Abstract
CRISPR‐Cas is RNA‐based prokaryotic immune systems that defend against exogenous genetic elements such as plasmids and viruses. Cas1 and Cas2 are highly conserved components that play an essential part in the adaptation stage of all CRISPR‐Cas systems. Characterization of CRISPR‐Cas genes in Thermococcus onnurineus reveals the association of the Cas2 gene with the putative type IV system that lacks Cas1 or its homologous genes. Here, we present a crystal structure of T. onnurineus Cas2 (Ton_Cas2) that exhibits a deep and wide cleft at an interface lined with positive residues (Arg16, Lys18, Lys19, Arg22, and Arg23). The obvious DNA recognizing loops in Cas2 from E. coli (Eco_Cas2) are absent in Ton_Cas2 and have significantly different shapes and electrostatic potential distributions around the putative nucleotide binding region. Furthermore, Ton_Cas2 lacks the hairpin motif at the C‐terminus that is responsible for Cas1 binding in Eco_Cas2. These structural features could be a unique signature and indicate an altered functional mechanism in the adaptation stage of Cas2 in type IV CRISPR‐Cas systems.
Keywords: CRISPR‐Cas system, Thermococcus onnurineus, Cas2, protein structure, type IV system
Introduction
The CRISPR‐Cas (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR Associated gene) system has been widely studied in microbial and archaeal antiviral immune systems.1 The CRISPR system protects the host organism from foreign genetic material via a three stage process of adaptation, processing, and interference. Cas proteins are thought to involve, in function or through various modifications, cleavage or binding activities to facilitate RNA or DNA biogenesis and cleavage.2, 3 CRISPR–Cas systems are typically encoded by a predicted single operon that encompasses the Cas1, Cas2 genes together with the genes for various RNA biogenesis and interference functions. Cas1 and Cas2 function as a complex in vivo and in vitro, and the Cas1–Cas2 complex exhibits DNA acquisition activity during the CRISPR‐Cas adaptive stage.4, 5 The current nomenclature classifies the CRISPR‐Cas systems of bacteria and archaea into five types (I–V) and 16 subtypes, based on phylogenetic and functional studies.6
Previous studies on Cas2 proteins report various activities and substrate specificities depending on the sources of the microorganisms. For example, Cas2 from Sulfolobus solfataricus (Sso_Cas2) that belongs to both type I‐A and type III‐B is characterized as a metal‐dependent single strand specific endoribonuclease with preference for U‐rich ss‐RNA.7 Cas2 from Bacillus halodurans (Bha_Cas2) that belongs to both type I‐C and type III‐B shows endonuclease activity toward double strand DNA substrates.8 Cas2 from D. vulgaris is known to exhibit neither nuclease nor ribonuclease activity.9 Contrary to expectation, recent structure studies have shown that the enzymatic activity of Cas2 is not required for spacer acquisition, suggesting a simple structural platform function in binding to Cas1.4, 10 The complex structure of Cas1‐Cas2‐DNA from E. coli of a type I‐E system indicates that the main function of Cas2 may be to form a scaffold between two dimeric Cas1 proteins and assist DNA substrate binding with the dsDNA substrate placed on one side of the Cas2 dimeric surface. Despite numerous biochemical characterization and structural analysis, the complete functional mechanism of Cas2 still remains elusive. To date, five structures of Cas2 from different organisms and CRISPR‐Cas systems have been reported in the PDB databank (Sso_Cas2:2I8E, Dvu_Cas2:3OQ2, Tth_Cas2:1ZPW, Pf_Cas2:2I0X, and Bha_Cas2:4ES1).7, 8, 9, 11 In this study, we report the association of Cas2 with the type IV CRISPR‐Cas system in Thermococcus onnurineus NA1 and analyze the structure of the protein—the first report of Cas2 in type IV CRISPR‐Cas systems to date.
Results
Characterization of CRISPR‐Cas system in T. onnurineus NA1
The hyperthermophilic archaeon, T. onnurineus NA1, has six CRISPR loci in the circular genome(1847607nt) based on the CRISPR database.12 The CRISPR locus in T. onnurineus contains 28‐30nt of repeat sequences and 7‐43nt of spacers [Fig. 1(A)]. Six repeats share similar nucleotide sequences and lengths, which are predicted to form a palindrome secondary structure, with an identical repeat sequence among loci 1, 5, and 6 and marginal differences between loci 2, 3, and 4. Repeat sequences are highly conserved in the beginning of the four nucleotides (GTTT) and in the terminal region (GXAAX) for all six regions. When ORFs near the CRISPR loci were analyzed, 16 Cas proteins were identified with two major cas gene cassettes: the subtype III‐A and the putative IV system. The subtype III‐A was located between loci 3 and 4 with similar individual cas genes and cassettes to those of P. yayanosii, a recently discovered thermophilic archea13, 14 [Fig. 1(B)]. The type IV system, previously described as a csf module, is known to form a minimal effector complex with four subunits.6 The putative type IV system in T. onnurineus was located on the downstream of locus 1 in that the signature proteins of Csf1 and Csf2 were identified, but the other two subunits could not be assigned, possibly due to the low sequence identity to known cas genes (<10%).
Figure 1.
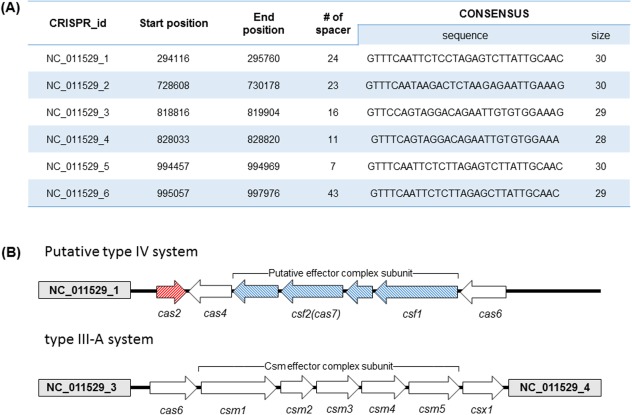
The CRISPR‐Cas system of T. onnurineus. A: Six CRISPR loci identified within the genome of T. onnurineus NA1, based on the CRISPR database (http://crispr.u-psud.fr/). B: Gene organization of the cas locus in type III and putative type IV. White arrows indicate ORFs present in T. onnurineus gene, and CRISPR loci 1, 3, and 4 are displayed (gray box). Red arrows represent the cas2 gene, and blue arrows represent the putative effector complex genes specific to type IV.
Interestingly, the cas2 gene is associated with the putative type IV array in the absence of the cas1 gene, located in the opposite direction to type IV array between CRISPR locus 1 and cas4. Genes in adaptation stage are often located externally cas gene array involved in RNA biogenesis or interference15 [Fig. 1(B)]. The CRISPR‐Cas systems are typically encoded by a single operon that encompasses the cas1 and the cas2 genes together because Cas1 and Cas2 form a complex and function with DNA acquisition activity during the adaptive stage.16 However, the CRISPR‐Cas system of T. onnurineus lacks the cas1 gene, and extensive BLAST searches for a Cas1 sequence could not detect any similar homologs, questioning the functional role of Cas2 in T. onnurineus. Cas4, known to be involved in the adaptation stage,17 is identified next to the cas2 loci in the type IV array. To investigate the function of Cas2, we purified the protein and analyzed the enzymatic property in a wide range of pH with various metal cofactors. However, the Cas2 protein did not exhibit any cleavage activity for dsDNA, ssDNA or ssRNA substrates at all.
Overall structure of Ton_Cas2
The structure of Ton_Cas2 was determined by the molecular replacement method using structure of the hypothetical protein of Pyrococcus furiosus (PDB code: 2I0X).18 Although the structure of the hypothetical proteins was available in PDB database, the function or characterization of the protein was not reported to date. The detailed statistics for the structure are described in Table 1. The overall structure of Ton_Cas2 showed a typical βαββαβ ferredoxin fold of double‐split α‐helices, and the antiparallel β‐sheet for each protomer was composed of 2 α‐helices and 5 β‐strands. The flexible C‐terminal region (residues Glu77‐Ilu85) in each protomer was not modeled in the final structure due to insufficient electron density maps. Cas2 is a dimer in crystal as well as in solution consistent with the reported Cas2 oligomerization.8, 11 The C‐terminal region of β5 (residues Arg71‐Lue74) from each protomer interacted with the β4 (residues Ser59‐Leu65) in the other molecule to form a β‐sheet of five strands in both subunits. Intensive hydrophobic interactions (Val4, Val6, Val28, Val32, Val61, Tyr63, and Leu65) produced a total buried surface of 1353.4 Å2 (24.6% of the total surface) at the dimer interface that were strengthened by 24 hydrogen bonds and four salt bridges (Lys46/Glu72, and Lys64/Glu72) [Fig. 2(A)]. The Dali server found the closest structure homologue to be initial search model of Cas2 from P. furiosus (Pf_Cas2; sequence identity, 79%) with a Z‐score of 14.8 and a 0.6 Å r.m.s.d. of over 76 Ca atoms. The Ton_Cas2 structure also aligns with Sso_Cas2 from S. solfataricus, which has ssRNase activity, and Bha_Cas2 from B. halodurans, which has dsDNase activity with r.m.s.d. of 2.3 and 1.7 Å, respectively. The dimer arrangements and overall conformations of these Cas2 homologues are similar, but the previously reported putative substrate binding region (loop1; β1‐α1, loop4; α2‐β4) around the conserved residue Asp8 is markedly different.7, 8, 11, 19 The invariant residue Asp8 is positioned at the end of the β1, together facing one another to form a cleft in the dimer. These Asp pairs are thought to play a catalytic function by coordinating a divalent metal ion for the substrate cleavage in Bha_Cas2. The distance between Asp pairs was reported to vary among Cas2 structures, raising the possibility of open‐close conformational change.8 The distance between the two Asp8 residues in Ton_Cas2 is 13.6Å, which is 7Å and 4Å longer than Sso_Cas2 and Bha_Cas2, respectively [Fig. 2(B)]. The wider distance between them may suggest an open state conformation of the current structure in that coordination of any metal cofactor between the Asp pair is not possible at the distance.
Table 1.
Data Collection and Structure Solution Parameters
| Crystal type | Overlapped plate |
|---|---|
| Unit cell parameters (Å) | a = 29.52 |
| b = 32.80 | |
| c = 83.16 | |
| Resolution (Å) | 50.00 – 1.7 |
| Space group | P2(1)2(1)2 |
| Completeness (%) | 99.18 (99.82)a |
| R sym (%)b | 4.62 (9.64) |
| I/σ (I) | 27.64 (17.23) |
| No. of refined atoms: protein/water | 12,582/66 |
| R factor/R free (%)c | 21.50/24.07 |
| r.m.s.d. bond length (Å) | 0.006 |
| r.m.s.d. bond angle | 0.814 |
| Ramachandran plot (%) | |
| Most favored region | 97. 4% |
| Additionally allowed region | 2.6% |
| Outlier region | 0% |
Numbers in parentheses are statistics from the highest resolution shell.
R sym = Σ|I obs – I avg|/Σ I obs, where I obs is the observed individual reflection, and I avg is the average over symmetry equivalents.
R factor = ΣǁF 0|–|F cǁ/Σ|F 0|, where |F 0| and |F c| are the observed and calculated structure factor amplitudes, respectively. R free was calculated using 5% of the data.
Figure 2.
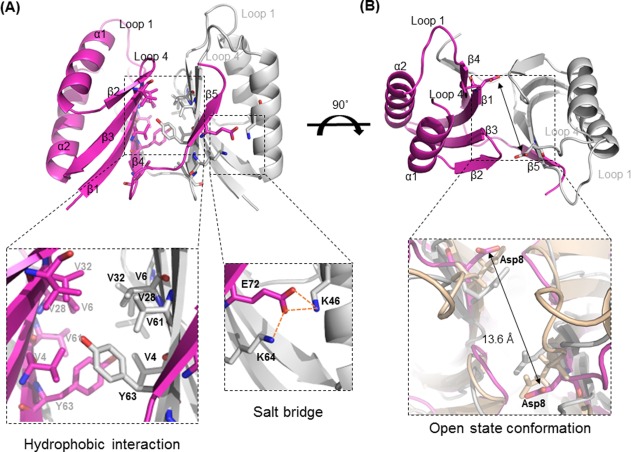
Overall structure properties of Ton_Cas2. A: Each protomer is represented by light magenta and white cartoons. Hydrophobic interactions and charge interactions are enlarged in the black dotted boxes, and interaction residues are indicated by sticks. B: Wide and open conformation of Ton_Cas2. The distance between the two Asp8 residues in dimer are enlarged in the black dotted box in superposed structures of promotor A with Sso_Cas2 (white) and Bha_Cas2 (wheat).
Structural comparison of Ton_Cas2
The structure of Ton_Cas2 reveals a few different features when compared to that of Eco_Cas2 in type I‐E, the most studied Cas2 in context with DNA substrate complex formation.5, 10 The significant difference between Ton_Cas2 and Eco_Cas2 is in the region of the C‐terminus of the protomer, a.a. 66‐76, that extends from β4 of the β‐sheet and forms a separate β‐strand in the other protomer. The main body of the βαββαβ ferredoxin fold is similar between protomer structures (r.m.s.d. of 0.31 Å at 51Cα positions) of Eco_Cas2 and Ton_Cas2, but the C‐terminal region of Ton_Cas2 is much shorter, lacking the β‐strand hairpin motif and notably folded into the other side of the protomer subunit in dimer to form a complete β‐sheet. In Eco_cas2, this region continues to complete the β‐sheet in the same side of the protomer and extends to yield a protruding loop with an additional hairpin structure in the end. The longer loop of the protruding architecture in Eco_Cas2 is responsible for binding together the dsDNA substrates with residues of Arg77 and Arg78 in addition to Arg14, Arg16 on α1 [Fig. 3(A)]. However, due to the shortened length of the loop and the absence of corresponding arginine residues, the shape and electrostatic surface distribution of the groove region is significantly altered in Ton_Cas2. Negative residues such as Glu12 on loop1 and Asp54, Glu55, Asp56, and Glu57 on loop4 are exposed in Ton_Cas2 with Arg16 positioned in opposing direction to the putative substrate binding site. Ton_Cas2 lacks the obvious arginine or other corresponding positive residues on the same surface responsible for DNA binding in Eco_Cas210 [Fig. 3(B)]. In addition, the angle of the dimeric arrangement of the two protomer subunits is different, resulting in wider geometry on the surface. Furthermore, Ton_Cas2 lacks the hairpin motif at the C‐terminus, which provides an essential binding surface to Cas1 molecules in Eco_Cas2, without any equivalent structural element in the position. In the absence of the hairpin motif, helix α1 in the main ferredoxin fold domain, of which the region in Eco_Cas2 provides an additional binding surface to Cas1 such as residue Ile21 and residue Trp22, is mainly constituted by hydrophilic residues without the conserved hydrophobic interaction residues in Ton_Cas2. Taken together, Ton_Cas2 shows very different DNA binding features on the surface and lacks the apparent Cas1 binding elements observed in Eco_Cas2, suggesting an altered function or property of the molecule.
Figure 3.
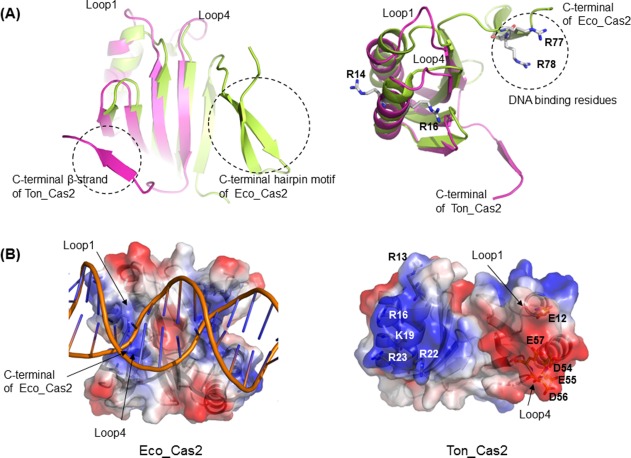
Structural comparison of Ton_Cas2. A: Superposition of Ton_Cas2 protomer (light magenta) on Eco_Cas2 protomer (limon) highlights the difference in C‐terminus (left) and the altered shape and orientation in the DNA binding region (right). B: Surface diagram of the putative substrate‐binding groove and electrostatic potential for Eco_Cas2 (left) and Ton_Cas2 (right). Positively charged regions of the molecular surface are shown in blue and negatively charged regions in red. DNA substrate is shown in cartoon for the complex structure of Eco_Cas2 (PDB code: 5DQT).
Discussion
Previously contradicting results on the enzymatic activity of Cas2 proteins were reported, including ribonuclease activity for Sso_Cas2 and DNase activity for Bha_Cas2, with no activity for Dvu_Cas2.7, 8, 9 Despite extensive investigation and analysis, we were not able to detect any DNA or RNA cleavage activity for Ton_Cas2. The lack of DNA or RNA cleavage activity in this study may be consistent with the previous notion that the main role of Cas2 is to form a scaffold between two dimeric Cas1 proteins and assist DNA substrate binding as determined by the Eco_Cas2 protein.4 However, the significantly altered architecture in the DNA binding surface of the dimeric molecule of Ton_Cas2 and the absence of Cas1 binding features observed in Eco_Cas2 may indicate that detailed DNA binding and the related functional mechanism could be different in Ton_Cas2 from that of Eco_Cas2. DNA binding property and specificity would determine its functional role, and thus correct analysis of the DNA interaction is important to understand the adaptation stage in the CRISPR‐Cas system. The DNA binding region of loop1 in Eco_Cas2 constitutes a suitable architecture by dimerization with the obvious positive electrostatic surface providing essential interaction with the double strand DNA substrate.5, 10 In contrast, the surface of Ton_Cas2 lacks these unique binding residues due to significantly altered geometry. The multiple alignments of Cas2 proteins among 25 sequences and analysis of this region indicate that Cas2 might be classified into several groups depending on the length of the loop and concomitant charge property in this region. For example, Cas2 proteins including Ton_Cas2 in this study contain a short loop1 and a short loop4 with charged residues of DEDED a highly conserved motif among this group. Cas2 proteins such as Bha_Cas2 showed an extended loop1 and short loop4 in which about 12 residues are posed on loop1, and <40% are invariant with Lys18 at end of loop1. With the conserved lysine residue at the end of the loop4, much wider positive electrostatic potential around helix1 and the loop regions were observed in this group. Cas2, including Sso_Cas2, showed a short loop1 and a long loop4 with electrostatic potential of both acidic and basic properties, different from the other two groups (Fig. 4).
Figure 4.
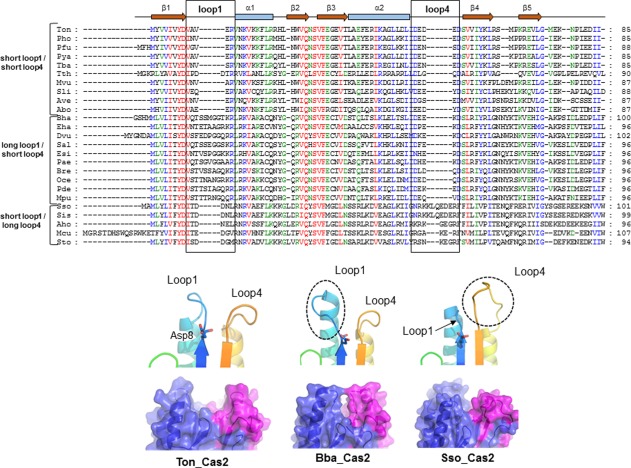
Multiple sequence alignments for Cas2 proteins. Upper: A sequence alignment of members of Cas2 compared with the sequence and secondary structure of Ton_Cas2. The numbering is shown on the right side of the alignment. Red, green, and blue highlighting indicates the residues conserved in 100, 80, and 60% of the proteins, respectively. Down: protomer architecture around the catalytic residue Asp8 (stick) and the putative active site groove between two protomer subunits (blue and pink). The additional loop regions found in other homologues are indicated by dotted circles. * Pho; P. horikoshii OT3, Pfu; P. furiosus, Pya; P. yayanosii CH1, Tth; T. Thermophilus, Tba; T. barophilus MP, Mvu; M. vulcanius M7, Sli; S. lipocalidus DSM 12680, Ave; A. veneficus SNP6, Abo; A. boonei T469, Sal; Salimicrobium sp. MJ3, Mpu; M. plutonius ATCC 35311, Eha; E. harbinense YUAN‐3, Esi; E. sibiricum 255‐15, Bha; B. Halodurans, Dvu; D. Vulgaris, Pae; Paenibacillus sp. oral taxon 786, Bre; Brevibacillus sp. CF112, Oce; Oceanobacillus sp. Ndiop, Pde; P. dendritiformis C454, Sso; S. solfataricus P2, Sis; S. islandicus M.14.25, Aho; A. hospitalis W1, Mcu; M. cuprina Ar‐4, Sto; S. tokodaii str. 7
Notably, the lack of the apparent feature for Cas1 interactions in Ton_Cas2 suggests an alternative functional mechanism or structural association in the adaptation stage in the CRISPR‐Cas system. This analysis might be related to the observation that the CRISPR‐Cas system in T. onnurineus lacks cas1 or its homologous genes. In some bacteria and archaea, cas1 gene is separately located from CRISPR system or lacked in the genome. The cas1 gene is considered to be a mobile element and thus the absence of cas1 gene could be resulted from gene transfer. Among the Thermococclaes, T. gammatolerans, and P. abyssi also showed similar CRISPR‐Cas system lacking the cas1 gene in their genome.15 The cas4 gene is located downstream, just next to cas2, in this putative type IV system of T. onnurineus, supporting the notion that Cas4 is involved in the adaptation stage. Type IV is not previously described in detail, and it is the first report of Cas2 in the putative type IV system that lacks the signature protein of Cas1 required for DNA acquisition. These observations point to the significant diversity of the CRISPR‐Cas system even within Cas2 proteins.
Conclusions
Cas1 and Cas2 are nearly universally conserved proteins in CRISR‐Cas system with a spacer acquisition function. However, the CRISPR‐Cas system of T. onnurineus lacks Cas1 or its homologue. The structural comparison showed a wider substrate binding groove and charged distribution, different from previously reported Cas2 structures. This could be a unique signature within some Cas2 proteins and provide a molecular basis in the adaptation stage for type IV CRISPR‐Cas systems.
Materials and Methods
Cloning and protein expression and purification
The gene encoding Ton_Cas2 (GenBank: YP_002306702.1, locus name: TON_0320) was amplified by PCR from T. onnurineus NA1 genomic DNA. The Ton_Cas2 specific primers 5′‐CGG CCG CTT ATG TAC GTG GTC ATC‐3′ and 5′‐GGT ACC CTA AAT GAT GTC CTC‐3′ were designed to introduce NotI and KpnI recognition sites. PCR was performed using Pfu KOD DNA polymerase (Novagen). The pPROEX‐HTa vector (Invitrogen) was prepared using a MiniPrep kit (Bioneer) and ligated (T4 DNA ligase, NEB) into vector treated with the restriction enzymes to create pPROEX‐Cas2. Escherichia coli MC1061 [F‐7, araD139, D(ara‐leu)7696, galE15, galK16, D(lac)X74, rpsL, (Strr), hsdR2, (rk 2mk1), mcrA, mcrB1] harboring pPROEX‐Cas2 was cultured in 6L of LB (Luria‐Bertani) medium containing ampicillin (50 μg/mL) to an OD600 of 0.65, and induced by adding 1 mM isopropyl β‐d−1‐thiogalactopyranoside and incubating at 18°C for 18 h. The cells were collected by centrifugation (8000g, 30 min), resuspended in 400 mL of lysis buffer (1× PBS), and sonicated in an ice bath (VC‐600 sonicator; Sonics & Materials, Newtown, CT). The supernatant was clarified by centrifugation (10,000g, 30 min, 4°C) and purified using a Histrap HP (GE Healthcare, lysis buffer; 1× PBS, elution buffer; 1× PBS, 250 mM imidazole), Resource Q (GE Healthcare, lysis buffer; 50 mM Tris‐HCl, pH 8.0, 50 mM NaCl, 1 mM dithiothreitol, elution buffer; 50 mM Tris‐HCl, pH 8.0, 1M NaCl, 1 mM dithiothreitol) and Hiload 16/60 superdex 200pg column (GE Healthcare) with an AKTA FPLC system (GE Healthcare). The final active fractions in elution buffer were dialyzed against 50 mM Tris‐HCl, pH 8.0, 150 mM NaCl, 1 mM dithiothreitol (DTT) buffer. The protein concentration was determined by the Bradford method using bovine serum albumin as a standard. The purity and apparent molecular mass of the protein were analyzed by discontinuous sodium dodecyl sulfate polyacrylamide gel electrophoresis.20
Crystallization and data collection
Ton_Cas2 crystallization trials were conducted using the sitting drop method at 18°C. We mixed a 1.5 uL 15 mg/mL Ton_Cas2 solution with an equal volume of crystallization reservoir solution containing 30% MPD, 100 mM Sodium acetate pH 4.6, 20 mM Calcium chloride. Before data collection, a rod‐type crystal was cryocooled to 95 K using a cryoprotectant consisting of the mother liquor. The data set was collected at Spring‐8, Japan. A total of 360 diffraction images were collected. The diffraction data set was integrated, processed, and scaled by HKL2000.21
Structure determination and refinement
The structure was determined by the molecular replacement method using the Phaser CCP4 suite22 and the structure of the Cas2 of P. furiosus (PDB Code 2I0X) as a searching model. The resultant model was refined in conjunction with model rebuilding using CNS.23 COOT24 was used for stereographic manual refinement and model building. After the Rcryst and Rfree values decreased to approximately 25%, the model structure was further refined using Phenix,25 and the structure was validated with PROCHECK.26 Sequence alignments and structure‐based sequence alignments were performed using ClustalW.27 Molecular images, including cartoon and stick representations, were produced using PyMOL.28 Detailed statistics for the data collection and refinement are listed in Table 1.
Atomic Coordinates
The atomic coordinates and structure factors (PDB code: 5G4D) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ, USA (http://www. rcsb.org/).
References
- 1. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- 2. Haft DH, Selengut J, Mongodin EF, Nelson KE (2005) A guild of 45 CRISPR‐associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, van der Oost J, Koonin EV (2011) Evolution and classification of the CRISPR‐Cas systems. Nat Rev Microbiol 9:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA (2014) Cas1‐Cas2 complex formation mediates spacer acquisition during CRISPR‐Cas adaptive immunity. Nat Struct Mol Biol 21:528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nunez JK, Harrington LB, Kranzusch PJ, Engelman AN, Doudna JA (2015) Foreign DNA capture during CRISPR‐Cas adaptive immunity. Nature 527:535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV (2015) An updated evolutionary classification of CRISPR‐Cas systems. Nat Rev Microbiol 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, Koonin EV, Edwards AM, Savchenko A, Yakunin AF (2008) A novel family of sequence‐specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem 283:20361–20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nam KH, Ding F, Haitjema C, Huang Q, DeLisa MP, Ke A (2012) Double‐stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)‐associated Cas2 protein. J Biol Chem 287:35943–35952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Samai P, Smith P, Shuman S (2010) Structure of a CRISPR‐associated protein Cas2 from Desulfovibrio vulgaris. Acta Cryst F66:1552–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J, Li J, Zhao H, Sheng G, Wang M, Yin M, Wang Y (2015) Structural and mechanistic basis of PAM‐dependent spacer acquisition in CRISPR‐Cas systems. Cell 163:840–853. [DOI] [PubMed] [Google Scholar]
- 11. Ka D, Kim D, Baek G, Bae E (2014) Structural and functional characterization of Streptococcus pyogenes Cas2 protein under different pH conditions. Biochem Biophys Res Commun 451:152–157. [DOI] [PubMed] [Google Scholar]
- 12. Grissa I, Vergnaud G, Pourcel C (2007) The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jun X, Lupeng L, Minjuan X, Oger P, Fengping W, Jebbar M, Xiang X (2011) Complete genome sequence of the obligate piezophilic hyperthermophilic archaeon Pyrococcus yayanosii CH1. J Bacteriol 193:4297–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jung TY, An Y, Park KH, Lee MH, Oh BH, Woo E (2015) Crystal structure of the Csm1 subunit of the Csm complex and its single‐stranded DNA‐specific nuclease activity. Structure 23:782–790. [DOI] [PubMed] [Google Scholar]
- 15. Koonin EV, Krupovic M (2015) Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat Rev Genet 16:184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nunez JK, Lee AS, Engelman A, Doudna JA (2015) Integrase‐mediated spacer acquisition during CRISPR‐Cas adaptive immunity. Nature 519:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lemak S, Nocek B, Beloglazova N, Skarina T, Flick R, Brown G, Joachimiak A, Savchenko A, Yakunin AF (2014) The CRISPR‐associated Cas4 protein Pcal_0546 from Pyrobaculum calidifontis contains a [2Fe‐2S] cluster: crystal structure and nuclease activity. Nucleic Acids Res 42:11144–11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen LQ, Fu Z‐Q, Liu Z‐J, Rose JP, Wang BC (2006) Southeast Collaboratory for Structural Genomics (SECSG). PDB ID: 2I0X, Hypothetical protein PF1117 from Pyrococcus Furiosus [Google Scholar]
- 19. Lintner NG, Frankel KA, Tsutakawa SE, Alsbury DL, Copie V, Young MJ, Tainer JA, Lawrence CM (2011) The structure of the CRISPR‐associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system. J Mol Biol 405:939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 21. Otwinowski Z, Minor W (1997) Processing of X‐ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed] [Google Scholar]
- 22. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Cryst D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse‐Kunstleve RW, Jiang J‐S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Cryst D54:905–921. [DOI] [PubMed] [Google Scholar]
- 24. Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Cryst D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 25. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L‐W, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Cryst D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291. [Google Scholar]
- 27. Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit 2 3. [DOI] [PubMed] [Google Scholar]
- 28. DeLano WL (2002) The PyMOL Molecular Graphic System. DeLano Scientific, San Carlos, CA, USA: (http://wwwpymolorg). [Google Scholar]


