Abstract
Objective
RNA-binding protein with multiple splicing (RBPMS) has been shown to physically interact with Smads and enhance transforming growth factor-β (TGF-β)–mediated Smad2/3 transcriptional activity in mammalian cells. Objective of this study was to examine whether expression of RBPMS is regulated by interleukin-1β (IL)-1β and TGF-β superfamily growth factors and whether expression of RBPMS is altered during aging and experimental osteoarthritis.
Methods
Expression of RBPMS protein was investigated in chondrocyte cell lines of murine (H4) and human (G6) origin using Western blot analysis. Regulation of RBPMS expression in H4 chondrocytes at mRNA level was done by reverse transcriptase–quantitative polymerase chain reaction. Furthermore, characterization of Smad signaling pathways regulating RBPMS expression was performed by blocking studies using small molecule inhibitors or by transfection studies with adenoviral vector constructs (constitutive-active ALK1 and constitutive-active ALK5). Expression of RBPMS in cartilage of different age groups of C57BL/6N mice (6 months and 20 months) and in a surgically induced osteoarthritis (OA) mouse model was analyzed using immunohistochemistry.
Results
RBPMS was shown to be expressed in chondrocytes and cartilage of murine, human, and bovine origin. TGF-β inhibited RBPMS expression while BMP2 and IL-1β increased its expression. TGF-β-induced inhibition was blocked by ALK5 inhibitor. Overexpression of ca-ALK1 stimulated RBPMS expression. Moreover, RBPMS expression was found to be reduced with ageing and in OA pathogenesis.
Conclusions
Expression of RBPMS in chondrocytes is regulated by TGF-β superfamily members and IL-1β, indicating a counter-regulatory mechanism. Expression of RBPMS, in cartilage and its reduction during ageing and OA might suggest its potential role in the maintenance of normal articular cartilage.
Keywords: RBPMS, Smad 1/5/8, Smad2/3, TGF-β, osteoarthritis
Introduction
Articular cartilage is an avascular tissue that covers bone ends in a diarthrodial joint and is underlined by a subchondral bone plate.1 During osteoarthritis (OA), degradation of cartilage matrix proteins exceeds synthesis resulting in net matrix loss.2 Transforming growth factor-β (TGF-β) is involved in the maintenance of articular cartilage via ALK5/Smad2/3 signaling pathway.3 During canonical TGF-β signaling, TGF-β binds to the type II receptor (TGFBR2), recruits and phosphorylates the TGF-β type I receptor (TGFBR1), activin receptor-like kinase-5 (ALK5). ALK5 subsequently phosphorylates Smad2 or Smad3. Thereafter, phosphorylated Smad2/3 forms a complex with Smad-4 and translocates to the nucleus and modulates transcription of TGF-β-responsive genes, such as aggrecan and collagen type II (Col2a1). Moreover, TGF-β has also been shown to counteract the catabolic effects induced by IL-1β in chondrocytes, including suppression of aggrecan synthesis and increased synthesis of cartilage degrading enzymes like matrix metalloproteinases (MMPs).4
Research in the field of cartilage biology, has identified recently the “double edged sword” effect of TGF-β in cartilage, like it has been reported in endothelial cells.5 Besides being a protective factor for cartilage, TGF-β has also been found to contribute to chondrocyte phenotypic alterations associated with OA development6 and progression by signaling via the ALK1/Smad1/5/8 pathway,7,8 thought to be involved in terminal differentiation.9,10 Moreover, age-dependent alterations in the relative expression of ALK5 and activin receptor-like kinase-1 (ALK1) receptors shifting toward a dominant Smad1/5/8 signaling pathway has been suggested to contribute to the pathogenesis of OA.8 So far various transcription factors and co-activators, like AP-1 family proteins, CBP/p300,11 Ski,12 and Sp113 have been reported to regulate TGF-β signaling by interacting with Smads. Recently, a protein called RNA-binding protein with multiple splicing (RBPMS) has been shown to physically interact with Smads and to enhance C-terminal phosphorylation of Smad2 and Smad3, further enhancing transcriptional activity.14
Since TGF-β signaling is crucial for maintaining healthy cartilage in a Smad-dependent manner, we have studied whether RBPMS is expressed in articular chondrocytes and whether this expression is modulated by growth factors such as TGF-β itself and BMP2, and by an inflammatory mediator (interleukin-1β [IL-1β]). In addition, we analyzed RBPMS expression in different age groups of C57BL/6N mice (6 months and 20 months) and in a model of experimental OA in mice to explore a putative role in maintenance of articular cartilage.
Methods
Chondrocyte Cell Line Culture
The murine chondrocyte cell line (H4)15 was cultured in 24-well plates in Dulbecco’s modified Eagle medium (DMEM)/F12 (1:1) medium with 5% fetal calf serum, 5% pyruvate and gentamycin (4.8 μg/μl). 20,000 cells were seeded in each well and cultured for 24 hours in a CO2 incubator at 37°C. After 24 hours, the medium was replaced with serum-free medium and culturing was continued for 24 hours. Subsequently, total cellular RNA was prepared by guanidinium isothiocyanate method, using TRIzol (Life Technologies) and evaluated for the expression of RBPMS mRNA transcripts using reverse transcriptase–quantitative polymerase chain reaction (RT-qPCR).
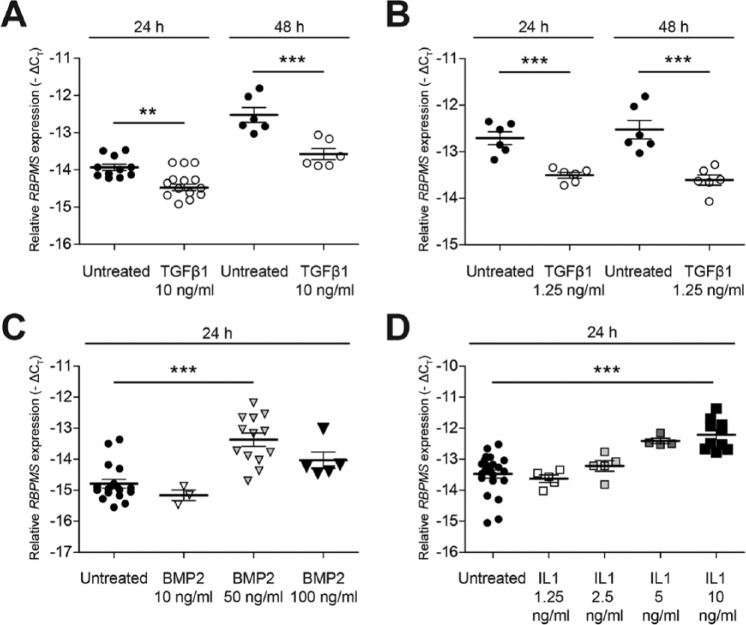
To evaluate the effect of TGF-β, IL-1β, and BMP2 on the expression of RBPMS mRNA transcripts, H4 chondrocytes were stimulated with recombinant human TGF-β1 (10 ng/mL or 1.25 ng/mL) (Biolegend), or recombinant murine IL-1β (10 ng/mL) (R&D Systems) or recombinant human BMP2 (50 ng/mL) (R&D Systems) in serum free conditions for 24 and 48 hours. Subsequently, mRNA was extracted and evaluated for the expression of RBPMS mRNA transcripts using RT-qPCR. These in vitro experiments were repeated 3 times ( Fig. 2A and 2B ), except for treatment with BMP2 at 10 ng/mL ( Fig. 2C ), which was done only twice. Each condition was done in duplicates or triplicates (biological replicates) and measured twice on qPCR.
Figure 2.
Expression of RBPMS (RNA-binding protein with multiple splicing) mRNA in H4 chondrocytes is differently influenced by members of TGF-β (transforming growth factor-β) superfamily ligands (TGF-β and BMP2) and IL-1β. H4 chondrocytes were either stimulated with TGF-β (10 ng/mL or 1.25 ng/mL) for 24 hours and 48 hours or BMP2 (50 ng/mL) for 24 hours in serum-free conditions and evaluated for the expression of RBPMS mRNA transcripts using RT-qPCR (reverse transcriptase–quantitative polymerase chain reaction). Expression of RBPMS mRNA transcripts was decreased significantly, whereas expression of RBPMS mRNA transcripts was increased significantly, when H4 chondrocytes were treated with either TGF-β (10 ng/mL or 1.25 ng/mL) (A and B) or BMP2 (50 ng/mL) (C), respectively. Expression of RBPMS in H4 chondrocytes was upregulated by IL-1β (D). H4 chondrocytes were stimulated with different concentration of IL-1β (1.25-10 ng/mL) in serum-free conditions and cultured for 24 hours and evaluated for the expression of RBPMS mRNA transcripts using RT-qPCR. Treatment of H4 chondrocytes with IL-1β (10 ng/mL) results in increased expression of RBPMS mRNA significantly. In all the experiments, each individual data point represents the mean −ΔCt of a sample analyzed in duplicates (*P < 0.05; **P < 0.005; ***P < 0.001).
Cartilage Tissue Samples
Samples of murine cartilage obtained from tibio-femoral joints of C57BL/6N mice and explants of cartilage from bovine metacarpal joints were used to analyze the expression of RBPMS mRNA transcripts. Cartilage tissue or explants were instantly frozen in liquid nitrogen and crushed using a micro-dismembrator (B. Braun). The crushed material was dissolved in RLT buffer supplied with the RNeasy Mini kit (Qiagen). Samples were treated with proteinase K, and RNA was isolated with the RNeasy Mini kit according to the manufacturer’s protocol. Complementary DNA (cDNA) was prepared by RT-PCR and was subsequently subjected to qPCR.
Quantitative Real-Time PCR
Although RBPMS has been previously shown to be expressed in various human cell lines and different tissues of xenopus, chick, rat, and human,16-18 it has not been previously investigated in cartilage tissue or chondrocyte cell lines. Therefore, expression of RBPMS mRNA was investigated in the murine chondrocyte cell line H415 and murine cartilage tissues by RT-qPCR, using primers designed to specifically amplify murine RBPMS mRNA transcript ( Table 1 ). The efficiency of this primer pair was validated using cDNA obtained from kidney tissue of mice, which has been reported to express RBPMS.19 mGapdh was used as a reference gene for relative quantification of genes of our interest (mId1, mPai1, and mRbpms). Quantitative PCR was performed using SYBR green on a Step One plus Real Time PCR system (Applied Biosystems) according to manufacturer’s protocol. Similarly, RBPMS mRNA expression was investigated in the human chondrocyte cell line G6,20 bovine primary chondrocytes and bovine cartilage explants by RT-qPCR, using the primers designed to specifically amplify human and bovine RBPMS mRNA transcripts, respectively. Sequences of all primer sets are given in Table 1 .
Table 1.
Primers used for RT-qPCR analysis.
| Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| mGapdh | GGCAAATTCAACGGCACA | GTTAGTGGGGTCTCGCTCCTG |
| mPai1 | ACGTCGTGGAACTGCCCTAC | CAGCGATGAACATGCTGAGG |
| mld1 | ACGACATGAACGGCTGCTAC | CAGGATCTCCACCTTGCTCAC |
| mRbpms | CAGTACTCCTCTGCCCAACAC | CTACTGGGGTAAAGTGCAGGTA |
| hGapdh | ATCTTCTTTTGCGTCGCCAG | TTCCCCATGGTGTCTGAGC |
| hALK1 | CCATCGTGAATGGCATCGT | GGTCATTGGGCACCACATC |
| hALK5 | CGACGG CGTTACAGTGTTTCT | CCCATCTGTCACACAAGTAAAATTG |
| hRbpms | CATTGCCAGAGAGCCATATGAG | AGGTGAAAGCAGGAGGAGGTA |
| bGapdh | CACCCACGGCAAGTTCAAC | TCTCGCTCCTGGAAGATGGT |
| bRbpms | ACCATTTAAGGGCTACGAAGGT | TGCGG GATTTCAG GATC GAA |
| bPai1 | CGAGCCAGGCGGACTTC | TGCGACACGTACAGAAACTCTTGA |
| bld1 | GCTCCGCTCAGCACTCTCAA | GATCGTCCGCTGGAACACA |
Relative quantification of all the target genes by qPCR was done either using comparative −ΔCt (Ct = cycle threshold) or ΔΔCt method. Predominantly, −ΔCt method was used to compare expression of gene of interest to that of the reference gene, and is calculated by subtracting the Ct value of a gene of interest from the Ct value of the reference gene (e.g., Ct Gapdh − Ct RBPMS). ΔΔCt method was used to calculate the effect size of adenoviral over expression by subtracting the −ΔCt values of ALK1 and ALK5 in LacZ-transduced cells from ALK1/5 −ΔCt levels in caALK1 or caALK5 transduced cells (e.g., −ΔCt caALK1 − −ΔCt caLacZ).
Transduction with Adenoviral Vector Constructs (Ad-caALK1/Ad-caALK5)
H4 chondrocytes were transduced with adenoviral vectors containing either constitutively active ALK1 (Ad-caALK1), constitutively active ALK5 (Ad-caALK5)8 or LacZ (Ad-LacZ) as a control at multiplicity of infection of 50 pfu/cell. The transfection experiment with H4 chondrocytes was done three times. Each time, each condition was tested in triplicate. H4 chondrocytes cultured as per the above mentioned protocol were subsequently washed with 0.9% saline, and incubated for 2 to 3 hours with adenoviral constructs. Then the transfected cells were cultured for 48 hours after which the expression pattern of RBPMS at mRNA level was evaluated by RT-qPCR. This experiment was repeated in bovine primary chondrocytes to ensure that the chondrocyte cell line responded similarly to primary cells. The primary bovine chondrocytes were isolated from bovine metacarpophalangeal joints as previously described21 from cows aged 9 months (n = 4) or 4 years (n = 2). These cells were transduced with either adenovirus overexpressing constitutive active ALK1 (Ad-caALK1) or Ad-caALK5 or Ad-LacZ control (multiplicity of infection of 50) for 2 hours and cultured for 48 hours thereafter to evaluate the expression pattern of RBPMS at the mRNA level. The transfection experiment with bovine primary chondrocytes was done twice, each time with chondrocytes of 3 cows.
Blocking of ALK1/Smad1/5/8 and ALK5/Smad2/3 Signaling Pathways
To analyze the effect of ALK1/Smad1/5/8 and ALK5/Smad2/3 signaling on expression of RBPMS mRNA transcripts, the H4 chondrocytes were treated with blockers of ALK1, 2, 3, 6 (pSmad1/5/8) and ALK4, 5, 7 (pSmad2/3) signaling pathways: The small molecule inhibitors, dorsomorphin (DM) and SB-505124 (SB5), respectively. H4 chondrocytes were treated with DM or SB5 (both 5 μM/mL) with or without TGF-β (10 ng/mL or 1.25 ng/mL) for 24 and 48 hours. Dimethyl sulfoxide (DMSO) was used as a control as the chemicals used were suspended in DMSO. After mRNA isolation, RBPMS mRNA expression was analyzed using RT-qPCR. These experiments were repeated 3 times.
Western Blot
Cells were lysed using lysis buffer (20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4) with freshly added protease inhibitor cocktail (cOmplete, Roche). Expression of RBPMS was assayed in H4 chondrocytes, G6 chondrocytes and mouse fibroblasts. One hundred micrograms of protein lysate was loaded on a 12% polyacrylamide gel with sodium dodecyl sulfate. 293T cell line (mammalian embryonic kidney cell line) and mouse liver tissue lysates were used as a positive control. Proteins were transferred onto a nitrocellulose membrane by wet blot transfer system using Tris–glycine transfer buffer (Tris 25 mM, glycine 200 mM, 20% methanol, pH 8.5) for 2 hours at 200 to 300 mA. The membrane was incubated with rabbit polyclonal antibodies (Abs) against RBPMS protein (1/2000) (Proteintech) overnight at 4°C. Fluorophore-labeled antibody Alexa Fluor 680 Donkey Anti-Rabbit IgG antibodies (1/10,000) (Life technologies) were used as secondary antibody. On these blots RBPMS should have a size between 28 and 31 kD(RBPMS).
For detection of phosphorylated Smad2/3, 20 µg of protein was loaded on a 7.5% polyacrylamide gel with sodium dodecyl sulfate. Proteins were transferred onto a nitrocellulose membrane by wet blot transfer system. The membrane was incubated with rabbit polyclonal Abs against pSmad2 (1/1000) (Cell Signaling Technology) overnight at 4°C. The fluorophore-labeled secondary antibody from Life Technologies was used (Alexa Fluor 680 Donkey Anti-Rabbit IgG antibodies (1/10,000). The membrane was imaged using Odyssey infrared imaging system (LI-COR).
Animals
C57BL/6N mice aged 6 months (n = 5) and 20 months (n = 5) were used for analyzing the pattern of RBPMS expression in relation to aging using immunohistochemistry. Similarly, C57BL/6N mice aged between 6 and 12 months (n = 6) and between 16 and 20 months (n = 4) were used for studying the expression of RBPMS mRNA levels in relation to aging using RT-qPCR.
C57BL/6N mice aged 10 to 12 weeks (n = 24) were used to induce OA by transecting the anterior attachment of the medial meniscus to the tibia of a right knee joint (destabilization of the medial meniscus model, DMM) as previously described by Glasson et al.22 Following DMM operation, mice were sacrificed at different time points (days 7, 14, 28, and 56) after surgery including at least 5 to 6 mice at each time point. Knee joints of sacrificed mice were isolated for histology. OA was confirmed by histological evaluation as previously described.23 The corresponding left knee joint of each mice was used as a control for comparison of DMM operated versus nonoperated knee joints. To observe any changes in the pattern of expression of RBPMS protein in DMM operated right knee joint of C57Bl/6 mice, immunohistochemistry was performed using anti-RBPMS antibody in knee joint sections of mice.
All animal procedures were carried out after obtaining approval from the Animal Ethics Committee of the Radboud University Nijmegen.
Immunohistochemistry
Knee joints were fixed, processed and sectioned using the standard protocol of the laboratory.8 Immunohistochemistry was performed as previously described.24 Sections were incubated with a specific primary Abs against RBPMS (1/100) for 1 hour at room temperature (Proteintech). Rabbit IgG (DAKO) was used as a negative control. A biotin-streptavidin detection system was used according to the manufacturer’s protocol (Vector Laboratories). Sections were counterstained with heamatoxylin and mounted with Permount.
Image Analysis
The number of RBPMS positive cells of articular chondrocytes in the tibial knee joints were scored using the image analysis system (Leica Imaging Systems) and a Leica DC 300F digital camera as previously described.25 For each knee joint, at least 3 tissue sections were measured and thereafter averaged per joint. All scoring was done in a blinded manner. The number of RBPMS positive cells were corrected for the average number of chondrocytes in consecutive sections stained with hematoxylin.
Statistical Analysis
Statistical analyses were performed using either a Kruskal-Wallis nonparametric test for nonnormally distributed data, and analysis of variance with Bonferroni’s multiple comparison test or Student’s t test were used for normally distributed data. All the analyses, including Bonferroni’s multiple comparison tests were done with the use of GraphPad Prism software 5. The Bonferroni correction was made by dividing the critical P value by the number of comparisons being made on a single data set in order to reduce the chances of obtaining false-positive results when multiple comparisons are performed on a single set of data. Results were considered significant, based on this modified P value, if P value was <0.05.
Results
Chondrocytes Express RBPMS at Both mRNA and Protein Levels
RBPMS has been previously shown to be expressed in various human cell lines and different tissues of xenopus, chick, rat, and human.16-18 Nevertheless, it was not known whether RBPMS is expressed in chondrocytes. Therefore, expression of RBPMS in chondrocytes was investigated with RT-qPCR ( Fig. 1A ). Our results showed that RBPMS was expressed in murine cartilage (mean Δ − ΔCt = −6.2), H4 chondrocytes (mean Δ − ΔCt = −13.8), G6 chondrocytes (mean Δ − ΔCt = −3.7), bovine cartilage explants (mean Δ − ΔCt = −11.0) and bovine primary chondrocytes (mean Δ − ΔCt = −13.7) all compared with GAPDH. The mRNA expression of RBPMS was clearly higher in murine cartilage tissue compared with H4 chondrocytes. Expression of RBPMS protein in H4 and G6 chondrocytes was confirmed by Western blot analysis ( Fig. 1B ). Instead of a single band ranging between ~28 and 32 kD, we observed at least 2 bands with molecular weight ranging between ~28 and 32 kD or higher. These bands could represent the existence of posttranslationally modified forms of the RBPMS protein or translated products of different mRNA splice variants that have been reported previously.16
Figure 1.
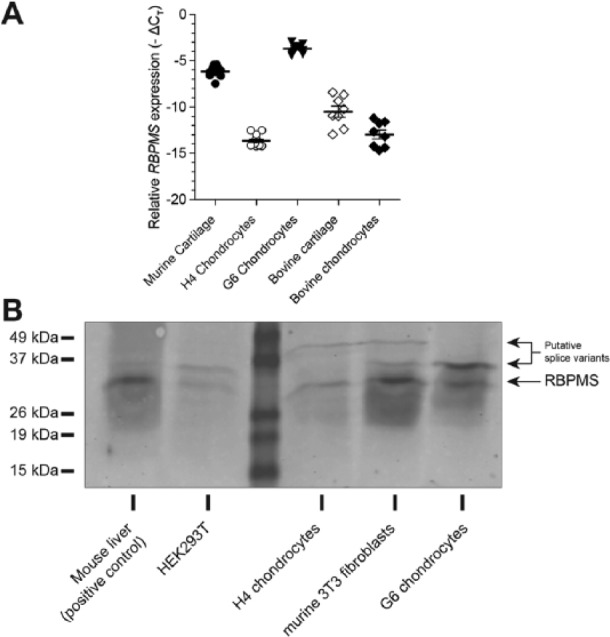
Expression of RBPMS (RNA-binding protein with multiple splicing) in primary chondrocytes, chondrocyte cell lines and articular cartilage. Expression of RBPMS at mRNA level was measured in isolated bovine chondrocytes, chondrocyte cell lines of murine (H4) and human origin (G6) and in murine and bovine articular cartilage by RT-qPCR (reverse transcriptase–quantitative polymerase chain reaction) (A). Expression of RBPMS protein (molecular weight range between 28 and 32 kD) was confirmed by Western blot analysis using polyclonal rabbit anti-RBPMS antibody in H4 chondrocytes (murine) and G6 chondrocytes (human) (B).
RBPMS Expression Is Regulated by TGF- β Superfamily Ligands and Inflammatory Factor IL-1β
We clearly confirmed the expression of RBPMS in chondrocyte cells and cartilage tissues at both mRNA and protein level. However, it was unknown whether its expression is regulated by growth factors of the TGF-β superfamily in chondrocytes. Therefore we investigated whether expression of RBPMS was regulated by TGF-β and BMP2. Stimulation of H4 chondrocytes with TGF-β (1.25 ng/mL or 10 ng/mL) led to a significant decrease in the levels of RBPMS mRNA transcripts at both 24 and 48 hours ( Fig. 2A and B ). In contrast, an increase in the levels of RBPMS mRNA transcript was observed when H4 chondrocytes were treated with BMP-2 (50 ng/mL) compared with that of untreated cells at 24 hours ( Fig. 2C ). No effect was found on the expression of RBPMS at lower concentration of BMP2 (10 ng/mL) and no significant effects at a concentration of 100 ng/mL.
Furthermore, in view of the observation that TGF-β possesses the ability to counteract the effects of IL-1β in chondrocytes, a major catabolic activator in chondrocytes, we investigated whether RBPMS expression is altered by IL-1β. Stimulation of H4 chondrocytes with increasing concentrations of IL-1β from 1.25 to 10 ng/mL showed an increase in the expression of RBPMS mRNA transcript (1.26 cycles), reaching a significant upregulation at an IL-1β concentration of 10 ng/mL ( Fig. 2D ).
RBPMS Expression Is Regulated via Smad Signaling
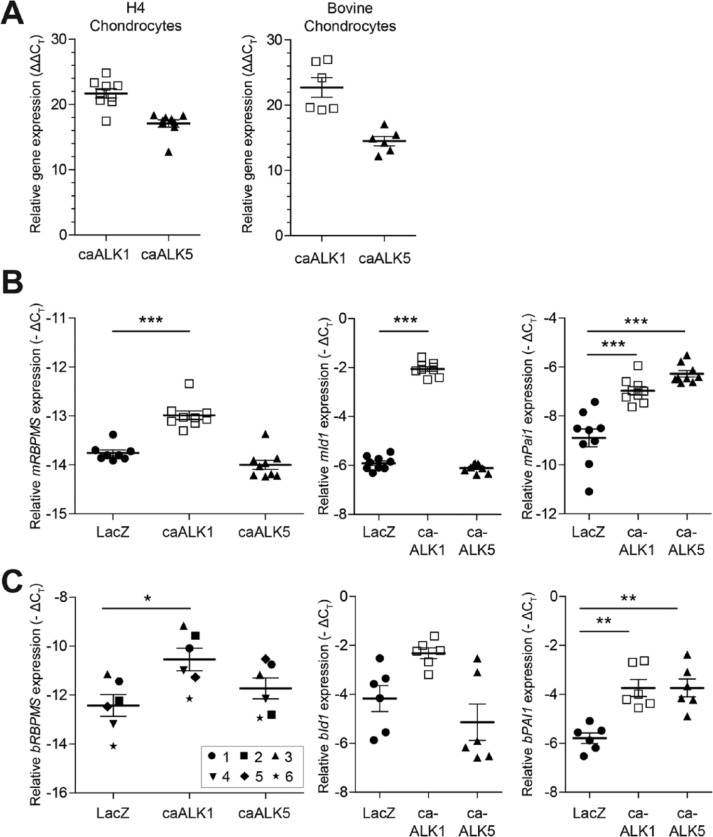
The opposite effect of TGF-β and BMP2 on RBPMS expression, prompted us to further explore the regulation of RBPMS expression by Smad signaling pathways in chondrocytes. Therefore, we transfected H4 chondrocytes with Ad-caALK1, Ad-caALK5, or Ad-LacZ. Efficiency of transfection was confirmed by checking the expression levels of human ALK1 and ALK5 in both murine H4 chondrocyte cell line and bovine primary chondrocytes ( Fig. 3A ). The mean ΔΔCT values of H4 chondrocytes transfected with Ad-caALK1 and Ad-caALK5 was 22.9 for ALK1 and 17.4 for ALK5 compared with H4 chondrocytes transfected with Ad-LacZ. Similarly, the mean ΔΔCT values of hALK1 and hALK5 in transfected bovine primary chondrocytes with Ad-caALK1 and Ad-caALK5 was 22.7 and 14.5, respectively compared with bovine primary chondrocytes transfected with Ad-LacZ.
Figure 3.
RBPMS (RNA-binding protein with multiple splicing) expression is regulated via Smad signaling. Effect on expression of RBPMS mRNA transcript on activation of the pSmad1/5/8 and pSmad2/3 pathway was done in H4 chondrocytes and bovine primary chondrocytes obtained from cartilage of different cows (n = 6). Efficiency of transfection was confirmed by checking the expression levels of human ALK1 and ALK5 in transfected H4 chondrocytes and primary bovine chondrocytes. Each individual data point in the figure represents the mean ΔΔCt of a sample analyzed in duplicates (A). Expression of RBPMS mRNA transcript was found to increase significantly when H4 chondrocytes were transfected with Ad-caALK1. Whereas, no significant effect was observed when H4 chondrocytes were transfected with Ad-caALK5 (B). Similar effect that was observed in H4 chondrocyte was also seen in primary chondrocytes obtained from bovine of different age groups (n = 6) (C). Transfection of Ad-caALK1 has resulted in increased expression of Id1 gene, the corresponding response gene of ALK1/smad1/5/8 signaling pathway and transfection of Ad-caALK5 has resulted in increased expression of Pai1 gene, the corresponding response gene of ALK5/smad2/3 signaling pathway in both H4 chondrocytes (B) and bovine primary chondrocytes (C). Each individual data point in figures B and C represents the mean −ΔCt of a sample analyzed in duplicates (*P < 0.05; **P < 0.005; ***P < 0.001).
Transfection of H4 chondrocytes with Ad-caALK1 increased the RBPMS mRNA levels significantly by 0.77 cycles, whereas no effect on the levels of RBPMS mRNA was found when they were transfected with Ad-caALK5 ( Fig. 3B ). To determine, whether bovine primary chondrocytes respond similarly as H4 chondrocytes, we transfected them with Ad-caALK1 and Ad-caALK5. This showed that AdcaALK1 increased RBPMS mRNA transcript levels by 1.9 cycles as was found in H4 chondrocytes ( Fig. 3C ). These results clearly show that activation of signaling via ALK1/Smad1/5/8 specifically lead to an increase in the expression of RBPMS. Further activation of ALK1/Smad1/5/8 and ALK5/Smad2/3 signaling pathways were checked by measuring the downstream markers of the respective signaling pathways. Both, H4 chondrocytes ( Fig. 3B ) and bovine primary chondrocytes ( Fig. 3C ) showed increased expression of Id1 when transfected with Ad-caALK1 and increased expression of Pai1 when transfected with Ad-caALK5, thus confirming the activation of ALK1/Smad1/5/8 and ALK5/Smad2/3 signaling pathways, respectively.
Opposite Effect of pSmad1/5/8 and pSmad2/3 Signaling on RBPMS Expression
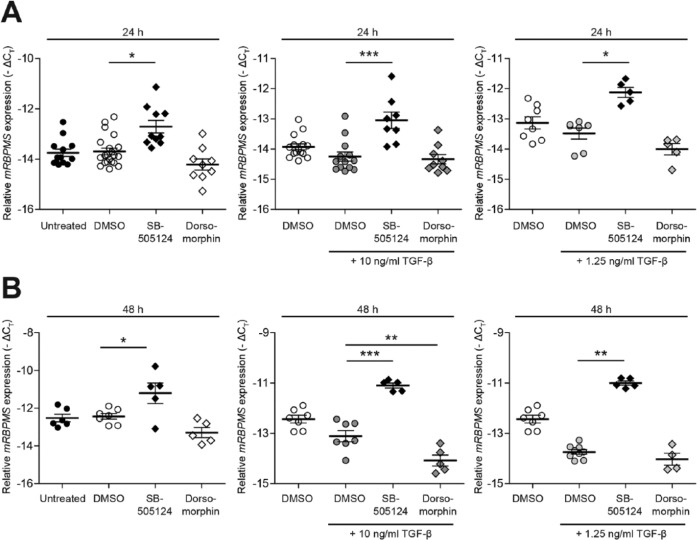
To validate our findings with regard to the different Smad signaling pathways that are involved in regulation of RBPMS expression, we analyzed the effect of blocking the Smad1/5/8 pathway or the Smad2/3 pathway. Treatment of H4 chondrocytes with DMSO (vehicle control) showed no significant effect on the expression of RBPMS when compared with untreated cells. Treatment of H4 chondrocytes with SB5 alone or in combination with TGF-β (10 ng/mL or 1.25 ng/mL) showed a significant upregulation of RBPMS mRNA transcripts at 24 hours ( Fig. 4A ) and 48 hours ( Fig. 4B ). Similarly the expression of RBPMS mRNA transcript was checked by blocking the pSmad1/5/8. Treatment of H4 chondrocytes with DM alone or with TGF-β (10 ng/mL or 1.25 ng/mL) led to downregulation of RBPMS mRNA expression at both 24 ( Fig. 4A ) and 48 hours ( Fig. 4B ); however, statistical significance was only reached when cells were treated with DM and TGF-β (10 ng/mL) together at 48 hours ( Fig. 4B ). These results indicate that the expression of RBPMS is regulated by pSmads, and that pSmad2/3 and pSmad1/5/8 have an opposite effect on RBPMS expression.
Figure 4.
Opposite effect of pSmad1/5/8 and pSmad2/3 signaling on RBPMS (RNA-binding protein with multiple splicing) expression. Effect of blocking pSmad1/5/8 and pSmad2/3 signaling on the expression of RBPMS mRNA transcript using small molecule kinase inhibitors was done in H4 chondrocytes. Blocking of pSmad2/3 signaling pathway with SB5 significantly increased the expression of RBPMS mRNA in H4 chondrocytes with or without TGF-β (transforming growth factor-β) (10 ng/mL or 1.25 ng/mL) at 24 hours (A) and 48 hours (B). Blocking of pSmad1/5/8 signaling pathway showed no effect on the expression of RBPMS mRNA without TGF-β (10 ng/mL) at both 24 hours (A) and 48 hours (B). Whereas, it resulted in downregulation of RBPMS mRNA expression when treated with TGF-β (10 ng/mL) at 48 hours (B). Each individual data point in all the figures represents the mean −ΔCt value of a sample analyzed in duplicates. (*P<0.05; **P<0.005; ***P<0.001)
Reduction in the Expression of RBPMS in Old Mice
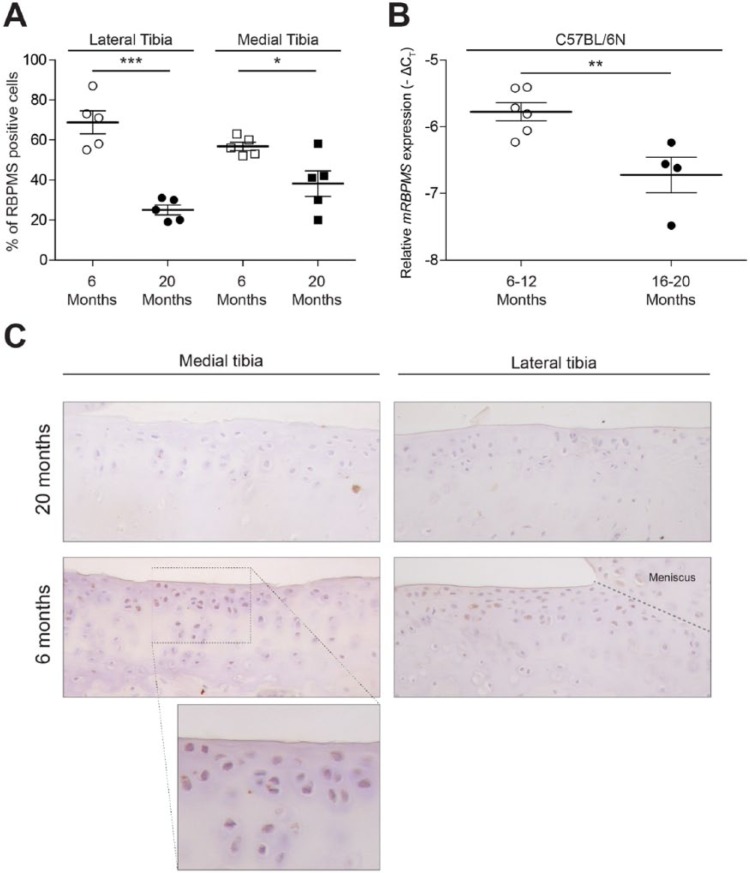
Although various risk factors have been identified, ageing is considered to be the major risk factor of primary OA.26 Since TGF-β signaling via ALK5/Smad/2/3 has been found to be reduced during ageing and RBPMS has also been found to be regulated by TGF-β signaling via ALK5/Smad/2/3, we investigated if there was a change in the expression pattern of RBPMS with aging. We performed immunohistochemistry using a specific anti-RBPMS antibody in knee joint sections of C57BL/6N mice of different age groups (6 and 20 months). The number of cells expressing RBPMS was found to be significantly reduced by 42% in lateral and 36% in medial tibial cartilage of mice in the 20-month age group compared with those in the 6-month age group ( Fig. 5A ). Furthermore, RBPMS mRNA transcripts quantified from the cartilage tissues of 16- to 20 month-old mice group was found to be reduced by 1.3 cycles compared with 6-month-old mice group ( Fig. 5B ).
Figure 5.
Reduction in the expression of RBPMS (RNA-binding protein with multiple splicing) in aging mice. Immunohistochemistry for RBPMS was performed in knee joints of 20 months and 6 months aged mice. The number of cells positive for RBPMS in lateral and medial tibia were measured with a computerized imaging system and corrected for the total number of cells in hematoxylin and eosin–stained sections. Expression of RBPMS-positive cells in knee joints of C57BL/6N mice were reduced by 42% in lateral and 36% in medial tibial cartilage of 20 months aged mice group compared with 6-month-old mice group (A). RBPMS mRNA transcripts was found to be reduced by 1.3 cycles in 16 to 20-month-old mice group compared with 6-month-old mice group (B). Representative sections are displayed in C (*P < 0.05; **P < 0.005; ***P < 0.001).
Reduction in the Expression of RBPMS in Experimental OA
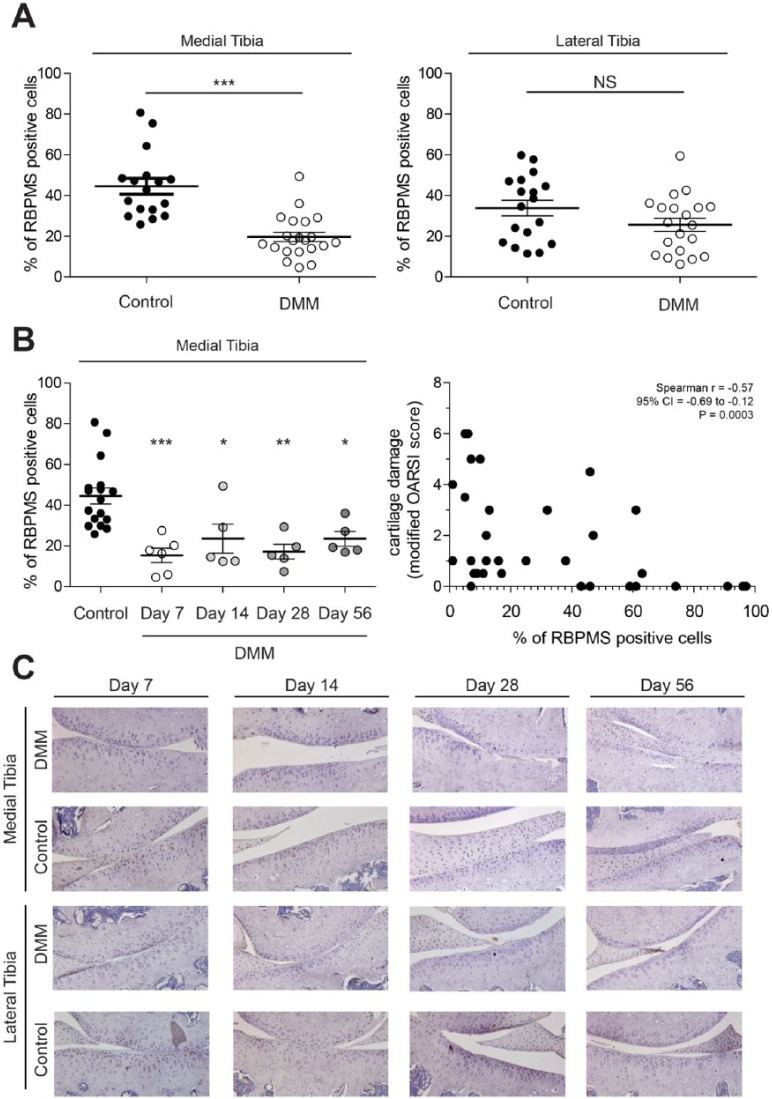
Reduction in RBPMS expression associated with advancing age, a major risk factor of primary OA, provoked us to investigate the impact of cartilage damage on RBPMS expression. Considerable reduction in the expression of RBPMS was seen in the cartilage of DMM-operated right knee joints compared to control left knee joints on the medial side of the tibia. The mean percentage of cells expressing RBPMS at medial knee joint was found to be reduced significantly by 25% whereas this was not significant at the lateral side ( Fig. 6A , left and right). The number of cells expressing RBPMS was found to be significantly reduced by 29%, 21%, 27%, and 21% on days 7, 14, 28, and 56, respectively, in medial tibial cartilage ( Fig. 6B , left). These data suggest that cartilage damage is associated with lowered expression of RBPMS. This is supported by the observation that the percentage of positive RBPMS cells correlates negatively with cartilage damage (Spearman r = −0.57, P = ***) in the DMM model of OA ( Fig. 6B , right).
Figure 6.
Reduction in the expression of RBPMS (RNA-binding protein with multiple splicing) in osteoarthritis (OA)-induced mice models. Expression of RBPMS was determined in the knee joints with the DMM (destabilization of the medial meniscus) model of experimental OA. Immunohistochemistry was performed for RBPMS using rabbit polyclonal anti-RBPMS Abs. The number of cells in medial and lateral tibia staining positive were measured with a computerized imaging system and corrected for the total number of cells counted in the respective section or cells counted in hematoxylin and eosin–stained adjacent sections. In the DMM model, the number of RBPMS-positive cells of the medial tibia was found to be reduced compared with that of control left knee joints. No significant reduction was observed in the expression of RBPMS-positive cells in lateral tibia of DMM operated right knee compared with lateral tibia of control left knee joint (A). Percentage of RBPMS-positive cells of medial tibia at different time points in comparison with control knee joint (B left), and the negative correlation between cartilage damage and percentage of positive RBPMS cells (B right). Representative sections are displayed in C. (*P < 0.05; **P < 0.005; ***P < 0.001).
Discussion
In this study, we investigated the expression and regulation of RBMPS mRNA transcripts in chondrocytes in vitro and RBMPS protein in murine aged and OA cartilage in vivo. RBPMS is a member of one of the largest families of RNA-binding proteins, containing a single RNA recognition motif (RRM) domain with 2 RNA-binding motifs (RNP1 and RNP2) at the N-terminus.16 To our knowledge, this is the first study to show the expression of RBPMS in chondrocytes and cartilage, although it has been previously shown to be expressed in various human cell lines and different tissues of xenopus, chick, rat, and human.16-18 Similar to other RNA-binding proteins, ribonucleoprotein domain (RNP) of mammalian RBPMS and Hermes, the Xenopus homologue of RBPMS has been identified to interact directly with mRNA, presuming its regulatory role in gene expression at the posttranscriptional level. However, specific mRNA targets interacting with RNP domain of mammalian RBPMS and hermes have not been identified yet.
From the time since the discovery of human RBPMS gene in 1998 by Shimamoto et al.,16 only few studies emphasizing its probable functional role have been published. Recently, RBPMS has been identified to interact with Smad proteins,14 the key mediators of TGF-β signaling, suggesting a role in transcriptional regulation of gene expression. Unlike, some of the cofactors, that have been found to interact with Smads and suppress TGF-β/Smad-induced transcription,12,27,28 RBPMS have been shown to activate TGF-β/Smad-induced transcription possibly by enhancing phosphorylation of Smad2 and Smad3.14
Among various growth factors, TGF-β has been found to be essential to maintain healthy cartilage3 and has also been suggested to be a protective factor that counteracts the effects of IL-1β in chondrocytes.29 However, it was not known whether TGF-β superfamily signaling regulates RBPMS expression in chondrocytes and whether this is regulated via the Smad1/5/8 or Smad2/3 signaling pathways. Therefore, to study the role of Smad1/5/8 and Smad/2/3 signaling in the regulation of RBPMS expression in chondrocytes, we carried out studies by blocking phosphorylation of either Smads (Smad2/3 and Smad 1/5/8) using small molecule inhibitors or by constitutively inducing phosphorylation of either Smads by transfection studies using adenoviral vectors (Ad-caALK1 and Ad-caALK5). RBPMS expression was found to be increased if phosphorylation of Smad2/3 was blocked or when Smad1/5/8 signaling was activated. This indicates that either reduced levels of pSmad2/3 or increased levels of pSmad1/5/8 regulate expression of RBPMS. Although we only showed indirect evidence for the role of these Smads in RBPMS regulation, we propose a counter-regulating mechanism of these Smads on RBPMS expression as depicted in Figure 7 .
Figure 7.
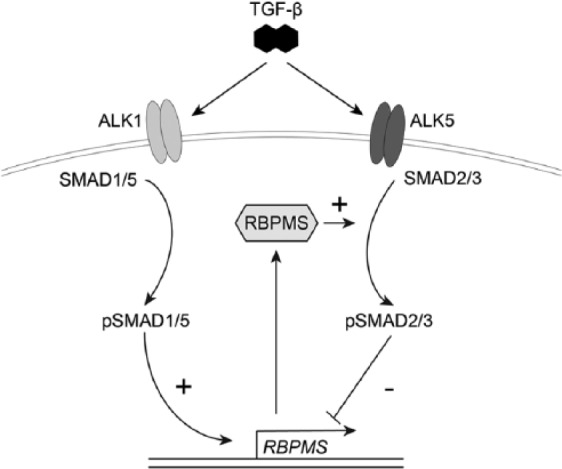
Counter-regulating mechanism on the effects of RBPMS (RNA-binding protein with multiple splicing) on Smad2/3 signaling. The scheme shows that RBPMS is regulated by TGF-β (transforming growth factor-β) signaling by pSmads, that is, negatively regulated by phosphorylated Smad2/3 and positively regulated by phosphorylated Smad1/5/8 and it also shows that RBPMS may be regulating TGF-β signaling by acting as linker molecule that might enhance phosphorylation of Smad2 and Smad3 by forming a complex with TGFBR (TGF-β receptor) and unphosphorylated Smad2/3.
Besides, IL-1β, a major catabolic activator in chondrocytes increased the expression of RBPMS mRNA transcripts. Also, this indicates a possible counter-regulation mechanism; in which Smad2/3 signaling is ultimately enhanced by IL-1β via upregulation of RBPMS expression. The underlying mechanism through which IL-1β regulates RBPMS expression is not known. However, it could be presumed to be mediated by suppression of phosphorylation of Smad2/3 through activation of TGF-β activated kinase 1 (TAK1) by IL-1β.31
Although the etiology of OA is not known clearly, OA is strongly correlated with increase in age.26 Previously, our group has also shown a reduction in Smad2/3 phosphorylation with age, which has been found to be associated with reduction in the expression of type I TGF-β receptors in cartilage of old mice, compromising TGF-β signaling capacity in chondrocytes of old mice.25 Therefore, we studied the expression of RBPMS in relation with ageing and OA using mouse models. Our data revealed a significant reduction in the expression of RBPMS with aging and OA, which might contribute to the observed reduced Smad2/3 signaling in old animals.25
Conversely, repeated injections of TGF-β into the murine knee joint have also been found to induce OA-like changes, suggesting a pathogenic role for excessive local TGF-β production in osteoarthritis.32 Our observation on reduced expression of the TGF-β signaling regulator RBPMS during aging and OA indicates that imbalance or functional loss of factors regulating TGF-β responsiveness and TGF-β-mediated signaling in cartilage might contribute to development of OA during ageing, emphasizing a possible role for RBPMS in the development of OA. Furthermore, the negative correlation found between cartilage damage and the percentage of positive RBPMS cells indicates that cartilage damage itself might affect RBPMS expression, and by doing so negatively affect pSmad2/3 phosphorylation. Our present results along with the finding of RBPMS associating with unphosphorylated rather than phosphorylated states of Smad2 and Smad3 reported in the previous study14 suggest that RBPMS might have a potential role in the maintenance of normal articular cartilage.
However, whether RBPMS is a significant functional factor that is involved in the regulation of TGF-β responsiveness and TGF-β mediated signaling in cartilage requires further studies by overexpressing or blocking RBPMS in chondrocytes and in OA mice models.
Footnotes
Acknowledgments and Funding: This study was supported in part through the Cutting-edge Research Enhancement and Scientific Training (DBT-CREST) award from the Department of Biotechnology, Ministry of Science and Technology, Government of India (Grant No. F.No.BT/HRD/03/01/2002-Vol.III).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: Ethical approval for this study was obtained from the Institutional Review Board (Animal Experiment Committee, Radboud University Nijmegen).
Animal Welfare: The present study followed international, national, and/or institutional guidelines for humane animal treatment and complied with relevant legislation.
References
- 1. Poole AR, Kojima T, Yasuda T, Mwale F, Kobayashi M, Laverty S. Composition and structure of articular cartilage: a template for tissue repair. Clin Orthop Relat Res. 2001;(391 Suppl):S26-33. [DOI] [PubMed] [Google Scholar]
- 2. Aigner T, McKenna L. Molecular pathology and pathobiology of osteoarthritic cartilage. Cell Mol Life Sci. 2002;59(1):5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang X, Chen L, Xu X, Li C, Huang C, Deng CX. TGF-β/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol. 2001;153(1):35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takahashi N1, Rieneck K, van der Kraan PM, van Beuningen HM, Vitters EL, Bendtzen K, et al. Elucidation of IL-1/TGF-β interactions in mouse chondrocyte cell line by genome-wide gene expression. Osteoarthritis Cartilage. 2005;13(5):426-38. [DOI] [PubMed] [Google Scholar]
- 5. Goumans MJ, Valdimarsdottir G, Itoh S, Lebrin F, Larsson J, Mummery C, et al. Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGFβ/ALK5 signaling. Mol Cell. 2003;12(4):817-28. [DOI] [PubMed] [Google Scholar]
- 6. van der Kraan PM, Blaney Davidson EN, Blom A, van den Berg WB. TGF-β signaling in chondrocyte terminal differentiation and osteoarthritis: modulation and integration of signaling pathways through receptor-Smads. Osteoarthritis Cartilage. 2009;17(12):1539-45. [DOI] [PubMed] [Google Scholar]
- 7. Finnson KW, Parker WL, ten Dijke P, Thorikay M, Philip A. ALK1 opposes ALK5/Smad3 signaling and expression of extracellular matrix components in human chondrocytes. J Bone Miner Res. 2008;23(6):896-906. [DOI] [PubMed] [Google Scholar]
- 8. Blaney Davidson EN, Remst DF, Vitters EL, van Beuningen HM, Blom AB, Goumans MJ, et al. Increase in ALK1/ALK5 ratio as a cause for elevated MMP-13 expression in osteoarthritis in humans and mice. J Immunol. 2009;182(12):7937-45. [DOI] [PubMed] [Google Scholar]
- 9. Yoon BS, Lyons KM. Multiple functions of BMPs in chondrogenesis. J Cell Biochem. 2004;93(1):93-103. [DOI] [PubMed] [Google Scholar]
- 10. Bonor J, Adams EL, Bragdon B, Moseychuk O, Czymmek KJ, Nohe A. Initiation of BMP2 signaling in domains on the plasma membrane. J Cell Physiol. 2012;227(7):2880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feng XH, Zhang Y, Wu RY, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-β-induced transcriptional activation. Genes Dev. 1998;12(14):2153-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luo K, Stroschein SL, Wang W, Chen D, Martens E, Zhou S, et al. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 1999;13(17):2196-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng XH, Lin X, Derynck R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-β. EMBO J. 2000;19(19):5178-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun Y, Ding L, Zhang H, Han J, Yang X, Yan J, et al. Potentiation of Smad-mediated transcriptional activation by the RNA-binding protein RBPMS. Nucleic Acids Res. 2006;34(21):6314-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Beuningen HM, Stoop R, Buma P, Takahashi N, van der Kraan PM, van den Berg WB. Phenotypic differences in murine chondrocyte cell lines derived from mature articular cartilage. Osteoarthritis Cartilage. 2002;10(12):977-86. [DOI] [PubMed] [Google Scholar]
- 16. Shimamoto A, Kitao S, Ichikawa K, Suzuki N, Yamabe Y, Imamura O, et al. A unique human gene that spans over 230 kb in the human chromosome 8p11-12 and codes multiple family proteins sharing RNA-binding motifs. Proc Natl Acad Sci U S A. 1996;93(20):10913-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerber WV, Yatskievych TA, Antin PB, Correia KM, Conlon RA, Krieg PA. The RNA-binding protein gene, hermes, is expressed at high levels in the developing heart. Mech Dev. 1999;80(1):77-86. [DOI] [PubMed] [Google Scholar]
- 18. Wilmore HP, McClive PJ, Smith CA, Sinclair AH. Expression profile of the RNA-binding protein gene hermes during chicken embryonic development. Dev Dyn. 2005;233(3):1045-51. [DOI] [PubMed] [Google Scholar]
- 19. Gerber WV, Vokes SA, Zearfoss NR, Krieg PA. A role for the RNA-binding protein, hermes, in the regulation of heart development. Dev Biol. 2002;247(1):116-26. [DOI] [PubMed] [Google Scholar]
- 20. van de Loo FA, Veenbergen S, van den Brand B, Bennink MB, Blaney-Davidson E, Arntz OJ, et al. Enhanced suppressor of cytokine signaling 3 in arthritic cartilage dysregulates human chondrocyte function. Arthritis Rheum. 2012;64(10):3313-23. [DOI] [PubMed] [Google Scholar]
- 21. Glansbeek HL, van der Kraan PM, Vitters EL, van den Berg WB. Correlation of the size of type II transforming growth factor β (TGF-β) receptor with TGF-β responses of isolated bovine articular chondrocytes. Ann Rheum Dis. 1993;52(11):812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15(9):1061-9. [DOI] [PubMed] [Google Scholar]
- 23. Pritzker KP, Gay S, Jimenez SA, Ostergaard K, Pelletier JP, Revell PA, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage. 2006;14(1):13-29. [DOI] [PubMed] [Google Scholar]
- 24. Blaney Davidson EN, Vitters EL, van der Kraan PM, van den Berg WB. Expression of transforming growth factor-β (TGFβ) and the TGFβ signalling molecule SMAD-2P in spontaneous and instability-induced osteoarthritis: role in cartilage degradation, chondrogenesis and osteophyte formation. Ann Rheum Dis. 2006;65(11):1414-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blaney Davidson EN, Scharstuhl A, Vitters EL, van der Kraan PM, van den Berg WB. Reduced transforming growth factor-beta signaling in cartilage of old mice: role in impaired repair capacity. Arthritis Res Ther. 2005;7(6):R1338-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van der Kraan PM, Blaney Davidson EN, van den Berg WB. A role for age-related changes in TGFβ signaling in aberrant chondrocyte differentiation and osteoarthritis. Arthritis Res Ther. 2010;12(1):201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-β signaling by the SnoN oncoprotein. Science. 1999;286(5440):771-4. [DOI] [PubMed] [Google Scholar]
- 28. Kim RH, Wang D, Tsang M, Martin J, Huff C, de Caestecker MP, et al. A novel smad nuclear interacting protein, SNIP1, suppresses p300-dependent TGF-beta signal transduction. Genes Dev. 2000;14(13):1605-16. [PMC free article] [PubMed] [Google Scholar]
- 29. van Beuningen HM, van der Kraan PM, Arntz OJ, van den Berg WB. In vivo protection against interleukin-1-induced articular cartilage damage by transforming growth factor-β 1: age-related differences. Ann Rheum Dis. 1994;53(9):593-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Remst DF, Blaney Davidson EN, Vitters EL, Bank RA, van den Berg WB, van der Kraan PM. TGF-ss induces Lysyl hydroxylase 2b in human synovial osteoarthritic fibroblasts through ALK5 signaling. Cell Tissue Res. 2014;355(1):163-71. [DOI] [PubMed] [Google Scholar]
- 31. Benus GF, Wierenga AT, de Gorter DJ, Schuringa JJ, van Bennekum AM, Drenth-Diephuis L, et al. Inhibition of the transforming growth factor β (TGFβ) pathway by interleukin-1β is mediated through TGFβ-activated kinase 1 phosphorylation of SMAD3. Mol Biol Cell. 2005;16(8):3501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Beuningen HM, Glansbeek HL, van der Kraan PM, van den Berg WB. Osteoarthritis-like changes in the murine knee joint resulting from intra-articular transforming growth factor-beta injections. Osteoarthritis Cartilage. 2000;8(1):25-33. [DOI] [PubMed] [Google Scholar]