of Natural Gas Sold to Commercial Consumers, Monthly. [(accessed on 20 November 2015)]; Available online: http://www.eia.gov/opendata/qb.cfm?sdid=NG.N3020US3.M.
- 27.Wendlandt K.D., Jechorek M., Helm J., Stottmeister U. Producing poly-3-hydroxybutyrate with a high molecular mass from methane. J. Biotechnol. 2001;86:127–133. doi: 10.1016/S0168-1656(00)00408-9. [DOI] [PubMed] [Google Scholar]
- 28.Levett I., Birkett G., Davies N., Bell A., Langford A., Laycock B., Lant P., Pratt S. Techno-economic assessment of poly-3-hydroxybutyrate (PHB) production from methane. Eng. Res. Design. 2016 under review. [Google Scholar]
- 29.Kracke F., Kromer J.O. Identifying target processes for microbial electrosynthesis by elementary mode analysis. BMC Bioinform. 2014 doi: 10.1186/s12859-014-0410-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pratt S. Ph.D. Thesis. The University of Queensland; Queensland, Australia: 2003. The Development of the TOGA Sensor for the Study of Biological Wastewater Treatment Systems. [Google Scholar]
- 31.Hanson R.S., Hanson T.E. Methanotrophic bacteria. Microbiol. Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalyuzhnaya M.G., Puri A.W., Lidstrom M.E. Metabolic engineering in methanotrophic bacteria. Metab. Eng. 2015;29:142–152. doi: 10.1016/j.ymben.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Strong P.J., Xie S., Clarke W.P. Methane as a resource: Can the methanotrophs add value? Environ. Sci. Technol. 2015;49:4001–4018. doi: 10.1021/es504242n. [DOI] [PubMed] [Google Scholar]
- 34.Whittenbury R., Phillips K.C., Wilkinson J.F. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 35.Vecherskaya M., Dijkema C., Stams A.J.M. Intracellular PHB conversion in a Type II methanotroph studied by 13c NMR. J. Ind. Microbiol. Biotechnol. 2001;26:15–21. doi: 10.1038/sj.jim.7000086. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y.X., Xin J.Y., Chen L.L., Song H., Xia C.U. Biosynthesis of poly-3-hydroxybutyrate with a high molecular weight by methanotroph from methane and methanol. J. Nat. Gas. Chem. 2008;17:103–109. doi: 10.1016/S1003-9953(08)60034-1. [DOI] [Google Scholar]
- 37.Helm J., Wendlandt K.D., Jechorek M., Stottmeister U. Potassium deficiency results in accumulation of ultra-high molecular weight poly-beta-hydroxybutyrate in a methane-utilizing mixed culture. J. Appl. Microbiol. 2008;105:1054–1061. doi: 10.1111/j.1365-2672.2008.03831.x. [DOI] [PubMed] [Google Scholar]
- 38.Asenjo J.A., Suk J.S. Microbial conversion of methane into poly-β-hydroxybutyrate (PHB): Growth and intracellular product accumulation in a type-II methanotroph. J. Ferment. Bioeng. 1986;64:271–278. doi: 10.1016/0385-6380(86)90118-4. [DOI] [Google Scholar]
- 39.Fei Q., Guarnieri M.T., Tao L., Laurens L.M.L., Dowe N., Pienkos P.T. Bioconversion of natural gas to liquid fuel: Opportunities and challenges. Biotechnol. Adv. 2014;32:596–614. doi: 10.1016/j.biotechadv.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Shiemke A.K., Cook S.A., Miley T., Singleton P. Detergent solubilization of membrane-bound methane monooxygenase requires plastoquinol analogs as electron donors. Arch. Biochem. Biophys. 1995;321:421–428. doi: 10.1006/abbi.1995.1413. [DOI] [PubMed] [Google Scholar]
- 41.Anthony C. The quinoprotein dehydrogenases for methanol and glucose. Arch. Biochem. Biophys. 2004;428:2–9. doi: 10.1016/j.abb.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 42.Culpepper M.A., Rosenzweig A.C. Structure and protein-protein interactions of methanol dehydrogenase from Methylococcus capsulatus (Bath) Biochemistry. 2014;53:6211–6219. doi: 10.1021/bi500850j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fassel T.A., Buchholz L.A., Collins M.L., Remsen C.C. Localization of methanol dehydrogenase in two strains of methylotrophic bacteria detected by immunogold labeling. Appl. Environ. Microbiol. 1992;58:2302–2307. doi: 10.1128/aem.58.7.2302-2307.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitmitto A., Myronova N., Basu P., Dalton H. Characterization and structural analysis of an active particulate methane monooxygenase trimer from Methylococcus capsulatus (Bath) Biochemistry-US. 2005;44:10954–10965. doi: 10.1021/bi050820u. [DOI] [PubMed] [Google Scholar]
- 45.Myronova N., Kitmitto A., Collins R.F., Miyaji A., Dalton H. Three-dimensional structure determination of a protein supercomplex that oxidizes methane to formaldehyde in Methylococcus capsulatus (Bath) Biochemistry. 2006;45:11905–11914. doi: 10.1021/bi061294p. [DOI] [PubMed] [Google Scholar]
- 46.Kalyuzhnaya M.G., Yang S., Rozova O.N., Smalley N.E., Clubb J., Lamb A., Gowda G.A.N., Raftery D., Fu Y., Bringel F., et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat. Commun. 2013 doi: 10.1038/ncomms3785. [DOI] [PubMed] [Google Scholar]
- 47.Rahalkar M. Ph.D. Thesis. Universitat Konstanz; Konstanz, Germany: 2006. Aerobic Methanotrophic Bacterial Communities in Sediments of Lake Constance. [Google Scholar]
- 48.Anthony C. How half a century of research was required to understand bacterial growth on C1 and C2 compounds; the story of the serine cycle and the ethylmalonyl-CoA pathway. Sci. Prog. 2011;94:109–137. doi: 10.3184/003685011X13044430633960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vorholt J.A., Marx C.J., Lidstrom M.E., Thauer R.K. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 2000;182:6645–6650. doi: 10.1128/JB.182.23.6645-6650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vincenzini M., de Philippis R. Polyhydroxyalkanoates. In: Cohen Z., editor. Chemicals from Microalgae. Taylor and Francis; London, UK: 1999. pp. 290–312. [Google Scholar]
- 51.Sheu D.S., Wang Y.T., Lee C.Y. Rapid detection of polyhydroxyalkanoate-accumulating bacteria isolated from the environment by colony PCR. Microbiol. UK. 2000;146:2019–2025. doi: 10.1099/00221287-146-8-2019. [DOI] [PubMed] [Google Scholar]
- 52.Madison L.L., Huisman G.W. Metabolic engineering of poly(3-hydroxyalkanoates): From DNA to plastic. Microbiol. Mol. Biol. R. 1999;63:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.York G.M., Stubbe J., Sinskey A.J. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 2002;184:59–66. doi: 10.1128/JB.184.1.59-66.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pieja A.J., Sundstrom E.R., Criddle C.S. Poly-3-hydroxybutyrate metabolism in the type II methanotroph Methylocystis parvus OBBP. Appl. Environ. Microbiol. 2011;77:6012–6019. doi: 10.1128/AEM.00509-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bourque D., Pomerleau Y., Groleau D. High cell density production of poly-b-hydroxybutyrate (PHB) from methanol by Methylobacterium extorquens: Production of high-molecular-mass PHB. Appl. Microbiol. Biotechnol. 1995;44:367–376. doi: 10.1007/BF00169931. [DOI] [Google Scholar]
- 56.Korotkova N., Chistoserdova L., Lidstrom M.E. Poly-b-hydroxybutyrate biosynthesis in the facultative methylotroph Methylobacterium extorquens AM1: Identification and mutation of gap11, gap20, and phaR. J. Bacteriol. 2002;184:6174–6181. doi: 10.1128/JB.184.22.6174-6181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wendlandt K.D., Jechorek M., Helm J., Stottmeister U. Production of PHB with a high molecular mass from methane. Polym. Degrad. Stabil. 1998;59:191–194. doi: 10.1016/S0141-3910(97)00164-X. [DOI] [Google Scholar]
- 58.Helm J., Wendlandt K.D., Rogge G., Kappelmeyer U. Characterizing a stable methane-utilizing mixed culture used in the synthesis of a high-quality biopolymer in an open system. J. Appl. Microbiol. 2006;101:387–395. doi: 10.1111/j.1365-2672.2006.02960.x. [DOI] [PubMed] [Google Scholar]
- 59.Zúñiga C., Morales M., Le Borgne S., Revah S. Production of poly-β-hydroxybutyrate (PHB) by Methylobacterium organophilum isolated from a methanotrophic consortium in a two-phase partition bioreactor. J. Hazard Mater. 2011;190:876–882. doi: 10.1016/j.jhazmat.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 60.Bowman J. The methanotrophs—The families methylococcaceae and methylocystaceae. In: Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E., editors. The Prokaryotes. Springer; New York, NY, USA: 2006. pp. 266–289. [Google Scholar]
- 61.Bowman J.P., Jiménez L., Rosario I., Hazen T.C., Sayler G.S. Characterization of the methanotrophic bacterial community present in a trichloroethylene-contaminated subsurface groundwater site. Appl. Environ. Microbiol. 1993;59:2380–2387. doi: 10.1128/aem.59.8.2380-2387.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heyer J., Berger U., Hardt M., Dunfield P.F. Methylohalobius crimeensis gen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. Int. J. Syst. Evol. Microbiol. 2005;55:1817–1826. doi: 10.1099/ijs.0.63213-0. [DOI] [PubMed] [Google Scholar]
- 63.Pieja A.J., Rostkowski K.H., Criddle C.S. Distribution and selection of poly-3-hydroxybutyrate production capacity in methanotrophic proteobacteria. Microb. Ecol. 2011;62:564–573. doi: 10.1007/s00248-011-9873-0. [DOI] [PubMed] [Google Scholar]
- 64.Follner C.G., Babel W., Valentin H.E., Steinbuchel A. Expression of polyhydroxyalkanoic-acid-biosynthesis genes in methylotrophic bacteria relying on the ribulose monophosphate pathway. Appl. Microbiol. Biotechnol. 1993;40:284–291. doi: 10.1007/BF00170382. [DOI] [Google Scholar]
- 65.Babel W. Pecularities of methylotrophs concerning overflow metabolism, especially the synthesis of polyhydroxyalkanoates. Fems. Microbiol. Lett. 1992;103:141–148. doi: 10.1111/j.1574-6968.1992.tb05831.x. [DOI] [Google Scholar]
- 66.Khosravi-Darani K., Mokhtari Z.B., Amai T., Tanaka K. Microbial production of poly (hydroxybutyrate) from C-1 carbon sources. Appl. Microbiol. Biotechnol. 2013;97:1407–1424. doi: 10.1007/s00253-012-4649-0. [DOI] [PubMed] [Google Scholar]
- 67.Pfluger A.R., Wu W.M., Pieja A.J., Wan J., Rostkowski K.H., Criddle C.S. Selection of Type I and Type II methanotrophic proteobacteria in a fluidized bed reactor under non-sterile conditions. Bioresour. Technol. 2011;102:9919–9926. doi: 10.1016/j.biortech.2011.08.054. [DOI] [PubMed] [Google Scholar]
- 68.Myung J., Wang Z.Y., Yuan T., Zhang P., van Nostrand J.D., Zhou J.Z., Criddle C.S. Production of nitrous oxide from nitrite in stable Type II methanotrophic enrichments. Environ. Sci. Technol. 2015;49:10969–10975. doi: 10.1021/acs.est.5b03385. [DOI] [PubMed] [Google Scholar]
- 69.Myung J., Galega W.M., van Nostrand J.D., Yuan T., Zhou J.Z., Criddle C.S. Long-term cultivation of a stable Methylocystis-dominated methanotrophic enrichment enabling tailored production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) Bioresour. Technol. 2015;198:811–818. doi: 10.1016/j.biortech.2015.09.094. [DOI] [PubMed] [Google Scholar]
- 70.Chidambarampadmavathy K., Karthikeyan O.P., Heimann K. Biopolymers made from methane in bioreactors. Eng. Life Sci. 2015;15:689–699. doi: 10.1002/elsc.201400203. [DOI] [Google Scholar]
- 71.Karthikeyan O.P., Chidambarampadmavathy K., Nadarajan S., Lee P.K.H., Heimann K. Effect of CH4/O2 ratio on fatty acid profile and polyhydroxybutyrate content in a heterotrophic-methanotrophic consortium. Chemosphere. 2015;141:235–242. doi: 10.1016/j.chemosphere.2015.07.054. [DOI] [PubMed] [Google Scholar]
- 72.Scott D., Brannan J., Higgins I.J. The effect of growth conditions on intracytoplasmic membranes and methane mono-oxygenase activities in Methylosinus trichosporium OB3b. Microbiology. 1981;125:63–72. doi: 10.1099/00221287-125-1-63. [DOI] [Google Scholar]
- 73.Shah N.N., Hanna M.L., Taylor R.T. Batch cultivation of Methylosinus trichosporium OB3b: V. Characterization of poly-b-hydroxybutyrate production under methane-dependent growth conditions. Biotechnol. Bioeng. 1996;49:161–171. doi: 10.1002/(SICI)1097-0290(19960120)49:2<161::AID-BIT5>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 74.Doronina N.V., Ezhov V.A., Trotsenko Y.A. Growth of Methylosinus trichosporium OB3b on methane and poly-b-hydroxybutyrate biosynthesis. Appl. Biochem. Microbiol. 2008;44:182–185. doi: 10.1134/S0003683808020099. [DOI] [PubMed] [Google Scholar]
- 75.Rahnama F., Vasheghani-Farahani E., Yazdian F., Shojaosadati S.A. PHB production by Methylocystis hirsuta from natural gas in a bubble column and a vertical loop bioreactor. Biochem. Eng. J. 2012;65:51–56. doi: 10.1016/j.bej.2012.03.014. [DOI] [Google Scholar]
- 76.Rostkowski K.H., Pfluger A.R., Criddle C.S. Stoichiometry and kinetics of the PHB-producing Type II methanotrophs Methylosinus trichosporium OB3b and Methylocystis parvus OBBP. Bioresour. Technol. 2013;132:71–77. doi: 10.1016/j.biortech.2012.12.129. [DOI] [PubMed] [Google Scholar]
- 77.Sundstrom E.R., Criddle C.S. Optimization of methanotrophic growth and production of poly(3-hydroxybutyrate) in a high-throughput microbioreactor system. Appl. Environ. Microbiol. 2015;81:4767–4773. doi: 10.1128/AEM.00025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song H., Zhang Y.X., Kong W.B., Xia C.G. Activities of key enzymes in the biosynthesis of poly-3-hydroxybutyrate by Methylosinus trichosporium IMV3011. Chin. J. Catal. 2012;33:1754–1761. doi: 10.1016/S1872-2067(11)60443-9. [DOI] [Google Scholar]
- 79.Suzuki T., Yamane T., Shimizu S. Mass production of poly-β-hydroxybutyric acid by fully automatic fed-batch culture of methylotroph. Appl. Microbiol. Biotechnol. 1986;23:322–329. doi: 10.1007/BF00257027. [DOI] [Google Scholar]
- 80.Ueda S., Matsumoto S., Takagi A., Yamane T. Synthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) from methanol and n-amyl alcohol by the methylotrophic bacteria Paracoccus denitrificans and Methylobacterium extorquens. Appl. Environ. Microbiol. 1992;58:3574–3579. doi: 10.1128/aem.58.11.3574-3579.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Daniel M., Choi J.H., Kim J.H., Lebeault J.M. Effect of nutrient deficiency on accumulation and relative molecular weight of poly-β-hydroxybutyric acid by methylotrophic bacterium, Pseudomonas 135. Appl. Microbiol. Biotechnol. 1992;37:702–706. doi: 10.1007/BF00174831. [DOI] [Google Scholar]
- 82.Taidi B., Anderson A.J., Dawes E.A., Byrom D. Effect of carbon source and concentration on the molecular-mass of poly(3-hydroxybutyrate) produced by Methylobacterium extorquens and Alcaligenes eutrophus. Appl. Microbiol. Biotechnol. 1994;40:786–790. doi: 10.1007/BF00173975. [DOI] [Google Scholar]
- 83.Kim S.W., Kim P., Lee H.S., Kim J.H. High production of poly-b-hydroxybutyrate (PHB) from Methylobacterium organophilum under potassium limitation. Biotechnol. Lett. 1996;18:25–30. doi: 10.1007/BF00137805. [DOI] [Google Scholar]
- 84.Ghatnekar M.S., Pai J.S., Ganesh M. Production and recovery of poly-3-hydroxybutyrate from Methylobacterium sp V49. J. Chem. Technol. Biotechnol. 2002;77:444–448. doi: 10.1002/jctb.570. [DOI] [Google Scholar]
- 85.Korotkova N., Lidstrom M.E. Connection between poly-b-hydroxybutyrate biosynthesis and growth on C-1 and C-2 compounds in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 2001;183:1038–1046. doi: 10.1128/JB.183.3.1038-1046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yezza A., Fournier D., Halasz A., Hawari J. Production of polyhydroxyalkanoates from methanol by a new methylotrophic bacterium Methylobacterium sp GW2. Appl. Microbiol. Biotechnol. 2006;73:211–218. doi: 10.1007/s00253-006-0458-7. [DOI] [PubMed] [Google Scholar]
- 87.Mokhtari-Hosseini Z.B., Vasheghani-Farahani E., Shojaosadati S.A., Karimzadeh R., Heidarzadeh-Vazifekhoran A. Effect of feed composition on PHB production from methanol by HCDC of Methylobacterium extorquens (DSMZ 1340) J. Chem. Technol. Biotechnol. 2009;84:1136–1139. doi: 10.1002/jctb.2145. [DOI] [PubMed] [Google Scholar]
- 88.Xin J.Y., Zhang Y.X., Dong J., Song H., Xia C.G. An experimental study on molecular weight of poly-3-hydroxybutyrate (PHB) accumulated in Methylosinus trichosporium IMV 3011. Afr. J. Biotechnol. 2011;10:7078–7087. [Google Scholar]
- 89.Orita I., Nishikawa K., Nakamura S., Fukui T. Biosynthesis of polyhydroxyalkanoate copolymers from methanol by Methylobacterium extorquens AM1 and the engineered strains under cobalt-deficient conditions. Appl. Microbiol. Biotechnol. 2014;98:3715–3725. doi: 10.1007/s00253-013-5490-9. [DOI] [PubMed] [Google Scholar]
- 90.Poroshina M.N., Doronina N.V., Ezhov V.A., Trotsenko Y.A. Comparative characteristics of biosynthesis of polyhydroxybutyrate from methanol by Methylobacteria extorquens G10 and Methyloligella halotolerans C2. Appl. Biochem. Microbiol. 2014;50:253–258. doi: 10.1134/S0003683814030132. [DOI] [PubMed] [Google Scholar]
- 91.Pieja A.J., Sundstrom E.R., Criddle C.S. Cyclic, alternating methane and nitrogen limitation increases PHB production in a methanotrophic community. Bioresour. Technol. 2012;107:385–392. doi: 10.1016/j.biortech.2011.12.044. [DOI] [PubMed] [Google Scholar]
- 92.Listewnik H.F., Wendlandt K.D., Jechorek M., Mirschel G. Process design for the microbial synthesis of poly-b-hydroxybutyrate (PHB) from natural gas. Eng. Life Sci. 2007;7:278–282. doi: 10.1002/elsc.200620193. [DOI] [Google Scholar]
- 93.Johnson K., van Loosdrecht M.C.M., Kleerebezem R. Influence of ammonium on the accumulation of polyhydroxybutyrate (PHB) in aerobic open mixed cultures. J. Biotechnol. 2010;147:73–79. doi: 10.1016/j.jbiotec.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 94.Ho A., de Roy K., Thas O., de Neve J., Hoefman S., Vandamme P., Heylen K., Boon N. The more, the merrier: Heterotroph richness stimulates methanotrophic activity. ISME J. 2014;8:1945–1948. doi: 10.1038/ismej.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lopez J.C., Quijano G., Perez R., Munoz R. Assessing the influence of CH4 concentration during culture enrichment on the biodegradation kinetics and population structure. J. Environ. Manag. 2014;146:116–123. doi: 10.1016/j.jenvman.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 96.Wise M.G., McArthur J.V., Shimkets L.J. Methanotroph diversity in landfill soil: Isolation of novel type I and type II methanotrophs whose presence was suggested by culture-independent 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 1999;65:4887–4897. doi: 10.1128/aem.65.11.4887-4897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cebron A., Bodrossy L., Stralis-Pavese N., Singer A.C., Thompson I.P., Prosser J.I., Murrell J.C. Nutrient amendments in soil DNA stable isotope probing experiments reduce the observed methanotroph diversity. Appl. Environ. Microbiol. 2007;73:798–807. doi: 10.1128/AEM.01491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dedysh S.N. Methanotrophic bacteria of acidic Sphagnum peat bogs. Microbiology. 2002;71:638–650. doi: 10.1023/A:1021467520274. [DOI] [Google Scholar]
- 99.Henckel T., Roslev P., Conrad R. Effects of O-2 and CH4 on presence and activity of the indigenous methanotrophic community in rice field soil. Environ. Microbiol. 2000;2:666–679. doi: 10.1046/j.1462-2920.2000.00149.x. [DOI] [PubMed] [Google Scholar]
- 100.Park S., Hanna M.L., Taylor R.T., Droege M.W. Batch cultivation of Methylosinus trichosporium OB3b.1. Production of soluble methane monooxygenase. Biotechnol. Bioeng. 1991;38:423–433. doi: 10.1002/bit.260380412. [DOI] [PubMed] [Google Scholar]
- 101.Choi D.W., Kunz R.C., Boyd E.S., Semrau J.D., Antholine W.E., Han J.I., Zahn J.A., Boyd J.M., de la Mora A.M., DiSpirito A.A. The membrane-associated methane monooxygenase (pMMO) and pMMO-NADH: Quinone oxidoreductase complex from Methylococcus capsulatus bath. J. Bacteriol. 2003;185:5755–5764. doi: 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chidambarampadmavathy K., Obulisamy P.K., Heimann K. Role of copper and iron in methane oxidation and bacterial biopolymer accumulation. Eng. Life Sci. 2015;15:387–399. doi: 10.1002/elsc.201400127. [DOI] [Google Scholar]
- 103.Gupta G., Bhardwaj K.N., Choudhary C., Chandna P., Jain K.K., Kaur A., Kumar S., Shrivastava B., Ninawe S., Singh A., Singh A., et al. Biofuels: The environment-friendly energy carriers. In: Singh A., Kuhad R.C., editors. Biotechnology for Environmental Management and Resource Recovery. Springer India; New Delhi, India: 2013. pp. 125–148. [Google Scholar]
- 104.Semrau J.D., DiSpirito A.A., Yoon S. Methanotrophs and copper. FEMS Microbiol. Rev. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 105.Zúñiga C., Morales M., Revah S. Polyhydroxyalkanoates accumulation by Methylobacterium organophilum CZ-2 during methane degradation using citrate or propionate as cosubstrates. Bioresour. Technol. 2013;129:686–689. doi: 10.1016/j.biortech.2012.11.120. [DOI] [PubMed] [Google Scholar]
- 106.Siegel M.H., Robinson C.W. Application of airlift gas-liquid-solid reactors in biotechnology. Chem. Eng. Sci. 1992;47:3215–3229. doi: 10.1016/0009-2509(92)85030-F. [DOI] [Google Scholar]
- 107.Chisti Y., Kasper M., Moo-Young M. Mass transfer in external-loop airlift bioreactors using static mixers. Can. J. Chem. Eng. 1990;68:45–50. doi: 10.1002/cjce.5450680106. [DOI] [Google Scholar]
- 108.Newlight Technologies: From Greenhouse Gas to Plastic. [(accessed on 20 November 2015)]. Available online: http://newlight.com/
- 109.Mango Materials. [(accessed on 20 November 2015)]. Available online: http://mangomaterials.com/
- 110.Koller M., Niebelschutz H., Braunegg G. Strategies for recovery and purification of poly[(R)-3-hydroxyalkanoates] (PHA) biopolyesters from surrounding biomass. Eng. Life Sci. 2013;13:549–562. doi: 10.1002/elsc.201300021. [DOI] [Google Scholar]
- 111.Chen G.Q., Patel M.K. Plastics derived from biological sources: Present and future: A technical and environmental review. Chem. Rev. 2012;112:2082–2099. doi: 10.1021/cr200162d. [DOI] [PubMed] [Google Scholar]
- 112.Sudesh K., Abe H., Doi Y. Synthesis, structure and properties of polyhydroxyalkanoates: Biological polyesters. Prog. Polym. Sci. 2000;25:1503–1555. doi: 10.1016/S0079-6700(00)00035-6. [DOI] [Google Scholar]
- 113.Myung J. PHA copolymers from methane; Proceedings of the International Conference and Exhibition on Biopolymers and Bioplastics; San Francisco, CA, USA. 10–12 August 2015. [Google Scholar]
- 114.McChalicher C.W.J., Srienc F. Investigating the structure-property relationship of bacterial PHA block copolymers. J. Biotechnol. 2007;132:296–302. doi: 10.1016/j.jbiotec.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 115.Kalyuzhnaya M.G., Lapidus A., Ivanova N., Copeland A.C., McHardy A.C., Szeto E., Salamov A., Grigoriev I.V., Suciu D., Levine S.R., et al. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat. Biotechnol. 2008;26:1029–1034. doi: 10.1038/nbt.1488. [DOI] [PubMed] [Google Scholar]
- 116.Bothe H., Møller Jensen K., Mergel A., Larsen J., Jørgensen C., Bothe H., Jørgensen L. Heterotrophic bacteria growing in association with Methylococcus capsulatus (Bath) in a single cell protein production process. Appl. Microbiol. Biotechnol. 2002;59:33–39. doi: 10.1007/s00253-002-0964-1. [DOI] [PubMed] [Google Scholar]
- 117.Slater S., Houmiel K.L., Tran M., Mitsky T.A., Taylor N.B., Padgette S.R., Gruys K.J. Multiple b-ketothiolases mediate poly(b-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J. Bacteriol. 1998;180:1979–1987. doi: 10.1128/jb.180.8.1979-1987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang Y.P., Chen R.H., Cai J.Y., Liu Z.G., Zheng Y.M., Wang H.T., Li Q.B., He N. Biosynthesis and thermal properties of PHBV produced from levulinic acid by Ralstonia eutropha. PLoS ONE. 2013;8:11. doi: 10.1371/journal.pone.0060318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Luo R.C., Chen J.Y., Zhang L., Chen G.Q. Polyhydroxyalkanoate copolyesters produced by Ralstonia eutropha PHB(−)4 harboring a low-substrate-specificity PHA synthase PhaC2(Ps) from Pseudomonas stutzeri 1317. Biochem. Eng. J. 2006;32:218–225. doi: 10.1016/j.bej.2006.10.005. [DOI] [Google Scholar]
- 120.Torres M.G., Talavera J.R.R., Munoz S.V., Carreon-Castro M.D., Munoz E.M.R., Padron G.H., Guerrero F.Q. Crystalline and spectroscopic characterization of poly(2-aminoethyl methacrylate hydrochloride) chains grafted onto poly[(R)-3-hydroxybutyric acid] Vib. Spectrosc. 2015;76:55–62. doi: 10.1016/j.vibspec.2014.12.003. [DOI] [Google Scholar]
- 121.Nguyen S. Graft copolymers containing poly(3-hydroxyalkanoates)—A review on their synthesis, properties, and applications. Can. J. Chem. 2008;86:570–578. doi: 10.1139/v08-044. [DOI] [Google Scholar]
- 122.Hazer D.B., Kilicay E., Hazer B. Poly(3-hydroxyalkanoate)s: Diversification and biomedical applications, a state of the art review. Mat. Sci. Eng. 2012;32:637–647. doi: 10.1016/j.msec.2012.01.021. [DOI] [Google Scholar]
- 123.Pina S., Oliveira J.M., Reis R.L. Natural-based nanocomposites for bone tissue engineering and regenerative medicine: A review. Adv. Mater. 2015;27:1143–1169. doi: 10.1002/adma.201403354. [DOI] [PubMed] [Google Scholar]
- 124.Yu L., Dean K., Li L. Polymer blends and composites from renewable resources. Prog. Polym. Sci. 2006;31:576–602. doi: 10.1016/j.progpolymsci.2006.03.002. [DOI] [Google Scholar]
- 125.Orts W.J., Nobes G.A.R., Kawada J., Nguyen S., Yu G.E., Raveneile F. Poly(hydroxyalkanoates): Biorefinery polymers with a whole range of applications. The work of Robert H. Marchessault. Can. J. Chem. 2008;86:628–640. doi: 10.1139/v08-050. [DOI] [Google Scholar]
- 126.Nguyen S., Yu G.E., Marchessault R.H. Thermal degradation of poly(3-hydroxyalkanoates): Preparation of well-defined oligomers. Biomacromolecules. 2002;3:219–224. doi: 10.1021/bm0156274. [DOI] [PubMed] [Google Scholar]
- 127.Saville R.M., Lee S., Regitsky D.D., Resnick S.M., Silverman J. Compositions and Methods for Biological Production of Lactate From C1 Compounds Using Lactate Dehydrogenase Transformants. Patent WO 2014205146. 2014 Dec 24;
- 128.Choi D., Chipman D.C., Bents S.C., Brown R.C. A techno-economic analysis of polyhydroxyalkanoate and hydrogen production from syngas fermentation of gasified bimass. Appl. Biochem. Biotechnol. 2010;160:1032–1046. doi: 10.1007/s12010-009-8560-9. [DOI] [PubMed] [Google Scholar]
- 129.Aragno M., Schlegel H.G. The mesophilic hydrogen-oxidizing (knallgas) bacteria. In: Balows A.T., Truper H.G., Dworkin M., Harder W., Schleifer K.H., editors. The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications. Volume 1. Springer-Verlag; Berlin, Germany: 1992. pp. 344–384. [Google Scholar]
- 130.Sonnleitner B., Heinzle E., Braunegg G., Lafferty R.M. Formal kinetics of poly-b-hydroxybutyric acid (PHB) production in Alcaligenes eutrophus H16 and Mycoplana rubra R14 with respect to the dissolved-oxygen tension in ammonium-limited batch cultures. Eur. J. Appl. Microbiol. 1979;7:1–10. doi: 10.1007/BF00522473. [DOI] [Google Scholar]
- 131.Panda B., Mallick N. Enhanced poly-b-hydroxybutyrate accumulation in a unicellular cyanobacterium, Synechocystis sp PCC 6803. Lett. Appl. Microbiol. 2007;44:194–198. doi: 10.1111/j.1472-765X.2006.02048.x. [DOI] [PubMed] [Google Scholar]