Abstract
Purpose
Affinity proteomic approaches by antibody bead arrays enable multiplexed analysis of proteins in body fluids. In the presented study, we investigated blood plasma within osteoporosis to discovery differential protein profiles and to propose novel biomarkers candidates for subsequent studies.
Experimental design
Starting with 4608 antibodies and plasma samples from 22 women for an untargeted screening, a set of 72 proteins were suggested for further analysis. Complementing these with targets from literature and other studies, a targeted bead array of 180 antibodies was built to profile for 92 proteins in plasma samples of 180 women from two independent population‐based studies.
Results
Differential profiles between osteoporosis patients and matched controls were discovered for 12 proteins in at least one of the two study sets. Among these targets, the levels of autocrine motility factor receptor (AMFR) were concordantly lower in plasma of female osteoporosis patients. Subsequently, verification of anti‐AMFR antibody selectivity was conducted using high‐density peptide and protein arrays, and Western blotting.
Conclusions and clinical relevance
Further validation in additional study sets will be needed to determine the clinical value of the observed decrease in AMFR plasma levels in osteoporosis patients, but AMFR may aid our understanding of disease mechanisms and could support existing tools for diagnosis and monitoring of patient mobility within osteoporosis.
Keywords: Antibody bead arrays, Biomarker discovery, Osteoporosis, Plasma
Abbreviations
- ALPL
alkaline phosphatase liver/bone/kidney
- AMFR
autocrine motility factor receptor
- AU
arbitrary unit
- BGLAP
bone gamma‐carboxyglutamate protein
- HPA
human protein atlas
- MFI
median fluorescent intensity
- NOTO
homeobox protein notochord
- PAEP
progestagen‐associated endometrial protein
1. Introduction
Osteoporosis, a chronic noncommunicable disease characterized by reduced bone density and increased bone fragility leading to higher risk of fracture, represents a major public health problem that is set to increase in importance as the proportion of elderly women and men in the population is on the rise 1, 2. In the European Union alone, approximately 3.5 million new fragility fractures occur each year, but the great majority of individuals at risk are not identified or treated 3. Osteoporosis is estimated to affect over 200 million people worldwide 4 as primarily identified by bone mineral density scanning, so there is a need for a better understanding of the mechanisms behind osteoporosis in order to improve early diagnosis, patient mobility monitoring, and prevention of osteoporotic fractures 1.
Clinical Relevance
In populations with an increasing proportion of elderly people, there is a need to improve our understanding, assessment, and management of osteoporosis related bone fractures. In the presented study, we provide results from affinity proteomic analysis of blood plasma, where we screened patient samples to discover potential candidate proteins. Our findings could be of value for subsequent studies that aim to support existing tools for diagnosis and monitoring of osteoporosis patients.
The analysis of blood plasma offers a minimally invasive window to disease processes is a sample type well suited for affinity proteomic analysis 5. Approaches such as antibody bead arrays 6 allow profiling of a large number of proteins and samples in parallel even in an unbiased manner 7, thus offering a promising approach for the identification of biomarkers. With the increasing availability of validated affinity reagents toward human proteins 8, 9, new opportunities arise to conduct large‐scale proteomic investigations of many proteins in different sample types.
In this study, we profiled plasma samples for proteins of differential abundance using the suspension bead array approach to compare osteoporosis patients with matched healthy controls. The aim was to screen and discover protein targets and to illustrate the potential of such candidates to serve as potential biomarker candidates for future studies within skeletal diseases such as osteoporosis.
2. Materials and methods
2.1. Samples
Plasma samples were extracted from biobanks of two independent sample cohorts. Blood samples were collected and prepared as plasma heparin, within the Swedish mammography cohort—clinical (SMC‐C; Dnr 2006/1490 and 2002/472c) 1, 10. The second set was from the Prospective Investigation of the vasculature in Uppsala Seniors cohort (PIVUS; Dnr 00–419 and 02–551) collected during 2001–2004 11. A summary of the cases and controls is given in Table 1 and a detailed description is provided as Supporting Information.
Table 1.
Sample demographics
| Study set | Cohort | Groups | Samples (N) | Age |
|---|---|---|---|---|
| Discovery | SMC‐C | Cases | 16 | N/A |
| Controls | 6 | N/A | ||
| Set‐1 | SMC‐C | Cases | 46 | 65 (59–70) |
| Controls | 44 | 63 (59–69) | ||
| Set‐2 | PIVUS | Cases | 45 | 73 (71–74) |
| Controls | 45 | 72 (71–74) |
For this study, samples originated from the Swedish Mammography Cohort‐Clinical (SMC‐C), denoted as study set 1, and the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS), denoted study set‐2.
2.2. Antibody suspension bead array assays
The antibodies available at the Human Protein Atlas (HPA), two bone turnover markers bone gamma‐carboxyglutamate and alkaline phosphatase, rabbit IgG (Jackson ImmunoResearch Laboratories), rabbit anti‐human albumin (Dako) were coupled to magnetic color‐coded microspheres (MagPlex®, Luminex Corp.) as previously described 12. Multiplexed antibody suspension bead arrays also contained one bead population processed in protein‐free buffer. For sample analysis, plasma was labeled at a 1:10 dilution in PBS with activated biotin (Pierce), and processed in accordance to previous protocols 13, 14. Liquids were transferred between plates using a liquid handler (CyBi‐SELMA, CyBio), beads were washed using a plate washer (EL406, Biotek), and assay plates (Greiner) incubated at 23°C for 16 h on a plate rotator (Grant). Lastly, beads were measured using a dedicated flow cytometer (FlexMap 3D, Luminex Corp.).
2.3. Target verification via Western blot
Western blot analysis was conducted using IgG/HSA‐depleted human plasma, lysates from RT‐4 cells, U‐251 MG cells, liver, and tonsil tissue samples as described previously 15 and as shown for three anti‐AMFR (autocrine motility factor receptor) primary detection antibodies. A second Western blot protocol was performed with a plasma sample from a healthy donor pool depleted from human serum albumin and immunoglobulin G using Affibody molecules (Affibody AB) 16, lysate from AMFR overexpressing cells (LY420107, Origene) and provided negative control cells (lacking AMFR vector). Those samples were loaded onto an SDS‐PAGE gel (4–12% Bis Tris, Invitrogen) in the amounts 1000, 100, 10, and 1 ng protein (Supporting Information Fig. 1–4). Transfer was performed onto a PVDF membrane (Invitrolon PVDF, 0.45 μm, Invitrogen), followed by membrane blocking with 1× TBS buffer containing 5% milk powder and 0.5% Tween 20 at 23°C for 1 h and incubation with anti‐AMFR HPA029018 in blocking buffer overnight at 4°C. The membrane was washed using 1 × TBS with 0.05% Tween 20, followed by assay read‐out with swine anti‐rabbit IgG‐HRP (Dako) and a peroxidase substrate (Clarity™ Western ECL Substrate, Bio‐Rad).
2.4. Peptide epitope mapping
Custom high‐density peptide microarrays were obtained in collaboration with Roche‐Nimblegen, and applied for epitope mapping as described before 17. Peptides of 12 amino acid lengths with an 11‐residue overlap were designed to cover the antigen region of AMFR. Binding analysis of anti‐AMFR antibody HPA029018 on 175 000 peptides occurred via secondary DyLight649 conjugated anti‐rabbit IgG and slide scanning at 2 μm resolution (MS200, Roche NimbleGen Inc., Madison, WI). The median fluorescent intensity of each spot in relation to its’ local background was used for further data analysis.
2.5. Protein microarray analysis
Antibody selectivity was further investigated using an antigen microarray that contained 15 728 spots from 13 363 antigens representing 9882 unique Ensembl gene IDs. These antigens were protein fragments that were generated and verified within the Human Protein Atlas 9. The array was produced in‐house as previously described 18, 19 (Sjöberg et al., submitted for publication) and by printing antigens onto epoxy slides (CapitalBio) using a noncontact microarrayer (ArrayJet Marathon M025). The printed slide was dried, blocked, washed, and incubated with HPA029018 diluted to 0.2 μg/mL for 1 h RT. Secondary antibody anti‐rabbit IgG (Alexa Flour® 647) was applied to the array after washing and incubated for 1 h RT. The array was washed, spun dry, and scanned at 10 μm resolution using a microarray scanner (G2565BA, Agilent) and images were processed using software (GenePix 5.1, Molecular Devices) to collect the median intensities per spot at 633 nm.
2.6. Experimental study design and data analysis
The assays were designed by randomizing samples, replicates of samples as well as sample‐free controls across 96‐well plates. Study set 1 and 2 were hosted on separate plates for labeling and each plate included one sample aliquot in triplicate and three sample‐free wells. These assays were repeated on separate days, resulting in two experimental datasets for statistical analysis.
Data evaluation and statistical analysis was performed using R 20. Median fluorescent intensities (MFI) based on > 50 events per bead identity were collected, and after excluding data generated in sample‐free wells, outlying samples were identified via robust principal components analysis 21 and excluded from comparative analysis. Nonprocessed data were used for the calculation of CV per bead ID to estimate the technical variation across aliquot replicates within each assay. The MFI values for each of the 96‐well sample plates used for labeling were normalized using probabilistic quotient normalization 22 and secondly across labeling plates using multidimensional MA‐normalization (Hong, M‐G et al., submitted for publication). Aliquot replicates were finally excluded and the normalized and log transformed data (normalized MFI or norm. MFI) was used for statistical analysis. The association of age with protein profiles was tested with a generalized linear model and profiles with unadjusted p‐values < 0.05 in replicated experiments were considered to be associated. For targeted analysis, Student's t‐test was used to assess an association to disease status. Antibodies with concordant trends and significance (p‐values, p < 0.05) in replicated assays and both study sets together with assay correlation of Spearman's rho > 0.5 were highlighted. To identify multiprotein signatures, a logistic regression model with L1 penalization (Lasso, 23) was used for training. To predict the performance of such models, a logistic regression with its’ parameter estimates was applied to fit a test dataset. A receiver operating characteristic curve was then calculated to estimate the performance of a model in the test dataset. The study sets 1 and 2 served both as training and test datasets in replicated analysis, respectively.
3. Results
3.1. Initial discovery and study design
Here, we describe the results from protein profiling of plasma samples in the context of osteoporosis by using antibody suspension bead arrays. At first, an initial undirected discovery was performed with 4608 antibodies and 22 plasma samples from females to propose antibodies for further analysis. As shown in Fig. 1A, seven antibodies were found to detect differential intensity levels (p < 0.001). Selection criteria were extended to include protein profiles from antibodies that revealed less‐significant trends (dashed line Fig. 1A), and secondly by manual annotation of group trends and from Wilcoxon rank‐sum test analysis. This resulted in a discovery derived list of 142 antibodies targeting 72 unique proteins. This collection was further supplemented with 25 candidate proteins (34 antibodies) proposed by genome wide association study analysis 24, 25, and as shown in Fig. 1B, eight protein targets were found to overlap with the discovery derived list. Subsequently, 176 antibodies (and five controls) were available to be coupled to create one bead array (Supporting Information Table 1) to study plasma samples collected within two population‐based cohorts (Table 1). For both study sets 1 and 2, 46 and 45 female osteoporosis patients matched with 44 and 45 healthy controls, respectively, were analyzed for differential detection and target verification (Fig. 1C).
Figure 1.
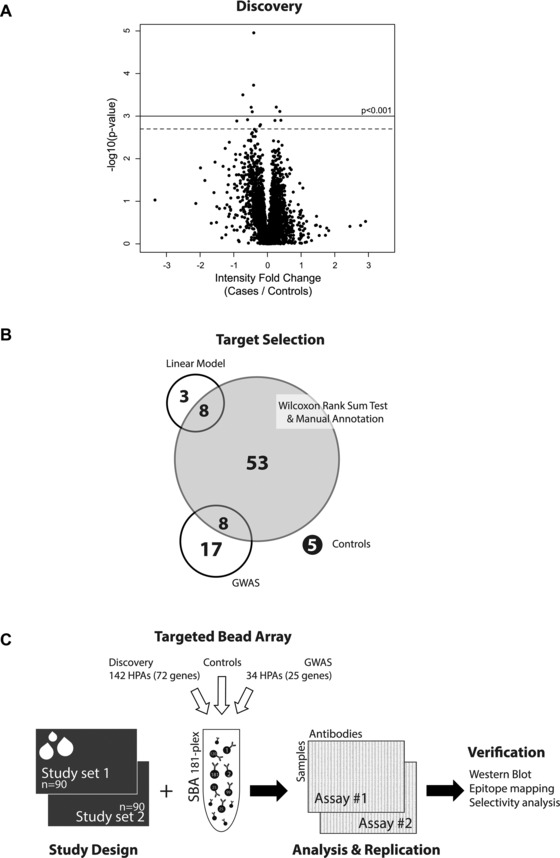
Study overview. (A) An initial discovery study on bead arrays was performed using 4608 antibodies on 16 cases and six controls. This provided seven antibodies with p < 0.001 using a linear model and 13 antibodies with extended selection (p < 0.002). Eleven HPA antibodies were available for generation of a new antibody suspension bead array. (B) To build an antibody array for subsequent targeted plasma analysis, statistical criteria (linear model and Wilcoxon rank sum test) as well as candidates from genome wide association studies were chosen. (C) Two study sets of 90 individuals were profiled with a bead array. Target verification was subsequently conducted for the most interesting finding using the listed methods. The numbers beside the selection criteria indicate unique antibodies (or genes).
3.2. Data quality assessment
The experimental study was designed to host the two study sets on two separate analysis plates and to repeat this analysis using the defined layout. The overall median technical CV based on intensities across aliquot replicates, ranged between 4–9%. For study set‐1 and its replicated assays, technical CVs across antibodies was 8% (0–21%) and 5% (0–23%), respectively. For study set‐2, technical CVs were 9% (0–67%) and 4% (0–16%). Intensity profiles between replicated assays were correlated with median Spearman's rho. Comparing sample profiles using all antibodies resulted in a median rho = 0.98 (0.94–0.99), and when correlating antibody profiles across samples; median of rho = 0.78 (–0.06–0.97). Out of 128 antibody pairs targeting the same protein, 2% correlated with a rho > 0.7 and 8% with rho > 0.5.
3.3. Comparative analysis in targeted discovery
The targeted plasma analysis was performed twice on both independent study sets. For 55 antibodies in study set‐1 and 75 antibodies in study set‐2, higher intensity levels were measured in cases compared to controls (Fig. 2A). There was an overlap of 20 out of 181 profiles (11%) that had the same trend in both study sets and assays. For another 16 antibodies, lower target levels were detected in osteoporosis patients. Further to this, profiles of 12 antibodies reached a statistically significant detection (p < 0.05) in one or both study sets and with a median assay correlation greater than rho = 0.5 (Fig. 2B, Supporting Information Table 2). Among these were antibodies for protein Autocrine Motility Factor Receptor (AMFR; HPA029018), progestagen‐associated endometrial protein (PAEP; HPA029473), and homeobox protein notochord (NOTO; HPA034660), as shown in Fig. 3A–C. For these binders, intensity levels between replicated assays were reproducible (median rho = 0.9) and in particularly AMFR reached statistical significance in all assays, as shown in Table 2. Noticeably, the detected levels of AMFR and PAEP were higher in controls as compared to the osteoporosis cases, while levels of NOTO were found to be higher in cases than in controls. In addition, the association of protein profiles with age was evaluated. A statistically significant association was found for the detected levels of KH domain containing, RNA binding, signal transduction associated 3 (HPA001369) and estrogen receptor 1, spectrin, beta, nonerythrocytic 1, speedy/RINGO cell‐cycle regulator family memberA, and AMFR (HPA002081, HPA013149, HPA035161, and HPA029019). Antibodies for bone turnover markers bone gamma‐carboxyglutamate protein (BGLAP) and alkaline phosphatase liver/bone/kidney (ALPL) did not reveal differential profiles when comparing osteoporosis cases and controls.
Figure 2.
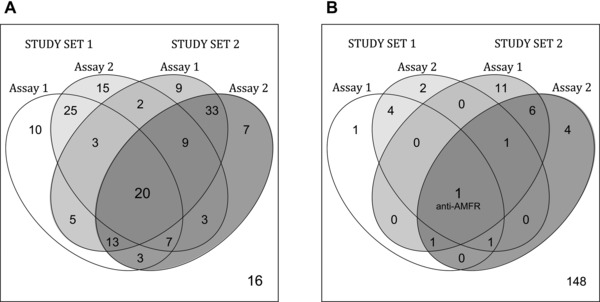
Summary of sample analysis in two study sets. The overlap within (A) trends and (B) statistical significance across both assays and study sets is shown. (A) As listed within the Venn diagram, 20 antibodies revealed a congruent higher detection level in osteoporosis cases than in controls, while 16 antibodies revealed congruent lower levels. (B) Protein targets of 11 antibodies were associated with osteoporosis status in both assays and one of the study sets, while only anti‐AMFR antibody HPA029018 was supportive in both study sets and assays.
Figure 3.
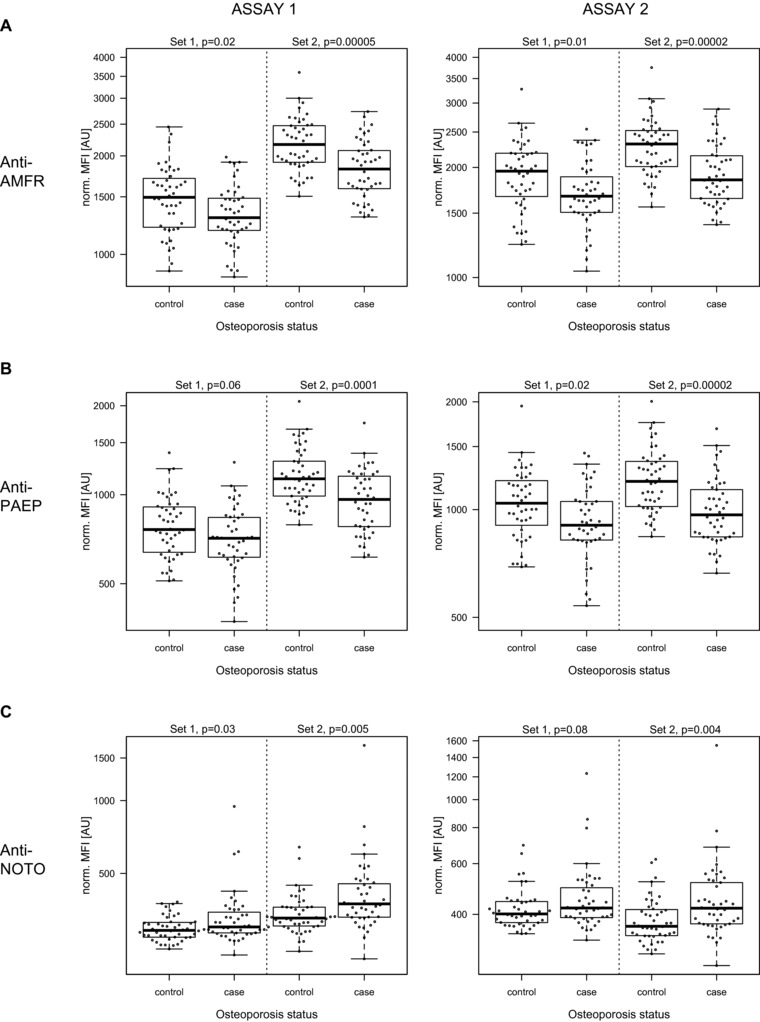
Differential profiles of candidate proteins. Boxplots from (A) anti‐AMFR HPA029018, (B) anti‐PAEP HPA029473, and (C) anti‐NOTO HPA034660 are shown as normalized data.
Table 2.
Performance of candidate proteins
| Antibody | Gene | Assay | Correlation (Rho) | p‐value | Technical CV | Trend | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Set‐1 | Set‐2 | Set‐1 | Set‐2 | Set‐1 | Set‐2 | Set‐1 | Set‐2 | |||
| HPA029018 | AMFR | 1 | 0.95 | 0.93 | 0.02 | 0.00005 | 5% | 2% | − | − |
| 2 | 0.01 | 0.00002 | 3% | 2% | − | − | ||||
| HPA029473 | PAEP | 1 | 0.94 | 0.95 | 0.06 | 0.0001 | 3% | 5% | − | − |
| 2 | 0.02 | 0.00002 | <1% | 5% | − | − | ||||
| HPA034660 | NOTO | 1 | 0.85 | 0.80 | 0.03 | 0.005 | 7% | 9% | + | + |
| 2 | 0.08 | 0.004 | 7% | 4% | + | + | ||||
Antibodies discriminating between cases and controls are summarized for repeated assays, including the trends of increased (+) or decreased (−) intensity levels in relation to the osteoporosis patient group.
Next, the performance of multivariate analyses was assessed. Thereto, one study set was applied to identify potential multiprotein signatures, which were then evaluated using the other set of samples. Models from the training set contained between two and six antibodies (Supporting Information Table 3), however when the performance of these models were estimated by calculating receiver operating characteristic curves in the test sets (Supporting Information Fig. 1) the areas under the curve remained low (< 0.63). These models did include anti‐PEAP and anti‐NOTO antibodies when training in study set‐2, while anti‐AMFR had not been selected by Lasso.
3.4. Target verification for AMFR using Western blot
Among the antibodies providing differential profiles, anti‐AMFR was found most supportive as concordant and significant trends were observed in all conducted assays for HPA029018. Thus, HPA029018 was chosen for further investigations with Western blot analysis, which is part of the routine antibody analysis within the Human Protein Atlas and uses lysates from cell lines and tissues as well as plasma 15. In Fig. 4A, which was taken from the Human Protein Atlas portal for HPA029018, the reactivity of the anti‐AMFR antibody resulted in two bands in the range of the Mr predicted for AMFR (73 kDa), predominantly in a lysate from tonsil. In plasma, one weak band was found in between the two mentioned bands for the tissues. Additionally, anti‐AMFR antibodies HPA028996 and HPA029019 detected a protein band of predicted Mr in whole tissue lysates from human liver and tonsil (Supporting Information Fig. 2). Of note, none of these additional antibodies provided supportive evidence in plasma analysis. Further evaluation of antibody HPA029018 with Western blot included a lysate from an AMFR‐overexpressed cell line. As seen in Fig. 4B, this revealed protein bands in a similar Mr range as the plasma pool.
Figure 4.
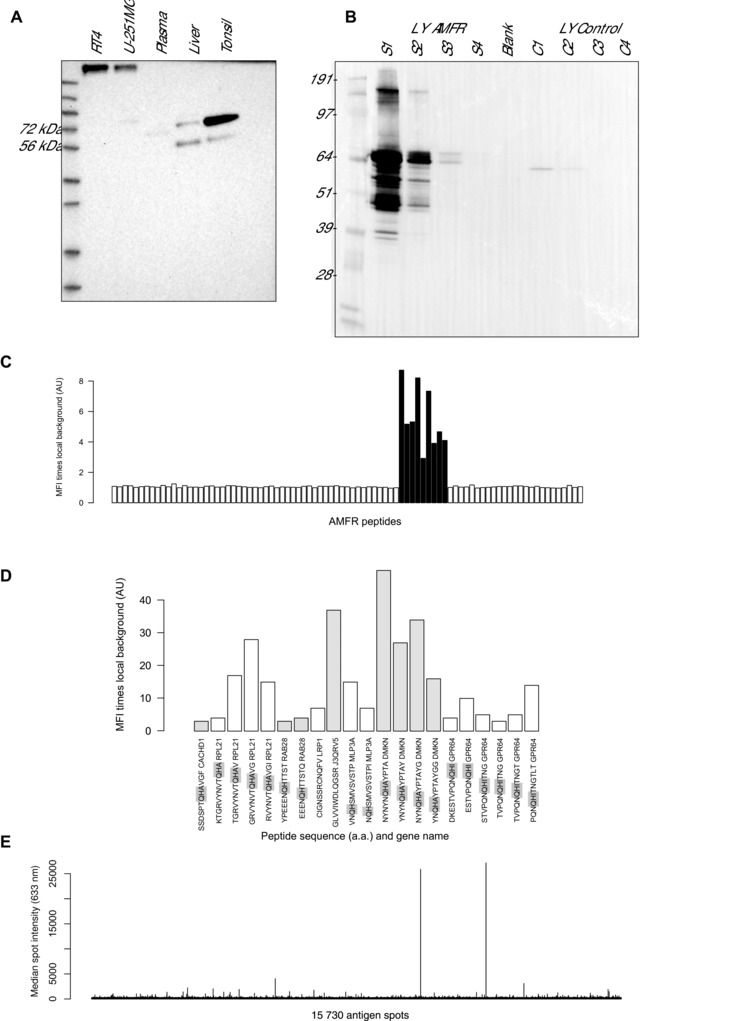
Verification of anti‐AMFR antibody. (A) Western blot analysis of tissue and cell lysates and plasma as taken from the Human Protein Atlas portal for HPA029018. (B) Western blot with different dilution of cell lysates (LY) overexpressing AMFR (Supporting Information Fig. 1–4) and nontransfected reference cells (C1–C4). (C) Epitope mapping of anti‐AMFR HPA029018 using a peptide array, and displaying reactive AMFR peptides with common motif (highlighted) and (D) identified peptides representing other proteins. (E) Interaction analysis of HPA029018 using antigen microarray built with 13 363 unique antigens.
3.5. Identification of anti‐AMFR peptide epitopes
Epitope mapping was performed using an ultra‐dense peptide array and revealed one, potentially two adjacent binding epitopes within the AMFR sequence in position 455–475 (Fig. 4C). A common motif consisting of AHQ was identified in the epitope region of anti‐AMFR (Supporting Information Table 4). The reverse sequence QHA, also discussed as retro‐peptide, was present in the sequences of nine out of the 21 peptides reactive with HPA029018 (Fig. 4D). Four out of the peptides contained QH and 6 peptides contained QHI, differing from epitope motif QHA with another hydrophobic residue. Only two of the 21 peptides detected above background levels among the 175 000 other peptides did not contain the identified full or similar version of the epitope motif.
3.6. Selectivity analysis of anti‐AMFR on antigen array of protein fragments
Further to using short peptides for selectivity confirmation of anti‐AMFR, we used a microarray that contained more than 13 000 unique antigens in forms of protein fragments, which represented almost 10 000 unique proteins. The analysis of anti‐AMFR antibody HPA029018 (Fig. 4E) resulted in a median intensity across all antigens of 238 AU (arbitrary unit), compared to the detection of the AMFR antigen at 26641 AU (average from five replicates; Supporting Information Fig. 3). Besides the protein fragment of AMFR, 15 out of 15 728 antigen spots (∼ 0.1%) were detected by anti‐AMFR but signal intensities did not exceed 15% of the AMFR protein fragment (Supporting Information Table 5). The peptide epitope motif QHA was present in three of these antigens, namely proteins encoded by the NUTM1, RFX6 and BRD4 genes. None of the other eight proteins identified during peptide‐based epitope mapping using the high‐density peptide array above, were among the protein fragments spotted on the antigen array.
4. Discussion
In the presented study, we used antibodies to profile proteins in plasma in relation to osteoporosis. After an initial discovery, a total of 180 plasma samples from females collected within two Swedish cohorts were analyzed using the suspension bead array assay, which identified an anti‐AMFR antibody revealing concordant and reproducible differences between osteoporosis cases and controls in both study sets. Further to this, we used Western blot, ultradense peptide array and antigen array analyses to validate the selectivity of the highlighted antibody.
We initiated this study by using an unbiased discovery approach, where we neither chose antibodies based on previously known relation to proteins important for the disease nor if the antibodies would be able to detect respective targets in the chosen assay system and sample type. In other words, the assay was not designed for known markers of disease and consequently such candidates might have been missed in our analysis. For this first screening phase, we used a small set of 22 samples (16 with osteoporosis diagnosis and six controls). At the time of the study, 4608 antibodies were available and included due to a concentration suitable for immobilization onto beads (> 0.05 mg/mL), as previously presented within multiple sclerosis 7. Further investigations in 180 plasma samples from females then revealed three candidates with discriminative power between osteoporosis and healthy controls, confirmed in the two study sets. These were proteins AMFR (autocrine motility factor receptor or E3 ubiquitin protein ligase), NOTO (notochord homeobox), and PAEP (progestagen‐associated endometrial protein). All these three antibodies had been included in the described protein profiling study because they were among the candidates found in the initial undirected, thus hypothesis generating, biomarker screening. The trend observed in the discovery set with anti‐PAEP was concordant with those for study sets 1 and 2. Besides AMFR, PAEP, and NOTO additional candidates such as cyclin D1 were detected at a differential level in osteoporosis cases and controls, but in only one of the study sets. Even though further investigation will be needed to reveal the potential of the candidates highlighted in our study, the presented findings illustrate the possibilities of using undirected and affinity‐based approaches for the discovery of novel and disease‐related candidate proteins within osteoporosis. A challenge of this strategy remains that the assay and context dependent antibody performance may hinder the confirmation of known markers of disease in parallel to the identified candidates. Antibodies for bone turnover markers BGLAP and ALPL illustrate this.
In the follow‐up and targeted analysis, the two study sets differed slightly in mean age (64 versus 72 years). Knowing that ageing may contribute to differences observed between the two study sets, as shown for AMFR, we chose to only focus on candidate proteins that revealed concordant results with samples from both collections. Our analysis found eight out of the 12 candidates in the older study set‐2, but further investigations would be needed to relate this trend to progression of osteoporosis or years post disease on‐set. Efforts to combine protein profiles in multivariate analysis did currently not yield a powerful prediction model for osteoporosis when being evaluated on these two independently collected study sets. As mentioned above, the nonoverlapping age ranges of patients connected to the two study sets could explain the weak performance of such multivariate models.
For the technical verification of antibody selectivity we conducted Western blot, epitope mapping, and cross‐reactivity analysis on microarray systems (Fig. 4). The anti‐AMFR antibody HPA029018 detected two protein bands in the range of the predicted molecular mass (73 kDa; Supporting Information Table 6) when using Western blot analysis with lysates from tissue and an AMFR‐overexpressing cell line, as well as a weak, single band in plasma. In particular the analysis using different amounts of lysate from cells overexpression AMFR show that a challenge with Western blot analysis is the local concentration (related to total protein load per lane). This calls for a careful interpretation of the data, as local enrichment of epitopes and prolonged image exposure may lead to the appearance of additional bands (off‐target). In addition, we conducted epitope mapping of the same antibody using an ultradense peptide array containing approximately 175 000 peptides, revealing one (or two adjacent) epitope(s) covering the sequence LNAMAHQIQEMFPQVPYHLV. Checking sequences beyond those representing AMFR peptides, additional peptides were recognized that contained the AHQ sequence motif found in the AMFR epitope region, while some of these sequences were related to an inverse order of residues (often called retro‐peptides). Cross‐reactivity was further investigated using an array containing more than 13 000 unique antigens to provide insight on possible interaction sites in targets of 50–150 residues length. Here, no other reactivity of the anti‐AMFR antibody was detected exceeding 15% of the intensity measured for the AMFR antigen, thus providing evidence of a selective AMFR binding.
In addition to affinity proteomics plasma data, investigations for transcript abundance were conducted within the Human Protein Atlas using RNA sequencing of 44 cell lines and 27 tissues 26. High transcript abundance was reported for AMFR and PAEP, while none was detected for NOTO. Expression of PAEP was group enriched with high abundance in the placenta and uterus, while AMFR was expressed in all tissues (Supporting Information Table 6). Transcripts of AMFR were found highly expressed in skeletal muscle and testis according to the GTEx portal (Genotype‐Tissue Expression project) 27. Again, no transcripts were found for NOTO but for PAEP, which were observed in the fallopian tubes, skin and cervix (and testis). In addition to this, immunohistochemistry had been conducted within the Human Protein Atlas on several tissues, where the anti‐PAEP antibody revealed high protein expression in the placenta and uterus, more specifically in decidual cells and a subset of the uterine glands, all in concordance with transcriptomic data. Immunohistochemistry analysis with anti‐AMFR revealed medium or high expression in a majority of the 82 analyzed normal tissues while anti‐NOTO revealed a mixed protein expression profile, with high expression in tissues of the reproductive and respiratory system.
To our knowledge, AMFR has not been associated with osteoporosis before. Links between AMFR and tumor progression as well as cell motility have been reported, with increased protein levels for many cancer types to suggest a role as a predictor of disease prognosis 28, 29. A decreased gene and protein expression of AMFR has further been observed in giant cell tumors of the bone, with association to a recurrent form of the tumor 30. The function of AMFR is related to polyubiquitination of proteins for e.g. proteasomal degradation. Decreased levels of AMFR found in plasma from osteoporosis patients may reflect a lower level of physical activity in osteoporotic patients, when considering that transcripts were abundant in skeletal muscle and mirroring a reduced turnaround in muscle proteins. In other recent studies on neuromuscular dystrophy 31 and childhood malaria 32, other muscle proteins have been detected in plasma as indicators of disease.
In conclusion, an affinity proteomics approach identified the protein AMFR through an unbiased screening as a potential marker in plasma to differentiate women diagnosed with osteoporosis compared to controls. Together with further validation, AMFR may aid future approaches and assist clinical tools for osteoporosis management and provide an additional route to increase our understanding of molecular mechanisms underlying osteoporosis.
The authors have declared no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Figure S1.
Figure S2.
Figure S3.
Table S1.
Table S2.
Table S3.
Table S4.
Table S5.
Table S6.
Acknowledgement
We thank the entire staff of the Human Protein Atlas and the whole Biobank Profiling group at SciLifeLab in Stockholm for their great efforts. We also thank the clinical teams and biobank staffs who helped with sample collection. The KTH Center for Applied Proteomics funded by the Erling‐Persson Family Foundation is acknowledged for financial support. This work was supported by grants for Science for Life Laboratory and the Knut and Alice Wallenberg Foundation.
5 References
- 1. Warensjo, E. , Byberg, L. , Melhus, H. , Gedeborg, R. et al., Dietary calcium intake and risk of fracture and osteoporosis: prospective longitudinal cohort study. BMJ 2011, 342, d1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhattacharyya, S. , Siegel, E. R. , Achenbach, S. J. , Khosla, S. , Suva, L. J. , Serum biomarker profile associated with high bone turnover and BMD in postmenopausal women. J. Bone Min. Res. 2008, 23, 1106–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hernlund, E. , Svedbom, A. , Ivergard, M. , Compston, J. et al., Osteoporosis in the European Union: medical management, epidemiology and economic burden. A report prepared in collaboration with the International Osteoporosis Foundation (IOF) and the European Federation of Pharmaceutical Industry Associations (EFPIA). Arch. Osteoporosis 2013, 8, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aaseth, J. , Boivin, G. , Andersen, O. , Osteoporosis and trace elements—an overview. J. Trace Elements Med. Biol. 2012, 26, 149–152. [DOI] [PubMed] [Google Scholar]
- 5. Landegren, U. , Vanelid, J. , Hammond, M. , Nong, R. Y. et al., Opportunities for sensitive plasma proteome analysis. Anal. Chem. 2012, 84, 1824–1830. [DOI] [PubMed] [Google Scholar]
- 6. Ayoglu, B. , Haggmark, A. , Neiman, M. , Igel, U. et al., Systematic antibody and antigen‐based proteomic profiling with microarrays. Exp. Rev. Mol. Diagn. 2011, 11, 219–234. [DOI] [PubMed] [Google Scholar]
- 7. Bystrom, S. , Ayoglu, B. , Haggmark, A. , Mitsios, N. et al., Affinity proteomic profiling of plasma, cerebrospinal fluid, and brain tissue within multiple sclerosis. J. Proteome Res. 2014, 13, 4607–4619. [DOI] [PubMed] [Google Scholar]
- 8. Stoevesandt, O. , Taussig, M. J. , Affinity proteomics: the role of specific binding reagents in human proteome analysis. Exp. Rev. Proteomics 2012, 9, 401–414. [DOI] [PubMed] [Google Scholar]
- 9. Uhlen, M. , Oksvold, P. , Fagerberg, L. , Lundberg, E. et al., Towards a knowledge‐based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [DOI] [PubMed] [Google Scholar]
- 10. Snellman, G. , Byberg, L. , Lemming, E. W. , Melhus, H. et al., Long‐term dietary vitamin D intake and risk of fracture and osteoporosis: a longitudinal cohort study of Swedish middle‐aged and elderly women. J. Clin. Endocrinol. Metab. 2014, 99, 781–790. [DOI] [PubMed] [Google Scholar]
- 11. Michaeelsson, K. , Lind, L. , Frystyk, J. , Flyvbjerg, A. et al., Serum adiponectin in elderly men does not correlate with fracture risk. J. Clin. Endocr. Metab. 2008, 93, 4041–4047. [DOI] [PubMed] [Google Scholar]
- 12. Haggmark, A. , Bystrom, S. , Ayoglu, B. , Qundos, U. et al., Antibody‐based profiling of cerebrospinal fluid within multiple sclerosis. Proteomics 2013, 13, 2256–2267. [DOI] [PubMed] [Google Scholar]
- 13. Schwenk, J. M. , Igel, U. , Neiman, M. , Langen, H. et al., Toward next generation plasma profiling via heat‐induced epitope retrieval and array‐based assays. Mol. Cell Proteomics 2010, 9, 2497–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Drobin, K. , Nilsson, P. , Schwenk, J. M. , Highly multiplexed antibody suspension bead arrays for plasma protein profiling. Methods Mol. Biol. 2013, 1023, 137–145. [DOI] [PubMed] [Google Scholar]
- 15. Algenas, C. , Agaton, C. , Fagerberg, L. , Asplund, A. et al., Antibody performance in western blot applications is context dependent. Biotechnol. J. 2014, 9, 435–445. [DOI] [PubMed] [Google Scholar]
- 16. Eriksson, C. , Schwenk, J. M. , Sjoberg, A. , Hober, S. , Affibody molecule‐mediated depletion of HSA and IgG using different buffer compositions: a 15 min protocol for parallel processing of 1–48 samples. Biotechnol. Appl. Bioc. 2010, 56, 49–57. [DOI] [PubMed] [Google Scholar]
- 17. Forsstrom, B. , Axnas, B. B. , Stengele, K. P. , Buhler, J. et al., Proteome‐wide epitope mapping of antibodies using ultra‐dense peptide arrays. Mol. Cell Proteomics 2014, 13, 1585–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ayoglu, B. , Haggmark, A. , Khademi, M. , Olsson, T. et al., Autoantibody profiling in multiple sclerosis using arrays of human protein fragments. Mol. Cell Proteomics 2013, 12, 2657–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sjoberg, R. , Sundberg, M. , Gundberg, A. , Sivertsson, A. et al., Validation of affinity reagents using antigen microarrays. N. Biotechnol. 2012, 29, 555–563. [DOI] [PubMed] [Google Scholar]
- 20. Ihaka, R. , Gentleman, R. , R: a language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar]
- 21. Hubert, M. , Rousseeuw, P. J. , Vanden Branden, K. , ROBPCA: a new approach to robust principal component analysis. Technometrics 2005, 47, 64–79. [Google Scholar]
- 22. Kato, B. S. , Nicholson, G. , Neiman, M. , Rantalainen, M. et al., Variance decomposition of protein profiles from antibody arrays using a longitudinal twin model. Proteome Sci. 2011, 9, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tibshirani, R. , Regression shrinkage and selection via the Lasso. J. Roy. Stat. Soc. B Met. 1996, 58, 267–288. [Google Scholar]
- 24. Estrada, K. , Styrkarsdottir, U. , Evangelou, E. , Hsu, Y. H. et al., Genome‐wide meta‐analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012, 44, 491‐+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rivadeneira, F. , Styrkarsdottir, U. , Estrada, K. , Halldorsson, B. V. et al., Twenty bone‐mineral‐density loci identified by large‐scale meta‐analysis of genome‐wide association studies. Nat. Genet. 2009, 41, 1199–U1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fagerberg, L. , Hallstrom, B. M. , Oksvold, P. , Kampf, C. et al., Analysis of the human tissue‐specific expression by genome‐wide integration of transcriptomics and antibody‐based proteomics. Mol. Cell Proteomics 2014, 13, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lonsdale, J. , Thomas, J. , Salvatore, M. , Phillips, R. et al., The genotype‐tissue expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nabi, I. R. , Watanabe, H. , Raz, A. , Identification of B16‐F1 melanoma autocrine motility‐like factor receptor. Cancer Res. 1990, 50, 409–414. [PubMed] [Google Scholar]
- 29. Chiu, C. G. , St‐Pierre, P. , Nabi, I. R. , Wiseman, S. M. , Autocrine motility factor receptor: a clinical review. Exp. Rev. Anticancer 2008, 8, 207–217. [DOI] [PubMed] [Google Scholar]
- 30. Guenther, R. , Krenn, V. , Morawietz, L. , Dankof, A. et al., Giant cell tumors of the bone: Molecular profiling and expression analysis of Ephrin A1 receptor, Claudin 7, CD52, FGFR3 and AMFR. Pathol. Res. Pract. 2005, 201, 649–663. [DOI] [PubMed] [Google Scholar]
- 31. Ayoglu, B. , Chaouch, A. , Lochmuller, H. , Politano, L. et al., Affinity proteomics within rare diseases: a BIO‐NMD study for blood biomarkers of muscular dystrophies. EMBO Mol. Med. 2014, 6, 918–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bachmann, J. , Burte, F. , Pramana, S. , Conte, I. et al., Affinity proteomics reveals elevated muscle proteins in plasma of children with cerebral malaria. PLoS Pathog. 2014, 10, e1004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Figure S1.
Figure S2.
Figure S3.
Table S1.
Table S2.
Table S3.
Table S4.
Table S5.
Table S6.


