Abstract
The interest about Staphylococcus aureus (S. aureus) and methicillin resistant S. aureus (MRSA) in livestock, and domestic and wild animals has significantly increased. The spread of different clonal complexes related to livestock animals, mainly CC398, and the recent description of the new mecC gene, make it necessary to know more about the epidemiology and population structure of this microorganism all over the world. Nowadays, there are several descriptions about the presence of S. aureus and/or MRSA in different animal species (dogs, sheep, donkeys, bats, pigs, and monkeys), and in food of animal origin in African countries. In this continent, there is a high diversity of ethnicities, cultures or religions, as well as a high number of wild animal species and close contact between humans and animals, which can have a relevant impact in the epidemiology of this microorganism. This review shows that some clonal lineages associated with humans (CC1, CC15, CC72, CC80, CC101, and CC152) and animals (CC398, CC130 and CC133) are present in this continent in animal isolates, although the mecC gene has not been detected yet. However, available studies are limited to a few countries, very often with incomplete information, and many more studies are necessary to cover a larger number of African countries.
Keywords: MRSA, MSSA, CC398, CC130, CC133, Africa
1. Introduction
Staphylococcus aureus (S. aureus) is a microorganism that is present as a commensal on the skin, the nose and mucous membranes of healthy humans and animals. However, it is also an opportunistic pathogen that can cause multiple infectious diseases of diverse severity. The epidemiology of this microorganism in animals has gained interest in the last years, not only because of their importance in veterinary medicine due to the increment of infectious processes caused by this pathogen (especially by methicillin-resistant S. aureus (MRSA) strains), but also because of the emergence of some clonal lineages associated with animals and their increasingly evidenced zoonotic potential. This is the case of the Sequence Type 398 (ST398), which has been identified as colonizer or infectious agent in pigs, cattle, horses, and poultry, as well as in people in contact with these animals (farmers, veterinarians, and slaughterhouse workers) [1,2,3,4,5,6]. Moreover, livestock associated (LA) MRSA infections have also been detected in relatives of farmers and some cases of MRSA of Clonal Complex 398 (CC398) have been identified in people without contact with animals [7]. These strains frequently exhibit multiresistance phenotypes. There are other clonal lineages (CC1, CC5, CC9, CC97, and CC130, among others) of LA-MRSA that are emerging, and whose importance is increasing in the last years. It should be pointed out that pets and wild animals can also act as reservoirs of MRSA strains, and play an important role in the epidemiology of this microorganism [5,8,9,10]. Recently, there has been growing interest not only in the study of MRSA strains but also of methicillin susceptible S. aureus (MSSA) strains, since these strains play an essential role in the evolution of different genetic lineages.
The number of studies focused on the antibiotic resistance problem in the African Continent has grown in last decade and they suggest that in this continent, as in other parts of the world, this problem is increasing; however, its real extent is currently unknown since surveillance of drug resistance is only carried out in a few countries [11]. The misuse of antibiotics due to poor control policies is promoting this resistance development [12]. Despite limited resources, during the last years in many of these countries, there are important efforts to establish good control measures to avoid this worrisome situation [13].
The study of S. aureus prevalence, antimicrobial resistance and clonal lineages in humans, animals and food in Africa has great relevance, taking into consideration the high diversity of ethnicities, cultures and religions that determine the lifestyle of African people. Most studies about MSSA and MRSA in the African continent are focused on human clinical isolates; nevertheless, the number of reviews focused on this topic is very scarce [14,15,16]. As would be expected, a higher diversity of clonal lineages is found among MSSA strains in comparison with MRSA strains, however, some clones (CC5 or CC8) have been found in methicillin resistant and susceptible strains [15]; a predominance of some clonal lineages (CC8 (ST239 and ST612), CC5, CC30, CC80, and CC88) has been identified in MRSA strains [15,16]. In many cases, CC88 is the dominant clonal lineage (24% to 83%) detected among MRSA strains in humans, and it has been named the “African clone” [15].
In this review, the objective is to report the situation of S. aureus in animals and food in Africa. The different African food habits highly influence the livestock industry of this continent. Moreover, there is a huge density and a high number of wild animal species that can be an important reservoir of this microorganism and of emerging antibiotic resistance mechanisms. These characteristics, and the close contact among humans, livestock, and domestic and wild animals, can have a relevant impact on the epidemiology of MSSA and MRSA. Therefore, it is essential to know what is happening, not only in strains from humans, but also in those of animal and food origin.
2. S. aureus in Animals in Africa
2.1. S. aureus Prevalence in Animals
Studies focused on the presence, prevalence and/or molecular typing of MSSA and MRSA strains from animals in Africa are rather limited and there is only information about certain countries (Table 1, Figure 1 and Figure 2) [17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38]. Until the moment when this review was performed, S. aureus strains had been reported in sick and healthy animals in 12 countries and MRSA strains had been only identified in seven of them (Côte d’Ivoire, Egypt, Nigeria, Senegal, South Africa, Sudan, and Tunisia). Most studies in animals have been performed in recent years, indicating an increased awareness of the role of animals in the evolution, epidemiology and dissemination of this microorganism.
Table 1.
Distribution and clonal lineages of S. aureus detected in animals in the African continent.
| Country | Tested Animals | Animals from Which S. aureus Was Detected | Detection of MRSA | Animals from Which MRSA Was Detected | Sampling Date | Healthy/Sick | Samples | Lineages of MRSA | Lineages of MSSA b | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Côte d’Ivoire | Domestic and wild animals | Goats, cats, dogs, sheep, poultry, primates | yes | Sheep | 2010–2013 | Healthy | Nasal and pharyngeal | CC88 | CC5, CC6, CC8, CC15, CC121, CC152, ST567, ST1472, ST2946, among others | [17] |
| Primates | Chimpanzees | no | - | 2007–2012 | Healthy | Mucosal, feces, oral, genital, fruit wedges | - | CC1, CC45, ST601, ST1928, ST2603, ST2621 | [18] | |
| Primates | Monkeys | no | - | - | Healthy | Nasal and fruit wedges | - | CC1, CC9, CC45, ST601, ST1782, ST1928, ST2023, ST2058, ST2059, ST2072, ST2603, ST2621 | [19] | |
| Democratic Republic of Congo | Domestic and wild animals | Civet, primates | no | - | 2010–2013 | Healthy | Nasal and pharyngeal | - | CC5, ST2473-ST2478, among others | [17] |
| Egypt | Dogs and cats | Dogs | yes | Dogs | - | Healthy and sick | Nasal, oral, ear, wound | HA-MRSA and CA-MRSA | - | [20] |
| Cattle, dogs, buffaloes, poultry | Cattle, dogs, buffaloes, poultry | yes | No specified | - | Sick | Milk, wounds, abscesses, internal organs, urine and nasal | ND a | - | [21] | |
| Gabon | Primates | Gorillas, chimpanzees | no | - | 2011 | Healthy and sick | Nasal, oral, vaginal, rectal | - | CC72, CC101 | [22] |
| Primates | Monkeys, gorillas, chimpanzees | no | - | - | Healthy | Nasal and fruit wedges | - | CC1, CC80, ST1851-ST1854, ST1856, ST1857, ST1872 ST1874, ST1928, ST2022, ST2023, ST2067, ST2071, ST2074 | [19] | |
| Domestic and wild animals | Sheep, primates | no | - | 2010–2013 | Healthy | Nasal and pharyngeal | - | CC101, CC80, ST1838, ST1851-ST1854, ST1857, ST1872-ST1874, ST1925, ST2022, ST2067, ST2071, ST2074, ST2295, ST2296, ST2721, among others | [17] | |
| Madagascar | Primates | Lemurs | no | - | 2007–2012 | Healthy | Mucosal, feces, oral, genital, fruit wedges | - | CC1, CC182, CC188, ST2435, ST2436 | [18] |
| Nigeria | Dogs, cats, chickens, pigs, horses, sheep, cattle, goats | Dogs, cats, chickens, pigs, horses, sheep, cattle, goats | - | - | - | Healthy and sick | Skin lesions, nasal, cloacal, milk | - | - | [23] |
| Bats | Bats | no | - | 2008–2010 | Healthy | Fecal | - | CC15, ST1725-ST1727, ST2463-ST2467, ST2470 | [24] | |
| Camels, sheep, cattle, goats | Camels, sheep, cattle, goats | yes | Camels, sheep, cattle, goats | 2012 | Healthy | Nasal and milk | ND | - | [25] | |
| Chickens | Chickens | no | - | - | Healthy | - | - | - | [26] | |
| Senegal | Pigs | Pigs | yes | Pigs | 2009–2011 | Healthy | Nasal | CC5, CC88 | CC1, CC5, CC8, CC15, CC72, CC97, CC121, CC152 | [27] |
| South Africa | Pigs, cattle, goats, chickens | Pigs | yes | Pigs | - | Healthy | Nasal, mouth wash, ear | ND | - | [28] |
| Chimpanzees | Chimpanzees | no | - | 2007, 2010, 2011 | Healthy | Nasal and oral | - | CC15, CC6, CC30, CC80, CC101 | [29] | |
| Cattle and pigs | Cattle and pigs | yes | Cattle and pigs | - | Healthy | Rump, flank, brisket, neck | ND | - | [30] | |
| Sudan | Sheep | Sheep | no | - | 2007–2008 | Sick | Abscesses | - | ST1464 | [31] |
| Sheep | Sheep | no | - | 2003–2005 | Sick | Pus samples | - | - | [32] | |
| Horse | Horse | yes | Horse | - | Sick | Lungs and peritoneum | ND | - | [33] | |
| Tunisia | Sheep | Sheep | yes | Sheep | 2010 | Healthy | Nasal | CC80 | CC8, CC130, CC522, ST1476, ST2076 | [34] |
| Donkeys | Donkeys | no | - | 2010 | Healthy | Nasal | - | CC1, CC6, CC7, CC15, CC22, CC72, CC133, CC522 | [35] | |
| Cattle, goats, dogs, cats | Cattle, goats, dogs, cats | no | - | 2010–2011 | Healthy | Nasal | - | CC6, CC15, CC30, CC45, CC130, CC133, CC188, CC522 | [36] | |
| Uganda | Chimpanzees | Chimpanzees | no | - | 2007, 2010, 2011 | Healthy | Nasal and oral | - | CC15, CC6, CC30, CC80, CC101 | [29] |
| Primates | Chimpanzees | no | - | 2007–2012 | Healthy | Mucosal, feces, oral, genital, fruit wedges | - | CC6, CC9, CC15, CC30, CC80, CC152, CC188 | [18] | |
| Zambia | Zebra | Zebra | - | - | Sick | Tissue | - | - | [37] | |
| Chimpanzees | Chimpanzees | no | - | 2007, 2010, 2011 | Healthy | Nasal and oral | - | CC15, CC6, CC30, CC80, CC101 | [29] | |
| Dogs and cats | Dogs | no | - | 2012 | Sick | Skin, ear, wound | - | CC398, CC5, CC15, CC152 | [38] |
a ND, non-determined in the study. b The clonal complexes were determined by e-BURST when sequence types were indicated in the study and were presumptively assumed according to the spa-types when the sequences types were not indicated in the study. Sequence Types instead of Clonal Complexes were indicated when they were not enclosed in any Clonal Complexes.
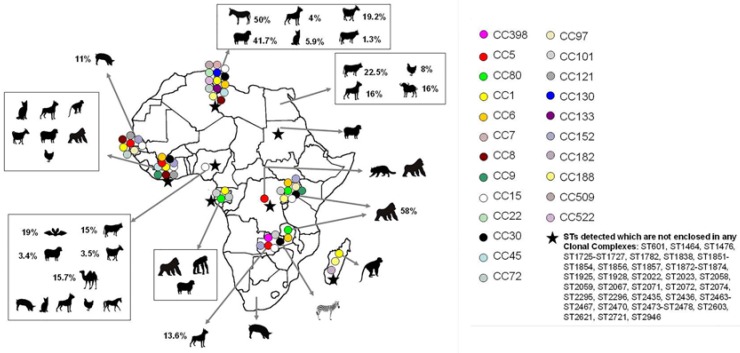
Figure 1.
Animal species, clonal lineages and prevalence of MSSA identified in the Africa continent. Prevalence (%) is calculated considering the total number of samples of each animal species included in the different studies and is indicated when this estimation is possible with the data shown in each publication. Moreover, in those cases, the number of samples studied is also indicated (%/number of samples). Clonal complexes detected in more than one country are indicated as a triangle. The clonal complexes were determined by e-BURST when sequence types were indicated in the study and were presumptively assumed according to the spa-types when the sequences types were not indicated in the study.
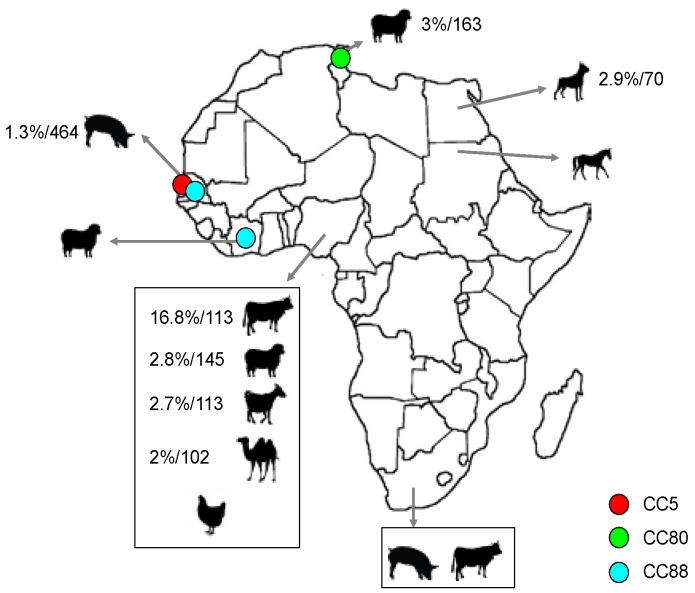
Figure 2.
Animal species, clonal lineages and prevalence of MRSA identified in the Africa continent. Prevalence (%) is calculated considering the total number of samples of each animal species included in the different studies and is indicated when this estimation is possible with the data shown in each publication. Moreover, in those cases, the number of samples studied is also indicated (%/number of samples). The clonal complexes were determined by e-BURST when sequence types were indicated in the study and were presumptively assumed according to the spa-types when the sequences types were not indicated in the study.
The comparison of MSSA and MRSA prevalence data in the different studies carried out in the African continent is difficult due to the different methodologies that have been employed. In some studies, this prevalence is calculated taking into consideration the number of total samples, in others the number of each species included and in others the number of staphylococcal strains isolated. In Figure 1 and Figure 2, the prevalence was estimated considering the total number of samples of each species tested when these data were included in each publication. In this way, the MSSA prevalence identified in the different countries was highly variable (from 3% to 58%) (Figure 1). Some clonal lineages seem to be better adapted to some animal species, and S. aureus rate might be higher in these animals. This could be the case, for example, of CC130 and CC133 lineages, which are highly associated with ruminants, as detected in studies performed on other continents [39,40]. On the African continent, there might be other clonal lineages associated with certain animal species, but most of these clones are still unknown. There is a specific subclade (ST1874, ST2058, and ST2071) that seems to be related to monkeys according to one study carried out in sub-Saharan Africa [19]. Results shown in Table 1 can be influenced by the methodology of sampling, and thus, the oro-pharyngeal S. aureus colonization rate was higher (72%) than the rectal prevalence (8.7%) in lemurs [18]. Prevalence rates were also very different depending on the animal analyzed, being 11% in lemurs and 50%–80% in chimpanzees (vaginal samples in both cases). In addition, some studies were performed including healthy [34,35,36] and/or sick animals [21] (Table 1).
In general, the MRSA colonization of animals detected in the African continent was very low (from 0% to 3%) [20,25,27,34], except for one study carried out in Nigeria [25], in which a colonization rate of 16.8% was observed in cattle samples (Figure 2). In countries on other continents, MRSA prevalence in healthy pets is usually lower than 1%, being between 9% and 20% in animals admitted to veterinary hospitals [41]. In one study performed in Egypt, MRSA isolates were identified in 2.9% of the analyzed dog samples and MRSA were not identified among the tested cat samples [20]. In livestock animals (especially in pigs), higher MRSA prevalence has been identified in European countries (4%–80%) [42]. In other farm animals, such as poultry, cattle and horses, the detected rates are normally lower than 13% [11,43,44]. On the African continent, MRSA has been found in variable rates in different livestock animals (cattle, sheep, pigs, goats, horses and camels) (Figure 2).
2.2. Population Structure of MSSA in Animals
As can be seen in Table 1 and Figure 1, a high diversity of clonal lineages has been identified among MSSA strains from animals in Africa. Twenty-three Clonal Complexes (CCs), 47 Sequence Types (STs) (which are not enclosed in any CC), and a many different spa-types were identified among these MSSA strains. Moreover, in these studies, numerous new STs [24,34,35,36,38] and spa-types [34,35] were detected. The most frequently found clonal lineages were CC1 and CC15, which were detected in the following countries: Côte d’Ivoire, Gabon, Madagascar, Nigeria, Senegal, Tunisia, Uganda and/or Zambia (Table 1 and Figure 1). In addition to CC1 and CC15, other clonal lineages (CC5, CC6, CC8, CC30, CC80, CC101, CC121, CC152, and CC188) were also identified in several African countries (Figure 1). Alternatively, some CCs were only observed in animals in one country. This is the case of CC398 in Zambia [38], CC130 and CC133 in Tunisia [34,35,36], and CC182 in Madagascar [18], among others.
Regarding clonal lineages associated with animals, there is only one description of MSSA CC398 in Africa, detected in the skin sample of one dog [38]. No other descriptions of MSSA or MRSA CC398 have been performed in other pets, in livestock animals or in wild animals in Africa. However, this clonal lineage has recently been identified in one MRSA clinical isolate in a hospital of Tunisia [45], and in MRSA and MSSA isolates from food samples, also in Tunisia, as will be detailed later [46]. Remarkably, other livestock associated CCs of high relevance have also been found in animals in Africa (Table 1 and Figure 1). MSSA CC130 strains were identified in Tunisia in sheep, goats, and one cow [34,36]. Monitoring of this lineage is very important since the new gene mecC has been identified mainly in CC130 strains in Europe [47,48]. However, this gene has not yet been found in any African country. CC133 is frequently found in ruminants [49,50,51] and this clonal lineage was identified in healthy donkeys in one study performed in Tunisia, and was the predominant CC found in 44% of the recovered isolates [35].
Other clonal lineages (CC1, CC5, CC8, CC9, CC30, CC97, and CC121) detected in animals in Africa have also been identified in livestock animals in other continents. CC1 has been previously found in pigs, cattle, poultry and horses in other studies [52,53,54], and identified in pigs, donkeys and non-human primates in Côte d’Ivoire, Gabon, Madagascar, Senegal, and Tunisia [17,18,19,27,35]. S. aureus strains belonging to CC5 are able to cause important infections in poultry [52]. Few studies about the presence of S. aureus in poultry have been performed in the Africa continent. Thus, MSSA strains have only been identified in poultry in Côte d’Ivoire, Egypt and Nigeria [17,21,23]. In one of these studies, molecular typing was performed and CC152 (and not CC5) was identified [16]. Although CC5 has not been found in poultry strains in Africa, this clone has been identified in MRSA and MSSA strains from other animal species (pigs, civets, dogs and goats) [17,27,38]. In Asian countries (China, Malaysia, and Thailand), the most common MRSA clone found in pigs is CC9 [55,56] and in Portugal CC30 (in addition to CC398) [57]. These CCs were identified in MSSA strains from animal species, except for pigs, in Côte d'Ivoire, Tunisia, Uganda, and/or Zambia [18,19,29,36]. However, it must be taken into consideration that the presence of this microorganism in pigs has only been studied for four works [23,27,28,30], and only in one of them the strains have been characterized [27]. In that study, the CCs identified in MSSA strains from pigs were CC1, CC5, CC72, CC97, CC121, CC15, CC152 and CC8 [27]. Regarding CC8, CC97 and CC121, these clonal lineages have been identified on other continents in cattle, horses, pigs, and rabbits [53,58,59,60,61].
2.3. Population Structure of MRSA in Animals
There are few studies in which MRSA strains have been identified in animals in Africa and only in three of them there is information about the ST or the CC detected [17,27,34] (Table 1 and Figure 2). The CCs identified were CC5 in pigs [27], CC80 in sheep [34] and CC88 in pigs and sheep [17,27]. CC5 and CC80 were also identified in MSSA strains in these and/or other studies [17,18,27,29,38]. Moreover, the three CCs found in MRSA of animals in Africa have been frequently detected among human clinical MRSA isolates in this continent [16,17]. MRSA isolates with the mecC gene have not yet been reported in Africa.
2.4. S. aureus Interspecies Transmission
MSSA and MRSA human-to-animal transmission has been suggested in some African studies [17,22,29]. Human related clonal lineages (CC15, CC72, CC80, CC101, and CC152) have been identified in MSSA strains from non-human primates, goats, sheep, poultry and pets [17,29]. Moreover, MRSA CC88 strains with the same spa-type (t189) were identified in humans and sheep in Côte d’Ivoire. In that study, samples were taken from domestic animals that lived in the same villages where the tested humans lived [17]. Another human-to-animal case transmission was identified in a sanctuary in Africa in which a veterinarian and a chimpanzee showed MSSA strains with the same spa-type t279 [29]. In addition, in the study of Nagel et al. [22], interspecies transmission of a widely spread human associated S. aureus CC72 strain (spa-type t148) was observed; this strain was found as colonizer agent in three gorillas and caused infection in one of them. Strains with the same spa-type were identified in chimpanzees in contact with the infected gorilla. These strains presented only one different band in Pulsed-Field Gel Electrophoresis (PFGE) compared with the strains obtained from gorillas [22].
On the other hand, in one study performed in Tunisia, nasal swabs of healthy people with different levels of interaction with animals were analyzed, and animal associated clonal lineages (CC30 and CC121) were found in some MSSA strains from people with frequent contact with animals [62]. In these cases, animal-to-human transmission might have happened.
3. S. aureus in Food in Africa
There are a high number of African studies focused on the microbiological analyses of food products (milk, meat, ready-to-eat, fish and eggs, among others). However, in most of them the main objective was usually to analyze the presence of different pathogens (among them, S. aureus), and to count CFU (Colony Forming Units) in order to determine the rate of contamination of the tested food [63,64,65,66,67,68]. In other studies, milk samples of sick animals were analyzed with the objective of detecting the presence of S. aureus as the mastitis-causing agent [69,70,71,72,73,74,75,76,77]. There were also a few papers in which the presence of S. aureus and/or MRSA was studied in food products from healthy animals. However, clonal lineages were determined only in a few of them [46,78].
3.1. MSSA Detection in Food Samples
MSSA strains have been identified in very diverse types of food in Africa in very different percentages (Table 2) [79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106]. The rates detected in raw meat, meat products and cooked meat have been from 3% to 81.8%. Cooked and uncooked samples were analyzed in one study carried out in Libya [91], and the prevalence was higher in raw chicken (29.6%), than in cooked meat (3.12%). In this case, it was demonstrated that the cooking process reduced the presence of this microorganism. However, the highest prevalence (81.8%) in meat samples was detected in one study performed in Cameroon in which cooked pork samples were analyzed [79]. In this case, human contamination during processing of these foods could be the most probable explanation. Some explanations of why this microorganism is present in food samples are given in the different publications. The fact that animals are kept in kitchens where food is prepared; direct contamination by the food handlers through coughing and sneezing; storage of food at high temperature; and/or some processed foods, which constitute a good culture medium for bacteria, are some of the possible reasons [88,97]. In addition, in the case of raw meat samples the source of contamination could also originate in the animal.
Table 2.
Detection of S. aureus in food from healthy animals in the African continent.
| Country | Samples | Number of Samples Studied | Date of Sampling | Raw/Cooked | Detection of MRSA a | S. aureus Prevalence b | Reference |
|---|---|---|---|---|---|---|---|
| Cameroon | Pork | 11 | - | Cooked | ND a | 81.8% | [79] |
| Côte d’Ivoire | Beef, chickens, pork | 240 | 2010 | Cooked | Yes | 7.9% | [80] |
| Democratic Republic of Congo | Beef | - | - | Raw | ND | - | [81] |
| Egypt | Sausage, hamburger | 60 | - | Raw | ND | - | [65] |
| Liver, meat | 60 | - | Cooked | ND | - | [63] | |
| Fish (sardine, feseikh, molouha) | 60 | - | Cooked | Yes | - | [82] | |
| Milk | 150 | - | Raw | ND | 41.2% | [83] | |
| Goat (milk and meat) | 100 | - | Raw | ND | 58% milk 18% goat meat | [84] | |
| Ethiopia | Milk | - | 2011–2012 | Raw | yes | 100% | [85] |
| Meat samples | 100 | - | Raw | ND | 21.2% | [86] | |
| Gabon | Chicken | 151 | 2011–2012 | Raw | no | 3.3% | [78] |
| Kenya | Milk | - | - | Raw and cooked | no | - | [87] |
| Milk, minced meat | 96 | - | - | yes | - | [88] | |
| Milk | - | 2001–2002 | Raw | yes | - | [89] | |
| Lesotho | Cattle, pigs, sheep | 237 | - | Raw | ND | 5% | [90] |
| Libya | Chicken burger | 120 | - | Raw and cooked | ND | 29.6% raw 3.12% cooked | [91] |
| Malawi | Home cooked food | 132 | - | Cooked | ND | 61% (63% maize flour porridge, 51% fish, 75% vegetables, 69% beans, 38% others) | [92] |
| Morocco | Turkey | 96 | 2011–2012 | Raw | no | - | [93] |
| Milk, lben, jben | - | 2005–2006 | Raw and cooked | yes | - | [94] | |
| Meat and beef offal | 156 | 2002–2004 | Raw | ND | 16% | [95] | |
| Namibia | Milk | 15 | 1995–1996 | Cooked | ND | - | [96] |
| Nigeria | Ready-to-eat food | 168 | - | Raw and cooked | ND | 33.3% (57.1% salad, 19.1% meat pie, 14.3% fish roll, 9.6% doughnut) | [97] |
| Milk | 510 | 2012 | Raw | yes | 30.4% | [25] | |
| Suya, balangu, kilishi, dambunnama, raw beef | 300 | - | Raw and cooked | yes | 9.7% | [98] | |
| Chicken | 400 | - | Raw | yes | - | [26] | |
| Ready to eat food (meat, fish, vegetable) | 880 | - | Raw and cooked | ND | 62% | [99] | |
| Somalia | Milk | - | - | Raw and cooked | no | - | [87] |
| South Africa | Milk | 28 | - | Raw | yes | 100% | [100] |
| Milk | 156 | 1995–1996 | Cooked | ND | - | [96] | |
| Poultry | - | - | Raw | ND | 24.1% | [64] | |
| Street food vending (beef, chicken, salad, gravy) | 132 | - | Raw and cooked | ND | 3% | [101] | |
| Sudan | Sausage | 40 | - | Raw | ND | - | [68] |
| Milk | 320 | - | Raw | ND | 8.8% | [102] | |
| Milk | 90 | - | Raw | ND | - | [103] | |
| Tanzania | Milk | 128 | 2003 | Raw | ND | 6.3% | [104] |
| Tunisia | Chicken, horse, sheep, veal | 164 | 2010–2011 | Raw | yes | 26.2% | [46] |
| Uganda | Eggs | 171 | - | Raw | ND | 18% surface 4% inside | [105] |
| Zimbabwe | Milk | 140 | 2009–2010 | Raw and cooked | ND | - | [106] |
a ND: non determined (methicillin resistance was not tested). b Prevalence is calculated considering the total number of samples included in the different studies when this estimation is possible with the data shown in each publication.
There are some methods such as molecular typing or determination of Immune Evasion Cluster (IEC) genes that could help us to know if the origin of S. aureus strains in meat samples might be human or animal [107,108]. However, clonal lineages were only determined in two studies [46,78]. In one of them, CC8, CC22, and CC398 were identified among chicken, sheep, and veal samples [46]. As previously noted, this was the first study in which CC398 has been found in food samples in the African continent [46]. In this study, twenty different spa-types were identified among MSSA strains. One of these spa-types (t1166) has been associated with CC133, and was detected in one strain isolated from a horse sample [46]. In another study carried out in Gabon, five MSSA strains were obtained from chicken samples [78]; three of them presented the spa-type t002 and belonged to ST5 (CC5), one showed the spa-type t386 and belonged to the singleton ST2622, and the remaining one had spa-type t591 and was non typeable by Multilocus Sequence Typing (MLST). Notably, the spa-type t002 was also identified in humans in Gabon [78]. It is important to mention that in this study, food samples were imported from industrialized countries (Brazil, Spain, USA, and Turkey), and it would be interesting to know if these clonal lineages are frequent in those countries. Until now, there is very scarce information about the spa-types t386 and t591. On the other hand, the spa-type t002 is widely spread all over the world [108].
Regarding milk samples from healthy animals, S. aureus prevalence was between 6.3% and 100% (Table 2). Most of the studies included raw milk samples from cattle or camels [83,85,104], although some of them also analyzed dairy products typical of the African continent, such as lben or jben [94]. One study carried out in Uganda analyzed the presence of different microorganisms in egg samples, and detected higher prevalence of S. aureus on the outer shell surfaces (18%) than inside the eggs (4%) [105]. Other types of food that have been analyzed include the following: beans, corn flour, doughnut, fish roll, salted fish, maize flour porridge, mangoes, meat pie, salad, pawpaw, and cassava [82,92,97,99,101] (Table 2).
3.2. MRSA Detection in Food Samples
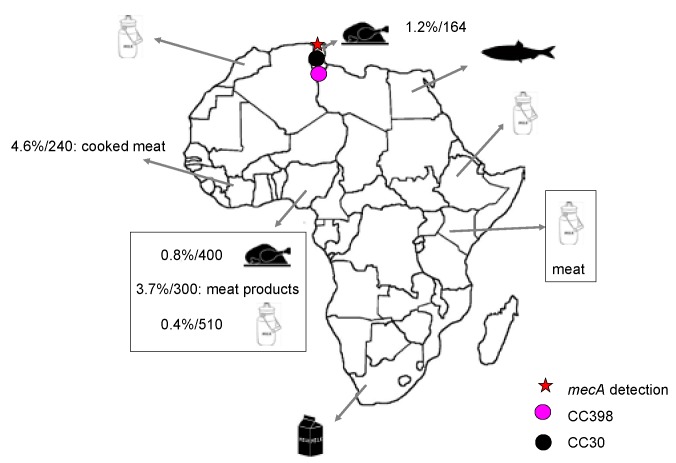
MRSA strains have been identified in meat, fish and milk samples from healthy animals in some studies performed in Africa (Table 2) (Figure 3). However, most of these MRSA strains have been identified by phenotypic methods, and the presence of the mecA gene was either not studied or not detected in many of them. MRSA strains have been found in raw meat, in meat products, and in cooked meat in Côte d’Ivoire, Nigeria, and Tunisia [26,46,80,98]. The presence of the mecA gene was analyzed in two of these four studies [46,98], and in only one of them this gene was found [46]. Molecular typing techniques were used in this last study [46], and two clonal lineages were identified (CC30 and CC398) in the two MRSA strains isolated from raw chicken samples.
Figure 3.
Food products, clonal lineages and prevalence of MRSA identified in the Africa continent. Prevalence (%) is calculated considering the total number of samples of each animal species included in the different studies and is indicated when this estimation is possible with the data shown in each publication. Moreover, in those cases, the number of samples studied is also indicated (%/number of samples). The clonal complexes were determined by e-BURST.
MRSA prevalence identified in meat samples in the African studies was in the range 0.8%–4.6% [26,46,80,98]. Interestingly, the highest percentage was identified in a unique study in which MRSA strains were found in cooked meat samples [80]. Salted fish samples were analyzed in Egypt and methicillin resistance was observed in 12 of the 95 S. aureus strains tested (12.6%) [82]. In five studies [25,85,89,94,100], the prevalence was calculated regarding the total S. aureus strains isolated, and the obtained values were variable. In the study performed in South Africa, the prevalence was 81.2%–93.2% in milk samples from communal farms and 5.7%–7% in those from commercial farms [100]. The percentages obtained in the remaining studies were 60.3% in Ethiopia [85], 28.57% in Nigeria [25], 15% in Morocco [94], and 7.8% in Kenya [89]. The presence of the mecA gene was not studied in any of them, and data regarding the clonal lineages of these MRSA strains were also not available.
4. Other Important Characteristics of S. aureus from Animals and Food in Africa
4.1. Antimicrobial Resistance of S. aureus
In some of the papers included in this review, in addition to methicillin resistance, antimicrobial resistance patterns to other agents were shown [17,18,19,20,21,22,24,25,28,29,30,31,34,35,36]. In general, MRSA isolates presented resistance to other non beta-lactam agents in addition to methicillin resistance, while MSSA isolates showed susceptibility to most of the antimicrobials tested. This situation is similar in MRSA and MSSA isolates from humans, animals and food in other parts of the world. In the studies in which MRSA isolates were obtained, resistance to tetracycline (5%–84%), erythromycin (1.7%–100%), clindamycin (9%–97%), trimethoprim-sulfametoxazole (1.9%–78%), tobramycin (0%–36%), ciprofloxacin (0%–42%) or vancomycin (9%–46%) were identified in different percentages [17,20,21,25,28,30,34,35,36,80,82]. Remarkably, in Nigeria all MRSA isolates obtained from camels, sheep and cattle showed resistance to mupirocin and fusidic acid and these antimicrobials are not routinely used in veterinary medicine in this zone [25]. Regarding MSSA isolates, in most of the studies, these isolates were susceptible to other non-beta-lactam antimicrobials. Only some of these isolates showed resistance to penicillin, tetracycline, erythromycin or clindamycin [17,18,19,22,24,25,29,31]. Nevertheless, in some cases, penicillin resistance was high among MSSA isolates [36,46,62], as occurs in other parts of the world; in the case of remote African regions, this phenotype is very rare, not only in animals but also in humans [29]. It must be taken into consideration that there are many factors that could be influencing the phenotypes detected. For example, it has been observed that MSSA isolates from chimpanzees in the wild were less resistant to penicillin, than isolates from chimpanzees living in captivity [18].
4.2. Virulence Determinants
Africa is considered endemic for Panton-Valentine-Leukocidin (PVL)-positive MSSA isolates [15,109]. Worryingly, this leukocidin has been identified in some MRSA from animals in Côte d’Ivoire, Gabon, Democratic Republic of Congo, Senegal and Tunisia [17,27,34], and in MSSA isolates in Côte d’Ivoire, Senegal and Tunisia [18,27,29,36]. In one study, PVL was significantly more frequent in isolates from chimpanzees than from humans (28% vs. 10%) [19]. According to these data, the possible role of animals as reservoir of this important virulence factor in this continent must be considered.
Other relevant virulence genes such as tst, eta, etb or etd have also been identified in animal isolates in Africa [18,23,27,34,35]. Moreover, the presence of genes encoding staphylococcal enterotoxins (SEs) responsible for food poisoning was studied in some articles. Some of these genes, such as sea, seb, sei, seh or seg, have been identified in isolates from different animal species in Africa [27,29,34,35]. Remarkably, these genes have also been found in isolates from food samples. In Egypt, SE genes were identified in 20.7% of raw goat milk samples and 11.1% of meat samples [84]. In Nigeria, 269 strains of 552 (48%) isolated from ready to eat food were enterotoxigenic, enterotoxin A being the most commonly found toxin [99]. However, in another study performed in Kenya, enterotoxin C was the most frequently produced type [88]; in this study, the highest percentage of enterotoxigenic strains was detected among chicken samples [88]. However, in one study performed in raw camel milk samples in Sudan, only three strains of 25 tested presented the enterotoxin C (the variant sec2) and the egc cluster [102].
5. Conclusions
The number of articles about the antibiotic resistance problem in African countries, and in particular about prevalence and clonal lineages of S. aureus strains in this continent, has increased in recent years. However, the available information is limited to a few countries, and is generally incomplete. Most of these studies are focused on clinical isolates, but there are some papers in which strains from various animal species (non-human primates, cows, pigs, donkeys, sheep, pets, bats, and camels) are analyzed. As in other parts of the world, animal MSSA strains present higher genetic diversity than MRSA strains. Clonal lineages associated with animals have been identified in several African countries, and the detection of MSSA CC398, CC130 and CC133 strains stand out. However, there is very scarce information about potential reservoirs and ways of dissemination of these clones in Africa. Relevantly, numerous new sequence types and spa-types have been identified in isolates of animals on this continent. Until now, the new mecC gene has not been detected in African countries, and further studies searching for its possible presence are required. On the other hand, there are several studies in which MSSA and MRSA strains have been found in food samples from healthy animals. However, in only two of them molecular typing of the S. aureus strains was performed. Therefore, the data in this regard are still insufficient. It is essential to know more about the current situation in these countries to assess the role of the food chain in the transmission of MRSA. Surveillance of MSSA and MRSA in humans, animals (pets, livestock and wild animals), and food in Africa can be a powerful tool for a better understanding of the epidemiology of this microorganism and for establishing appropriate control measures.
Acknowledgments
Carmen Lozano has a contract associated with Project SAF2012-35474. We thank Fernanda Ruiz-Larrea for critical review of the manuscript.
Author Contributions
Carmen Lozano contributed to the search of articles and to their tabulation and classification into different categories. She also contributed to the general design and the analysis of the review and to the writing of the paper. Haythem Gharsa and Karim Ben Slama helped in the general review of the manuscript. Myriam Zarazaga contributed to the search for articles and in the general review of the manuscript. Carmen Torres contributed to the general design of the manuscript and reviewed the manuscript. She also contributed to the writing of the paper and supervised all the work performed.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Golding G.R., Bryden L., Levett P.N., McDonald R.R., Wong A., Wylie J., Graham M.R., Tyler S., van Domselaar G., Simor A.E., et al. Livestock-associated methicillin-resistant Staphylococcus aureus sequence type 398 in humans, Canada. Emerg. Infect. Dis. 2010;16:587–594. doi: 10.3201/eid1604.091435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mediavilla J.R., Chen L., Uhlemann A.C., Hanson B.M., Rosenthal M., Stanak K., Koll B., Fries B.C., Armellino D., Schilling M.E., et al. Methicillin-susceptible Staphylococcus aureus ST398, New York and New Jersey, USA. Emerg. Infect. Dis. 2012;18:700–702. doi: 10.3201/eid1804.111419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuny C., Layer F., Köck R., Werner G., Witte W. Methicillin susceptible Staphylococcus aureus (MSSA) of clonal complex CC398, t571 from infections in humans are still rare in Germany. PLoS ONE. 2013;8:12. doi: 10.1371/journal.pone.0083165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witte W., Strommenger B., Stanek C., Cuny C. Methicillin resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg. Infect. Dis. 2007;13:255–258. doi: 10.3201/eid1302.060924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gómez P., Lozano C., Camacho M.C., Lima-Barbero J.F., Hernández J.M., Zarazaga M., Höfle U., Torres C. Detection of MRSA ST3061-t843-mecC and ST398-t011-mecA in white stork nestlings exposed to human residues. J. Antimicrob. Chemother. 2016;71:53–57. doi: 10.1093/jac/dkv314. [DOI] [PubMed] [Google Scholar]
- 6.Gómez-Sanz E., Simón C., Ortega C., Gómez P., Lozano C., Zarazaga M., Torres C. First detection of methicillin-resistant Staphylococcus aureus ST398 and Staphylococcus pseudintermedius ST68 from hospitalized equines in Spain. Zoonoses Public Health. 2014;61:192–201. doi: 10.1111/zph.12059. [DOI] [PubMed] [Google Scholar]
- 7.Benito D., Lozano C., Rezusta A., Ferrer I., Vasquez M.A., Ceballos S., Zarazaga M., Revillo M.J., Torres C. Characterization of tetracycline and methicillin resistant Staphylococcus aureus strains in a Spanish hospital: Is livestock-contact a risk factor in infections caused by MRSA CC398? Int. J. Med. Microbiol. 2014;304:1226–1232. doi: 10.1016/j.ijmm.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Gómez P., González-Barrio D., Benito D., García J.T., Viñuela J., Zarazaga M., Ruiz-Fons F., Torres C. Detection of methicillin-resistant S. aureus carrying the mecC gene in wild small mammals in Spain. J. Antimicrob. Chemother. 2014;69:2061–2064. doi: 10.1093/jac/dku100. [DOI] [PubMed] [Google Scholar]
- 9.Strommenger B., Kehrenberg C., Kettlitz C., Cuny C., Verspohl J., Witte W., Schwarz S. Molecular characterization of methicillin-resistant Staphylococcus aureus strains from pet animals and their relationship to human isolates. J. Antimicrob. Chemother. 2006;57:461–465. doi: 10.1093/jac/dki471. [DOI] [PubMed] [Google Scholar]
- 10.Baptiste K.E., Williams K., Willams N.J., Wattret A., Clegg P.D., Dawson S., Corkill J.E., O’Neill T., Hart C.A. Methicillin-resistant staphylococci in companion animals. Emerg. Infect. Dis. 2005;11:1942–1944. doi: 10.3201/eid1112.050241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geneva: World Health Organization Antimicrobial Resistance: Global Report on Surveillance 2014. [(accessed on 30 November 2015)]. Available online: http://www.who.int/drugresistance/documents/surveillancereport/en/
- 12.Kimang’a A.N. A situational analysis of antimicrobial drug resistance in Africa: Are we losing the battle? Ethiop. J. Health Sci. 2012;22:135–143. [PMC free article] [PubMed] [Google Scholar]
- 13.BoscoNdihokubwayo J., Ahmed Yahaya A., Tamer Desta A., Ki-Zerbo G., AssamoahOdei E., Keita B., PanaAssimawe P., Nkhoma W. Antimicrobial Resistance in the African Region: Issues, Challenges and Actions Proposed. [(accessed on 19 August 2015)]. Available online: http://www.afro.who.int/en/clusters-a-programmes/hss/blood-safety-laboratories-a-health-technology/blt-highlights/3861-antimicrobial-resistance-in-the-african-region-issues-challenges-and-actions-proposed.html.
- 14.Falagas M.E., Karageorgopoulos D.E., Leptidis J., Korbila I.P. MRSA in Africa: Filling the global map of antimicrobial resistance. PLoS ONE. 2013;8:12. doi: 10.1371/journal.pone.0068024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaumburg F., Alabi A.S., Peters G., Becker K. New epidemiology of Staphylococcus aureus infection in Africa. Clin. Microbiol. Infect. 2014;20:589–596. doi: 10.1111/1469-0691.12690. [DOI] [PubMed] [Google Scholar]
- 16.Abdulgader S.M., Shittu A.O., Nicol M.P., Kaba M. Molecular epidemiology of Methicillin-resistant Staphylococcus aureus in Africa: A systematic review. Front. Microbiol. 2015 doi: 10.3389/fmicb.2015.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaumburg F., Pauly M., Anoh E., Mossoun A., Wiersma L., Schubert G., Flammen A., Alabi A.S., Muyembe-Tamfum J.J., Grobusch M.P., et al. Staphylococcus aureus complex from animals and humans in three remote African regions. Clin. Microbiol. Infect. 2015;21:e1–e8. doi: 10.1016/j.cmi.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Schaumburg F., Mugisha L., Kappeller P., Fichtel C., Köck R., Köndgen S., Becker K., Boesch C., Peters G., Leendertz F. Evaluation of non-invasive biological samples to monitor Staphylococcus aureus colonization in great apes and lemurs. PLoS ONE. 2013;8:12. doi: 10.1371/annotation/c2148f4d-866d-479a-b0e6-97aa6ab931f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaumburg F., Alabi A.S., Köck R., Mellmann A., Kremsner P.G., Boesch C., Becker K., Leendertz F.H., Peters G. Highly divergent Staphylococcus aureus isolates from African non-human primates. Environ. Microbiol. Rep. 2012;4:141–146. doi: 10.1111/j.1758-2229.2011.00316.x. [DOI] [PubMed] [Google Scholar]
- 20.Abdel-moein K.A., El-Hariri M., Samir A. Methicillin-resistant Staphylococcus aureus: An emerging pathogen of pets in Egypt with a public health burden. Transbound. Emerg. Dis. 2012;59:331–335. doi: 10.1111/j.1865-1682.2011.01273.x. [DOI] [PubMed] [Google Scholar]
- 21.El-Jakee J., Nagwa A.S., Bakry M., Zouelfakar S.A., Elgabry E., Gad El-Said W.A. Characteristics of Staphylococcus aureus Strains Isolated from Human and Animal Sources. Am. Eur. J. Agric. Environ. Sci. 2008;4:221–229. [Google Scholar]
- 22.Nagel M., Dischinger J., Türck M., Verrier D., Oedenkoven M., Ngoubangoye B., Le Flohic G., Drexler J.F., Bierbaum G., Gonzalez J.P. Human-associated Staphylococcus aureus strains within great ape populations in Central Africa (Gabon) Clin. Microbiol. Infect. 2013;19:1072–1077. doi: 10.1111/1469-0691.12119. [DOI] [PubMed] [Google Scholar]
- 23.Adesiyun A.A., Lenz W., Schaal K.P. Production of toxic shock syndrome toxin-1 (TSST-1) by Staphylococcus aureus strains isolated from humans, animals and foods in Nigeria. Microbiologica. 1992;15:125–133. [PubMed] [Google Scholar]
- 24.Akobi B., Aboderin O., Sasaki T., Shittu A. Characterization of Staphylococcus aureus isolates from faecal samples of the Straw-Coloured Fruit Bat (Eidolon helvum) in Obafemi Awolowo University (OAU), Nigeria. BMC Microbiol. 2012 doi: 10.1186/1471-2180-12-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mai-siyama I.B., Okon K.O., Adamu N.B., Askira U.M., Isyaka T.M., Adamu S.G., Mohammed A. Methicllin-resistant Staphylococcus aureus (MRSA) colonization rate among ruminant animals slaughtered for human consumption and contact persons in Maiduguri, Nigeria. Afr.J. Microbiol. Res. 2014;8:2643–2649. doi: 10.5897/AJMR2014.6855. [DOI] [Google Scholar]
- 26.Otalu O., Junaidu K., Chukwudi O.E., Jarlath U.V. Multi-drug resistant coagulase positive Staphylococcus aureus from live and slaughtered chickens in Zaria, Nigeria. Int. J. Poult. Sci. 2011;10:871–875. doi: 10.3923/ijps.2011.871.875. [DOI] [Google Scholar]
- 27.Fall C., Seck A., Richard V., Ndour M., Sembene M., Laurent F., Breurec S. Epidemiology of Staphylococcus aureus in pigs and farmers in the largest farm in Dakar, Senegal. Foodborne Pathog. Dis. 2012;9:962–965. doi: 10.1089/fpd.2012.1197. [DOI] [PubMed] [Google Scholar]
- 28.Adegoke A.A., Okoh A.I. Species diversity and antibiotic resistance properties of Staphylococcus of farm animal origin in Nkonkobe Municipality, South Africa. Folia Microbiol. Praha. 2014;59:133–140. doi: 10.1007/s12223-013-0275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaumburg F., Mugisha L., Peck B., Becker K., Gillespie T.R., Peters G., Leendertz F.H. Drug-resistant human Staphylococcus aureus in sanctuary apes pose a threat to endangered wild ape populations. Am. J. Primatol. 2012;74:1071–1075. doi: 10.1002/ajp.22067. [DOI] [PubMed] [Google Scholar]
- 30.Tanih N.F., Sekwadi E., Ndip R.N., Bessong P.O. Detection of pathogenic Escherichia coli and Staphylococcus aureus from cattle and pigs slaughtered in abattoirs in Vhembe District, South Africa. Sci. World J. 2015 doi: 10.1155/2015/195972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elbir H., Feil E.J., Drancourt M., Roux V., El Sanousi S.M., Eshag M., Colque-Navarro P., Kühn I., Flock J.I. Ovine clone ST1464: A predominant genotype of Staphylococcus aureus subsp. anaerobius isolated from sheep in Sudan. J. Infect. Dev. Ctries. 2010;4:235–238. doi: 10.3855/jidc.632. [DOI] [PubMed] [Google Scholar]
- 32.Musa N.O., Babiker A., Eltom K., Rodwan K., El Sanousi S.M. Prevalence of Staphylococcus aureus subsp. anaerobius in Sub-Clinical Abscess Cases of Sheep. Br. Microbiol. Res. J. 2012;2:131–136. [Google Scholar]
- 33.Omer M.M., Abusalab S., Gumaa M.M., Mulla S.A., Osman H.M., Sabiel Y.A., Ahmed A.M. Staphylococcus aureus isolated from a horse in a sudden death condition in Kassala state, eastern Sudan. Pak. J. Biol. Sci. 2008;11:2028–2031. doi: 10.3923/pjbs.2008.2028.2031. [DOI] [PubMed] [Google Scholar]
- 34.Gharsa H., Ben Slama K., Lozano C., Gómez-Sanz E., Klibi N., Ben Sallem R., Gómez P., Zarazaga M., Boudabous A., Torres C. Prevalence, antibiotic resistance, virulence traits and genetic lineages of Staphylococcus aureus in healthy sheep in Tunisia. Vet. Microbiol. 2012;156:367–373. doi: 10.1016/j.vetmic.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Gharsa H., Ben Sallem R., Ben Slama K., Gómez-Sanz E., Lozano C., Jouini A., Klibi N., Zarazaga M., Boudabous A., Torres C. High diversity of genetic lineages and virulence genes in nasal Staphylococcus aureus isolates from donkeys destined to food consumption in Tunisia with predominance of the ruminant associated CC133 lineage. BMC Vet. Res. 2012 doi: 10.1186/1746-6148-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gharsa H., Ben Slama K., Gómez-Sanz E., Lozano C., Zarazaga M., Messadi L., Boudabous A., Torres C. Molecular characterization of Staphylococcus aureus from nasal samples of healthy farm animals and pets in Tunisia. Vector Borne Zoonotic Dis. 2015;15:109–115. doi: 10.1089/vbz.2014.1655. [DOI] [PubMed] [Google Scholar]
- 37.Pandey G.S., Nomura Y., Kobayashi K., Fujise H., Yamada T. Cutaneous staphylococcal granuloma in a free living zebra (Equus burchelli) in Zambia. J. Vet. Med. Sci. 1998;60:137–138. doi: 10.1292/jvms.60.137. [DOI] [PubMed] [Google Scholar]
- 38.Youn J.H., Park Y.H., Hang’ombe B., Sugimoto C. Prevalence and characterization of Staphylococcus aureus and Staphylococcus pseudintermedius isolated from companion animals and environment in the veterinary teaching hospital in Zambia, Africa. Comp. Immunol. Microbiol. Infect. Dis. 2014;37:123–130. doi: 10.1016/j.cimid.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Guinane C.M., Ben Zakour N.L., Tormo-Mas M.A., Weinert L.A., Lowder B.V., Cartwright R.A., Smyth D.S., Smyth C.J., Lindsay J.A., Gould K.A., et al. Evolutionary genomics of S. aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol. Evol. 2010;2:454–466. doi: 10.1093/gbe/evq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.García-Álvarez L., Holden M.T., Lindsay H., Webb C.R., Brown D.F., Curran M.D., Walpole E., Brooks K., Pickard D.J., Teale C., et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loeffler A., Lloyd D.H. Companion animals: A reservoir for methicillin-resistant Staphylococcus aureus in the community? Epidemiol. Infect. 2010;138:595–605. doi: 10.1017/S0950268809991476. [DOI] [PubMed] [Google Scholar]
- 42.Crombé F., Argudín M.A., Vanderhaeghen W., Hermans K., Haesebrouck F., Butaye P. Transmission dynamics of Methicillin-Resistant Staphylococcus aureus in pigs. Front. Microbiol. 2013 doi: 10.3389/fmicb.2013.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persoons D., van Hoorebeke S., Hermans K., Butaye P., de Kruif A., Haesebrouck F., Dewulf J. Methicillin-resistant Staphylococcus aureus in poultry. Emerg. Infect. Dis. 2009;15:452–453. doi: 10.3201/eid1503.080696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huber H., Koller S., Giezendanner N., Stephan R., Zweifel C. Prevalence and characteristics of meticillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. [(accessed on 30 November 2015)];Euro Surveill. 2010 15:19542. Available online: http://www.eurosurveillance.org/images/dynamic/ee/v15n16/art19542.pdf. [PubMed] [Google Scholar]
- 45.Elhani D., Gharsa H., Kalai D., Lozano C., Gomez P., Boutheina J., Aouni M., Barguellil F., Torres C., Ben Slama K. Clonal lineages detected among tetracycline resistant MRSA isolates of a Tunisian Hospital, with detection of lineage ST398. J. Med. Microbiol. 2015 doi: 10.1099/jmm.0.000066. [DOI] [PubMed] [Google Scholar]
- 46.Chairat S., Gharsa H., Lozano C., Gómez-Sanz E., Gómez P., Zarazaga M., Boudabous A., Torres C., Ben Slama K. Characterization of Staphylococcus aureus from raw meat samples in tunisia: Detection of clonal lineage ST398 from the African Continent. Foodborne Pathog. Dis. 2015;12:686–692. doi: 10.1089/fpd.2015.1958. [DOI] [PubMed] [Google Scholar]
- 47.Paterson G.K., Harrison E.M., Holmes M.A. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014;22:42–47. doi: 10.1016/j.tim.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paterson G.K., Morgan F.J., Harrison E.M., Cartwright E.J., Török M.E., Zadoks R.N., Parkhill J., Peacock S.J., Holmes M.A. Prevalence and characterisation of human mecC methicillin-resistant S. aureus isolates in England. J. Antimicrob. Chemother. 2014;69:907–910. doi: 10.1093/jac/dkt462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rabello R.F., Moreira B.M., Lopes R.M.M., Teixeira L.M., Riley L.W., Castro A.C.D. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. J. Med. Microbiol. 2007;56:1505–1511. doi: 10.1099/jmm.0.47357-0. [DOI] [PubMed] [Google Scholar]
- 50.Hasman H., Moodley A., Guardabassi L., Stegger M., Skov R.L., Aarestrup F.M. spa type distribution in Staphylococcus aureus originating from pigs, cattle and poultry. Vet. Microbiol. 2010;141:326–331. doi: 10.1016/j.vetmic.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 51.Porrero M.C., Hasman H., Vela A.I., Fernández-Garayzábal J.F., Domínguez L., Aarestrup F.M. Clonal diversity of Staphylococcus aureus originating from the small ruminants goats and sheep. Vet. Microbiol. 2012;156:157–161. doi: 10.1016/j.vetmic.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 52.Lowder B.V., Guinane C.M., Ben Zakour N.L., Weinert L.A., Conway-Morris A., Cartwright R.A., Simpson A.J., Rambaut A., Nübel U., Fitzgerald J.R. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA. 2009;106:19545–19550. doi: 10.1073/pnas.0909285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smyth D.S., Feil E.J., Meaney W.J., Hartigan P.J., Tollersrud T., Fitzgerald J.R., Enright M.C., Smyth C.J. Molecular genetic typing reveals further insights into the diversity of animal-associated Staphylococcus aureus. J. Med. Microbiol. 2009;58:1343–1353. doi: 10.1099/jmm.0.009837-0. [DOI] [PubMed] [Google Scholar]
- 54.Franco A., Hasman H., Iurescia M., Lorenzetti R., Stegger M., Pantosti A., Feltrin F., Ianzano A., Porrero M.C., Liapi M., et al. Molecular characterization of spa type t127, sequence type 1 methicillin-resistant Staphylococcus aureus from pigs. J. Antimicrob. Chemother. 2011;66:1231–1235. doi: 10.1093/jac/dkr115. [DOI] [PubMed] [Google Scholar]
- 55.Cui S., Li J., Hu C., Jin S., Li F., Guo Y., Ran L., Ma Y. Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J. Antimicrob. Chemother. 2009;64:680–683. doi: 10.1093/jac/dkp275. [DOI] [PubMed] [Google Scholar]
- 56.Vestergaard M., Cavaco L.M., Sirichote P., Unahalekhaka A., Dangsakul W., Svendsen C.A., Aarestrup F.M., Hendriksen R.S. SCCmec Type IX element in methicillin resistant Staphylococcus aureus spa type t337 (CC9) isolated from pigs and pork in Thailand. Front. Microbiol. 2012;3:1–4. doi: 10.3389/fmicb.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pomba C., Hasman H., Cavaco L.M., da Fonseca J.D., Aarestrup F.M. First description of meticillin-resistant Staphylococcus aureus (MRSA) CC30 and CC398 from swine in Portugal. Int. J. Antimicrob. Agents. 2009;34:193–194. doi: 10.1016/j.ijantimicag.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 58.Vancraeynest D., Haesebrouck F., Deplano A., Denis O., Godard C., Wildemauwe C., Hermans K. International dissemination of a high virulence rabbit Staphylococcus aureus clone. J. Vet. Med. B Infect. Dis. Vet. Public Health. 2006;53:418–422. doi: 10.1111/j.1439-0450.2006.00977.x. [DOI] [PubMed] [Google Scholar]
- 59.Walther B., Monecke S., Ruscher C., Friedrich A.W., Ehricht R., Slickers P., Soba A., Wleklinski C.G., Wieler L.H., Lübke-Becker A. Comparative molecular analysis substantiates zoonotic potential of equine methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2009;47:704–710. doi: 10.1128/JCM.01626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gómez-Sanz E., Torres C., Lozano C., Fernández-Pérez R., Aspiroz C., Ruiz-Larrea F., Zarazaga M. Detection, molecular characterization, and clonal diversity of methicillin-resistant Staphylococcus aureus CC398 and CC97 in Spanish slaughter pigs of different age groups. Foodborne Pathog. Dis. 2010;7:1269–1277. doi: 10.1089/fpd.2010.0610. [DOI] [PubMed] [Google Scholar]
- 61.Sakwinska O., Giddey M., Moreillon M., Morisset D., Waldvogel A., Moreillon P. Staphylococcus aureus host range and human-bovine host shift. Appl. Environ. Microbiol. 2011;77:5908–5915. doi: 10.1128/AEM.00238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ben Slama K., Gharsa H., Klibi N., Jouini A., Lozano C., Gómez-Sanz E., Zarazaga M., Boudabous A., Torres C. Nasal carriage of Staphylococcus aureus in healthy humans with different levels of contact with animals in Tunisia: genetic lineages, methicillin resistance, and virulence factors. Eur. J. Clin. Microbiol. Infect. Dis. 2011;30:499–508. doi: 10.1007/s10096-010-1109-6. [DOI] [PubMed] [Google Scholar]
- 63.Abou E.A. Bacteriological quality of ready to eat meals. J. Egypt Public Health Assoc. 1995;70:627–641. [PubMed] [Google Scholar]
- 64.Geornaras I., de Jesus A., van Zyl E., von Holy A. Microbiological survey of a South African poultry processing plant. J. Basic Microbiol. 1995;35:73–82. doi: 10.1002/jobm.3620350204. [DOI] [PubMed] [Google Scholar]
- 65.Abd El Aziz T. Screening of some food poisoning bacteria in sausage and hamburger meat. J. Egypt Public Health Assoc. 1996;71:47–61. [PubMed] [Google Scholar]
- 66.Karama M., de Jesus A.E., Veary C.M. Microbial quality of ostrich carcasses produced at an export-approved South African abattoir. J. Food Prot. 2003;66:878–881. doi: 10.4315/0362-028x-66.5.878. [DOI] [PubMed] [Google Scholar]
- 67.Nel S., Lues J.F., Buys E.M., Venter P. Bacterial populations associated with meat from the deboning room of a high throughput red meat abattoir. Meat Sci. 2004;66:667–674. doi: 10.1016/S0309-1740(03)00187-6. [DOI] [PubMed] [Google Scholar]
- 68.Elhag N.B., Babiker E.R.B., Mahdi A.A. Microbial profile of sausages in Khartoum State. J. Agri. Food Appl. Sci. 2014;2:206–219. [Google Scholar]
- 69.Kivaria F.M., Noordhuizen J.P., Nielen M. Interpretation of California mastitis test scores using Staphylococcus aureus culture results for screening of subclinical mastitis in low yielding smallholder dairy cows in the Dar es Salaam region of Tanzania. Prev. Vet. Med. 2007;78:274–285. doi: 10.1016/j.prevetmed.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 70.Almaw G., Zerihun A., Asfaw Y. Bovine mastitis and its association with selected risk factors in smallholder dairy farms in and around Bahir Dar, Ethiopia. Trop. Anim. Health Prod. 2008;40:427–432. doi: 10.1007/s11250-007-9115-0. [DOI] [PubMed] [Google Scholar]
- 71.Abera M., Demie B., Aragaw K., Regassa F.G., Regassa A. Isolation and identification of Staphylococcus aureus from bovine mastitic milk and their drug resistance patterns in Adama town, Ethiopia. J. Vet. Med. Anim. Health. 2010;2:29–34. [Google Scholar]
- 72.Mengistu F., Molla B., Ali A.I. Camel mastitis, associated bacterial pathogens and its impact on milk quality in Gewane district, afar regional state, Northeastern Ethiopia. Bull. Anim. Health Prod. Afr. 2010;58:241–247. [Google Scholar]
- 73.Kadja M.C., Kpodekon M., Kane Y., Tchassou K., Kaboret Y., Mainil J., Taminiau B. Typing of Staphylococcus aureus strains isolated from milk cows with subclinical mastitis in Dakar, Senegal. Bull. Anim. Health Prod. Afr. 2010;58:201–209. doi: 10.4314/bahpa.v58i3.64207. [DOI] [Google Scholar]
- 74.Haftu R., Taddele H., Gugsa G., Kalayou S. Prevalence, bacterial causes, and antimicrobial susceptibility profile of mastitis isolates from cows in large-scale dairy farms of Northern Ethiopia. Trop Anim Health Prod. 2012;44:1765–1771. doi: 10.1007/s11250-012-0135-z. [DOI] [PubMed] [Google Scholar]
- 75.Alemu G., Almaw G., Abera M. Incidence rate of Staphylococcus aureus and Streptococcus agalactiae in subclinical mastitis at smallholder dairy cattle farms in Hawassa, Ethiopia. Afr. J. Microbiol. Res. 2014;8:252–256. [Google Scholar]
- 76.Ibrahim A.I., Duprez J., Bada-Alambedji R., Moula N., Mainil J.G., Bardiau M. Antibiotic resistance trend of Staphylococcus aureus isolated between 2010 and 2012 from mastitis cases in Azawak zebu in Niger. Afr. J. Microbiol. Res. 2014;35:3271–3275. [Google Scholar]
- 77.Iceland Kasozi K., BoscoTingiira J., Vudriko P. High prevalence of subclinical mastitis and multidrug resistant Staphylococcus aureus are a threat to dairy cattle production in Kiboga District (Uganda) Open J. Vet. Med. 2014;4:35–43. doi: 10.4236/ojvm.2014.44005. [DOI] [Google Scholar]
- 78.Schaumburg F., Alabi A.S., Frielinghaus L., Grobusch M.P., Köck R., Becker K., Issifou S., Kremsner P.G., Peters G., Mellmann A. The risk to import ESBL-producing Enterobacteriaceae and Staphylococcus aureus through chicken meat trade in Gabon. BMC Microbiol. 2014 doi: 10.1186/s12866-014-0286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yannick N., Rawlings N., Emmanuela A. Assessment of bacteriological quality of cooked pork meat sold along the commercial streets of Nkwen through Bambili Metropolis, Cameroon. Afr. J. Food Sci. 2013;7:441–445. doi: 10.5897/AJFS2013.1108. [DOI] [Google Scholar]
- 80.Attien P.S., Moussaoui W., Dadié T., Chabi Sika K., Djéni T., Bankole H.S., Kotchoni S.O., Edoh V., Prévost G., Djè M., et al. Prevalence and antibiotic resistance of Staphylococcus strains isolated from meat products sold in Abidjan streets (Ivory Coast) Afr. J. Microbiol. Res. 2013;7:3285–3293. [Google Scholar]
- 81.Mathieu A., Isigidi B.K., Devriese L.A., Godard C., Vanhoof R. Characterization of Staphylococcus aureus and Salmonella species strains isolated from bovine meat in Zaire. Int. J. Food Microbiol. 1991;14:119–126. doi: 10.1016/0168-1605(91)90098-A. [DOI] [PubMed] [Google Scholar]
- 82.Ezzeldeen N.A., Mansour H.A., Ahmed A.A. Phenotypic and molecular identification of Staphylococcus aureus isolated from some egyptian salted fish. World Appl. Sci. J. 2011;15:1703–1712. [Google Scholar]
- 83.Gwida M.M., EL-Gohary F.A. Zoonotic bacterial pathogens isolated from raw milk with special reference to Escherichia coli and Staphylococcus aureus in Dakahlia Governorate, Egypt. Open Access Sci. Rep. 2013 doi: 10.4172/scientificreports705. [DOI] [Google Scholar]
- 84.Khalifa N.O., Elhofy F.I., Fahmy H.A., Barakat A.M.A. Epidemiological and genetic studies of enterotoxigenic Staphylococcus aureus isolated from goat and human. Am. J. Infect. Dis. Microbiol. 2015;3:32–37. [Google Scholar]
- 85.Daka D., Silassie S., Yihdego D. Antibiotic-resistance Staphylococcus aureus isolated from cow’s milk in the Hawassa area, South Ethiopia. Ann. Clin. Microbiol. Antimicrob. 2012 doi: 10.1186/1476-0711-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haileselassie M., Taddele H., Adhana K., Kalayou S. Food safety knowledge and practices of abattoir and butchery shops and the microbial profile of meat in Mekelle city, Ethiopia. Asian Pac. J. Trop. Biomed. 2013;3:407–412. doi: 10.1016/S2221-1691(13)60085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Njage P.M.K., Dolci S., Jans C., Wangoh J., Lacroix C., Meile L. Phenotypic and genotypic antibiotic resistance patterns of Staphylococcus aureus from raw and spontaneously fermented camel milk. Eur. J. Food Res. Rev. 2013;3:87–98. doi: 10.9734/EJNFS/2013/2853. [DOI] [Google Scholar]
- 88.Ombui J.N., Kimotho A.M., Nduhiu J.G. Antimicrobial resistance patterns and plasmid profiles of Staphylococcus aureus isolated from milk and meat. East Afr. Med. J. 2000;77:463–467. doi: 10.4314/eamj.v77i9.46688. [DOI] [PubMed] [Google Scholar]
- 89.Shitandi A., Mwangi M. Occurrence of miltiple antimicrobial resistance among Staphylococcus aureus isolates from Kenyan milk. J. Food Technol. Afr. 2004;9:23–25. [Google Scholar]
- 90.Seeiso T.M., McCrindle C.M. An investigation of the quality of meat sold in Lesotho. J.S. Afr. Vet. Assoc. 2009;80:237–242. doi: 10.4102/jsava.v80i4.215. [DOI] [PubMed] [Google Scholar]
- 91.El Shrek Y.M., Ali M.R. Microbiological study of spiced chicken burgers in Tripoli City, Libya. East Mediterr. Health J. 2012;18:653–662. doi: 10.26719/2012.18.6.653. [DOI] [PubMed] [Google Scholar]
- 92.Taulo S., Wetlesen A., Abrahamsen R., Kululanga G., Mkakosya R., Grimason A. Microbiological hazard identification and exposure assessment of food prepared and served in rural households of Lungwena, Malawi. Int. J. Food Microbiol. 2008;125:111–116. doi: 10.1016/j.ijfoodmicro.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 93.Abdellah E.A., Fouzia R.F., Bouchra O. Prevalence and antibiogram study of Escherichia coli and Staphylococcus aureus in turkey meat in Morocco. Pharm. Anal. Acta. 2013 doi: 10.4172/2153-2435.1000270. [DOI] [Google Scholar]
- 94.Bendahou A., Lebbadi M., Ennanei L., Essadqui F.Z., Abid M. Characterization of Staphylococcus species isolated from raw milk and milk products (lben and jben) in North Morocco. J. Infect. Dev. Ctries. 2008;2:218–225. doi: 10.3855/jidc.266. [DOI] [PubMed] [Google Scholar]
- 95.Cohen N., Ennaji H., Hassar M., Karib H. The bacterial quality of red meat and offal in Casablanca (Morocco) Mol. Nutr. Food Res. 2006;50:557–562. doi: 10.1002/mnfr.200500180. [DOI] [PubMed] [Google Scholar]
- 96.Beukes E.M., Bester B.H., Mostert J.F. The microbiology of South African traditional fermented milks. Int. J. Food Microbiol. 2001;63:189–197. doi: 10.1016/S0168-1605(00)00417-7. [DOI] [PubMed] [Google Scholar]
- 97.Isara A.R., Isah E.C., Lofor P.V., Ojide C.K. Food contamination in fast food restaurants in Benin City, Edo State, Nigeria: implications for food hygiene and safety. Public Health. 2010;124:467–471. doi: 10.1016/j.puhe.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 98.Ndahi M.D., Kwaga J.K., Bello M., Kabir J., Umoh V.J., Yakubu S.E., Nok A.J. Prevalence and antimicrobial susceptibility of Listeria monocytogenes and methicillin-resistant Staphylococcus aureus strains from raw meat and meat products in Zaria, Nigeria. Lett. Appl. Microbiol. 2014;58:262–269. doi: 10.1111/lam.12183. [DOI] [PubMed] [Google Scholar]
- 99.Sokari T. Distribution of enterotoxigenic Staphylococcus aureus in ready-to-eat foods in eastern Nigeria. Int. J. Food Microbiol. 1991;12:275–279. doi: 10.1016/0168-1605(91)90079-5. [DOI] [PubMed] [Google Scholar]
- 100.Ateba C.N., Mbewe M., Moneoang M.S., Bezuidenhout C.C. Antibiotic-resistant Staphylococcus aureus isolated from milk in the Mafikeng Area, North West province, South Africa. S. Afr. J. Sci. 2010;106:1–6. doi: 10.4102/sajs.v106i11/12.243. [DOI] [Google Scholar]
- 101.Mosupye F.M., von Holy A. Microbiological hazard identification and exposure assessment of street food vending in Johannesburg, South Africa. Int. J. Food Microbiol. 2000;61:137–145. doi: 10.1016/S0168-1605(00)00264-6. [DOI] [PubMed] [Google Scholar]
- 102.Shuiep E.S., Kanbar T., Eissa N., Alber J., Lämmler C., Zschöck M., El Zubeir I.E., Weiss R. Phenotypic and genotypic characterization of Staphylococcus aureus isolated from raw camel milk samples. Res. Vet. Sci. 2009;86:211–215. doi: 10.1016/j.rvsc.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 103.Yagoub S.O., Awadalla N.E., El Zubeir I.E.M. Incidence of some potential pathogens in raw milk in Khartoum North (Sudan) and their susceptinility to antimicrobial agents. J. Anim. Vet. Adv. 2005;4:341–344. [Google Scholar]
- 104.Kivaria F.M., Noordhuizen J.P., Kapaga A.M. Evaluation of the hygienic quality and associated public health hazards of raw milk marketed by smallholder dairy producers in the Dar es Salaam region, Tanzania. Trop. Anim. Health Prod. 2006;38:185–194. doi: 10.1007/s11250-006-4339-y. [DOI] [PubMed] [Google Scholar]
- 105.Higenyi J., Kabasa J.D. Microbial contamination load of hatching eggs in Butaleja, eastern Uganda. Anim. Vet. Sci. 2014;2:22–30. doi: 10.11648/j.avs.20140202.12. [DOI] [Google Scholar]
- 106.Mhone T.A., Matope G., Saidi P.T. Aerobic bacterial, coliform, Escherichia coli and Staphylococcus aureus counts of raw and processed milk from selected smallholder dairy farms of Zimbabwe. Int. J. Food Microbiol. 2011;151:223–228. doi: 10.1016/j.ijfoodmicro.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 107.Lozano C., López M., Gómez-Sanz E., Ruiz-Larrea F., Torres C., Zarazaga M. Detection of methicillin-resistant Staphylococcus aureus ST398 in food samples of animal origin in Spain. J. Antimicrob. Chemother. 2009;64:1325–1326. doi: 10.1093/jac/dkp378. [DOI] [PubMed] [Google Scholar]
- 108.Benito D., Gómez P., Lozano C., Estepa V., Gómez-Sanz E., Zarazaga M., Torres C. Genetic lineages, antimicrobial resistance, and virulence in Staphylococcus aureus of meat samples in Spain: Analysis of immune evasion cluster (IEC) genes. Foodborne Pathog. Dis. 2014;11:354–356. doi: 10.1089/fpd.2013.1689. [DOI] [PubMed] [Google Scholar]
- 109.Stegger M., Wirth T., Andersen P.S., Skov R.L., de Grassi A., Simões P.M., Tristan A., Petersen A., Aziz M., Kiil K., et al. Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus. MBio. 2014 doi: 10.1128/mBio.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]