Summary
The Pold3 gene encodes a subunit of the Polδ DNA polymerase complex. Pold3 orthologues are not essential in Saccharomyces cerevisiae or chicken DT40 cells, but the Schizzosaccharomyces pombe orthologue is essential. POLD3 also has a specialized role in the repair of broken replication forks, suggesting that POLD3 activity could be particularly relevant for cancer cells enduring high levels of DNA replication stress. We report here that POLD3 is essential for mouse development and is also required for viability in adult animals. Strikingly, even Pold3+/- mice were born at sub-Mendelian ratios and, of those born, some presented hydrocephaly and had a reduced lifespan. In cells, POLD3 deficiency led to replication stress and cell death, which were aggravated by expression of activated oncogenes. Finally, we show that Pold3 deletion destabilizes all members of the Polδ complex, explaining its major role in DNA replication and the severe impact of its deficiency.
Introduction
The bulk of DNA replication in eukaryotes is carried by two major DNA polymerase complexes, Polδ and Polε, in charge of replicating the lagging and leading strands, respectively (Johansson and Dixon, 2013). Each complex contains several subunits, whose roles are not completely defined. The Polδ complex was originally isolated as a dimer of p125 (POLD1) and p50 (POLD2) with POLD1 bearing the polymerase catalytic activity and POLD2 serving an essential structural role (Lee et al., 1984; Ng et al., 1991). Later studies identified two additional subunits (POLD3 and POLD4), which in vitro stimulated the catalytic activity of the complex (Hughes et al., 1999; Liu et al., 2000; Mo et al., 2000). Recent studies in yeast and mammals have attracted specific attention onto POLD3 (Pol32, in S. cerevisiae), since it has been shown to regulate the repair of broken replication forks through break-induced replication (BIR) and to promote repair-associated DNA synthesis in mitosis, pathways that might be particularly relevant in the context of replication stress (RS) and therefore for cancer cells (Costantino et al., 2014; Lydeard et al., 2007; Mayle et al., 2015; Minocherhomji et al., 2015).
POLD3 was first isolated from calf thymus, and shown to have two important roles within the POLD complex. It stabilizes the POLD1-POLD2 interaction and, through a C-terminal PIP box, it facilitates the interaction of the POLD complex with PCNA (Ducoux et al., 2001; Shikata et al., 2001). In addition, POLD3 is necessary for the recruitment of POLD1 to sites of UV-light induced DNA damage (Ogi et al., 2010). Moreover, POLD3 presents additional functions that are independent of genome duplication during S-phase. First, a complex of POLD3-POLD2 interacts with Rev3 and Rev7, increasing the activity of the translesion (TLS) polymerase Pol ζ (Lee et al., 2014). Deletion of the chicken POLD3 orthologue in DT40 cells further supported a role in TLS (Hirota et al., 2015). And second, a recent study described that POLD3 is essential for BIR in human cells, a pathway that is particularly important for cancer cells (Costantino et al., 2014). However, to what extent POLD3 is essential for normal cells is still unclear. In S. cerevisiae, where the Polδ complex is only made of 3 subunits (Pol3: POLD1; Pol31: POLD2; Pol32: POLD3), deletion of the POLD3 orthologue Pol32 is not lethal, but sensitizes cells to replication challenges such as hydroxyurea (HU) or alkylating agents (Gerik et al., 1998). However, the POLD3 orthologue Cdc27 is essential in S. pombe, where the Polδ complex is built of 4 subunits (Pol3: POLD1; Cdc1: POLD2; Cdc27: POLD3; Cdm1: POLD4) (MacNeill et al., 1996; Zuo et al., 1997). Nevertheless, POLD3 is not essential for chicken DT40 cells, and human cells tolerate its depletion by siRNA (Costantino et al., 2014; Hirota et al., 2015).
Given the growing interest on POLD3, we here sought to clarify whether it is essential in mammalian cells, and to specifically interrogate how an acute depletion of POLD3 impacts the Polδ complex. By developing a conditional knockout strain, we found that POLD3 is essential in mice both during embryonic development but also when deleted postnatally. Furthermore, Pold3 heterozygous mice are born at sub-Mendelian ratios and present a reduced lifespan. These severe phenotypes are mechanistically explained by the discovery that the absence of POLD3 destabilizes all members of the Polδ complex, which leads to a severe replication defect and widespread replication stress in mammalian cells.
Results
POLD3 is essential for mouse development
To investigate the physiological roles of POLD3 we generated a conditional knockout (cKO) strain by using a gene-trap targeting construct that can be converted into a cKO vector (Pold3GT:lox; see Methods) (Figure 1A). Southern blot analysis confirmed the correct integration of the targeting construct in embryonic stem (ES) cells (Figure 1B), from which mouse chimaeras were subsequently generated. After verifying the transmission of the mutant allele by PCR, we crossed the strain with mice that ubiquitously express the FLP recombinase from the β-ACTIN promoter (Rodriguez et al., 2000). This eliminated the gene-trap and neomycin resistance cassette and led to the generation of the Pold3 cKO strain (Pold3lox).
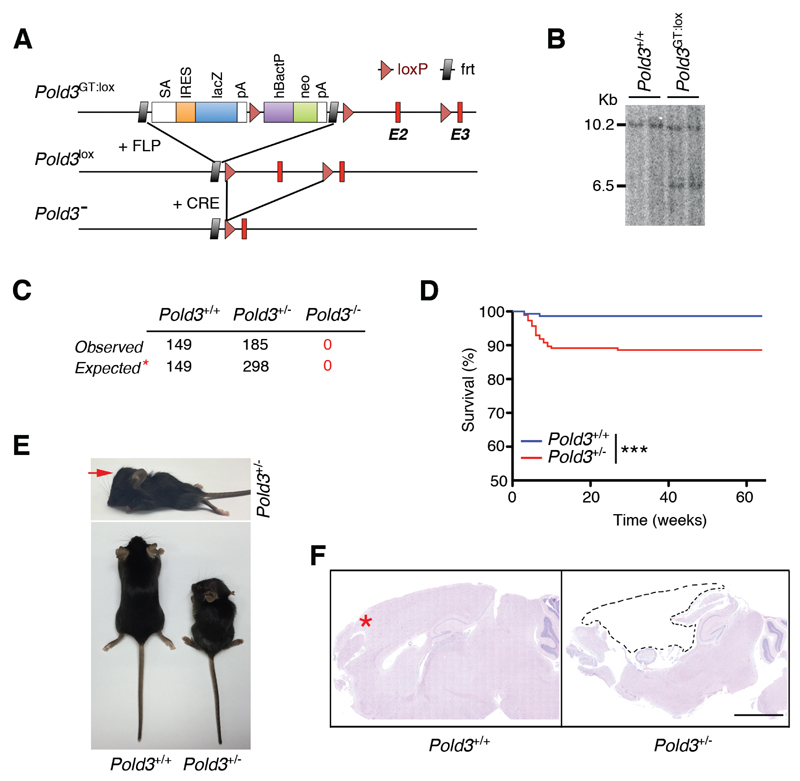
Figure 1. POLD3 is essential for mouse development.
(A) Scheme illustrating the targeting construct used for the generation of Pold3 mutant mice (top), and how the Pold3lox and Pold3- alleles are obtained upon recombination with Flp or Cre recombinases, respectively.
(B) Southern blot illustrating the presence of ES clones harboring the correct integration of the Pold3GT:lox allele that were subsequently used for the generation of mutant mice. The 10.2 Kb band corresponds to the endogenous Pold3 gene, and the 6.5 Kb band to the targeted allele.
(C) Table representing the genotypes observed from inter-crossing Pold3 heterozygous mice. The expected values are those estimated from the offspring of a homozygous-lethal cross. Only 62% (185/298) of the expected Pold3+/- mice are born.
(D) Kaplan-Meyer curves of Pold3+/+ (n = 149) and Pold3+/- (n = 185) mice. The p value was calculated with the Mantel-Cox log rank test. ***P<0.001.
(E) Representative pictures of 2 month-old Pold3+/+ and Pold3+/- littermate mice illustrating the enlarged head morphology (red arrow) and overall dwarfism found in a subset of Pold3 heterozygous mice.
(F) Brain sections of Pold3+/+ and Pold3+/- mice. Note that the forebrain (red arrow) is almost inexistent on Pold3 heterozygous brain (dashed lines). The skull cavity of the mutant mouse was found full of liquid upon necropsy.
To determine whether POLD3 is essential in mice, we crossed Pold3lox/+ mice with an EIIA-Cre strain that ubiquitously expresses the Cre recombinase (Lakso et al., 1996). This generated mice heterozygous for a Pold3 null allele. Pold3+/- mice were subsequently inter-crossed to explore whether POLD3-deficient mice could be born. Out of 334 mice born from heterozygous crosses, no Pold3-/- mice were obtained. We also failed to obtain Pold3-knockout mouse embryo fibroblasts (MEFs) as early as 10.5 dpc. Strikingly, only 62% of the Pold3 heterozygous animals expected from a homozygous-lethal cross were born (185 out of 298; Figure 1C). Moreover, 11% of the Pold3+/- mice that were born died prematurely within the first 10 weeks (Figure 1D). Pold3 heterozygous mice that died prematurely were dwarf from birth and presented an enlarged skull size reminiscent of hydrocephaly (Figure 1E). Pathological analyses confirmed this diagnosis, which was most evident in moribund mice and thus, likely causative of their death (Figure 1E,F). The fact that the phenotype was more penetrant in a fraction of the mice and the preferential targeting of the brain are reminiscent of syndromes associated with embryonic replication stress (RS), such as, for example, the Seckel syndrome, caused by reduced levels of the ATR kinase (Murga et al., 2009). Given that brain development is dependent on rapid cell proliferation at specific developmental stages, genetic defects that limit the capacity for DNA replication, such as Pold3 heterozygosity or Atr hypomorphism, would render brain development very sensitive to any factor, environmental or genetic, that further compromises cell proliferation during critical developmental stages.
Pold3 deletion leads to lethality in adult mice
To further investigate the physiological consequences of Pold3 deletion, we used a previously developed transgenic strain that ubiquitously expresses a tamoxifen-inducible version of the Cre recombinase (UbCre) (Ruzankina et al., 2007). Cre activation and the subsequent deletion of the Pold3lox allele in adult mice was accomplished by supplementing the diet with 4-hydroxy-tamoxifen (4-OHT), starting at 5 weeks of age (weaning). This strategy has previously enabled the deletion of other essential genes, such as the SMC5/6 complex member Nsmce2 or Atr, which in both cases led to premature ageing but with mice surviving for up to 1 or 2 years of 4-OHT treatment, respectively (Jacome et al., 2015; Ruzankina et al., 2007). In striking contrast, UbCre/Pold3lox/lox mice started to show symptoms of severe distress only days after starting the diet and all died within 15 weeks (Figure 2A). These effects were dependent on Pold3 deletion, as they were not observed in Pold3lox/lox mice lacking the UbCre gene or in UbCre/Pold3+/+ mice, all of which were administered the same 4-OHT-containing diet.
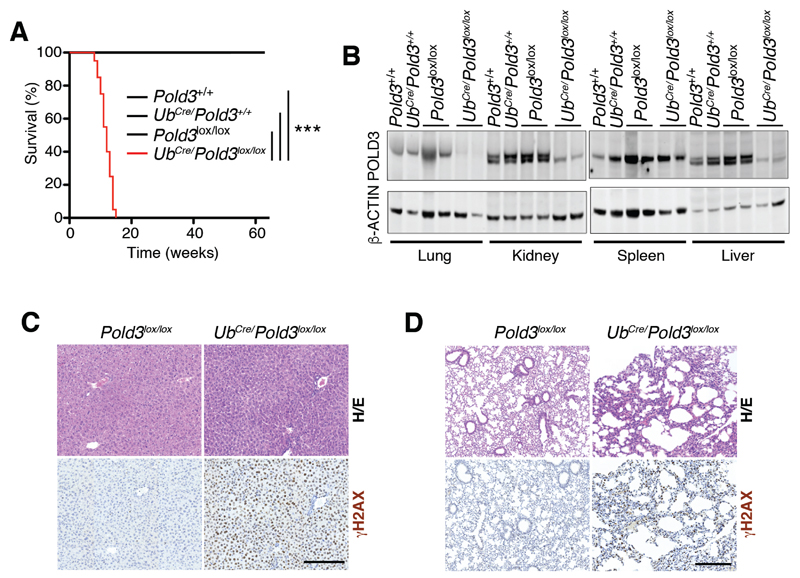
Figure 2. Pold3 deletion is severely toxic in adult mice.
(A) Kaplan-Meyer curves of mice from the indicated genotypes that were exposed to a 4-OHT containing diet that started at week 5. The p value was calculated with the Mantel-Cox log rank test. ***P<0.001.
(B) WB illustrating the depletion of POLD3 levels that is observed on organs from UbCre/Pold3lox/lox mice fed the 4-OHT diet for 1 month. Note that no reduction in POLD3 levels is observed in the spleen.
(C,D) γH2AX immunohistochemistry from the liver (C) and lung (D) of Pold3lox/lox and UbCre/Pold3lox/lox mice fed the 4-OHT diet for 1 month. H/E: Hematoxylin/Eosin. Scale bar (black) indicates 200 μm.
To determine the efficiency of Pold3 deletion, we monitored POLD3 protein levels by western blotting (WB) in organs from UbCre/Pold3lox/lox and control mice fed for 1 month with the tamoxifen-containing diet (Figure 2B). 4-OHT induced a significant depletion of POLD3 in the lung, kidney and liver. In contrast, no clear reduction was observed in the spleen, consistent with previous studies targeting essential genes in adult mice (Jacome et al., 2015; Ruzankina et al., 2007) and which can be explained by the selection for cells that did not undergo Cre-mediated recombination in organs with a high turnover rate. Next, to determine whether Pold3 deletion led to RS, we performed an immunohistochemical analysis for phosphorylated histone H2AX (γH2AX), a marker of DNA damage and RS. In all organs tested, Pold3 deletion led to the widespread accumulation of cells with pan-nuclear γH2AX staining, indicative of RS (Figure 2C,D). This overall accumulation of DNA damage was associated with a noticeable decrease in tissue cellularity, as shown, for example, for the lungs (Figure 2D). In summary, Pold3 deletion in adult mice a generalized accumulation of DNA damage, leading to a severe pathology and ultimately the death of the animal.
POLD3 is essential for DNA replication in B cells
As noted above, we did not observe a decrease in POLD3 protein levels in the spleens of UbCre/Pold3lox/lox mice treated with 4-OHT, suggesting that the spleen is quickly replenished by cells that retained intact Pold3 alleles. To specifically investigate the role of POLD3 in splenic lymphocytes, we crossed Pold3lox/lox mice with a CD19Cre knock-in strain that drives the deletion of the floxed alleles in B cells (Rickert et al., 1997). Compared to wild-type (wt) or CD19Cre-negative Pold3lox/lox B cells, Pold3-deleted B cells showed very limited expansion following stimulation with lipopolysaccharide in vitro (Figure 3A), which correlated with a significant reduction in DNA replication in CD19Cre-positive Pold3lox/lox B cells, as measured by BrdU incorporation (Figure 3B).
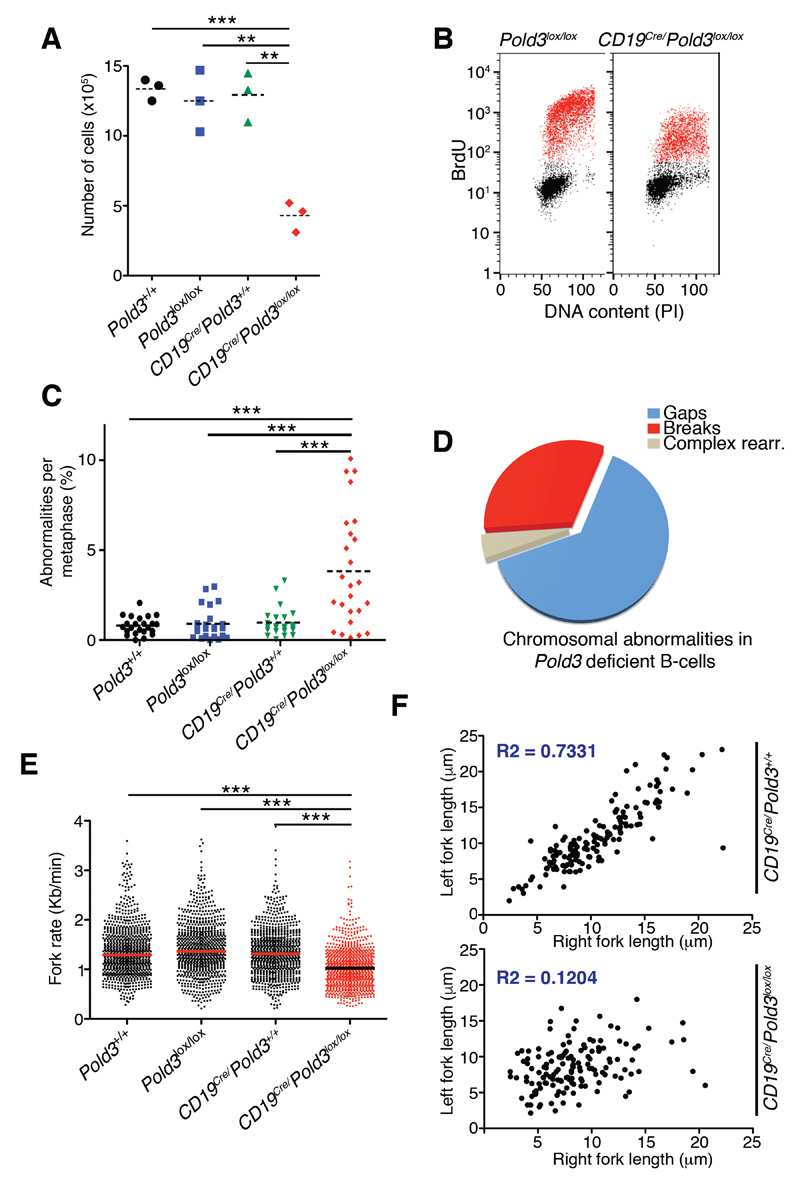
Figure 3. POLD3 sustains DNA replication and prevents RS in B cells.
(A) Number of cells after stimulating 2.5x105 freshly isolated splenic B-lymphocytes for 72 hrs with LPS. Dashed lines indicate mean values. **P<0.01; ***P<0.001.
(B) FACS analysis of DNA content (propidium iodide, PI) versus 5’-bromo-2’-deoxyuridine (BrdU) incorporation in Pold3lox/lox and CD19Cre/Pold3lox/lox B cells, 48 hrs after stimulation with LPS (25 μg/ml). BrdU was used at 10μM for 30 min.
(C) Chromosomal abnormalities found per metaphase in B cells of the indicated genotypes 2 days after stimulation with LPS (25 μg/ml). Dashed lines indicate mean values. ***P<0.001.
(D) Distribution of the different types of chromosomal rearrangements observed in metaphases from CD19Cre/Pold3lox/lox B cells.
(E) Fork rates were measured in stretched DNA fibers (see Methods) prepared from B cells of the indicated genotypes. Data are representative of three indicative experiments. Approximately 900 tracks were measured to estimate fork rates. ***P<0.001.
(F) Fork asymmetry (represented as left vs right fork lengths) in DNA fibers prepared from B cells of the indicated genotypes. The determination coefficient is indicated in blue (R2).
See also Figure S1.
We next explored whether deletion of Pold3 in CD19Cre-positive Pold3lox/lox B cells led to RS and genomic instability, using wt, CD19Cre-negative Pold3lox/lox and CD19Cre-positive Pold3+/+ cells as controls. Indeed, by western blotting, we observed higher levels of H2AX and CHK1 phosphorylation in the CD19Cre-positive Pold3lox/lox B cells (Figure S1A), as well as decreased cell viability in response to inhibition of the RS-response kinase ATR (Figure S1B). Moreover, analysis of metaphase chromosomes from Pold3-deleted B cells revealed evidences of genomic instability, such as chromosome and chromatid breaks, cruciform structures and, most frequently, gaps indicative of incomplete DNA replication (Figure 3C,D). Finally, to directly evaluate DNA replication, we performed analyses on stretched DNA fibers. These experiments revealed that POLD3-deficiency led to a slower progression of replication forks (Figure 3E and S1C) and demonstrated the presence of RS, as evidenced by a significant increase in fork stalling and asymmetry (Figure 3F and S1D-F). Importantly, the slower rates were confirmed in origin-driven forks; i.e. forks progressing around newly fired origins (“green-red-green” tracks) or in two proximal “red-green” tracks moving in opposite directions within the same fiber, as these forks are less likely to arise from BIR or repair-related processes (Figure S1C). Collectively, these results reveal that POLD3 is essential to sustain overall DNA replication during S phase and to suppress RS in B cells.
POLD3 is haploinsufficient for stabilizing the Polδ complex
To determine the mechanism by which POLD3 is essential in mammalian cells we immunoprecipitated POLD3 from nuclear extracts of wt and Pold3-deleted B lymphocytes, and subjected the precipitates to Mass Spectrometry (MS) (Table S1). Ranking the immunoprecipitated proteins by the difference in their abundance between wt and POLD3-deficient extracts, led to POLD1 and POLD2 ranking first from the entire list, whereas POLD4 ranked seventh. This not only validated the POLD3 immunoprecipitation (since it efficiently pulled down the whole Polδ complex), but also revealed that in addition to POLD3 all other members of the Polδ complex (POLD1, POLD2 and POLD4) were present at reduced levels in extracts lacking POLD3. Western blotting confirmed the overall low levels of POLD1-3 in POLD3-deficient B lymphocytes, while levels of the POLE subunit from the leading strand polymerase complex Polε remained unaltered (Figure 4A, see also Figure S2). Of note, and as mentioned above when deleting Pold3 in adult tissues, the reduction of POLD1-3 levels found in POLD3 deficient B-lymphocytes is likely more severe than observed due to the selection for wt cells that escaped Cre-mediated recombination. Accordingly, the reduction of POLD1-3 levels is more severe in slowly proliferating cell types such as mouse embryonic fibroblasts (MEFs) (see below).
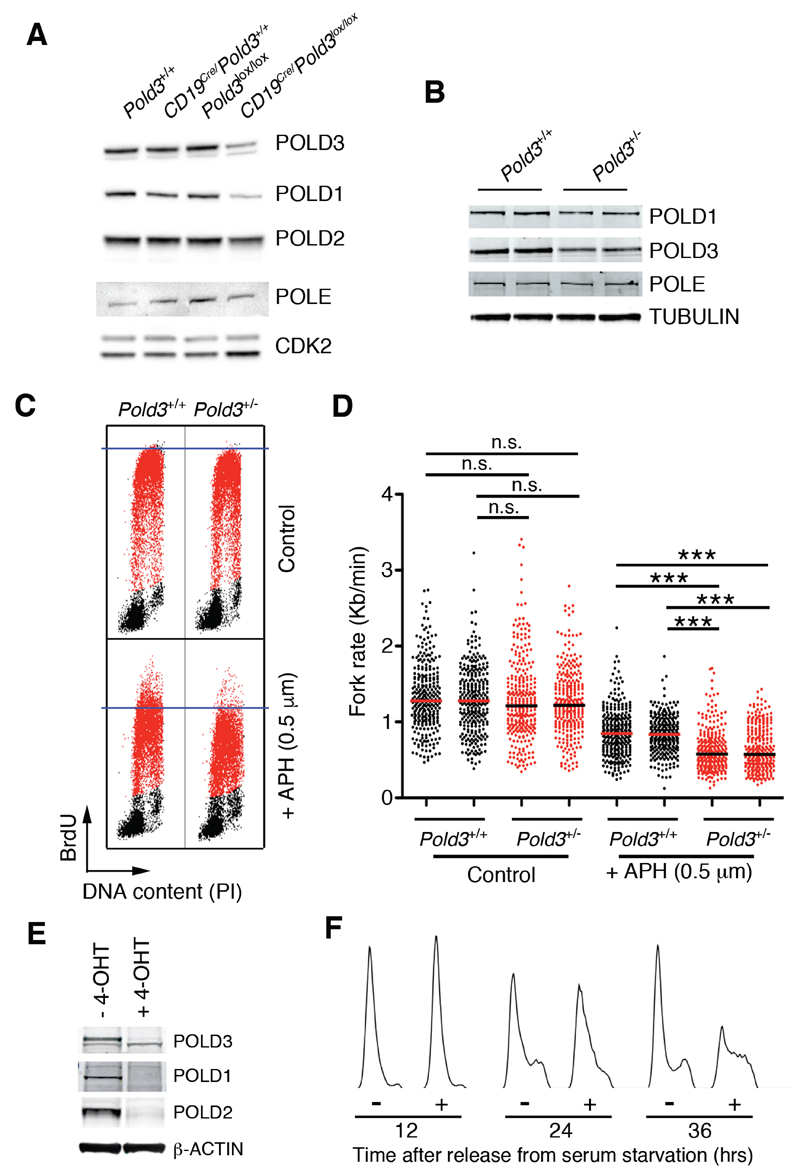
Figure 4. POLD3 maintains the stability of the POLD complex.
(A) WB illustrating the levels of POLD1, POLD2, POLD3 and POLE in B cell cultures from the indicated genotypes, 48 hrs after stimulation with LPS (25 μg/ml). CDK2 was used as a loading control.
(B) WB of POLD1, POLD3 and POLE levels from Pold3+/+ and Pold3+/- B cells. TUBULIN is shown as a loading control.
(C) Representative FACS analysis of DNA content (PI) versus BrdU incorporation in Pold3+/+ and Pold3+/- B cells. 48 hrs after stimulation with LPS (25 μg/ml) cells were exposed (or not) to 0.5 μM aphidicolin (APH) for 4 hrs, and then to BrdU (10μM) for another 30 min. Note that the APH treatment had a bigger impact on in limiting DNA replication on Pold3 heterozygous cells.
(D) Fork rates were measured in stretched DNA fibers prepared from B cells of the indicated genotypes treated as in (C). Data from two independent B cell cultures of each genotype are shown. Approximately 300 tracks were measured to estimate fork rates in each condition. ***P<0.001.
(E) WB illustrating the levels of POLD1, POLD2 and POLD3 in UbCre/Pold3lox/lox MEF exposed (or not) to 4-OHT (1 μM) and that were serum starved (0.1% serum) for 72 hrs and subsequently released in 15% serum-containing media for 24 hrs.
(F) FACS analyses of DNA content (PI) from UbCre/Pold3lox/lox MEF exposed (or not) to 4-OHT (1 μM) and that were serum starved (0.1% serum) for 72 hrs and subsequently released in 15% serum-containing media for the indicated times. In (E) and (F) 4-OHT was present throughout the entire experiment where indicated.
See also Figures S2, S3 and Table S1.
Next, and given the phenotypes observed in Pold3 heterozygous mice, we analyzed B cell extracts from wt and Pold3+/- mice. These experiments revealed lower levels of POLD3, but also of the catalytic subunit POLD1 in Pold3 heterozygous B cells (Figure 4B). To determine whether the partial reduction in POLD1 found in Pold3+/- cells had functional consequences we evaluated the impact of the DNA polymerase inhibitor aphidicolin (APH) in wt and Pold3 heterozygous cells. Since the dose of a chemical inhibitor needed to inhibit its target correlates with the levels of the target protein, we reasoned that the lower levels of the catalytic subunit POLD1 could increase the sensitivity of Pold3 heterozygous cells to APH. Indeed, FACS analyses showed that whereas overall DNA replication rates were similar in both cell types, a low dose of APH had a bigger impact in reducing BrdU incorporation rates in Pold3 heterozygous cells (Figure 4C). In addition, analyses of fork rate progression in stretched DNA fibers confirmed a higher sensitivity of Pold3+/- cells to APH (Figure 4D). Altogether, these data reveal that POLD3 is haploinsufficient for stabilizing the Polδ complex, which could explain the severe effects of its deficiency in mammalian cells.
POLD3 deficiency sensitizes MEFs to oncogene expression
To extend our findings to other cell types and to explore the impact of Pold3 deletion in the context of oncogenes, we generated mouse embryonic fibroblasts (MEFs) from UbCre/Pold3lox/lox mice. Treatment of these MEFs with 4-OHT led to the depletion of POLD3, POLD1 and POLD2 (with no effect on POLE) in all cellular compartments analysed (cytoplasm, nucleus and chromatin) (Figure S2A). Moreover, a 2-day treatment with 4-OHT led to a partial reduction in BrdU incorporation rates and slower S-phase progression UbCre/Pold3lox/lox mice (Figure S2B). Since residual POLD3 protein levels could explain the partial reduction in DNA replication observed upon a 2-day 4-OHT treatment, we took two independent approaches to further reduce POLD3. First, we exposed UbCre/Pold3lox/lox MEFs for up to 7 days of continuous 4-OHT treatment. This experiment revealed a progressive reduction in POLD1, POLD2 and POLD3 levels, which correlated with a stepwise decrease in DNA replication rates (measured by EdU incorporation) in these cells (Figure S2C,D). In addition to this experiment, UbCre/Pold3lox/lox MEFs were serumstarved in the presence of 4-OHT for 72 hrs and subsequently released in the presence of serum. This approach further reduced POLD1-3 levels, and revealed a severe deficiency of Pold3-deleted cells in completing DNA replication with a concomitant accumulation of cells in S phase (Figure 4E,F). In order to evaluate the impact of POLD3 deficiency in the context of oncogenes, we first needed to define a faster way to deplete the protein. As an alternative way of depleting POLD3 in asynchronous cultures, we infected Pold3lox/lox MEFs with adenoviruses expressing Cre (AdCre), which is more efficient than the 4-OHT treatment. Consistent with our observations with the UbCre allele, exposure to AdCre led to a severe reduction in DNA replication in Pold3lox/lox MEFs, together with an accumulation of γH2AX as measured by High-Throughput Microscopy (HTM), which was aggravated by the expression of RASG12V or MYC oncogenes (Figure S3A,B). Moreover, while expression of RASG12V together with E1A oncogenes transforms primary MEFs, it was highly toxic in the context of POLD3-deficient MEFs (Figure S3C). Of note, we were unable to obtain any RASG12V/E1A-transformed clones lacking POLD3 expression, suggesting that oncogenic transformation does not bypass the essential nature of Pold3.
Discussion
Recent studies have indicated a unique role for POLD3 in DNA synthesis occurring during the repair of RS-induced DNA breaks (Costantino et al., 2014; Mayle et al., 2015; Minocherhomji et al., 2015). Given that oncogene-induced RS is a major cause of genomic instability in cancer, targeting POLD3 could thus offer an opportunity to preferentially target cancer cells. While our research confirms that POLD3 deficiency is particularly toxic for cells expressing oncogenes, it has also identified a haploinsufficient role of Pold3 in stabilizing the Polδ complex and in safeguarding genomic integrity during DNA replication during S phase. Nevertheless, and besides the role of POLD3 in origin-associated DNA synthesis, other functions of POLD3 in BIR or TLS could also contribute to the dramatic phenotypes observed upon Pold3 deletion in mice. The role of POLD3 in TLS has been mostly associated as being an integral member of the TLS-specific DNA polymerase zeta (Polζ) complex (Johnson et al., 2015; Makarova et al., 2012). In this regard, our proteomic analysis in B cells did not show changes on members of the Polζ complex in POLD3 deficient cells (Table S1), although the constitutive levels of this complex might be low in the absence of sufficient lesions that need to be dealt by TLS. Interestingly, recent work in chicken DT40 cells has revealed that POLD3 could also operate in TLS independently of Polζ and as part of the Polδ complex (Hirota et al., 2016; Hirota et al., 2015), a function that could in principle be conserved in mammals. Nevertheless, and given the severe defects of Pold3 nullyzygosity in overall DNA replication, alternative genetic approaches will be needed to perform a dedicated analysis of the roles of POLD3 in BIR or TLS in mammalian cells. In what regards to the idea of targeting POLD3 as a way to preferentially kill cancer cells, our results predict a broad deleterious effect of POLD3 inhibition although the phenotypes of depleting versus inhibiting a protein are often not the same. The identification of separation of function mutants with impaired repair-associated DNA synthesis, but with an intact origin-linked DNA replication function (if these functions of POLD3 can be separated), should be an interesting alternative to further explore the potential for targeting POLD3 in cancer therapy.
Experimental Procedures
Mouse models
For the generation of Pold3 mutant mice, a linearized construct targeting Pold3 (EUCOMM) was electroporated into ES cells. Upon identification of properly recombined ES clones, these were used for the generation of chimaeras using standard procedures. CD19Cre, UbCre, EIIA-Cre and actin-FLP strains have been described before (Lakso et al., 1996; Rickert et al., 1997; Rodriguez et al., 2000; Ruzankina et al., 2007). Mice were kept under standard conditions at specific-pathogen free facility of the Spanish National Cancer Centre in a mixed C57BL/6-129/Sv background. All mouse work was performed in accordance with the Guidelines for Humane Endpoints for Animals Used in Biomedical Research, and under the supervision of the Ethics Committee for Animal Research of the “Instituto de Salud Carlos III”.
DNA fiber analyses
The analysis of stretched DNA fibers was performed as described before (Jacome et al., 2015). Briefly, B cell cultures were pulse-labeled with 50 μM CldU (20 min) followed by 250 μM IdU (20 min). Labeled cells were collected and DNA fibers were spread in buffer containing 0.5% SDS, 200 mM Tris pH 7.4 and 50 mM EDTA. For immunodetection of labeled tracks, fibers were incubated with primary antibodies against CldU and BrdU and developed with the corresponding secondary antibodies conjugated to Alexa dyes. A mouse anti-ssDNA antibody was used to assess fiber integrity.
POLD3 immunoprecipipation
500 μg of nuclear extracts from B cells were immunoprecipitated with Protein-G Dynabeads that were previously coated with an anti-POLD3 antibody (Sigma). Proteins were eluted in 8M urea in 0.1M triethylammonium bicarbonate, and digested with trypsin for subsequent analyses. Proteomics was performed by NanoLC-MS/MS using an LTQ-Orbitrap Velos (Thermo Scientific) coupled to a nanoLC Ultra system (Eksigent), equipped with a nanoelecroespray ion source (Proxeon Biosystems). Raw data was processed using Proteome Discoverer 1.4 (Thermo) and filtered for frequent contaminants.
See the Supplemental Information for a full list of antibodies, as well as other methods used in this study.
Supplementary Material
Acknowledgements
Research was funded by Fundación Botín, Banco Santander, through its Santander Universities Global Division and by grants from the Spanish Ministry of Economy and Competitiveness (MINECO) (SAF2014-59498-R; SAF2014-57791-REDC), Fundació La Marato de TV3, Howard Hughes Medical Institute and the European Research Council (ERC-617840) to OF; by a Marie-Curie International Outgoing Fellowshp (IOF) from the FP7 Marie Curie Actions and a grant from MINECO (BFU2014-55168-JIN) that was co-funded by European Regional Development Funds (FEDER) to EL; by a grant from MINECO (BFU2013-49153) to JM; and by the European Commission (ERC grant ONIDDAC) to TDH.
Footnotes
Author Contributions
M.M. participated in the generation of POLD3 deficient mice and in most experiments of the manuscript. E.L. and M.E.A. helped with biochemistry and proteomic analyses. M.D. and J.M. helped with stretched DNA fiber analyses. I.K., N.G., S.K.S. and T.D. helped in the analyses of POLD3 deficiency in mice and in experiments of DNA replication in MEFs. O.F. coordinated the study and wrote the manuscript.
The authors declare no competing financial interests.
References
- Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducoux M, Urbach S, Baldacci G, Hubscher U, Koundrioukoff S, Christensen J, Hughes P. Mediation of proliferating cell nuclear antigen (PCNA)-dependent DNA replication through a conserved p21(Cip1)-like PCNA-binding motif present in the third subunit of human DNA polymerase delta. J Biol Chem. 2001;276:49258–49266. doi: 10.1074/jbc.M106990200. [DOI] [PubMed] [Google Scholar]
- Gerik KJ, Li X, Pautz A, Burgers PM. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- Hirota K, Tsuda M, Mohiuddin, Tsurimoto T, Cohen IS, Livneh Z, Kobayashi K, Narita T, Nishihara K, Murai J, et al. In vivo evidence for translesion synthesis by the replicative DNA polymerase delta. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Yoshikiyo K, Guilbaud G, Tsurimoto T, Murai J, Tsuda M, Phillips LG, Narita T, Nishihara K, Kobayashi K, et al. The POLD3 subunit of DNA polymerase delta can promote translesion synthesis independently of DNA polymerase zeta. Nucleic Acids Res. 2015;43:1671–1683. doi: 10.1093/nar/gkv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes P, Tratner I, Ducoux M, Piard K, Baldacci G. Isolation and identification of the third subunit of mammalian DNA polymerase delta by PCNA-affinity chromatography of mouse FM3A cell extracts. Nucleic Acids Res. 1999;27:2108–2114. doi: 10.1093/nar/27.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacome A, Gutierrez-Martinez P, Schiavoni F, Tenaglia E, Martinez P, Rodriguez-Acebes S, Lecona E, Murga M, Mendez J, Blasco MA, et al. NSMCE2 suppresses cancer and aging in mice independently of its SUMO ligase activity. EMBO J. 2015;34:2604–2619. doi: 10.15252/embj.201591829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E, Dixon N. Replicative DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Klassen R, Prakash L, Prakash S. A Major Role of DNA Polymerase delta in Replication of Both the Leading and Lagging DNA Strands. Mol Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Tan CK, Downey KM, So AG. Further studies on calf thymus DNA polymerase delta purified to homogeneity by a new procedure. Biochemistry. 1984;23:1906–1913. doi: 10.1021/bi00304a003. [DOI] [PubMed] [Google Scholar]
- Lee YS, Gregory MT, Yang W. Human Pol zeta purified with accessory subunits is active in translesion DNA synthesis and complements Pol eta in cisplatin bypass. Proc Natl Acad Sci U S A. 2014;111:2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Mo J, Rodriguez-Belmonte EM, Lee MY. Identification of a fourth subunit of mammalian DNA polymerase delta. J Biol Chem. 2000;275:18739–18744. doi: 10.1074/jbc.M001217200. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- MacNeill SA, Moreno S, Reynolds N, Nurse P, Fantes PA. The fission yeast Cdc1 protein, a homologue of the small subunit of DNA polymerase delta, binds to Pol3 and Cdc27. EMBO J. 1996;15:4613–4628. [PMC free article] [PubMed] [Google Scholar]
- Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, Bjergbaek L, Lupski JR, Ira G. DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349:742–747. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y, Hickson ID. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015;528:286–290. doi: 10.1038/nature16139. [DOI] [PubMed] [Google Scholar]
- Mo J, Liu L, Leon A, Mazloum N, Lee MY. Evidence that DNA polymerase delta isolated by immunoaffinity chromatography exhibits high-molecular weight characteristics and is associated with the KIAA0039 protein and RPA. Biochemistry. 2000;39:7245–7254. doi: 10.1021/bi0000871. [DOI] [PubMed] [Google Scholar]
- Murga M, Bunting S, Montana MF, Soria R, Mulero F, Canamero M, Lee Y, McKinnon PJ, Nussenzweig A, Fernandez-Capetillo O. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Tan CK, Downey KM, Fisher PA. Enzymologic mechanism of calf thymus DNA polymerase delta. J Biol Chem. 1991;266:11699–11704. [PubMed] [Google Scholar]
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata K, Ohta S, Yamada K, Obuse C, Yoshikawa H, Tsurimoto T. The human homologue of fission Yeast cdc27, p66, is a component of active human DNA polymerase delta. J Biochem. 2001;129:699–708. doi: 10.1093/oxfordjournals.jbchem.a002909. [DOI] [PubMed] [Google Scholar]
- Zuo S, Gibbs E, Kelman Z, Wang TS, O'Donnell M, MacNeill SA, Hurwitz J. DNA polymerase delta isolated from Schizosaccharomyces pombe contains five subunits. Proc Natl Acad Sci U S A. 1997;94:11244–11249. doi: 10.1073/pnas.94.21.11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.