Abstract
Scripts are used in PyMOL to exert precise control over the appearance of the output and to ease remaking similar images at a later time. We developed horizontal scripts to ease script development. A horizontal script makes a complete scene in PyMOL like a traditional vertical script. The commands in a horizontal script are separated by semicolons. These scripts are edited interactively on the command line with no need for an external text editor. This simpler workflow accelerates script development. In using PyMOL, the illustration of a molecular scene requires an 18‐element matrix of view port settings. The default format spans several lines and is laborious to manually reformat for one line. This default format prevents the fast assembly of horizontal scripts that can reproduce a molecular scene. We solved this problem by writing a function that displays the settings on one line in a compact format suitable for horizontal scripts. We also demonstrate the mapping of aliases to horizontal scripts. Many aliases can be defined in a single script file, which can be useful for applying costume molecular representations to any structure. We also redefined horizontal scripts as Python functions to enable the use of the help function to print documentation about an alias to the command history window. We discuss how these methods of using horizontal scripts both simplify and enhance the use of PyMOL in research and education.
Keywords: molecular graphics, molecular visualization, molecular artwork, scientific illustration, scientific communication, Python scripting, molecular literacy, molecular orientation, reproducible research
Introduction
The uploading and viewing of protein structures in various molecular graphics viewers is generally intuitive, but the use of these programs for structure analysis and figure making requires knowledge of the options available in gui pulldowns, plugins, and commands. Many users have trouble recalling this knowledge because they use these programs infrequently. One program that is difficult to master is PyMOL. In spite of this difficulty, PyMOL is used widely in structural biology.1, 2 PyMOL is often used for molecular visualization by crystallographic, molecular dynamics simulation, and protein modeling software packages.3, 4, 5, 6, 7, 8, 9, 10, 11 PyMOL provides exquisite control of the display of images. For this reason, PyMOL is also favored for making molecular images for journal covers. New users of PyMOL with little training in computing have trouble mastering PyMOL because the agile use of PyMOL ultimately requires the use of multiple commands in scripts. Most beginners are reluctant to use script files for many reasons. The traditional script file has each command listed on a single line in a vertical array. These vertical scripts are tedious to use for three reasons. First, an external editor is needed to change the parameter values. Next, the script has to be saved, reloaded into PyMOL, rerun, and then the parameters often need to be edited again. This cycle of four steps is slow and tiring. Second, extra mouse movements are required to move the mouse cursor from line to line while editing the vertical script. Third, the molecular scene settings are required to set the molecule with a specific orientation, zoom, and slab thickness. These settings are printed to the command history window with the get_view command. The default settings span several lines and are prone to error in copying from the command history window and pasting into the script file. An easier approach to using scripts in PyMOL would empower a wider range biologists to do advanced structure analyses and thereby gain deeper scientific insights into a protein's function from its structure.
We tested with first‐year graduate students in biomedicine the idea that horizontal scripts combined with a compact format for the molecular scene settings would be easier for the students to master than vertical scripts. The horizontal script has all of the commands on one line. The script can be edited on the command line without needing to use an external text editor between each cycle of edits. This interactive approach removes the saving and reloading steps in the four‐step cycle of editing vertical scripts. Read line commands move the mouse cursor swiftly between commands. This new approach to script writing accelerates script development and reduces user fatigue and frustration. After explaining the assembly of horizontal scripts, we demonstrate four ways in which they can be used to ease and enhance the use of PyMOL. Our students had no trouble using horizontal scripts when the four methods were introduced gradually. One of the goals of structural biology is to enable other biologists to use molecular structures to think about their experimental systems and to plan their next experiments. Horizontal scripting advances the field toward this goal because it eases the use of molecular graphics with scripts by biologists with little training in computing.
Results
We present four methods of using horizontal scripts from the simplest to the most advanced (Fig. 1). We use this order to introduce trainees to scripting in PyMOL. In method one [Fig. 1(A)], the user copies a row of commands from a document. They paste the commands onto the command line in the external GUI of PyMOL (Fig. 2). In the second method [Fig. 1(B)], the commands are assigned to an alias. The user pastes the aliased commands onto the command line and hits the return key to activate the alias. The commands are executed by entering the name of the alias. In the third method [Fig. 1(C)], multiple aliased commands are listed in a PyMOL command language script file (i.e., a *.pml file). The user sources the script from inside PyMOL to activate the aliases for the rest of the session. In the fourth method [Fig. 1(D)], the aliased commands are included in a Python script. This script provides documentation that is accessible with PyMOL's help command. This documentation can include the aliased PyMOL commands. The printing of the commands to the command history window enables the copying and pasting of some or all of the displayed commands onto the command line during the assembly of new horizontal scripts. We introduce novice users to all four methods by the second or third training session. Expert users will find each method useful in different situations. The four methods introduce new users to commands, horizontal scripts, aliases, pml script files, and Python script files.
Figure 1.

Workflows of the four methods of using horizontal scripts. A: Copy and paste (method one). B: Paste aliased row of commands (method two). C: PyMOL command language script with multiple aliased rows of commands (method three). D: Python script with several aliased rows of commands and docstrings (method four).
Figure 2.
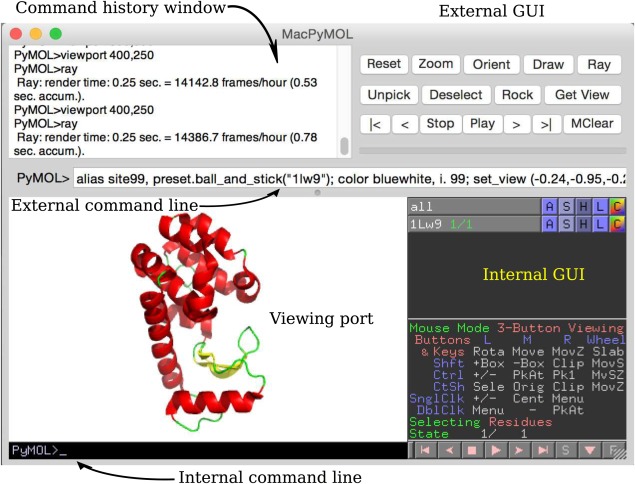
Image showing parts of the MacPyMOL GUI: the two command lines, the internal GUI, the external GUI, and the viewing port. The viewing port is the OpenGL window in which the molecular scene is viewed.
Method one: Pasting horizontal scripts
In method one, we copied a row of commands from a PDF or plain text file. The row had a whitespace at the end to avoid the copying of the newline character and then the subsequent need to delete this character from the command line in PyMOL. New users find the first method easy to apply because they are already skilled at copying and pasting text from daily computer use [Table 1, Figs. 1, 2, and Fig. 3(C)]. The row of commands is analogous to a horizontal script. PyMOL commands are usually listed vertically in a script file with one command per line. However, PyMOL will accept many commands on one line if semicolons separate the commands [Fig. 3(C)]. The pasting of one long line of commands was much easier than pasting and entering each command separately. An external text editor was not required for the editing of the horizontal script after the horizontal script was pasted onto the command line. We hit the up‐arrow key to recall a previous line of commands for editing on the command line. The cursor was moved to either end of the command line with Readline commands.12 Control‐a moved the cursor to the beginning of the line, and control‐e to moved it to the end of the line. Both of these commands work in either of PyMOL's command lines. On the external command line (the command line at the top of the PyMOL GUI), command‐f advanced the cursor to the end of a word, and command‐b moved the cursor to the beginning of a word. Also on the external command line, control‐f advanced the cursor by one character and control‐b moved it backwards by one character. The interactive editing of the commands on one line was fast because the user does not have to shift the cursor between lines. Several commands can be edited in one round of editing before they are submitted as a group. This efficient method of editing the horizontal scripts accelerated the development of molecular scenes.
Table 1.
The Advantages and Disadvantages of Four Methods of Using Horizontal Scripts in PyMOL
| Method | Advantages | Disadvantages |
|---|---|---|
|
Copy‐and‐paste (Method one) |
One paste operation No installation Easy to edit on command line |
Add one line at a time |
|
Paste with alias (Method two) |
One paste operation No installation Invoke with alias name Easy to edit on command line |
Add one alias at a time |
|
PyMOL script (Method three) |
Multiple aliases defined at once Invoke with alias name Need no knowledge of Python Easy to edit |
Must find the script Load with @filename.pml |
|
Python script (Method four) |
Multiple aliases defined at once Invoke with alias name On‐line alias description Easy to extend and adapt |
Must find script Load with “run filename.py” Need knowledge of Python |
Figure 3.

Scene settings and horizontal scripts. A: Molecular scene settings from the get_view() command with nine decimal places in the default format. B: Molecular scene settings from the function roundview() with one decimal place. C: A long row of PyMOL commands separated by semicolons on the PyMOL command line and the scene settings with the default of two decimal places from the function roundview(). D: The alias site99 mapped to the row of commands in (C) on the PyMOL command line. E: The alias site99 mapped to a long row of commands from (C) in a Python script.
The function roundview()
A matrix with 18 elements describes the molecular scene in the viewing port of the PyMOL GUI. This matrix enables the recording and the recreation of a molecular scene at a later time [Fig. 3(A)]. The matrix includes the center of rotation and orientation of the molecular scene. It also includes the positions of the front and rear clipping planes, and the orthoscopic flag. The built‐in PyMOL function get_ view() printed the matrix to the command history window as an array of three columns and six rows [Fig. 3(A)]. The set_view() command applied the matrix. This matrix is tedious to manually reformat to fit on one line. In addition, the matrix elements have nine decimal places. The last seven or eight decimal places can be removed without a discernible change in the scene.
We wrote a substitute for get_view() called roundview() [Fig. 3(B), Supporting Information S1_ File]. This new function enabled resetting of the number of decimal places [e.g., set to one in Fig. 3(B)]. The script wrote the matrix elements on one line as arguments of the set_view() command. We copied the returned set_view() command from the command history window to a text file or to the command line in PyMOL. A semicolon must separate the set_view() command from the other commands on the line. The roundview() function greatly eases the use of the set_view() command on the command line. This roundview() function also enables the use of compact scene settings in traditional vertical script files that have one command per line.
Our trainees welcomed the simplified viewing port settings from the roundview() function, the multiple commands linked by semicolons on one line, and the Readline commands for the fast movement of the cursor on the command line during interactive editing of the horizontal script. These three features eased their use of PyMOL. These features should also interest educators and lab directors who want to introduce molecular graphics to beginners, to improve their own expertise with PyMOL, or both. Users can develop horizontal scripts for use in other molecular graphics computer programs and with other GUI‐driven scientific software programs that have internal command lines.
Limits to row length
The width of the computer screen set the practical limit of the length of the horizontal script. The PyMOL command line showed 165 characters of 12pt font on a 12‐inch screen, 250 characters on a 15‐inch screen, and 350 characters on a 24‐inch screen. However, we worked with one horizontal script that was ten times wider; the NA alias in the startupAliases.py script had 57 commands with 535 words and 3581 characters. The plain text editor TextWrangler.app has a horizontal scroll‐bar that enabled the viewing of scripts wider than the screen when line wrapping was turned off. We selected and copied the single line from the document and pasted it onto the command line. Most laptop screens are 12 inches wide, so a command length of 165 characters was a practical limit for use in a workshop or classroom. This limit was still twice the 80 character width used for a letter document.
Navigation of the command line
A horizontal script can be copied and pasted onto the command line just below the command history window in PyMOL. The cursor needs to be moved swiftly on the command line to edit the horizontal script with agility. The cursor can be moved to the next site to be edited in the horizontal script by using a mouse. The end of long horizontal scripts will be out of view initially on computer screens that are too narrow to show the entire horizontal script. The end of the script can be revealed by moving the cursor to the end of the script with the command control‐e. The cursor can be moved back to the beginning of the script with the command control‐a. The up arrow key is used to recall previously entered horizontal scripts for editing and reuse. The movement of the cursor with the mouse and the last three commands are sufficient for most editing tasks. These commands are available in most versions of PyMOL.
Method two: paste aliased horizontal scripts
The second method used aliases to extend the use of horizontal scripts [Fig. 3(B)]. Aliases enabled the rapid recall and reapplication of horizontal scripts. PyMOL has an alias() function [Fig. 3(D)] that we used to assign an alias name to a row of commands. The alias name may have only two or three unique characters for easy use. The alias name was entered on the command line to execute the row of commands. Users can edit the commands in the alias definition as described in method one. Once defined during a PyMOL session, the alias remained available throughout the session. The aliases also survived the execution of the reinitialize command that deleted all of the current molecular objects. It was sometimes faster to enter the alias name than to find an old row of commands with the up‐arrow key during a long PyMOL session, so method two provides advantages over method one.
Method three: sourcing aliases from a PyMOL command script
The third method loaded a PyMOL script file that contained several aliases [Table 1, Fig. 3(C,D)]. We sourced the script from the current directory with the command @filename.pml. The names and descriptions of the aliases were printed to the command history window. The aliases remained active for the entire PyMOL session and were reused many times. The script of aliases can also be sourced from the .pymolrc startup file in the user's home directory by adding @filename.pml on a new line. The sourcing of a file of aliases was easier than defining aliases one at a time on the command line.
The aliases enabled easy navigation between different sites in a protein molecule. A series of aliases could be used to give guided tours of a molecule in seminars, workshops, and lectures. A series of aliases could also be used for self‐guided tours in collaborative editing of a series of alternative figures, journal articles, tutorials, and homework. The PyMOL commands in the script file were easy for new users to understand and edit. The disadvantage of the third method was the lack of on‐line documentation (i.e., docstrings, Table 1). Method four addressed this shortcoming [Fig. 1(D)].
Method four: sourcing aliases from a python script
PyMOL can also read Python script files. Python scripts can print statements in the command history window. We defined each alias as a Python function with its own documentation string. We printed these “docstrings” to the command history window by entering help alias name on the PyMOL command line. We describe below three Python scripts containing aliases. The docstrings in the last script contained the alias's PyMOL code as a vertical script for easy copying of single lines of commands during the recycling of small parts of the code by copying and pasting them onto the command line. Of course, the entire script can also be copied and pasted into an external text file for use as traditional vertical script. The docstring also contained the commands in a horizontal script. This horizontal script can be copied and pasted onto the command line to quickly edit the parameter values during customization of the script for a new molecular scene.
To make the first script, we converted the wtT4L.pml file into a Python script wtT4L.py [Fig. 3(E), Supporting Information S2_File]. We added print statements that described the alias. The wtT4L.py script generated molecular scenes of wild‐type phage T4 lysozyme (Fig. 4). We entered run wtT4L.py to load the script. The script set the background color to white, fetched the structure from the PDB, and printed the names of the aliases in the command history window. There was no need to open the script in an external text editor to find the names of the aliases. This script had simple Python syntax. It required modest knowledge of Python syntax to be modified for use with new molecules.
Figure 4.
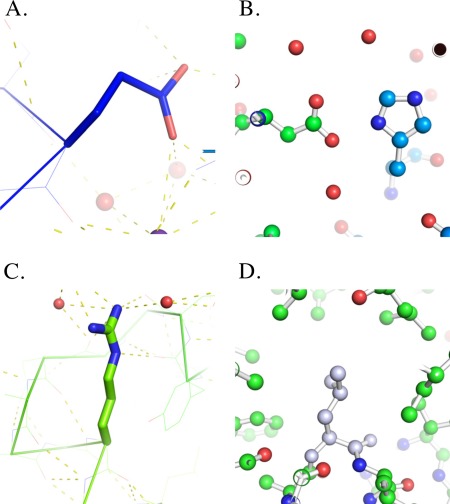
Four sites in T4 lysozyme (PDB‐ID 3fa0 [13]) mapped to four aliases in the script file wtT4L.py (see Supporting Information S2_ File). A: Site11, Glu11 in the active site.25 B: Site31, the His31‐Asp70 salt bridge that adds 3 − 5 kcal/mol of stability to T4 lysozyme.26 C: Site96, the site of the Arg96 →His mutant that suffers a large loss in stability.27 D: Site99, the site of a large cavity upon replacement of Leu99 with alanine.28
In the second Python script (exam2.py, Supporting Information S4_File), each alias was defined inside a Python function. Each function had a documentation string (or docstring) that explained the alias. For example, we entered help q1 to print the documentation string for the first alias q1. This alias was the first question of a take‐home exam in a first‐year graduate course.
In the third Python script (startupAliases.py, Supporting Information S5_File), we inclued a number of standard views and complex scenes. We included the PyMOL commands for the alias in the docstring for easy recycle of code fragments. Typing help alias name printed a detailed description and the commands mapped to the alias. The command history window sometimes became full of past commands. To reprint the list of aliases, we entered the alias SA (Show Aliases).
On‐line access to the commands in the third script enabled convenient reuse of past code. We copied the code from the command history window and pasted it onto the command line during assembly of a new horizontal script. The command parameter values were then edited on the PyMOL command line. The final horizontal script was copied to a separate plain text file for later reuse.
We put the aliases in startupAliases.py in three groups: standard views of proteins, complex scenes, and customized representations. An example of the first group was of phage T4 lysozyme in its standard view. An example of the second category included the commands to make electron density maps. An example of the third category included the commands to make ambient occlusion images.13 The startupAliases.py file was stored in ∼./Scripts/PyMOLScripts folder. This file was called by the ∼/.pymolrc startup file with the command run ∼./Scripts/PyMOLScripts/startupAliases.py. The display of the commands in the command history window reduced the need to search for old scripts for code to use as templates.
Access to documentation on‐line for aliased commands extended the uses of horizontal scripts. The printing of PyMOL code to the command history window eased the reuse of old commands. The access to functional chunks of code on‐line should also speed up the mastery of PyMOL.
Limits on the “reinitialize” and “label” functions in horizonal scripts
We found two limitations to the use of horizontal scripts. First, the reinitialize command did not work completely when run from a horizontal script in PyMOL versions 1.6 and more recent. To solve this problem, we substituted the reinitialize command with the command delete all that removes all molecular objects in the current PyMOL session. Second, only one label command was allowed in a horizontal script. The label command had to be the last command in a horizontal script, and it could not be followed by a semicolon. A series of label commands were put in a PyMOL script file called Labels.pml. This file was sourced from the horizontal script with the command @Labels.pml. These solutions to two limitations on using horizontal scripts in PyMOL were easy to find. Additional limitations on the use of horizontal scripts may arise in future versions of PyMOL or may be discovered by other users. These new limitations and their solutions will be posted on the EasyPyMOL GitHub website.
Discussion
We used horizontal scripts in PyMOL to make molecular scenes and to aid the recall of PyMOL commands. We developed four methods of horizontal scripting with rising levels of sophistication (Table 1, Fig. 1). The first method eases the interactive development of blocks of code during a PyMOL session. The last method is useful for presentations, self‐guided tours of a molecular structure, learning the PyMOL command syntax, and recycling code for standard views and complex scenes. These methods combine PyMOL's ability to accept horizontal scripts, the availability inside PyMOL of the Readline commands for agile editing on the command line, and the output of the roundview() function. This function gives the scene settings in a compact format for easy addition to a series of commands in a single row. The horizontal scripts complement and extend the approaches outlined earlier by others.1, 14 We discuss below how horizontal scripts enable new ways to use PyMOL.
Agile editing of many commands on the command line
The PyMOL wiki http://pymolwiki.org/ has examples of several commands on one line, but there are no examples of using many commands on one line. There are three possible reasons. First, the multiple commands in a traditional script file are often used with the molecular scene settings returned from PyMOL's get_view command. The default format of these settings is too long for easy use on one line. We solved this problem with the roundview() function that writes the settings on one line in a compact format. Second, many commands on one line are not so easy to edit by moving the cursor with repetitive use of the backspace key or spacebar. However, the Readline commands in PyMOL enable rapid movement of the cursor on the command line. Third, past examples of using several commands on one line often used two‐to‐four commands per line. We used up to 57 commands per line along with the compact settings from the function roundview(). This large number of commands exceeded by a factor of five the ten commands in a typical block of related PyMOL commands that are required to accomplish a complex task. Our extensions of the use of multiple commands in one row make horizontal scripts attractive to users at all levels of expertise.
Comparison to PyMOL session files
PyMOL session files are an alternative to using scripts. A PyMOL session file stores a molecular scene in a binary format. The creation of the session file can be done with the PyMOL GUI and without writing a script file. However, the session files are large and can be hard to send by e‐mail as attachments. In addition, the session files are not universal. They cannot be opened on different operating systems, and they are not always backward or forward compatible with different versions of PyMOL on the same computer. This means once a user creates a session file, they should retain the version of PyMOL that was used to make it. This measure can consume a lot of disk because PyMOL is updated several times a year.
Laboratory presets
Aliased horizontal scripts can be shared within a laboratory to make consistent figures for publication. Comparative studies often have a reference molecule such as of a wild‐type protein or an apoenzyme. The reference molecule is often shown in a standard orientation that minimizes the overlap of secondary structure elements. This standard orientation adds consistency between figures within and between manuscripts. The settings for the standard orientation of the reference structure can be stored in a horizontal script and then assigned to an alias for easy reuse. Standard orientations may also be valuable for ligand binding sites, active sites, interfaces between oligomers, and crystal lattice contacts. Likewise, a lab may have preferred styles of presenting molecular representations for specific purposes. These preferences can be encoded in horizontal scripts and then assigned aliases. All of the aliases could be stored in one script file for easy sharing among lab or project members. This script of template aliases could be installed permanently by sourcing it in the .pymolrc file in a user's home directory. This script could also be read by all computer accounts by sourcing it from the pymol_startup.py script file. The file of template scripts promotes consistent use of standard views and molecular representations by project members.
Library of horizontal scripts
Alternatively, the startupAliases.py script can be modified to include new aliases to the lab's presets and to commonly used blocks of code. By typing help alias name, the PyMOL code is printed to the command history window. This code can be copied to the command line in PyMOL and edited as required. The presence of the PyMOL code in the command history window helps new users learn the command syntax by example and helps old users by refreshing their faded memories of the command syntax. This approach addresses directly the problems of building skills with PyMOL by new users and of fading memories of the commands by experienced users. The on‐line source of code templates provided by the startupAliases.py file complements the existing high‐quality documentation that comes with PyMOL. This built‐in documentation explains the syntax but does not always illustrate it with examples and does not provide examples involving multiple commands. The startupAliases.py file addresses this gap with custom examples. By filling this gap in the documentation and by addressing the problems of skill building and memory fading that make PyMOL hard to learn and use, the horizontal scripts make PyMOL easier to use for both new and experienced users.
Science education
Instructors use molecular graphics to deepen the understanding of key concepts about molecular structure and biology15, 16, 17, 18 (see Refs. 17, 18 for examples using PyMOL). These concepts are usually introduced earlier with 2‐D images.19 Yet, many students struggle to understand 3D structures because their 3‐D molecular literacy has not been developed. We build their molecular literacy by spending two hours on a detailed tour in PyMOL of a human protein of importance in cysteine metabolism (human γ‐glutamyltranspeptidase) with beta sheets and alpha helices, post‐translational modifications, an autocleaving threonine, and ion binding sites.20 We discuss covalent structure, van der Waals surfaces, protein‐water interactions, and hydrogen bonding as we move from site to site. We use horizontal scripts to aid this instruction. We distribute by e‐mail the scripts in a plain text file without line wrapping. The students copy and paste the horizontal scripts into PyMOL (method one) to reproduce the scenes on their laptop computers. After one to two hours of using method one for horizontal scripting, we introduce the three other methods. These directed exercises build the students' understanding of how the commands and scripts work. We include horizontal scripts in homework assignments that test the students' understanding of molecular structure. These scripts display specific sites in a molecular structure. The above uses of horizontal scripts reduce the time spent by students navigating between sites and gives the students access to the results of applying advanced commands without needing to master the commands. They have a record of the commands that they can use during self‐study. The scripts appeared to reduce student anxiety about using the command line during the workshop and allowed them to focus on the principles of protein structure.
Materials and Methods
PyMOL
We used the incentive version of PyMOL (version 1.7.6.4) to develop the scripts.21 The scripts have been subsequently tested in PyMOL versions 1.5.0.5 and 1.8.0.5. Students used the education‐only versions of PyMOL on their personal laptop computers. We edited the scripts with the plain text editor TextWrangler on the Mac or Jedit on all operating systems. The coordinates of the crystal structure of bacteriophage T4 lysozyme were used in the examples (PDB ID: 3fa0.22). This protein's small size (164 residues) made it easy for students to view elaborate molecular representations in PyMOL on their personal laptop computers, many of which had modest computer memory.
Writing the viewport settings on one line
We developed a Python script, roundview.py, to reformat the viewing port settings from six lines to one line for easy pasting onto the command‐line. We tested the roundview() function in PyMOL on Windows 8 32‐bit, Ubuntu 14.04 64‐bit, and Mac OS X 10.10.5 64‐bit. The roundview() function does not depend on features that are limited to the incentive versions of PyMOL.
Installation and use of roundview.py
The roundview.py script and associated scripts were downloaded by viewing the “raw” code and saving it from https://github.com/MooersLab/EasyPyMOL.git. If the program git was installed, the scripts were downloaded by typing the command git clone https://github.com/MooersLab/EasyPyMOL.git. The installation of the roundview.py script followed three steps: create a subfolder called ∼/Scripts/PyMOLScripts, move roundview.py to ∼/Scripts/PyMOLScripts, and edit (may need to create with a plain text editor) the hidden ∼/.pymolrc file (or the visible ∼/pymolrc.pml file on Windows) by adding on a new line the command run∼/Scripts/PyMOLScripts/roundview.py. Upon restart, PyMOL automatically loaded roundview.py. The function was executed by typing roundview. This installation method is simple and robust because the subfolder is secure during removal or updating of PyMOL. The same steps can be used to install the other scripts mentioned in this paper. The roundview function was used multiple times without needing to reload it. The roundview function also remained active after the issuing of the reinitialize command. Each time the roundview command was issued, the scene settings were written to the last line of a plain text file called roundview.txt in the current working directory. The on‐line documentation was printed to the command history window by typing help roundview.
An alternate way to install the scripts was to use the plugin manager in the plugin pulldown (note: plugins are not available in MacPyMOL). The ∼/Scripts/PyMOLScripts path was added to the list of paths used to search for scripts upon restart of PyMOL. The roundview.py script was listed under the Installed Plugin tab. The “Load on startup” box can be checked to ensure that the roundview function is always available. Users of MacPyMOL do not have the plugin pulldown, so they should use the first method to install roundview.py. Links to the YouTube video showing these two methods of installing roundview.py are found on the EasyPyMOL GitHub website (https://github.com/MooersLab/EasyPyMOL).
Workshop setting
We tested the horizontal scripts in two classes of 22 − 25 first‐year graduate students in the biomedical sciences. The classroom had a projector for computerized slides, a screen for projected images, and power outlets for the laptop computers of all students. The room also had internet access via wifi. The internet was used to retrieve files of atomic coordinates from the Protein Data Bank23 and electron density maps from the Electron Density Server.24 The students were introduced to PyMOL in a workshop format. Before the workshop, we instructed the students in how to install PyMOL and asked each student to bring a laptop computer, power supply, and a three‐button mouse.
Script availability
The scripts mentioned in this paper are available in the supplementary materials and at https://github.com/MooersLab/EasyPymol. This website contains links to several YouTube videos that describe how to download, install, and use the scripts.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Acknowledgments
The authors thank 2 years of graduate students in the Molecular Systems course at OUHSC for using the horizontal scripts in class and assignments. They thank Chiedza Kanyumbu for comments on the paper and Kathy Kyler for editing the paper. They thank William Beasley for reviewing the EasyPyMOL GitHub website.
References
- 1. Mura C, McCrimmon CM, Vertrees J, Sawaya MR (2010) An introduction to biomolecular graphics. PLoS Comput Biol 6:e1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Craig PA, Michel LV, Bateman RC (2013) A survey of educational uses of molecular visualization freeware. Biochem Mol Biol Educ 41:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adams PD, Afonine PV, Bunkczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive python‐based system for macromolecular structure solution. Acta Crystallogr D: Biol Crystallogr 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Panjkovich A, Svergun DI (2016) SASpy: a PyMOL plugin for manipulation and refinement of hybrid models against small angle X‐ray scattering data. Bioinformatics 32:2062–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lill MA, Danielson ML (2011) Computer‐aided drug design platform using PyMOL. J Comput‐Aided Mol Des 25:13–19. [DOI] [PubMed] [Google Scholar]
- 6. Makarewicz T, Kaźmierkiewicz R (2016) Improvements in GROMACS plugin for PyMOL including implicit solvent simulations and displaying results of PCA analysis. J Mol Model 22:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Das R, Baker D (2008) Macromolecular modeling with rosetta. Annu Rev Biochem 77:363–382. [DOI] [PubMed] [Google Scholar]
- 8. Chaudhury S, Lyskov S, Gray JJ (2010) PyRosetta: a script‐based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26:689–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baugh EH, Lyskov S, Weitzner BD, Gray JJ (2011) Real‐time PyMOL visualization for Rosetta and PyRosetta. PLoS One 6:e21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adolf‐Bryfogle J, Dunbrack RL, Jr (2013) The PyRosetta Toolkit: a graphical user interface for the Rosetta software suite. PloS One 8:e66856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schenkelberg CD, Bystroff C (2015) InteractiveROSETTA: a graphical user interface for the PyRosetta protein modeling suite. Bioinformatics; 31:4023–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fox B, Ramey C. (2016) GNU readline library, edition 6.3, for readline library version 6.3. Free Software Foundation, Cambridge, MA.
- 13. Tarini M, Cignoni P, Montani C (2005) Ambient occlusion and edge cueing for enhancing real time molecular visualization. Visual Comput Graphics IEEE Trans 12:1237–1244. [DOI] [PubMed] [Google Scholar]
- 14. Via A, Rother K, Tramontano A (2014) Managing your biological data with Python. CRC Press, Boca Raton. [Google Scholar]
- 15. Ealy JB (2004) Students understanding is enhanced through molecular modeling. J Sci Educ Technol 13:461–471. [Google Scholar]
- 16. Appling JR, Peake LC (2004) Instructional technology and molecular visualization. J Sci Educ Technol 13:361–365. [Google Scholar]
- 17. Kramer IM, Dahmani HR, Delouche P, Bidabe M, Schneeberger P (2012) Education catching up with science: preparing students for three‐dimensional literacy in cell biology. CBE‐Life Sci Educ 437–447. (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simmons AD, Nguyen TK, Follis JL, Ribes‐Zamora A (2014) Using a PyMOL activity to reinforce the connection between genotype and phenotype in an undergraduate genetics laboratory. PloS One 9:e114257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richardson J, Richardson D, Sirochman R, Bateman RC, Jr , Booth D (2002) Teaching and assessing three‐dimensional molecular literacy in undergraduate biochemistry. J Chem Educ 79:551–552. [Google Scholar]
- 20. West MB, Chen Y, Wickham S, Heroux A, Cahill K, Hanigan MH, Mooers BHM (2013) Novel insights into eukaryotic γ‐glutamyltranspeptidase 1 from the crystal structure of the glutamate‐bound human enzyme. J Biol Chem 288:31902–31913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schrödinger LLC. (2016) The PyMOL molecular graphics system, version 1.8.2.0.
- 22. Mooers BH, Tronrud DE, Matthews BW (2009) Evaluation at atomic resolution of the role of strain in destabilizing the temperature‐sensitive T4 lysozyme mutant Arg 96 His. Protein Sci 18:863–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J (2000) The Protein Data Bank and the challenge of structural genomics. Nat Struct Mol Biol 7:957–959. [DOI] [PubMed] [Google Scholar]
- 24. Taylor T, Harris M, Kleywegt G, Wählby A, Zou J, Jones T (2004) The Uppsala electron‐density server. Acta Crystallogr, Sect D: Biol Crystallogr 60:2240–2249. [DOI] [PubMed] [Google Scholar]
- 25. Shoichet BK, Baase WA, Kuroki R, Matthews BW (1995) A relationship between protein stability and protein function. Proc Natl Acad Sci USA 92:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anderson DE, Becktel WJ, Dahlquist FW (1990) pH‐induced denaturation of proteins: a single salt bridge contributes 3‐5 kcal/mol to the free energy of folding of T4 lysozyme. Biochemistry 29:2403–2408. [DOI] [PubMed] [Google Scholar]
- 27. Grutter MG, Hawkes RB, Matthews BW (1979) Molecular basis of thermostability in the lysozyme from bacteriophage T4. Nature 277:667–669. [DOI] [PubMed] [Google Scholar]
- 28. Eriksson AE, Baase WA, Zhang XJ, Heinz DW, Blaber M, Baldwin EP, Matthews BW (1992) Response of a protein structure to cavity‐creating mutations and its relation to the hydrophobic effect. Science 255:178–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information


