Abstract
BECN1 (Beclin 1), a highly conserved eukaryotic protein, is a key regulator of autophagy, a cellular homeostasis pathway, and also participates in vacuolar protein sorting, endocytic trafficking, and apoptosis. BECN1 is important for embryonic development, the innate immune response, tumor suppression, and protection against neurodegenerative disorders, diabetes, and heart disease. BECN1 mediates autophagy as a core component of the class III phosphatidylinositol 3‐kinase complexes. However, the exact mechanism by which it regulates the activity of these complexes, or mediates its other diverse functions is unclear. BECN1 interacts with several diverse protein partners, perhaps serving as a scaffold or interaction hub for autophagy. Based on extensive structural, biophysical and bioinformatics analyses, BECN1 consists of an intrinsically disordered region (IDR), which includes a BH3 homology domain (BH3D); a flexible helical domain (FHD); a coiled‐coil domain (CCD); and a β‐α‐repeated autophagy‐specific domain (BARAD). Each of these BECN1 domains mediates multiple diverse interactions that involve concomitant conformational changes. Thus, BECN1 conformational flexibility likely plays a key role in facilitating diverse protein interactions. Further, BECN1 conformation and interactions are also modulated by numerous post‐translational modifications. A better structure‐based understanding of the interplay between different BECN1 conformational and binding states, and the impact of post‐translational modifications will be essential to elucidating the mechanism of its multiple biological roles.
Keywords: autophagy, BECN1/Beclin 1/ATG6/VPS30, conformational flexibility, intrinsically disordered protein, interaction hub, class III phosphatidylinositol 3‐kinase complexes
Abbreviations
- αMoRF
α‐molecular recognition features
- AMBRA1
autophagy/BECN1 regulator 1
- AMPK
AMP‐associated protein kinase
- ATG6
autophagy‐related protein 6
- BARAD
β‐α repeated autophagy‐specific domain
- BARKOR
Beclin 1‐associated autophagy‐related key regulator
- BATS
BARKOR/ATG14 autophagosome targeting sequence
- BCL2BD
BCL2‐binding domain
- BECN1
BCL2 interacting coiled‐coil protein or Beclin 1
- BH3D
BCL2 homology 3 domain
- BSA
buried surface area
- CC1
coiled‐coil 1
- CCD
coiled‐coil domain
- CD
circular dichroism
- CoIP
co‐immunoprecipitation
- CRM1
chromosomal maintenance protein 1
- Cyro‐EM
cryo‐electron microscopy
- ECD
evolutionarily conserved domain
- ELMs
eukaryotic linear motifs
- FHD
flexible helical domain
- GAPR1
Golgi‐associated plant pathogenesis‐related protein 1
- HCMV
human cytomegalovirus
- HDX‐MS
hydrogen‐deuterium exchange mass spectrometry
- HEAT
Huntington, elongation factor 3 protein phosphatase 2A, yeast kinase TOR1
- HV
herpesvirus
- HIV
human immunodeficiency virus
- HSV‐1
herpes simplex virus type 1
- IDR
intrinsically disordered region
- IRS1
internal repeat sequence 1
- ITC
isothermal titration calorimetry
- KSHV
Kaposi's sarcoma associated γHV
- LAMP1
lysosome associated membrane protein
- LC3
light chain 3
- NES
nuclear export signal
- NMR
nuclear magnetic resonance
- nPIST
neuronal isoform of protein‐interaction, specifically with TC10
- PI
phosphatidylinositol
- PI3KC3
class III phosphatidylinositol 3‐kinase
- PI3KR4
PI3K Ser/Thr kinase regulatory protein or p150
- PI3P
phosphatidylinositol 3‐phosphate
- PR
proline‐rich
- PTM
post‐translational modification
- RAB7
Ras‐associatd protein Rab‐7a
- SAXS
small‐angle X‐ray scattering
- SIRT1
sirtuin 1
- SLAMF1
signaling lymphocyte‐activation molecule family 1
- TAB2/3
TAK1‐binding proteins 2/3
- TAK1‐IKK
TAK1 protein kinase‐Iκk kinase
- TFE
2,2,2‐trifluoroethanol
- TRS1
terminal repeat sequence 1
- UVRAG
UV radiation resistance associated gene
- VMP1
vacuole membrane protein 1
- VPS30
vacuolar protein sorting protein 30
- WASH
Wiskott‐Aldrich syndrome protein and SCAR homolog.
Statement of Importance
BECN1/Beclin 1 is a highly conserved eukaryotic protein essential for autophagy, a cellular homeostasis pathway. BECN1 is important for embryonic development, innate immune responses, tumor suppression, and protection against neurodegenerative disorders, diabetes, and heart disease. BECN1 likely serves as an interaction hub or scaffold that targets proteins to specific membranes for autophagosome formation. Here we elucidate the BECN1 domain architecture, highlighting conformations observed in different interaction states that enable diverse signals to modulate autophagy.
Introduction
BECN homologs are highly conserved in all eukaryotes. BECN1 is also called Beclin 1 (BCL2‐interacting coiled‐coil protein) in mammals,1 BEC‐1 in worms and insects, Beclin 1 or ATG6 (autophagy‐related protein 6) in plants, and ATG6 or VPS30 (vacuolar protein sorting protein 30) in yeast. While most organisms have a single BECN gene, a second paralog, BECN2 or Beclin 2, has been recently identified in mammals.2
BECN1 was the first mammalian protein shown to be involved in macroautophagy3 (hereafter called autophagy), a cellular pathway first identified in yeast.4 Autophagy is a catabolic homeostasis process wherein cytoplasmic contents including damaged organelles, long‐lived or aggregated proteins, and pathogens, are surrounded by multilayered vesicles called autophagosomes, which fuse with lysosomes enabling degradation of the sequestered contents.5, 6, 7, 8, 9 Thus, autophagy is a survival mechanism that enables cells to withstand nutrient deprivation, environmental stressors, aging, and infection by recycling intracellular debris to generate metabolic precursors, such as amino acids and ATP. However, excessive or uncontrolled autophagy can cause cell death, often called autophagic cell death or autosis.10
BECN1, an essential gene required for embryonic survival and normal development, plays roles in phagocytosis and clearance of apoptotic cells during embryonic development.11, 12 Various diseases are associated with BECN1 mutations or altered expression levels. BECN1 is an important tumor suppressor in many cancers3, 13, 14, 15, 16 and is monoallelically deleted in 40%–75% of human breast, ovary, and prostate cancers.13 BECN1 deficiency and malfunction contributes to neurodegenerative disorders including Huntington's,17 Alzheimer's,18 Parkinson's, and Lewy body19 disease. Brain injury upregulates BECN1, suggesting increased autophagy is neuroprotective.20 Therapeutic BECN1 overexpression clears mutant ataxin‐3 to alleviate Machado‐Joseph disease.21 Type‐2 diabetes increases BECN1 activation and autophagy in human heart, promoting progressive loss of cardiac cells.22, 23 Ischemia triggers autophagy in hibernating myocardium, possibly to clear damaged organelles and unfolded proteins to support survival.24 However, during reperfusion BECN1:BCL2 interaction is increased and autophagy is attenuated, protecting the recovering myocardium from autophagic cell death.25, 26 Some viruses target BECN1 to evade autophagic degradation, including the human immunodeficiency virus (HIV),27 influenza A virus,28 African swine fever virus,29 foot and mouth disease virus30 α‐herpesvirus (αHV) herpes simplex virus type 1 (HSV‐1),31 βHV human cytomegalovirus (HCMV),32 and γHVs Kaposi's sarcoma associated HV (KSHV) and γHV68.33, 34, 35
Proteins that execute autophagy are conserved in all eukaryotes and many of these conserved autophagy‐related effectors are called ATG proteins.6, 7, 8, 9, 36 Stages of autophagy include autophagy initiation signaling, autophagosome nucleation, autophagosome expansion, and autophagosome maturation which involves docking and fusion with the lysosome. Multiprotein complexes mediate each stage.6, 7, 8, 9 BECN1 is a core component of the class III phosphatidylinositol 3‐kinase (PI3KC3 or VPS34 in yeast) complexes required for autophagosome nucleation and autophagosome maturation, which also include the PI3K Ser/Thr kinase regulatory protein (PI3KR4/p150/VPS15) and either ATG14/BARKOR or VPS38/UVRAG (UV radiation resistance associated gene).37 38, 39, 40 41
Beyond autophagy, BECN1 is implicated in several other important functions. Yeast VPS30 localizes to vacuolar membranes and endosomes for vacuole protein sorting.37 C. elegans 42 and Drosophila 43 BECN1 contributes to endocytic trafficking. Caspases‐3, 7 and 8, cleave BECN1 at D133 or D149 to generate autophagy‐inactive N‐ and C‐terminal fragments. The C‐terminal fragment translocates to mitochondria, triggering the release of pro‐apoptotic factors like cytochrome c to initiate apoptosis.44, 45 BECN1 also localizes to the nucleus where it reduces Transcription Factor EB activation, thereby regulating transcription of autophagosome maturation‐related proteins such as LC3 (light chain 3), LAMP1 (lysosome associated membrane protein), and RAB7 (Ras‐associated protein Rab‐7a).46, 47
BECN1 Domain Architecture
Human BECN1 is a 450‐residue protein that shares 29%–99% identity with homologs from diverse eukaryotes (Fig. 1). Based on several structural, biophysical and bioinformatics studies, including careful analysis of sequence conservation, we delineate four structurally‐distinct domains/regions common to all BECN1 homologs, with all numbering corresponding to human BECN1 [Figs. 1, 2]: (i) an intrinsically disordered region (IDR, residues 1‐140),49, 50 (ii) a flexible helical domain (FHD, residues 141‐171),51 also called coiled‐coil 1 (CC1),52 (iii) a coiled‐coil domain (CCD, residues 175‐265)53, 54 and (iv) a β‐α‐repeated autophagy‐specific domain (BARAD, residues 266‐450).55, 56 While the sequence within the IDR is poorly conserved, the FHD, CCD, and BARAD are all highly conserved; therefore residues 244‐337 that include the C‐terminus of the CCD and N‐terminus of the BARAD were previously called the evolutionarily conserved domain (ECD)57 (Fig. 1). However, the extensive conformational flexibility of BECN1 has resulted in significant ambiguity in these domain boundaries, as briefly summarized below, and more substantially discussed in subsequent sections.
Figure 1.
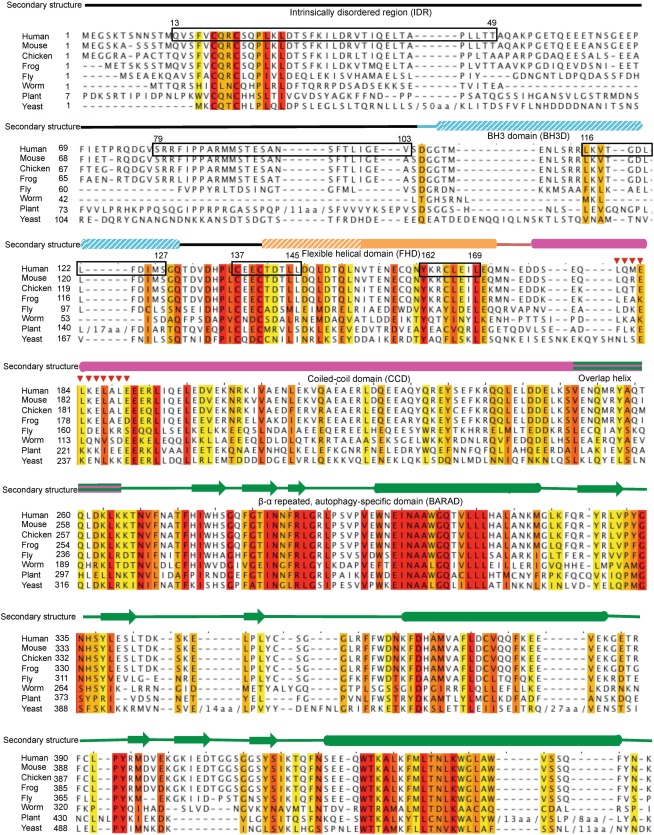
Sequence alignment of BECN1 orthologs from diverse organisms. Increasing background color intensity corresponds to increasing residue conservation with red corresponding to invariant residues. Experimentally determined secondary structure is displayed above the alignment, with cylinders representing helices, arrows representing strands and lines representing coil, color‐coded by domains as follows: IDR (black), BH3D (cyan), FHD (orange), CCD (magenta), and BARAD (green). Solid colors indicate natively folded stable structural elements, horizontal stripes indicate elements that may fold as part of two domains and diagonal stripes indicate binding‐induced secondary structure. Anchor regions are boxed in black. Red triangles indicate the human NES.
Figure 2.
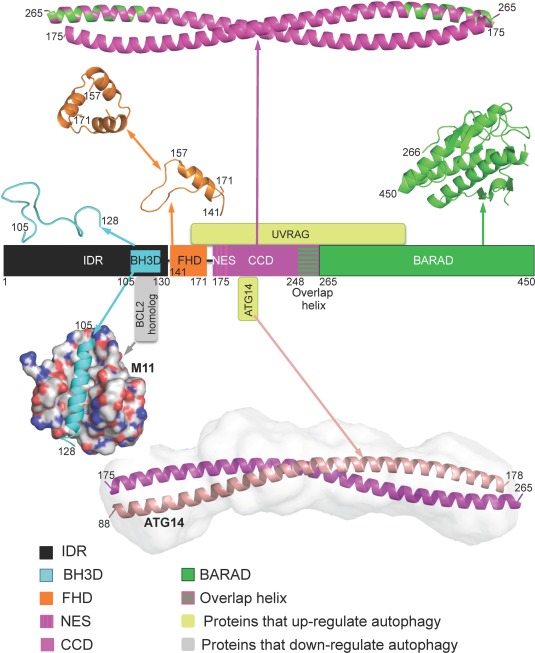
Domain architecture of BECN1 and selected interactions. Domains are as in Figure 1, with corresponding domain structures displayed in ribbon above the schematic. Two structurally characterized interactions are displayed below the schematic: (i) The BECN1 BH3D (cyan ribbon) bound to M11 (grey molecular surface) and (ii) The BECN1 CCD (magenta ribbon): ATG14 CCD (salmon ribbon) heterodimer is shown modeled into its SAXS‐derived molecular envelope. Yellow‐green boxes represent interacting proteins that up‐regulate autophagy while grey boxes represent interacting proteins that down‐regulate autophagy. All molecular figures were prepared using the program PyMOL.48
Neither the boundaries nor the function of the IDR are well delineated. Within the human BECN1 IDR, residues 105–130 have been identified as a BCL2 homology 3 domain (BH3D) that is required and sufficient for binding to BCL2s,34, 35, 58, 59, 60, 61 but are disordered in the absence of binding49, 50 [Figs. 1, 2]. BECN1 residues 144–269 were initially predicted to constitute a single CCD.62 However, biophysical and crystallographic studies have established residues 175‐265 as an independently folding CCD,53, 54 while residues 141‐171 constitute an independent structural domain, the FHD,51, 52 which is partly disordered51 [Figs. 1, 2]. Lastly, even within the well folded CCD53, 54 and BARAD,55, 56 residues 248‐265 adopt mutually exclusive conformations in different crystal structures (Fig. 3).
Figure 3.

Mutually exclusive packing of the “overlap helix”. The overlap helix‐containing BARAD (PDB 4DDP, green ribbon) is superimposed onto the CCD (PDB 3Q8T, magenta ribbon). Residues of the overlap helix in each structure, as well as the partner CCD helix, are colored in alternating pink and green.
Thus, conformational flexibility is an over‐riding feature of BECN1 and many BECN1 regions are now known to undergo binding‐associated conformational changes. Here we will review and summarize current information regarding BECN1 structure in the absence of interactions, conformational transitions mediated by binding of different partners, selected structurally‐uncharacterized interactions and post‐translational modifications.
Intrinsically disordered region (residues 1‐140)
BECN1 contains a long IDR comprising nearly one‐third of its sequence. BECN1 residues 42–115 were first defined as a consensus IDR, based on bioinformatics analysis using four different programs, as residues 13–41, 116–130, and 137–145 were predicted to have secondary structure.49 However, concurrent experimental analyses using circular dichroism (CD) spectroscopy and 1H‐nuclear magnetic resonance (NMR) spectroscopy indicated BECN1 residues 116–130 are disordered and this disorder persists in constructs including adjacent BECN1 domains. Thus, the BECN1 IDR extends beyond the conservative consensus definition, to encompass the poorly conserved residues 42–136.49
Disorder of the BECN1 IDR has now been confirmed by multiple structural and biophysical studies. No density corresponding to this region is visible in either the 28 Å cryo‐electron microscopy (Cryo‐EM) reconstructions63 or the 4.4 Å structure of full‐length BECN1 within PI3KC3 complexes.52 In the latter, a four‐turn helix unconnected to any polypeptide chain was proposed to constitute the BECN1 BH3D; however, no identifiable side‐chain electron density is visible, and other proteins within the complex are also missing many residues in the same region. Further, yeast do not encode BCL2 proteins, and VPS30 lacks a conserved BH3D sequence motif, raising questions about the assignment of this four‐turn helix as a BH3D.
More recently, 2D 1H‐15N heteronuclear single quantum coherence NMR showed that a BECN1 fragment comprising residues 1–150 is disordered when the four cysteines of invariant18C‐x‐x‐C21 and 137C‐x‐D/E‐C140 motifs were mutated to serines.50 However, as other studies show that residues 141‐171 constitute a distinct domain, the FHD/CC1,51, 52 we define the IDR as comprising of, at most, residues 1‐140. Further, these experiments do not preclude the possibility that interactions of the invariant cysteines mediate local structure, especially since residues 13–41 and 137–145, including these cysteines, are predicted to be structured. However, pending experimental evidence verifying structure in these regions, we include the entire region encompassing residues 1‐140 in the current definition of the IDR.
As is typical for IDRs,64 the BECN1 IDR lacks a well‐packed hydrophobic core, and is rich in disorder‐promoting residues, containing 51% polar and charged residues, 6% glycine, and 6% proline. Excluding short regions around the invariant CxxC motifs, the IDR is very poorly conserved amongst BECN1 homologs. The sequence and structural flexibility of IDRs is thought to enable their participation in diverse, and often multiple, interactions with high specificity and reversibility, allowing them to regulate functions such as protein recruitment, signaling, transcription, and translation.65, 66, 67, 68, 69, 70 Further, IDRs are often the sites of post‐translational modifications that regulate the functions of these proteins.71
Not surprisingly, the BECN1 IDR contains several binding motifs. These include “Anchor regions”, i.e. sequences flanking or overlapping IDRs that are predicted by the program ANCHOR to nucleate binding‐associated folding;72 and eukaryotic linear motifs (ELMs), which are short, evolutionarily plastic, linear sequence motifs experimentally shown to be key for various protein–protein interactions that were identified using the ELM server65, 73 (Table 1). The BECN1 IDR contains three complete Anchor regions, residues 13–49, 79–103, and 116–127, and a fourth Anchor region comprising residues 137–145 that extends into the FHD (Fig. 1).
Table 1.
Location of Predicted ELMs and Anchor Regions in BECN1
| Interacting protein/domain | BECN1 residues comprising | ||
|---|---|---|---|
| ELMs | ELM sequence | Overlapping Anchors | |
| GSK3 | 3–10 | GSKTSNNS | – |
| 4–11 | SKTSNNST | – | |
| 8–15 | NNSTMQVS | – | |
| 101–108 | GEVSDGGT | – | |
| CK1 | 4–10 | SKTSNNS | – |
| 7–13 | SNNSTMQ | – | |
| 90–96 | STESANS | 79‐103 | |
| 93–99 | SANSFTL | 79‐103 | |
| 127–133 | SGQTDVD | – | |
| N‐GLC | 7–12 | SNNSTM | – |
| 8–13 | NNSTMQ | – | |
| 110–115 | ENLSRR | – | |
| FHA1 (Forkhead‐associated domain 1) | 9–15 | NSTMQVS | – |
| 41–47 | ELTAPLL | 13–49 | |
| 117–123 | KVTGDLF | 116–127 | |
| 139–145 | ECTDTLL | 137–145 | |
| PIKK | 19–25 | QRCSQPL | 13–49 |
| 54–60 | PGETQEE | – | |
| PKA | 19–25 | QRCSQPL | 13–49 |
| 35–41 | DRVTIQE | 13–49 | |
| 14‐3‐3 protein | 26–31 | KLDTSF | 13–49 |
| PLK | 27–33 | LDTSFKI | 13–49 |
| 40–46 | QELTAPL | 13–49 | |
| FHA2 (Forkhead‐associated domain 2) | 36–42 | RVTIQEL | 13–49 |
| 55–61 | GETQEEE | – | |
| 128–134 | GQTDVDH | – | |
| TRAF2 | 38–41 | TIQE | 13–49 |
| 57–60 | TQEE | – | |
| 64–67 | SGEE | – | |
| TRAF6 | 52–60 | AKPGETQEE | – |
| GlcNHglycan | 63–66 | NSGE | – |
| 92–95 | ESAN | 79–103 | |
| 126–129 | MSGQ | – | |
| Pin1 (phospho‐specific PPIase) | 69–74 | FIETPR | – |
| ProDKin | 69–75 | FIETPRQ | – |
| BRCA1 | 78–82 | VSRRF | 79–103 |
| AMBRA1 (WD40 domain) | 80–82 | RRF | 79–103 |
| 114–116 | RRL | – | |
| NDR (N‐arginine dibasic convertase) | 80–82 | RRF | 79–103 |
| 114–116 | RRL | – | |
| Caspase3‐7 | 102–106 | EVSDG | – |
| Cyclin | 114–118 | RRLKV | – |
| SKI1 (Subtilisin/kexin isozyme‐1) | 114–118 | RRLKV | – |
| SH3 | 129–135 | QTDVDHP | – |
Two of the BECN1 IDR Anchor regions have been identified as part of α‐molecular recognition features (αMoRFs), i.e. regions that undergo disorder‐to‐helix transitions upon binding to partners.74, 75 The BH3D, comprising residues 105‐130 and encompassing the third Anchor region, was the first BECN1 αMoRF identified.49 The BH3D is disordered, even in BECN1 constructs including adjacent structured domains, but folds into a helix upon binding to various BCL2 homologs49, 61 or in the presence of 2,2,2‐trifluoroethanol (TFE), a chemical that induces helicity in αMoRFs even in the absence of their binding partners.76 Similar methodology shows that residues 76–105, which include the second Anchor region, is likely also an αMoRF.76
Flexible helical domain (residues 141–171)
Immediately following the IDR is a highly conserved BECN1 domain, the FHD or CC1, which was delineated by combined sequence and structural analyses (Fig. 1). It was named the FHD (flexible helical domain) because the X‐ray crystal structure of the isolated domain demonstrates that the N‐terminal half comprising residues 141–156 is disordered, while the C‐terminal half comprising residues 157–171 forms a 2.5‐turn α‐helix51 (Fig. 2). CD and small‐angle X‐ray scattering (SAXS) confirm this disorder and show that partial disorder persists in BECN1 fragments that include the flanking BH3D and CCD. Further, the FHD crystal structure and SAXS data indicate the BECN1 FHD trimerizes in the absence of other interactions (Fig. 2). Conformers generated by long time‐scale molecular dynamic simulations fitted to the experimental SAXS data also suggest that the FHD is a trimer, wherein each FHD has a helical C‐terminal part and an unstructured N‐terminal region that transiently samples helical conformations.51
The FHD has two invariant residues, Y162 and L169; and several highly conserved residues (Fig. 1). Importantly, these conserved FHD residues are critical for starvation‐induced upregulation of autophagy.51 Of these residues, Y162, L166, and L169 map to one face of the helix, while the remaining are disordered or in coil conformation. However, if the FHD were to become completely helical, all the conserved residues would map to the same helical face. In the crystal structure, the ordered conserved residues pack about the three‐fold to stabilize the FHD trimer (Fig. 2).
In addition to the Anchor region comprising residues 137‐145 that overlaps both the IDR and FHD (Fig. 1), the FHD contains another Anchor region comprising residues 162‐169.51 Like the BH3D and αMoRF‐containing IDR fragments from other proteins, TFE induces a marked increase in FHD helicity and decrease in disorder, suggesting that it is an αMoRF that undergoes a disorder‐to‐helix transition upon binding to appropriate partners.
Coiled‐coil domain (residues 175–265)
The BECN1 CCD, which is well conserved amongst homologs (Fig. 1), is sufficient for self‐interaction in cells.77, 78 BECN1 homo‐oligomers were detected in mammalian cells via co‐immunoprecipitation (CoIP) and immunoblotting, even during starvation and rapamycin‐induced autophagy.78 Further, while in vitro isothermal titration calorimetry (ITC) studies indicate that UVRAG, which heterodimerizes via the BECN1 CCD, disrupts the BECN1 homodimer;77 overexpression of the UVRAG CCD in cell culture diminishes, but does not abolish homo‐oligomerization.77, 78 These studies indicate that the BECN1 CCD also exists as a homodimer.
X‐ray crystal structures show that the 91‐residue BECN1 CCD forms a straight, ∼130 Å long, anti‐parallel, left‐handed coiled‐coil homodimer [Fig. 4(A)]. Each CCD helix has 13 heptad residue repeats (a‐b‐c‐d‐e‐f‐g), that stabilize the homodimer by interactions of residues at the “a” and “d” positions.53, 54 The 26 interacting pairs are comprised of thirteen unique pairs related by the homodimer two‐fold symmetry [Fig. 4(A)]. Six of these 13 unique pairs comprise of hydrophobic residues. However, residues with polar and/or bulky side‐chains occupy either the “a” or “d” positions of each of the remaining seven pairs, resulting in non‐ideal packing. Interestingly, while many of the non‐ideal or acceptable pairings are conserved amongst BECN homologs, the ideal pairings are not. The non‐ideal packing interface results in a metastable homodimer, with a relatively weak K d of 89 µM for rat BECN153 and 48 μM for human54 BECN1. CoIP and thermal stability experiments show that multi‐site alanine mutagenesis of hydrophobic CCD interface residues results in a monomeric CCD, while multi‐site mutagenesis of hydrophilic CCD interface residues to leucines increases the thermal stability of the BECN1 homodimer.53
Figure 4.
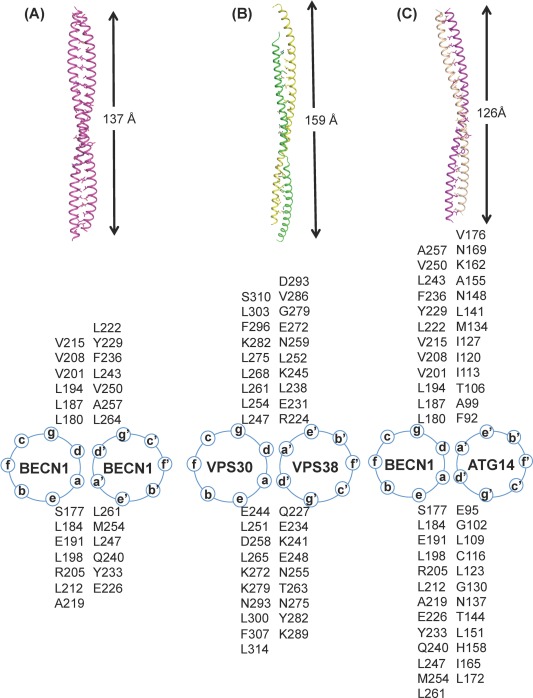
The BECN1 CCD dimers. (A) BECN1 CCD homodimer, (B) VPS30:VPS38 CCD heterodimer and (C) BECN1:ATG14 CCD heterodimer. The upper panel shows three dimers in ribbon with interface residues rendered in stick with atoms color‐coded by atom type: C, magenta for BECN1, wheat for ATG14, green for VPS38 and yellow for VPS30; O, red; N, blue and S, yellow. The length of each dimer is indicated. The lower panel shows the corresponding helical wheel of the dimers.
Almost half (43%) of the residues comprising the BECN1 CCD homodimer are charged, with a predominance of acidic residues resulting in a highly negatively charged surface.53, 54 Around 36% of these charged residues are conserved. Six pairs of interchain polar interactions mediated by polar interface residues, further stabilize the BECN1 homodimer. Furthermore, some of the solvent‐exposed, charged residues cluster in triplets, forming intrachain salt bridges that neutralize potential Coulombic repulsion to further stabilize the CCD structure.
β‐α Repeated, autophagy‐specific domain (residues 266–450)
The 1.6 Å crystal structure of the yeast VPS30 residues 320–557 (corresponding to human BECN1 residues 266‐450) reveals a novel protein fold, consisting of three repeats of a three‐stranded, anti‐parallel β‐sheet followed by a α‐helix (Fig. 1), with the three repeats arranged around an approximate central 3‐fold such that the helices form a central three‐helix bundle surrounded by the β‐sheets55 (Fig. 2). Largely conserved, hydrophobic residues (Fig. 1) stabilize the packing of the helices, as well as of the β‐sheets against the three‐helix bundle. This novel domain is required for autophagy but not vacuolar sorting, and was therefore named the β‐α repeated autophagy‐specific domain (BARAD).55
The BARAD fold is conserved in the crystal structure of human BECN1; however this structure includes residues 248–264 preceding the BARAD, which pack against the BARAD as an additional helix56 that we call the “overlap helix”. The overlap helix corresponds to the C‐terminal four turns of the CCD (Fig. 1). Overlap helix residues that pack against the BARAD56 also constitute the CCD homodimer interface.53, 54 Therefore, extensive steric clashes would prevent the overlap helix from simultaneously existing as part of both domains within a single molecule (Fig. 3). Thus, the crystallized human CCD and BARAD structures represent mutually exclusive conformations of BECN1, with the overlap helix able to exist as part of either domain under different physiological conditions. Indeed, hydrogen‐deuterium exchange mass spectrometry (HDX‐MS) experiments show that in the yeast PI3KC3 complex, VPS30 residues corresponding to the overlap helix undergo membrane binding‐induced changes, but do not directly bind membranes.52
Liposome binding assays reveal that BARAD co‐sedimentation with liposomes requires an “aromatic finger” comprising BARAD residues F259, F360 and W361.56 Further, cellular assays demonstrate that wild‐type BARAD, but not a F259D + F360D + W361D mutant, co‐localizes with lipid membranes. Loss of the aromatic finger reduces starvation‐induced autophagy. Thus, autophagy induction requires membrane binding by the BARAD aromatic finger. The aromatic finger is only partially conserved: the first aromatic residue is conserved amongst all homologs, while the second and third residues are not conserved in worm or yeast (Fig. 1). However, in yeast VPS30, basic residues that may also promote interactions with acidic lipid head‐groups occupy equivalent positions. Notably, a yeast VPS30 region found by HDX‐MS to interact directly with lipid membranes52 includes residues analogous to the aromatic finger.
Structurally‐Characterized BECN1 Interactions
BCL2 binding to the BECN1 BH3D
BECN1 was initially discovered as a BCL2‐interacting protein,1 with binding of the anti‐apoptotic BCL2 to BECN1 downregulating autophagy.33, 34, 35 BECN1 residues 88–150 were named the BCL2‐binding domain (BCL2BD), as this region was sufficient for binding BCL2 and BCL2L1 (BCL‐XL), and its deletion weakened BECN1 interaction with BCL2 in mammalian cells.1 Sequence analysis and structural studies demonstrated that the BECN1 BCL2BD contains a BH3D, comprising residues 105‐130.34, 35, 58, 59, 60 A minimal BH3D consists of a four‐turn, amphipatic α‐helix bearing the sequence motif: Hy‐X‐X‐X‐Hy‐K/R‐X‐X‐Sm‐D/E‐X‐Hy (where Hy: hydrophobic residues; Sm: small residues, typically glycine).61 The BH3D is conserved amongst pro‐apoptotic proteins and enables binding of anti‐apoptotic BCL2 proteins.61 The BH3D is also conserved among BECN1 homologs from humans to Drosophila, organisms that encode anti‐apoptotic BCL2 proteins; but not in yeast ATG6/VPS30 (Fig. 1), presumably because yeast do not encode BCL2 proteins.
Diverse BCL2 homologs bind the isolated BECN1 BH3D with moderate affinity.34, 35, 49, 59 Mutagenesis studies in combination with diverse binding assays show that the BH3D is both necessary and sufficient for BECN1 interaction with anti‐apoptotic BCL2 proteins. Structures of the BECN1 BH3D bound to human BCL2L159, 60 or the γHV68 BCL2 homolog, M11 (Fig. 2),34, 35 show that, like BH3Ds from pro‐apoptotic proteins, the BECN1 BH3D binds as a helix to a hydrophobic groove on the surface of the BCL2 proteins (Fig. 2). In each structure, the hydrophobic interaction interface buries conserved BECN1 BH3D hydrophobic residues: L112, L116, and F123. The BECN1 G120‐D121 pair, which is highly conserved in all BH3Ds, interacts with a Gly‐Arg pair (G86‐R87 of M11 and G138‐R139 of BCL2L1) that is highly conserved in anti‐apoptotic BCL2 homologs. While G120 packs tightly against the BCL2 glycine; D121 forms a bidentate salt bridge with the BCL2 arginine. Thus, anti‐apoptotic BCL2 proteins bind the BECN1 BH3D in a manner similar to the other BH3Ds.
It has now been conclusively shown that the BECN1 BH3D is part of a larger IDR.49, 50 Notably, residues toward the BH3D C‐terminus (residues 116‐123) comprise an Anchor region.49, 76 Mutation of these Anchor region residues abolishes BCL2 binding and abrogates binding‐induced helical transition within the BH3D.49 Thus, the BECN1 BH3D is an αMoRF that undergoes a binding‐dependent conformational transition to down‐regulate BECN1‐mediated autophagy. Conversely, conserved residues lining the hydrophobic groove of BCL2 proteins are critical for BECN1 binding.35
Despite the overall similarity in the mode of binding of different BCL2 homologs to BECN1, there are significant differences. Variations in residues lining the hydrophobic groove of each BCL2 protein dictate a differential binding affinity for BECN1. Thus, the BECN1 BH3D binds to M11 with 1.1 μM affinity, and to human and KSHV BCL2 with weaker affinities of 8.0 μM and 13.3 μM, respectively.35 Further, adjacent BECN1 regions differentially impact binding to diverse BCL2s,34, 35 although this has not yet been fully investigated. Two mechanisms regulate BCL2‐mediated inhibition of BECN1‐dependent autophagy: competitive binding of other BH3D‐containing proteins79 and modulation of BECN1:BCL2 interaction by phosphorylation of either partner.80, 81, 82, 83
The mechanism by which BCL2 proteins bind the BECN1 BH3D is well established; however, the mechanism by which this interaction downregulates BECN1‐mediated autophagy is not. BCL2 overexpression diminishes BECN1 CoIP with VPS34.33 Further, in the presence of BCL2L1 or KSHV BCL2, BECN1 binds UVRAG with 4‐fold lower affinity.77 ITC experiments indicate BCL2L1 and KSHV BCL2 bind the BECN1 homodimer.77 Therefore, BCL2 binding may stabilize the BECN1 homodimer and disrupt interactions between BECN1 and other components of the autophagy nucleation complex.
BECN1 in PI3KC3 complexes
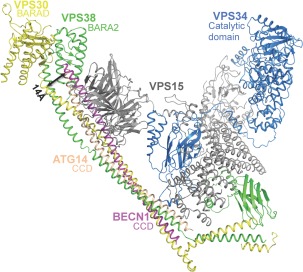
BECN1 is a core component of PI3KC3/VPS34 complexes involved in autophagy and vesicle trafficking.37, 62 BECN1 associates with either ATG14/BARKOR or UVRAG/VPS38 to form two mutually exclusive PI3KC3 complexes: (i) Complex I comprising BECN1:ATG14:PI3KC3:p150 or (ii) Complex II comprising BECN1:UVRAG:PI3KC3:p150.38, 40, 41 While p150/PI3KR4/VPS15 is an obligate partner of PIK3C3, association of the other proteins up‐regulates PI3KC3 activity to convert phosphatidylinositol (PI) to phosphatidylinositol 3‐phosphate (PI3P), an essential signal for autophagosome formation.84, 85, 86 Complex I nucleates autophagosomes while Complex II mediates autophagosome maturation.37, 38, 40, 41, 87
PI3KC3/VPS34 contains a N‐terminal C2 domain, a central helical domain, and C‐terminal lipid kinase domain.88, 89 VPS34 forms a heterodimer with N‐terminally myristoylated VPS1537, 90 (p150 in humans), a protein Ser/Thr kinase, which consists of a N‐terminal kinase domain, a central HEAT (Huntington, elongation factor 3 protein phosphatase 2A, yeast kinase TOR1) domain, and a C‐terminal WD40 domain.88 The VPS34 C‐terminal lipid kinase domain interacts with the VPS15 N‐terminal kinase domain (Fig. 5).52
Figure 5.
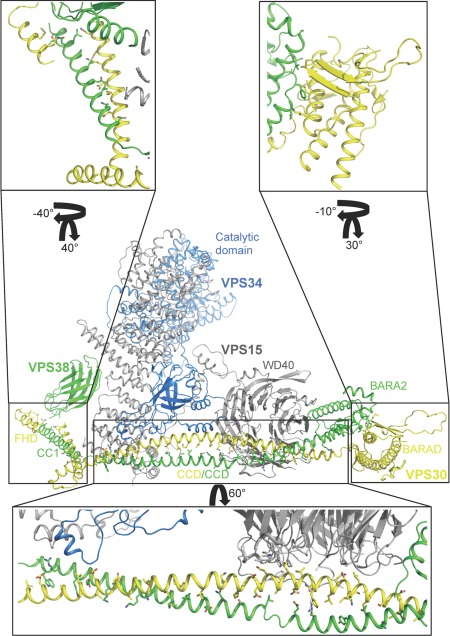
BECN1 interactions in the PI3KC3 complex II. All proteins are shown in ribbon, colored as: VPS30, yellow; VPS38, green; VPS15, grey, and VPS34, blue. Protein domains implicated in membrane interaction, the BECN1 BARAD, UVRAG BARA2 and PI3KC3 catalytic domain are labeled. Boxes indicate regions that were enlarged and rotated to demonstrate interactions. Interacting side chains, mutated to the VPS30 sequence from the 4.4 Å poly‐alanine structure, are displayed as sticks.
Human ATG14 is a 492‐residue protein comprised of a N‐terminal cysteine‐rich repeat region required for targeting ATG14 to the endoplasmic reticulum,91 a CCD comprising at least residues 88‐178, and a C‐terminal BATS (Barkor/Atg14 autophagosome targeting sequence) functional domain responsible for sensing and preferentially binding highly curved, PI3P‐rich membranes.92 ATG14 residues 88‐178 are sufficient for binding the BECN1 CCD.53, 54 Human UVRAG, a 699‐residue protein, consists of a proline‐rich (PR) sequence, a C2 domain, a CCD and a C‐terminal domain.62 Based on the recent structure of yeast VPS38 within Complex II,52 the UVRAG CCD comprises residues 230‐321.
Self‐association of the BECN1 CCD is 11‐27‐fold weaker than heterodimerization with human or rat ATG14 respectively, and 370‐fold weaker than heterodimerization with rat UVRAG.53, 54, 77 This likely facilitates homodimer dissociation and formation of BECN1:ATG14 or BECN1:UVRAG heterodimers upon induction of autophagy. Heterodimerization increases cellular stability of each protein.54
The 28 Å Cryo‐EM reconstructions of Complex I and II reveal nearly identical overall V‐shaped envelopes.63 PI3KC3 and VPS15 comprise one arm of each V‐envelope, while the other arm is comprised of BECN1 and either ATG14 or UVRAG. MBP‐tag mapping of the Cryo‐EM reconstructions reveals that the BECN1 BARAD is located at the tip of one arm of the V, with BECN1 in a parallel arrangement with either ATG14 or UVRAG, extending to the N‐termini of each protein located at the base of the V. Further, the VPS15 HEAT domain is positioned to interact with N‐terminal regions of BECN1 and ATG14 or UVRAG, at the base of the V‐envelope, while the VPS15 WD40 domain is positioned to interact with C‐terminal regions of BECN1 and ATG14 or UVRAG.
A recent 4.4 Å crystal structure of PI3KC3 Complex II52 confirms this overall architecture and also shows that, as expected, the BECN1/VPS30:UVRAG/VPS38 CCD heterodimer of each protein constitutes the largest component of the VPS30:VPS38 binding interface. VPS30 equivalents of BECN1 homodimer residues were modeled to contribute to the interface with VPS38. However, this heterodimer is more poorly packed than the BECN1 CCD homodimer as VPS30 CCD hydrophobic residues mostly pair with polar and charged VPS38 residues, while two hydrophobic VPS38 residues, L238 and L252, interact with charged VPS30 residues, D258 and K272, respectively [Figs. 4(B), 5, lower inset]. Lastly, the hydrophobic VPS38 V286 interacts with the bulky VPS30 F207.
Unexpectedly, this structure reveals that the C‐terminal VPS38 domain has a fold similar to the VPS30 BARAD (Fig. 5), albeit with two, rather than three, repeats of the three‐stranded, anti‐parallel β‐sheet and helix unit.52 Therefore, this domain was named the VPS38 BARA2 domain. Further, the VPS30:VPS38 interaction extends beyond the CCD, including the FHD/CC1 toward the N‐terminus, and the BARAD at the C‐terminus (Fig. 5). The BARAD interactions involve the first α‐helix and second β‐sheet of the VPS30 BARAD and the second α‐helix of the VPS38 BARA2. The VPS30 FHD/CC1 forms a coiled‐coil with a VPS38 helix (hence these helices were named CC1 in both VPS30 and VPS38),52 verifying that the BECN1/VPS30 FHD is an αMoRF.51 Strikingly however, conserved VPS30 FHD/CC1 residues are mostly solvent‐exposed and uninvolved in binding VPS38.52 This may either reflect inaccurate sequence assignment due to the low‐resolution of the structure; or indicate that these residues bind other autophagy regulators such as AMBRA1 (Autophagy/BECN1 regulator 1), which binds to a region that includes the FHD.
Within the yeast PI3KC3 Complex II, the VPS30 FHD/CC1 also interacts with the VPS15 WD40 domain and the CBR1 loop of the VPS34 C2 domain (Fig. 5).52 Unexpectedly, the PI3KC3 C2 domain does not interact with membranes, rather it is responsible for nucleating the complex by binding VPS15 and the BECN1:UVRAG CCD. Further, although the BECN1/VPS30 ECD (comprising parts of the CCD and BARAD) was previously shown to be required for PI3KC3/VPS34 interaction with VPS30,57 it does not directly interact with VPS34 in this crystal structure.52 Rather, the N‐terminal region of the VPS30 overlap helix, which lies within the ECD, interacts with the VPS15 WD40 domain, which in turn interacts with VPS34 (Fig. 5). Membrane phosphorylation requires both PI3KC3 catalytic activity and the membrane binding function of the BECN1 BARAD aromatic finger; domains located at the tips of each arm of the V‐shaped complex [Figs. 5, 6]. Therefore, complex formation may serve to position/anchor the PI3KC3 catalytic domain onto autophagosomal membranes for phosphorylation and serve as a scaffold for recruiting other proteins that modulate PI3KC3 activity.
Figure 6.
Displacement of the BECN1 BARAD domain because of the curved BECN1:ATG14 quaternary structure. All proteins are shown in ribbon, colored as in Figures 4 and 5. Arrows indicate altered positions of equivalent residues.
SAXS studies of the BECN1:ATG14 CCD heterodimer show that it has an extended but curved envelope with a radius of 15 nm54 [Figs. 2, 4(C))]. In comparison, the BECN1 homodimer is straight [Fig. 4(A)]; while in the PI3KC3 complex II X‐ray structure, the VPS30:VPS38 CCD heterodimer has a shallower radius of curvature of 17 nm [Fig. 4(B)]. Further, CD spectroscopy and SAXS indicate the BECN1:ATG14 CCD heterodimer is less structured than the BECN1 CCD homodimer.
A computational model, based on optimized interactions of the ATG14 and BECN1 CCDs, that also fits well to experimental SAXS data (Fig. 2), indicates that BECN1 CCD residues mediating homodimerization are also responsible for heterodimerization [Fig. 4(A,C)].54 The BECN1:ATG14 heterodimer model has a total buried surface area (BSA) of 4449 Å2, slightly less than the BECN1 homodimer which has a total BSA of 4849 Å2. In contrast, the VPS30 CCD and VPS38 CCD are shifted relative to each other within the PI3KC3 complex II,52 resulting in a total BSA of 3319 Å2, which is less than either the BECN1:ATG14 CCD heterodimer or BECN1 CCD homodimer. While VPS38/UVRAG contributes mostly polar and charged residues to the VPS30:VPS38 CCD interface, approximately half the residues contributed by ATG14 to the BECN1:ATG14 interface are hydrophobic (Fig. 4).
BECN1:ATG14 heterodimer association is 10‐folder tighter than BECN1 homodimerization, however only ten of 25 BECN1:ATG14 interface pairs are hydrophobic [Fig. 4(C)].54 Strikingly, many of the polar residues that form non‐ideal pairs in the BECN1 CCD homodimer, pair either with polar residues in the BECN1:ATG14 CCD heterodimer model, or are stabilized by additional hydrogen bonds or charged interactions with residues adjacent to their partner (Fig. 4). Many of these interface pairs are conserved amongst diverse eukaryotes. Thus, ATG14 and BECN1 CCD heterodimerization involves conserved, hydrophobic pairs, as well as conserved, polar ATG14 residues paired with conserved, polar BECN1 residues. ITC, CoIPs and cellular autophagy assays indicate that BECN1 and ATG14 interface residues identified from this model are required for BECN1:ATG14 heterodimerization and starvation‐induced autophagy.53, 54
The enhanced curvature of the BECN1:ATG14 CCD may position the BECN1 BARAD up to 14 Å away from the corresponding VPS30 BARAD position in the PI3KC3 complex II (Fig. 6), placing lipid‐binding domains of Complex I closer to each other than in Complex II, thereby modulating the association of these complexes with membranes of different curvature. Thus, differential incorporation of ATG14 or UVRAG may target PI3KC3 to different membranes based on membrane curvature and lipid composition.
Structurally Uncharacterized BECN1 Interactions
BECN1 appears to be a major interaction hub for autophagy, as it is implicated in binding over 25 other proteins,45, 49, 93, 94, 95 primarily based on CoIP and cellular pull‐down assays. Many of these interactions modulate cellular autophagy levels; however, some may also mediate BECN1 functions beyond autophagy. BECN1 domains that are responsible for specific interaction have been delineated for some of these interactions.
BECN1 IDR residues 88‐123 are required for binding VMP1 (Vacuole membrane protein 1), which results in dissociation of BCL2 and starvation‐induced up‐regulation of autophagy.96, 97, 98, 99, 100 The BECN1 FHD is required for binding AMBRA1, a highly disordered 1300‐residue protein.101, 102 AMBRA1 binding promotes BECN1 phosphorylation, PI3KC3 Complex I formation, and autophagy nucleation.103, 104, 105, 106
Several proteins interact with the BECN1 FHD‐CCD. TAB2 and TAB3 (TAK1‐binding proteins 2 and 3) bind this region and inhibit autophagy,107, 108 providing a node of crosstalk between autophagy and TAK1‐IKK (TAK1 protein kinase‐IκB kinase) signaling. Similarly, binding of the BECN1 FHD‐CCD to the pro‐apoptotic, BH3‐only protein Bim, results in mislocalization of BECN1 and autophagy suppression.109, 110, 111 Interestingly, the BECN1:BCL2L1 interaction is unaffected by BECN1:Bim binding, since the BECN1 BH3D binds BCL2/BCL2L1, while the BECN1 FHD‐CCD binds Bim. Lastly, SLAMF1 (signaling lymphocyte‐activation molecule family1), a microbial sensor that regulates bacterial phagocytosis, binds the BECN1 BH3D‐FHD‐CCD region within the PI3KC2 Complex II, resulting in VPS34 activation to produce PI3P, to regulate phagosomomal/endosomal membrane fusion.112
The BECN1 CCD also binds the nPIST (neuronal isoform of protein‐interaction, specifically with TC10) CCD to enable binding of the δ2 glutamate receptor and activate autophagy in lurcher Purkinje cells.113 WASH (Wiskott‐Aldrich syndrome protein and SCAR homolog) also binds the BECN1 CCD, abolishing BECN1 K437 ubiquitination, inactivating PI3KC3 and suppressing starvation‐induced autophagy; while being essential for endosome sorting.114, 115 The WASH:BECN1 interaction likely prevents excess autophagy during early embryonic development. The BECN1 CCD also serves as a interaction platform in the context of the BECN1:ATG14 heterodimer to recruit downstream autophagy effectors such as the ATG12‐ATG5‐ATG16 and LC3 complexes necessary for autophagosome elongation.116
The BECN1 CCD N‐terminal residues 180‐190 constitute a leucine‐rich nuclear export signal (NES) (Fig. 1) that binds the chromosomal maintenance protein 1 (CRM1) to enable nuclear export of BECN1.117 The NES L184 and L187 are essential for BECN1 nuclear export and for BECN1‐mediated autophagy and tumor suppression. In the BECN1 CCD homodimer, the C‐terminal overlap helix of one helix packs against the N‐terminus of the partner helix, which contains the BECN1 NES (Fig. 2). L184 and L187, which are required for NES function,117 participate in coiled‐coil pairings with residues within the overlap helix in the CCD homodimer [Fig. 4(A)]. Therefore, in order to bind CRM1, the BECN1 NES cannot be packed against the BECN1 overlap helix. In conditions where the NES is exposed to allow nuclear export, the overlap helix would be free to pack as part of the BARAD, as seen in one of the BARAD crystal structures.
Lastly, virus‐encoded proteins also target the BECN1 CCD. The βHV HCMV encodes two proteins, the tegument protein TRS1 (terminal repeat sequence 1) and IRS1 (internal repeat sequence 1), which share a common N‐terminal domain that binds the BECN1 FHD‐CCD to control autophagy to modulate viral replication and infection.118, 119
Several proteins target the BARAD to regulate autophagy. BARAD residues 267‐284 are essential for autophagy, and also necessary and sufficient for in vivo interaction with autophagy inhibitors such as the endogenous GAPR1 (Golgi‐associated plant pathogenesis‐related protein 1), and the HIV‐encoded Nef.120 HSV‐1‐encoded ICP34.5 also inhibits BECN1‐mediated autophagy.31 Loss of BECN1 residues 237‐450 abrogates the in vivo interaction with ICP34.5; therefore, the BECN1 BARAD, CCD, or some combination of both, likely mediate this interaction.
Post‐Translational Modifications (PTMs) of BECN1
PTMs play important roles in regulating BECN1 interactions and function. PTMs of BECN1 and its interaction partners include phosphorylation to regulate catalytic activity and protein‐protein interactions, ubiquitination to control signal degradation, and acetylation to impact gene expression and metabolism.
Phosphorylation
Phosphorylation is the most common BECN1 PTM. The poorly conserved BECN1 IDR contains several phosphorylation sites required for starvation‐induced activation of PI3KC3. BECN1 IDR phosphorylation affects binding and may result in conformational changes that impact autophagy or other cellular functions.
In C. elegans, BECN1 S14 (S15 in human) is phosphorylated by the Ser/Thr‐protein kinase ULK1 involved in autophagy initiation. This phosphorylation is crucial for VPS34 activation during amino acid starvation‐induced autophagy.104 Upon starvation, AMBRA1 also promotes association of AMPK (AMP‐activated protein kinase) and ULK1 to facilitate phosphorylation of AMPK at T172, and ULK1 at S317; which can then phosphorylate BECN1 at S14/S15, increasing PI3KC3 Complex I formation, PI3KC3 activation and autophagy.103, 104 ATG14 and UVRAG also stimulate this phosphorylation by promoting BECN1 association with ULK1.104 This phosphorylation site is conserved from human to worm BECN1, highlighting its importance in PI3KC3 activation and autophagy induction.
Starvation‐induced autophagy and tumor suppression in MCF7 cells requires phosphorylation of BECN1 S90 by the MAPK p38.121 BCL2 binding to the BECN1 BH3D prevents MAPK‐mediated phosphorylation at S90, likely because steric conflicts prevent MAPK binding.
AMPK phosphorylates BECN1 at S93/S96 to enable BECN1 interaction with PI3KC3 and formation of pro‐autophagic PI3KC3 complexes.122 Phosphorylation at these two sites, as well as at S15, is dependent on heterodimerization with either ATG14 or UVRAG. Drug‐induced BECN1 phosphorylation at S93/S96 by AMPK causes AMPK:BECN1:caspase 8 complex formation, resulting in caspase‐8 cleavage of BECN1 to down‐regulate autophagy and up‐regulate apoptosis.123 Therefore, S93/S96 phosphorylation also cross‐regulates autophagy and apoptosis.
MST1 phosphorylates BECN1 BH3D T108, enhancing BECN1 and BCL2/BCL2L1 interaction, which stabilizes the BECN1 homodimer and significantly impairs BECN1:ATG14 and BECN1:VPS34 interaction.82 In contrast, starvation‐dependent phosphorylation of T119 in the BECN1 BH3D Anchor region by ROCK1 or DAPK promotes BECN1:BCL2 complex dissociation, while ROCK1 inhibition increases BECN1:BCL2 interaction, downregulating starvation‐induced autophagy.83
Within the BECN1 CCD, S234 and S295 are phosphorylated by AKT, enhancing BECN1 interaction with 14‐3‐3 and intermediate filament proteins, whose depletion increases autophagy.124 Since intermediate filament proteins are markers of tumor initiation and progression, their interaction with 14‐3‐3 and BECN1 emphasizes the important regulatory role of autophagy in tumorigenesis inhibition.125 Cellular assays demonstrate that the EGFR tyrosine kinase binds and phosphorylates BECN1 at the CCD Y229, Y233 and BARAD Y352 to promote homodimer formation, while diminishing CoIP with PI3KC3, presumably due to decreased interaction with the VPS15 WD40 within PI3KC3 complexes.121 This inactivates the PI3KC3 kinase and suppresses autophagy.
Lastly, BECN1 phosphorylation also relates to other PTMs. BECN1 acetylation at K430 and K437 by p300 requires BARAD S409 phosphorylation by casein kinase 1.126 BECN1 phosphorylation at Y352 is important for NEDD4 binding, which ubiquitinates BECN1.127, 128
Acetylation
Acetylation involves the attachment of an acetyl group, normally at the N‐terminus of a protein. BECN1 is acetylated by p300, a lysine acetyltransferase, at K430 and K437 and is deacetylated by SIRT1 (sirtuin 1).126 K430 and K437 acetylation promotes the recruitment of RUBICON to the UVRAG:BECN1 complex to inhibit autophagosome maturation and endocytic trafficking.
Ubiquitination
BECN1 undergoes K11‐, K63‐ and K48‐linked ubiquitination. K63‐linked ubiquitination of BECN1 BH3D K117 by TRAF6 E3 ligase promotes binding to the PI3KC3 complex rather than to BCL2 proteins, to induce autophagy; and is crucial for TLR4‐triggered autophagy in macrophages.129 Similarly, K63‐linked ubiquitination of BARAD K437 by the AMBRA1‐DDB1‐Cul4‐Rbx1 E3 ligase augments PI3KC3 activity to induce autophagy.115 NEDD4 ubiquitinates BECN1 with K11‐linked polyubiquitin chains to increase proteasomal degradation of BECN1.128 This degradation is greatly enhanced upon PI3KC3 knock‐down in cells, suggesting that association within the PI3KC3 complex protects BECN1 from degradation.128 BECN1 is the first tumor suppressor shown to be regulated by K11‐linked polyubiquitination, although the BECN1 ubiquitination site is unidentified. Besides binding BECN1 within the PI3KC3 complex, AMBRA1 also triggers polyubiquitination and BECN1 stabilization during autophagy.115
Targeting BECN1‐Mediated Autophagy
Understanding the structural details of BECN1 and various BECN1‐mediated interactions is critical not only for understanding the mechanism of its various biological functions, but also for rational design of therapeutics targeting BECN1‐mediated autophagy.
A BH3D mimetic ABT737, competitively disrupts the interaction between BECN1 and BCL2L1 or BCL2, releasing BECN1 from BCL2 protein down‐regulation, thereby stimulating autophagy.79 Further, BH3D mimics can be designed to differentially bind and selectively inhibit BCL2 proteins for therapeutic purposes. Detailed structural and biophysical information regarding binding of the BECN1 BH3D to human BCL2 proteins and the γHV68 BCL2, M11, showed that mutation of a conserved BH3D GD pair, that typically stabilizes BH3D:BCL2 interactions, to a EA pair, abrogates binding to BCL2L1 or BCL2, but not to M11.49, 130 This enabled development of a cell‐permeable peptide inhibitor that exploits the more promiscuous binding of M11, relative to human BCL2 proteins, to selectively inhibit M11‐mediated down‐regulation of autophagy, without impacting cellular BCL2‐mediated down‐regulation of autophagy.130
Another cell‐permeable peptide, Tat‐beclin 1 was derived from residues 267‐284 of the BECN1 BARAD based upon CoIP and autophagy assays that demonstrate this region is crucial for binding to HIV‐encoded Nef, as well as for rescue of starvation‐induced autophagy. Tat‐beclin 1 treatment improves clearance of protein aggregates in mammalian cell culture, stimulates autophagy in mammalian cells and transgenic mice, and improves survival of Chikungunya and West Nile virus‐infected mice.120 Thus, a better understanding of the structural basis and mechanism of the many BECN1 interactions should permit the design of additional therapeutics to specifically modulate various stages of autophagy as well as other BECN1 functions.
Conclusion
Over the last decade, much has been learnt about BECN1 structure, conformational flexibility and interactions; however, numerous unanswered questions remain. BECN1 has now been trapped in diverse, mutually exclusive structural states. BECN1 homotrimers and homodimers, caused by oligomerization of the FHD and CCD respectively, cannot exist in the context of PI3KC3 complexes that mediate autophagy. However, BECN1 homooligomers exist in cells, even upon induction of autophagy. What then, is the biological function of these homooligomers? Does BECN1 simultaneously homodimerize and homotrimerize to form higher order oligomers?
The conformationally dynamic “overlap helix” within the CCD can exist as part of the CCD, such as within the PI3KC3 complexes, as well in a conformation where it packs against the BARAD. However, it is unclear what interactions or biological states involve the latter conformation, and how different overlap helix conformations influence BECN1 homo‐oligomerization. The interactions and conformational states that expose the BECN1 NES located at the CCD N‐terminus to enable nuclear export have also not yet been explored.
The mechanism by which BCL2 homologs bind BECN1 is well established, however the mechanisms by which this interaction down‐regulates autophagy and/or stabilizes the homodimer state of BECN1 remains unknown. Despite the substantial information provided by structural studies of the PI3KC3 complexes, it remains unclear precisely how complex formation modulates PI3KC3 activity. Notably, mutation of many of the highly conserved BECN1 residues that mediate interactions within the complex, abrogates starvation‐induced, rather than basal, autophagy.51, 54, 130 While membrane binding influences the structure of BECN1 and perhaps of other Complex I and II proteins, the precise conformational transitions have not been mapped. We are only now beginning to understand the differences in the structures and interactions of Complex I and II; elucidation of the mechanism by which each complex mediates different stages of autophagy will require much more research.
The mechanism by which diverse BECN1 interactions compete with, or complement each other, to modulate different BECN1 functions is largely unknown. The BECN1 IDR likely mediates many uncharacterized interactions, but as these have not yet been studied in detail, the effect of these diverse interactions on the conformation of the IDR and/or other BECN1 domains, and the mechanism by which these modulate BECN1 function is not understood. Similarly, autophagy suppression by diverse viral proteins is poorly understood. Further, the modulation of diverse BECN1 interactions, conformational states, and functions by various PTMs adds another layer of complexity to elucidating a complete structure‐based mechanism of BECN1 function. Clearly, much additional research is needed to develop a comprehensive structure‐based understanding of the interplay between different BECN1 conformational and binding states, and the impact of post‐translational modifications to elucidate the mechanism of its diverse biological roles. Ultimately, such an understanding would enable the selective targeting of diverse BECN1 functions/interactions for therapeutic benefit.
Conflict of interest statement
The authors declare no conflict of interest
Acknowledgments
The authors would like to thank Dr. Christopher Colbert for useful discussions. This work was supported by Doctoral Dissertation Awards for Y.M. and M.S.
References
- 1. Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B (1998) Protection against fatal Sindbis virus encephalitis by Beclin 1, a novel Bcl‐2‐interacting protein. J Virol 72:8586–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. He C, Wei Y, Sun K, Li B, Dong X, Zou Z, Liu Y, Kinch LN, Khan S, Sinha S, Xavier RJ, Grishin NV, Xiao G, Eskelinen E‐L, Scherer PE, Whistler JL, Levine B (2013) Beclin 2 functions in autophagy, degradation of G protein‐coupled receptors, and metabolism. Cell 154:1085–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B (1999) Induction of autophagy and inhibition of tumorigenesis by Beclin 1. Nature 402:672–676. [DOI] [PubMed] [Google Scholar]
- 4. De Duve C, Wattiaux R (1966) Functions of lysosomes. Annu Rev Physiol 28:435–492. [DOI] [PubMed] [Google Scholar]
- 5. Lawrence BP, Brown WJ (1992) Autophagic vacuoles rapidly fuse with pre‐existing lysosomes in cultured hepatocytes. J Cell Sci 102:515–526. [DOI] [PubMed] [Google Scholar]
- 6. Levine B, Klionsky DJ (2004) Development by self‐digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell 6:463–477. [DOI] [PubMed] [Google Scholar]
- 7. Klionsky DJ (2010) The autophagy connection. Dev Cell 19:11–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xie Z, Klionsky DJ (2007) Autophagosome formation: Core machinery and adaptations. Nat Cell Biol 9:1102–1109. [DOI] [PubMed] [Google Scholar]
- 9. Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27:107–132. [DOI] [PubMed] [Google Scholar]
- 10. Liu Y, Shoji‐Kawata S, Sumpter RM, Wei Y, Ginet V, Zhang L, Posner B, Tran KA, Green DR, Xavier RJ, Shaw SY, Clarke PG, Puyal J, Levine B (2013) Autosis is a Na+,K+‐ATPase‐regulated form of cell death triggered by autophagy‐inducing peptides, starvation, and hypoxia‐ischemia. Proc Natl Acad Sci USA 110:20364–20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yue Z, Jin S, Yang C, Levine AJ, Heintz N (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA 100:15077–15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qu X, Zou Z, Sun Q, Luby‐Phelps K, Cheng P, Hogan R, Gilpin C, Levine B (2007) Autophagy gene‐dependent clearance of apoptotic cells during embryonic development. Cell 128:833–836. [DOI] [PubMed] [Google Scholar]
- 13. Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen E‐L, Mizushima N, Ohsumi Y, Cattoretti G, Levine B (2003) Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112:1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koneri K, Goi T, Hirono Y, Katayama K, Yamaguchi A (2007) Beclin 1 gene inhibits tumor growth in colon cancer cell lines. Anticancer Res 27:1453–1457. [PubMed] [Google Scholar]
- 15. Miracco C, Cosci E, Oliveri G, Luzi P, Pacenti L, Monciatti I, Mannucci S, De Nisi MC, Toscano M, Malagnino V, Falzarano SM, Pirtoli L, Tosi P (2007) Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol 30:429–436. [PubMed] [Google Scholar]
- 16. Li Z, Chen B, Wu Y, Jin F, Xia Y, Liu X (2010) Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. BMC Cancer 10:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J (2006) Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem 281:14474–14485. [DOI] [PubMed] [Google Scholar]
- 18. Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger P, Small S, Spencer B, Rockenstein E, Levine B, Wyss‐Coray T (2008) The autophagy‐related protein Beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest 118:2190–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spencer B, Potkar R, Trejo M, Rockenstein E, Patrick C, Gindi R, Adame A, Wyss‐Coray T, Masliah E (2009) Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in alpha‐synuclein models of Parkinson's and Lewy body diseases. J Neurosci 29:13578–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diskin T, Tal‐Or P, Erlich S, Mizrachy L, Alexandrovich A, Shohami E, Pinkas‐Kramarski R (2005) Closed head injury induces upregulation of Beclin 1 at the cortical site of injury. J Neurotrauma 22:750–762. [DOI] [PubMed] [Google Scholar]
- 21. Nascimento‐Ferreira I, Santos‐Ferreira T, Sousa‐Ferreira L, Auregan G, Onofre I, Alves S, Dufour N, Colomer Gould VF, Koeppen A, Déglon N, Pereira de Almeida L (2011) Overexpression of the autophagic beclin‐1 protein clears mutant ataxin‐3 and alleviates Machado‐Joseph disease. Brain 134:1400–1415. [DOI] [PubMed] [Google Scholar]
- 22. Munasinghe PE, Riu F, Dixit P, Edamatsu M, Saxena P, Hamer NS, Galvin IF, Bunton RW, Lequeux S, Jones G, Lamberts RR, Emanueli C, Madeddu P, Katare R (2015) Data supporting the activation of autophagy genes in the diabetic heart. Data Brief 5:269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Munasinghe PE, Riu F, Dixit P, Edamatsu M, Saxena P, Hamer NS, Galvin IF, Bunton RW, Lequeux S, Jones G, Lamberts RR, Emanueli C, Madeddu P, Katare R (2016) Type‐2 diabetes increases autophagy in the human heart through promotion of Beclin‐1 mediated pathway. Int J Cardiol 202:13–20. [DOI] [PubMed] [Google Scholar]
- 24. Yan L, Vatner DE, Kim SJ, Ge H, Masurekar M, Massover WH, Yang G, Matsui Y, Sadoshima J, Vatner SF (2005) Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci USA 102:13807–13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peng W, Liu Y, Xu WJ, Xia QH (2013) Role of Beclin 1‐dependent autophagy in cardioprotection of ischemic preconditioning. J Huazhong Univ Sci Technol Med Sci 33:51–56. [DOI] [PubMed] [Google Scholar]
- 26. Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J (2007) Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP‐activated protein kinase and Beclin 1 in mediating autophagy. Circ Res 100:914–922. [DOI] [PubMed] [Google Scholar]
- 27. Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong CS, Wu L, Kominami E, Ueno T, Yamamoto A, Federico M, Panganiban A, Vergne I, Deretic V (2009) Autophagy pathway intersects with HIV‐1 biosynthesis and regulates viral yields in macrophages. J Cell Biol 186:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, Lee M, Strowig T, Arrey F, Conenello G, Pypaert M, Andersen J, Garcia‐Sastre A, Münz C (2009) Matrix Protein 2 of Influenza A Virus Blocks Autophagosome Fusion with Lysosomes. Cell Host & Microbe 6, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hernaez B, Cabezas M, Munoz‐Moreno R, Galindo I, Cuesta‐Geijo M, Alonso C (2013) A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Curr Mol Med 13:305–316. [PubMed] [Google Scholar]
- 30. Gladue DP, O'Donnell V, Baker‐Branstetter R, Holinka LG, Pacheco JM, Fernandez‐Sainz I, Lu Z, Brocchi E, Baxt B, Piccone ME, Rodriguez L, Borca MV (2012) Foot‐and‐mouth disease virus nonstructural protein 2C interacts with Beclin1, modulating virus replication. J Virol 86:12080–12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Orvedahl A, Alexander D, Talloczy Z, Sun QH, Wei YJ, Zhang W, Burns D, Leib DA, Levine B (2007) HSV‐1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1:23–35. [DOI] [PubMed] [Google Scholar]
- 32. Chaumorcel M, Lussignol M, Mouna L, Cavignac Y, Fahie K, Cotte‐Laffitte J, Geballe A, Brune W, Beau I, Codogno P,AE (2012) The human cytomegalovirus protein TRS1 inhibits autophagy via its interaction with Beclin 1. J Virol 86:2571–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B (2005) Bcl‐2 antiapoptotic proteins inhibit Beclin 1‐dependent autophagy. Cell 122:927–939. [DOI] [PubMed] [Google Scholar]
- 34. Ku B, Woo J‐S, Liang C, Lee K‐H, Hong H‐S, Xiaofei E, Kim K‐S, Jung JU, Oh B‐H (2008) Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral Bcl‐2 of murine γ‐Herpesvirus 68. PLoS Pathog 4:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sinha S, Colbert CL, Becker N, Wei Y, Levine B (2008) Molecular basis of the regulation of Beclin 1‐dependent autophagy by the γ‐herpesvirus 68 Bcl‐2 homolog M11. Autophagy 4:989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Legakis JE, Klionski DJ, Overview of autophagy In Deretic V, Ed. (2006) Autophagy in immunity and infection. Weinheim, Germany: Wiley‐VCH Verlag, pp 3–17. [Google Scholar]
- 37. Kihara A, Noda T, Ishihara N, Ohsumi Y (2001) Two distinct Vps34 phosphatidylinositol 3‐kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae . J Cell Biol 152:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Itakura E, Kishi C, Inoue K, Mizushima N (2008) Beclin 1 forms two distinct phosphatidylinositol 3‐kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19:5360–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q (2008) Identification of Barkor as a mammalian autophagy‐specific factor for Beclin 1 and class III phosphatidylinositol 3‐kinase. Proc Natl Acad Sci USA 105:19211–19216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Itakura E, Mizushima N (2009) Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1‐PI3K complexes. Autophagy 5:534–536. [DOI] [PubMed] [Google Scholar]
- 41. Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama‐Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T (2009) Two Beclin 1‐binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11:385–396. [DOI] [PubMed] [Google Scholar]
- 42. Ruck A, Attonito J, Garces KT, Nunez L, Palmisano NJ, Rubel Z, Bai Z, Nguyen KC, Sun L, Grant BD, Hall DH, Melendez A (2011) The Atg6/Vps30/Beclin 1 ortholog BEC‐1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy 7:386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shravage BV, Hill JH, Powers CM, Wu L, Baehrecke EH (2013) Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development 140:1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, Roelandt R, De Rycke R, Verspurten J, Declercq W, Agostinis P, Vanden Berghe T, Lippens S, Vandenabeele P (2010) Caspase‐mediated cleavage of Beclin‐1 inactivates Beclin‐1‐induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis 1:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kang R, Zeh H, Lotze M, Tang D (2011) The Beclin 1 network regulates autophagy and apoptosis. Cell Death Diff 18:571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma XC, Liu HY, Murphy JT, Foyil SR, Godar RJ, Abuirqeba H, Weinheimer CJ, Barger PM, Diwan A (2015) Regulation of the transcription factor EB‐PGC1alpha axis by beclin‐1 controls mitochondrial quality and cardiomyocyte death under stress. Mol Cell Biol 35:956–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koukourakis MI, Kalamida D, Giatromanolaki A, Zois CE, Sivridis E, Pouliliou S, Mitrakas A, Gatter KC, Harris AL (2015) Autophagosome proteins LC3A, LC3B and LC3C have distinct subcellular distribution kinetics and expression in cancer cell lines. PLoS ONE 10:e0137675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. DeLano WL (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA, USA.
- 49. Mei Y, Su M, Soni G, Salem S, Colbert C, Sinha S (2014) Intrinsically disordered regions in autophagy proteins. Proteins 82:565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee EF, Perugini MA, Pettikiriarachchi A, Evangelista M, Keizer DW, Yao S, Fairlie WD (2016) The BECN1 N‐terminal domain is intrinsically disordered. Autophagy 12:460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mei Y, Ramanathan A, Glover K, Christopher SC, Sanishvili R, Chakravarthy S, Yang Z, Colbert CL, Sinha SC (2016) Conformational flexibility enables function of a BECN1 region essential for starvation‐mediated autophagy. Biochemistry 55:1945–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, Williams RL (2015) Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350:178–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li X, He L, Che KH, Funderburk SF, Pan L, Pan N, Zhang M, Yue Z, Zhao Y (2012) Imperfect interface of Beclin1 coiled‐coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun 3:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mei Y, Su M, Sanishvili R, Chakravarthy S, Colbert CL, Sinha SC (2016) Identification of BECN1 and ATG14 coiled‐coil interface residues important for starvation‐induced autophagy. Biochemistry. DOI: 10.1021/acs.biochem.6b00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Noda NN, Kobayashi T, Adachi W, Fujioka Y, Ohsumi Y, Inagaki F (2012) Structure of the novel C‐terminal domain of vacuolar protein sorting 30/autophagy‐related protein 6 and its specific role in autophagy. J Biol Chem 287:16256–16266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang F‐L, Deng H, Liu L, Gao N, Yu L, Shi Y (2012) Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res 22:473–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Furuya N, Yu F, Byfield M, Pattingre S, Levine B (2005) The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 1:46–52. [DOI] [PubMed] [Google Scholar]
- 58. Maiuri M, Le Toumelin G, Criollo A, Rain J, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman J, Geneste O, Kroemer G (2007) Functional and physical interaction between Bcl‐XL and a BH3‐like domain in Beclin‐1. Embo J 26:2527–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Oberstein A, Jeffrey PD, Shi Y (2007) Crystal structure of the Bcl‐XL‐Beclin 1 peptide complex: Beclin 1 is a novel BH3 only protein. J Biol Chem 282:13123–13132. [DOI] [PubMed] [Google Scholar]
- 60. Feng W, Huang S, Wu H, Zhang M (2007) Molecular basis of Bcl‐XL's target recognition versatility revealed by the structure of Bcl‐XL in complex with the BH3 domain of Beclin‐1. J Mol Biol 372:223–235. [DOI] [PubMed] [Google Scholar]
- 61. Sinha S, Levine B (2009) The autophagy effector Beclin 1: A novel BH3‐only protein. Oncogene 27:S137–S148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU (2006) Autophagic and tumour suppressor activity of a novel Beclin1‐binding protein UVRAG. Nat Cell Biol 8:688–699. [DOI] [PubMed] [Google Scholar]
- 63. Baskaran S, Carlson L‐A, Stjepanovic G, Young LN, Kim DJ, Grob P, Stanley RE, Nogales E, Hurley JH (2014) Architecture and dynamics of the autophagic phosphatidylinositol 3‐kinase complex. eLife 3:e05115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z (2001) Intrinsically disordered proteins. J Mol Graph Model 19:26–59. [DOI] [PubMed] [Google Scholar]
- 65. Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ (2008) Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci 13:6580–6603. [DOI] [PubMed] [Google Scholar]
- 66. Jones S, Thornton JM (1996) Principles of protein‐protein interactions. Proc Natl Acad Sci USA 93:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wright P, Dyson H (1999) Intrinsically unstructured proteins: Re‐assessing the protein structure‐function paradigm. J Mol Biol 293:321–331. [DOI] [PubMed] [Google Scholar]
- 68. Dunker AK, Obradovic Z (2001) The protein trinity–linking function and disorder. Nat Biotechnol 19:805–806. [DOI] [PubMed] [Google Scholar]
- 69. Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208. [DOI] [PubMed] [Google Scholar]
- 70. Wright P, Dyson H (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gao J, Xu D (2012) Correlation between posttranslational modification and intrinsic disorder in protein. Pac Symp Biocomput 94–103. [PMC free article] [PubMed] [Google Scholar]
- 72. Dosztányi Z, Mészáros B, Simon I (2009) ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics 25:2745–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dinkel H, Michael S, Weatheritt RJ, Davey NE, Roey KV, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A, Jödicke L, Dammert MA, Schroeter C, Hammer M, Schmidt T, Jehl P, McGuigan C, Dymecka M, Chica C, Luck K, Via A, Chatraryamontri A, Haslam N, Grebnev G, Edwards RJ, Steinmetz MO, Meiselbach H, Diella F, Gibson TJ (2012) ELM—The database of eukaryotic linear motifs. Nucleic Acids Res 40:D242–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mohan A, Oldfield C, Radivojac P, Vacic V, Cortese M, Dunker A, Uversky V (2006) Analysis of molecular recognition features (MoRFs). J Mol Biol 362:1043–1059. [DOI] [PubMed] [Google Scholar]
- 75. Vacic V, Oldfield C, Mohan A, Radivojac P, Cortese M, Uversky V, Dunker A (2007) Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res 6:2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Glover K, Mei Y, Sinha S (2016) Identifying intrinsically disordered protein regions likely to undergo binding‐induced helical transitions. Biochim Biophys Acta. DOI:10.1016/j.bbapap.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Noble C, Dong J, Manser E, Song H (2008) Bcl‐xL and UVRAG cause a monomer‐dimer switch in Beclin1. J Biol Chem 283:26274–26282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Adi‐Harel S, Erlich S, Schmukler E, Cohen‐Kedar S, Segev O, Mizrachy L, Hirsch JA, Pinkas‐Kramarski R (2010) Beclin 1 self‐association is independent of autophagy induction by amino acid deprivation and rapamycin treatment. J Cell Biochem 110:1262–1271. [DOI] [PubMed] [Google Scholar]
- 79. Maiuri M, Criollo A, Tasdemir E, Vicencio J, Tajeddine N, Hickman J, Geneste O, Kroemer G (2007) BH3‐only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl‐2/Bcl‐XL. Autophagy 3:374–476. [DOI] [PubMed] [Google Scholar]
- 80. Wei Y, Pattingre S, Sinha S, Bassik M, Levine B (2008) JNK1‐mediated phosphorylation of Bcl‐2 regulates starvation‐induced autophagy. Mol Cell 30:678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wei Y, Sinha S, Levine B (2008) Dual role of JNK1‐mediated phosphorylation of Bcl‐2 in autophagy and apoptosis regulation. Autophagy 4:949–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, Sciarretta S, Del Re DP, Zablocki DK, Hsu C‐P, Lim D‐S, Isobe M, Sadoshima J (2013) Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl‐2. Nat Med 19:1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gurkar A, Chu K, Raj L, Bouley R, Lee S, Kim Y, Dunn S, Mandinova A, Lee S (2013) Identification of ROCK1 kinase as a critical regulator of Beclin1‐mediated autophagy during metabolic stress. Nat Commun 4:2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3‐phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182:685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Obara K, Noda T, Niimi K, Ohsumi Y (2008) Transport of phosphatidylinositol 3‐phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae . Genes Cells 13:537–547. [DOI] [PubMed] [Google Scholar]
- 86. Obara K, Ohsumi Y (2008) Dynamics and function of PtdIns(3)P in autophagy. Autophagy 4:952–954. [DOI] [PubMed] [Google Scholar]
- 87. Liang C, Lee JS, Inn K‐S, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU (2008) Beclin1‐binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 10:776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Backer JM (2008) The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J 410:1–17. [DOI] [PubMed] [Google Scholar]
- 89. Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman B, Shokat K, Williams R (2010) Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science 327:1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Stack JH, Herman PK, Schu PV, Emr SD (1993) A membrane‐associated complex containing the Vps15 protein kinase and the Vps34 PI 3‐kinase is essential for protein sorting to the yeast lysosome‐like vacuole. Embo J 12:2195–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T (2010) Autophagy requires endoplasmic reticulum targeting of the PI3‐kinase complex via Atg14L. J Cell Biol 190:511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Fan W, Nassiri A, Zhong Q (2011) Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc Natl Acad Sci USA 108:7769–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Behrends C, Sowa ME, Gygi SP, Harper JW (2010) Network organization of the human autophagy system. Nature 466:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. He C, Levine B (2010) The Beclin 1 interactome. Curr Opin Cell Biol 22:140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Maiuri MC, Criollo A, Kroemer G (2010) Crosstalk between apoptosis and autophagy within the Beclin 1 interactome. Embo J 29:515–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dusetti NJ, Jiang Y, Vaccaro MI, Tomasini R, Azizi Samir A, Calvo EL, Ropolo A, Fiedler F, Mallo GV, Dagorn JC, Iovanna JL (2002) Cloning and expression of the rat vacuole membrane protein 1 (VMP1), a new gene activated in pancreas with acute pancreatitis, which promotes vacuole formation. Biochem Biophys Res Commun 290:641–649. [DOI] [PubMed] [Google Scholar]
- 97. Jiang PH, Motoo Y, Vaccaro MI, Iovanna JL, Okada G, Sawabu N (2004) Expression of vacuole membrane protein 1 (VMP1) in spontaneous chronic pancreatitis in the WBN/Kob rat. Pancreas 29:225–230. [DOI] [PubMed] [Google Scholar]
- 98. Molejon MI, Ropolo A, Vaccaro MI (2013) VMP1 is a new player in the regulation of the autophagy‐specific phosphatidylinositol 3‐kinase complex activation. Autophagy 9:933–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ropolo A, Grasso D, Pardo R, Sacchetti ML, Archange C, Lo Re A, Seux M, Nowak J, Gonzalez CD, Iovanna JL, Vaccaro MI (2007) The pancreatitis‐induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J Biol Chem 282:37124–37133. [DOI] [PubMed] [Google Scholar]
- 100. Molejon MI, Ropolo A, Re AL, Boggio V, Vaccaro MI (2013) The VMP1‐Beclin 1 interaction regulates autophagy induction. Sci Rep 3:1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Strappazzon F, Vietri‐Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, Piacentini M, Levine B, Cecconi F (2011) Mitochondrial BCL‐2 inhibits AMBRA1‐induced autophagy. Embo J 30:1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gu W, Wan D, Qian Q, Yi B, He Z, Gu Y, Wang L, He S (2014) Ambra1 is an essential regulator of autophagy and apoptosis in SW620 cells: pro‐survival role of Ambra1. PLoS One 9:e90151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Shi CH, Wu J, Fu M, Zhang BH, Wang J, Yang X, Chi YP (2014) Ambra1 modulates starvation‐induced autophagy through AMPK signaling pathway in cardiomyocytes. Biochem Biophys Res Commun 452:308–314. [DOI] [PubMed] [Google Scholar]
- 104. Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL (2013) ULK1 induces autophagy by phosphorylating Beclin‐1 and activating VPS34 lipid kinase. Nat Cell Biol 15:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Stirnimann CU, Petsalaki E, Russell RB, Muller CW (2010) WD40 proteins propel cellular networks. Trends Biochem Sci 35:565–574. [DOI] [PubMed] [Google Scholar]
- 106. Cianfanelli V, De Zio D, Di Bartolomeo S, Nazio F, Strappazzon F, Cecconi F (2015) Ambra1 at a glance. J Cell Sci 128:2003–2008. [DOI] [PubMed] [Google Scholar]
- 107. Criollo A, Niso‐Santano M, Malik SA, Michaud M, Morselli E, Marino G, Lachkar S, Arkhipenko AV, Harper F, Pierron G, Rain JC, Ninomiya‐Tsuji J, Fuentes JM, Lavandero S, Galluzzi L, Maiuri MC, Kroemer G (2011) Inhibition of autophagy by TAB2 and TAB3. Embo J 30:4908–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Niso‐Santano M, Criollo A, Malik SA, Michaud M, Morselli E, Marino G, Lachkar S, Galluzzi L, Maiuri MC, Kroemer G (2012) Direct molecular interactions between Beclin 1 and the canonical NF kappa B activation pathway. Autophagy 8:268–270. [DOI] [PubMed] [Google Scholar]
- 109. Luo S, Garcia‐Arencibia M, Zhao R, Puri C, Toh PP, Sadiq O, Rubinsztein DC (2012) Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol Cell 47:359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A (1999) The proapoptotic activity of the Bcl‐2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell 3:287–296. [DOI] [PubMed] [Google Scholar]
- 111. Lei K, Davis RJ (2003) JNK phosphorylation of Bim‐related members of the Bcl2 family induces Bax‐dependent apoptosis. Proc Natl Acad Sci USA 100:2432–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ma CY, Wang NH, Detre C, Wang GX, O'Keeffe M, Terhorst C (2012) Receptor signaling lymphocyte‐activation molecule family 1 (Slamf1) regulates membrane fusion and NADPH oxidase 2 (NOX2) activity by recruiting a Beclin‐1/Vps34/ultraviolet radiation resistance‐associated gene (UVRAG) complex. J Biol Chem 287:18359–18365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Yue ZY, Horton A, Bravin M, DeJager PL, Selimi F, Heintz N (2002) A novel protein complex linking the delta 2 glutamate receptor and autophagy: Implications for neurodegeneration in Lurcher mice. Neuron 35:921–933. [DOI] [PubMed] [Google Scholar]
- 114. Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A (2009) The Arp2/3 activator WASH controls the fission ofeEndosomes through a large multiprotein complex. Dev Cell 17:712–723. [DOI] [PubMed] [Google Scholar]
- 115. Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, Huang G, Ye B, Li C, Dai Z, Hou N, Cheng X, Sun Q, Li L, Yang X, Fan Z (2013) WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. Embo J 32:2685–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhong Y, Wang QJ, Li XT, Yan Y, Backer JM, Chait BT, Heintz N, Yue ZY (2009) Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1‐phosphatidylinositol‐3‐kinase complex. Nat Cell Biol 11:468–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Liang XH, Yu J, Brown K, Levine B (2001) Beclin 1 contains a leucine‐rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res 61:3443–3449. [PubMed] [Google Scholar]
- 118. Romanowski MJ, Shenk T (1997) Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate‐early transcription unit within irs1 whose product antagonizes transcriptional activation. J Virol 71:1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mouna L, Hernandez E, Bonte D, Brost R, Amazit L, Delgui LR, Brune W, Geballe AP, Beau I, Esclatine A (2016) Analysis of the role of autophagy inhibition by two complementary human cytomegalovirus BECN1/Beclin 1‐binding proteins. Autophagy 12:327–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Shoji‐Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, Huerta C, Virgin HW, Helms JB, Eerland R, Tooze SA, Xavier R, Lenschow DJ, Yamamoto A, King D, Lichtarge O, Grishin NV, Spector SA, Kaloyanova DV, Levine B (2013) Identification of a candidate therapeutic autophagy‐inducing peptide. Nature 494:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wei Y, An Z, Zou Z, Sumpter R, Su M, Zang X, Sinha S, Gaestel M, Levine B (2015) The stress‐responsive kinases MAPKAPK2/MAPKAPK3 activate starvation‐induced autophagy through Beclin 1 phosphorylation. eLife 4:e05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL (2013) Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Song X, Kim SY, Zhang L, Tang D, Bartlett DL, Kwon YT, Lee YJ (2014) Role of AMP‐activated protein kinase in cross‐talk between apoptosis and autophagy in human colon cancer. Cell Death Dis 5:e1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wang R, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B (2012) Akt‐mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338:956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Altomare DA, Testa JR (2005) Perturbations of the AKT signaling pathway in human cancer. Oncogene 24:7455–7464. [DOI] [PubMed] [Google Scholar]
- 126. Sun T, Li X, Zhang P, Chen WD, Zhang HL, Li DD, Deng R, Qian XJ, Jiao L, Ji J, Li YT, Wu RY, Yu Y, Feng GK, Zhu XF (2015) Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun 6:7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137:133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Platta HW, Abrahamsen H, Thoresen SB, Stenmark H (2012) Nedd4‐dependent lysine‐11‐linked polyubiquitination of the tumour suppressor Beclin 1. Biochem J 441:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Shi CS, Kehrl JH (2010) Traf6 and A20 differentially regulate TLR4‐induced autophagy by affecting the ubiquitination of Beclin 1. Autophagy 6:986–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Su M, Mei Y, Sanishvili R, Levine B, Colbert CL, Sinha S (2014) Targeting γ‐herpesvirus 68 Bcl‐2–mediated down‐regulation of autophagy. J Biol Chem 289:8029–8050. [DOI] [PMC free article] [PubMed] [Google Scholar]


