Abstract
Background
EGFR, KRAS, and ALK alterations are major genetic changes found in non-small cell lung cancers (NSCLCs). Testing advanced lung adenocarcinoma tumors for these three genes is now standard care. The purpose of this study was to investigate the clinicopathologic expression pattern of these three genes in East Asian NSCLC patients.
Patients and methods
We conducted a retrospective study of all patients tested for mutations of these three genes at a single institute in Korea between 2006 and 2014. Study data were extracted from electronic medical records. Univariate and multivariate logistic regression analyses were used to measure associations between clinicopathologic features and alterations of EGFR, KRAS, and ALK.
Results
We detected 12 EGFR-mutated tumors with additional mutations in KRAS (N=6, 0.1%) or ALK (N=6, 0.1%). General clinicopathologic characteristics of tumors with EGFR, KRAS, or ALK mutations were similar to previous reports. Patients having EGFR L858R point mutations were older than patients having EGFR exon 19 deletions. EGFR G719X point mutations were more common in men and smokers than exon 19 deletions or L858R point mutations. Tumors having KRAS G12C mutations were less often of mucinous type than those with G12D or G12V, mutations.
Conclusions
This is the largest three gene molecular epidemiology study in East Asian NSCLC patients. Each genetic alteration was associated with distinct clinicopathologic characteristics. Furthermore, different age and sex are associated with different subtypes of EGFR and KRAS mutations.
Keywords: lung cancer, EGFR, KRAS, ALK, molecular epidemiology
INTRODUCTION
EGFR, KRAS, and ALK alterations are the major genetic changes in lung adenocarcinoma[1]. Drugs targeting EGFR and ALK have improved clinical outcomes in patients with mutations in those genes[2, 3]. Since targeted therapy was discovered, mutation testing has increased[4, 5]. Molecular testing of EGFR and ALK in lung adenocarcinoma is recommended by the guidelines from College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology[6].
EGFR mutation is associated with certain clinical and histologic factors, and is more prevalent in adenocarcinomas, women, Asians, and those who never smoked[7–9]. Despite differences between reports, histology is related to EGFR mutation status. Tumors with papillary, micropapillary, acinar, and lepidic (bronchioloalveolar) patterns more frequently have EGFR mutations than do tumors with a solid pattern[10–16]. EGFR mutation is rare in mucinous adenocarcinoma[17]. EGFR mutations tend to occur in older patients[15, 18–21]. Alternatively, KRAS mutation is associated with smokers, men, a solid pattern tumors, and mucinous adenocarcinoma[7, 15, 22–24]. ALK mutation is associated with non-smokers, younger patients, adenocarcinoma, a solid pattern tumors, and signet ring cell type tumors[25–34].
Genetic alterations of EGFR, KRAS, and ALK typically are mutually exclusive[35]. However, exceptional cases may have concurrent mutations of those genes[36–39]. Sometimes, mutations of other genes can occur after chemotherapy, which can cause resistance to targeted therapy[40–43].
In this study, we characterized the clinicopathologic features and genetic changes associated with EGFR, KRAS, and ALK in lung cancer.
RESULTS
EGFR tests
A total of 7,463 EGFR mutation tests were performed on samples from 6,878 patients. There were 55 failed tests due to insufficient biopsy materials. Test materials from 254 cases were not from lung cancer. Thus 7,154 tests and 6,583 patients remained (Figure S1). Of these, 545 patients were tested for EGFR mutation more than once. Among those patients, 11 had second primary tumors and 1 had a third primary tumor. Among the 6,595 tumors, 2,387 had EGFR mutations, and 60 had more than 2 EGFR mutations other than T790M.
EGFR tests were performed on 4,322 biopsy specimens, 2,548 resected specimens, and 115 cytology specimens. From 4,407 (62.8%) specimens obtained from lung, 4,344 tests were performed by PNA-clamping. Among these, 3,534 tests were confirmed by Sanger sequencing. Sanger sequencing alone was used to test 2,861 tumors. The tumor proportion ranged from 1 to 99% (Table S1). In univariate analysis, the EGFR mutation detection rate was low when the specimen was obtained by biopsy (OR[odds ratio]: 0.78, p<0.001), or from lymph node (OR: 0.56, P<0.001) or bronchus(OR: 0.67, P<0.001), when the tumor proportion was lower than 20% (OR: 0.71, P<0.001), or when the test was performed by Sanger sequencing only (OR: 0.81, P=0.003). However, in the multivariate analysis, there was no significant difference in mutation rates between biopsy and resection(OR: 1.17, P=0.020) or biopsy and cytology (OR: 1.08, P=0.874)(Table S2). There was a weak positive correlation between ΔCT-1 and tumor proportion (R2 = 0.0068). The ΔCT-1 of T790M was usually less than that of other EGFR mutations (Figure S2).
Association between EGFR mutation and clinicopathologic variables
All clinical and histopathologic variables are summarized in Tables S3 and S4. Adenocarcinoma accounted for a large proportion of cases (4,984 cases, 75.6%). The most frequent primary pattern observed was acinar pattern (65.5%). Of the adenocarcinomas, 2,295 (46%) tumors had EGFR mutations, 358 (9.2%) had KRAS mutations, and 270 (7.2%) had ALK rearrangements. 60 tumors (1.2%) had more than 2 EGFR mutations other than T790M.
In multivariate analysis, EGFR mutations were frequent in women (OR: 1.83, P<0.001), middle-aged patients (OR: 1.34, P<0.001), those who never smoked (OR: 2.04, P<0.001), adenocarcinomas (OR: 14.0, P<0.001), well (OR: 2.46, P<0.001) to moderately (OR: 2.73, P<0.001) differentiated tumors, small-sized tumors (OR for 1cm increase: 0.91, P=0.003), tumors of non-mucinous type (OR: 26.8, P<0.001), tumors without signet ring cells (OR: 17.2, P=0.007), and tumors with lepidic (OR: 2.18, P=0.003), acinar (OR: 3.38, P<0.001) and papillary (OR: 3.17, P<0.001) patterns (Table S5 and Figure S3). The relation between EGFR mutation and age was non-linear. In patients under 40, the EGFR mutation rate increased with increasing age, while in patients over 60, the EGFR mutation rate decreased with increasing age.
Differences between types of EGFR mutations
Deletions in exon 19 (N=1,262) and L858R point mutations (N=921) were the most common mutations. These two mutations accounted for approximately 90% of all EGFR mutations. Less common mutations included G719X point mutations (N=81), insertions in exon 20 (N=54), S768I point mutations (N=20), insertions in exon 19 (N=11), and L861Q point mutations (N=10) (Table S6).
Deletions in exon 19 frequently occurred in younger patients (OR for 1-year increase: 0.98, P<0.001). Conversely, L858R point mutations frequently occurred in older patients (OR for 1-year increase: 1.02, P<0.001). In multivariate analysis comparing EGFR mutation types, older patients were more likely to have L858R mutations than exon 19 deletions (OR for 1-year increase: 1.03, P<0.001) (Figure 1). Compared to exon 19 deletion, G719X mutation was more likely to occur in men(OR: 1.69, P=0.167) and smokers (OR: 2.04, P=0.058), but those factors were not independent in multivariate analysis (Table 1 and Figure 2).
Figure 1. Comparison between exon 19 deletion and L858R point mutation.
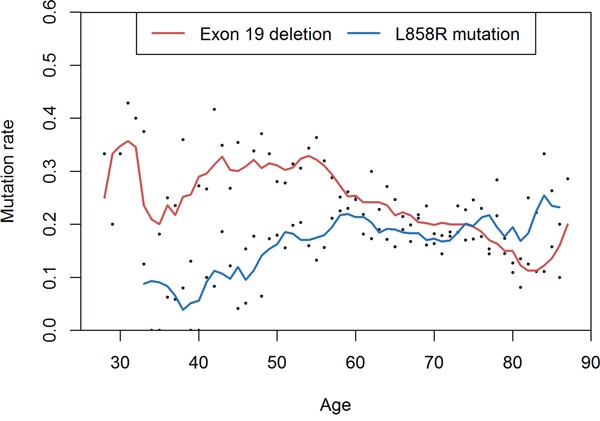
Deletions in exon 19 are frequent in younger patients and L858R mutations are frequent in older ages.
Table 1. Multivariate analysis of subtypes of EGFR mutation.
| vs. E19 | Age (per 1 year) | Sex (male vs. female) | Smoking (ever vs. never) | ||||
|---|---|---|---|---|---|---|---|
| OR | P-value | OR | P-value | OR | P-value | ||
| E19 vs. | L858R | 1.03 | <0.001 | 1.01 | 0.914 | 1.00 | 0.994 |
| L861Q | 1.07 | 0.050 | 0.38 | 0.389 | 1.68 | 0.645 | |
| G719X | 1.03 | 0.028 | 1.69 | 0.167a | 2.04 | 0.058a | |
| S768I | 0.97 | 0.418 | 1.19 | 0.889 | 4.59 | 0.227 | |
| E20 | 1.00 | 0.756 | 1.66 | 0.178 | 0.61 | 0.247 | |
p-value is less than 0.001 in univariate analysis
E19: exon 19 deletion, E20: exon 20 insertion
Figure 2. The proportion and subtypes of EGFR mutation.
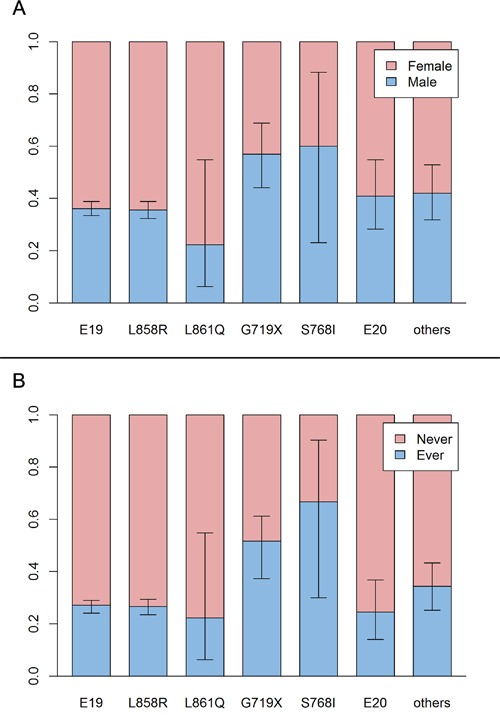
A. Sex proportion and subtypes of EGFR mutation. Deletion in exon 19 and L858R appears more often in women. However, G719X and S768I do not have this tendency. B. Proportion of smokers and subtypes of EGFR mutation. The trend is similar to the sex proportion. E19: deletion in exon 19, E20: insertion in exon 20.
Primary T790M mutation
There were 15 patients with a T790M EGFR mutation without history of previous targeted therapy (primary T790M mutation). One primary T790M mutation presented without other EGFR mutations. Eight of these patients were women and nine had never smoked. Their mean age was 65.3 years, and all patients had adenocarcinoma. The ΔCT-1 of secondary (patients who received targeted therapy) T790M was lower than the ΔCT-1 of coexisting EGFR mutations (average difference of ΔCT-1: 2.74). However, the ΔCT-1 of the primary T790M mutation was not very different from the ΔCT-1 of coexisting EGFR mutations (average difference of ΔCT-1: −0.20). Ten patients were treated with EGFR inhibitors. Tumor progressed in nine patients, while insufficient time has passed to assess the other patient (Table 2).
Table 2. Clinical Data of Patients Having Primary T790M mutation.
| Age | Sex | Smoking | Other EGFR mutation | ΔCT-1(other than T790M) | ΔCT-1 (T790M) | Targeted therapy | Response | |
|---|---|---|---|---|---|---|---|---|
| PT01 | 53 | M | Former | Positive | 3.9 | 4.59 | ||
| PT02 | 57 | F | Former | Positive | Gefitinib | PD | ||
| PT03 | 70 | F | Former | Positive | Gefitinib | PD | ||
| PT04 | 63 | F | Never | Positive | Gefitinib, Lapatinib | PD | ||
| PT05 | 83 | F | Never | Positive | ||||
| PT06 | 78 | M | Never | Negative | ||||
| PT07 | 57 | F | Never | Positive | Gefitinib | PD | ||
| PT08 | 65 | M | Former | Positive | Gefitinib, Afatinib | PD | ||
| PT09 | 53 | M | Never | Positive | 8.06 | 6.72 | ||
| PT10 | 41 | M | Former | Positive | 5.99 | 5.74 | Gefitinib | PD |
| PT11 | 78 | M | Former | Positive | 2.75 | 4.17 | Gefitinib | PD |
| PT12 | 77 | F | Never | Positive | 4.46 | 4.63 | Gefitinib | PD |
| PT13 | 69 | F | Never | Positive | 3.7 | 3.89 | ||
| PT14 | 75 | M | Never | Positive | 4.91 | 5.64 | Gefitinib | NA |
| PT15 | 61 | F | Never | Positive | 6.31 | 6.29 |
PD: progressed disease, NA: not accessible due to short follow up time
Association between KRAS mutation and clinicopathologic variables
In multivariate analysis, KRAS mutations were frequent in men (OR: 1.67, P=0.003), older patients (OR for 1-year increase: 1.03, P<0.001), smokers (OR: 1.78, P<0.001), adenocarcinomas (OR: 7.28, P<0.001), large-sized tumors (OR for 1cm increase: 1.17, P<0.001), poorly-differentiated tumors (vs. moderate differentiation, OR: 1.88, P=0.001), and mucinous type (OR: 9.09, P<0.001) and solid pattern (vs. acinar pattern, OR: 2.57, P<0.001) tumors (Table S7). Among those variables, mucinous type was the most distinguishing factor. There were three prevalent KRAS mutations: G12C (N=108, 27.2%), G12D (N=107, 27.0%), and G12V (N=89, 22.3%). G12C mutations were infrequent in mucinous type tumors compared to G12D (OR: 4.98, P=0.007) and G12V mutations (OR; 5.58, P=0.006) (Figure 3). In univariate analysis, G12C mutations were frequent in men and smokers compared to G12D and G12V mutations. However, those were not independent factors in multivariate analysis (Table 3 and Figure S4).
Figure 3. Comparison of proportion of mucinous type between subtypes of KRAS mutation.
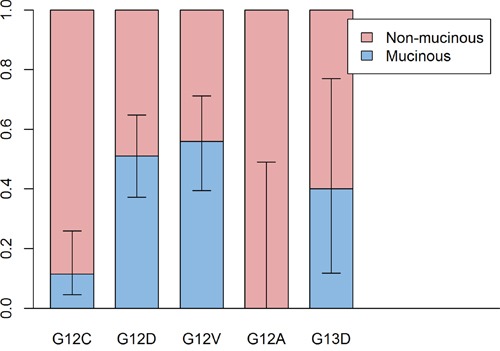
Proportion of mucinous type is higher in G12D and G12V subtypes than G12C subtype.
Table 3. Multivariate analysis of subtypes of KRAS mutations.
| Non-mucinous vs. Mucinous | Sex (Male vs. Female) | Smoking (Never vs. Ever) | |||||
|---|---|---|---|---|---|---|---|
| OR | P-value | OR | P-value | OR | P-value | ||
| vs. G12C | G12D | 4.98 | 0.007 | 0.96 | 0.968a | 0.18 | 0.044a |
| G12V | 5.58 | 0.006 | 1.02 | 0.988a | 0.18 | 0.060a | |
| G12A | 0.73 | 0.841 | 0.43 | 0.788 | 0.38 | 0.755 | |
| G13D | 7.22 | 0.082 | 1.04 | 0.983 | 0.85 | 0.945 | |
p-value is less than 0.01 in univariate analysis
Association between ALK rearrangement and clinicopathologic variables
In multivariate analysis, ALK rearrangements were frequent in younger patients (OR for 1 year increase: 0.95, P<0.001), those who never smoked (OR: 1.73, P=0.005), adenocarcinomas (OR: 6.99, P<0.001), poorly differentiated tumors (vs. moderate differentiation, OR: 2.54, P<0.001), signet ring cell types (OR: 20.3, P<0.001), cribriform (vs. acinar pattern, OR: 22.9, p<0.001) or solid patterns (vs. acinar pattern, OR: 2.96, P=0.002), tumors with lymph node metastasis (N2 vs N0, OR: 3.95, P<0.001), and tumors invading blood vessels (OR: 3.85, P<0.001), lymphatic vessels (OR: 2.13, P=0.004), or nerves (OR: 2.96, P=0.019) (Table S8 and Figure S5). Signet ring cell type and cribriform pattern were highly associated with ALK rearrangements.
Double mutations
Among tumors with EGFR mutations, 12 had additional mutations in KRAS (N= 6) or ALK (N=6) (Figure S6). Three of these EGFR mutations were detected only by PNA clamping and not by Sanger sequencing. Nine of these EGFR mutations were confirmed by Sanger sequencing, seven of which were major subtypes of EGFR mutation (deletion in exon 19 and L858R point mutation). Four were G719X point mutations, comprising 33% of the double mutants, which is a higher proportion than that observed in tumors having only EGFR mutations. The other EGFR mutation was an R803W point mutation, a very rare subtype. In five tumors, KRAS mutations presented at codon 12 or 13, and one tumor had two KRAS mutations, at codons 21 and 34. Among the six ALK alterations tested by immunohistochemistry, two were confirmed by FISH. The mean age of patients having both EGFR and ALK mutations was higher than that of patients having ALK rearrangements (P=0.012). Except for one tumor, tumors having both EGFR and KRAS mutations were moderately differentiated. Tumors having both EGFR and ALK mutation tended to be poorly differentiated. Nine of 15 patients had stages higher than III. One patient had a history of EGFR targeted therapy and ALK targeted therapy prior to mutation testing. The remaining patients had no history of targeted therapy prior to mutation tests. Six patients were treated with EGFR tyrosine kinase inhibitors and three were treated with ALK inhibitors. The follow-up period was insufficient to measure response (Table 4).
Table 4. Patients having EGFR mutation plus KRAS or ALK mutations.
| Age | Sex | Smoking | Pack-year | Stage | EGFR mutation | KRAS mutation | Differentiation | Targeted therapy | |
|---|---|---|---|---|---|---|---|---|---|
| DM01 | 77 | M | Current | 57 | IIA | L858R | I21S, P34S | Moderate | |
| DM02 | 79 | F | Never | IV | G719X | G12D | Moderate | ||
| DM03 | 51 | F | Never | IIIA | Exon 19 deletion | G12V | Moderate | erlotinib | |
| DM04 | 64 | F | Never | IV | L858R | G12D | Unknown | gefitinib | |
| DM05 | 64 | F | Never | IA | L858R | G13A | Moderate | ||
| DM06 | 58 | M | Former | 20 | IIIA | exon 19 deletion | G13C | Poor | gefitinib |
| Age | Sex | Smoking | Pack-year | Stage | EGFR mutation | ALK methods | Differentiation | Targeted therapy | |
| DM07 | 63 | F | Never | IIB | L858R | IHC | Poor | gefitinib | |
| DM08 | 67 | F | Never | IV | R803W | IHC | Poor | erlotinib | |
| DM09 | 69 | F | Never | IIIA | G719X | IHC | Moderate | ||
| DM10 | 57 | M | Former | 15 | IV | G719X | IHC | Poor | crizotinib |
| DM11 | 59 | F | Never | IV | Exon 19 deletion | IHC & FISH | Unknown | crizotinib | |
| DM12 | 63 | F | Never | IV | G719X | IHC & FISH | Moderate | gefitinib |
M: male, F: female, IHC: immunohistochemistry, FISH: fluorescence in situ hybridization
Double primary tumors
Among the 12 identified second or third primary tumors, 10 had genetic profiles that differed from their previous tumors. The histologic type was different in one second primary tumor. Another second primary tumor was histologically similar to the previous tumor, and had no mutations in EGFR, KRAS, or ALK. All second primary tumors arose at different sites from the prior tumors (Table S9).
DISCUSSION
We analyzed data from a large number of lung cancer patients from a single institution, assessing genetic alterations of EGFR, KRAS, and ALK. Most results were consistent with previous reports[7, 10, 16, 48]. However, contrary to previous reports[18, 19], EGFR mutations were more frequent in tumors from patients between 40 and 64 years of age than from other age groups. The relationship between age and EGFR mutation frequency was different with different mutation type. Exon 19 deletions occurred frequently in patients under 65, while L858R point mutations occurred frequently in patients over 40. Summing these data, the EGFR mutation frequency was highest in middle-aged patients. One report describes similar comparison of age between EGFR mutation subtypes[7]. Although it did not reach statistical significance in multivariate analysis, the G719X point mutation was frequent in men and smokers than other mutation subtypes. Of the 81 patients with G719X mutations, 44 (54%) were men and 39 (48%) smoked. This finding is similar to a previous report[39].
The T790M mutation is the most common cause of EGFR-targeted therapy resistance[49]. This mutation typically is detected after targeted therapy and is present as a minor clone prior to treatment[50]. In the 15 cases with primary T790M mutations here, the average difference in ΔCT-1 between T790M and other coexisting EGFR mutations was −0.20, whereas the average difference between T790M and other coexisting EGFR mutations was 2.73 in secondary T790M mutations. The ΔCT-1 of primary T790M was not very different from the ΔCT-1 of other coexisting EGFR mutations, indicating that the T790M mutation was present as a major clone in these cases. The T790M mutation may play an important role in this situation other than just resistance to EGFR tyrosine kinase inhibitors. There was no clinicopathologic difference in our analysis between patients with primary T790M mutations and patients without primary T790M mutations. A recent study with more patients with primary T790M mutations showed that primary T790M mutation is associated with never smoking and development of brain metastasis[51].
KRAS mutations were frequent in men, older patients, smokers, adenocarcinomas, mucinous tumor types, large-sized tumors, poorly differentiated tumors, and tumors with a solid pattern, consistent with previous reports[23, 24]. ALK rearrangements were frequent in younger patients, those who never smoked, adenocarcinomas, poorly differentiated tumors, signet ring cell types, and tumors with cribriform or solid patterns, also consistent with previous reports[33, 47]. All KRAS mutations were point mutations. Like the L858R point mutation of EGFR, the KRAS mutation rate increased as patient age increased. All ALK mutations were chromosomal rearrangements. Like ALK rearrangements in other tumors[52, 53], ALK rearrangements in lung cancer frequently occur in younger patients. G13C mutations were infrequent in mucinous types compared with G12D and G12V point mutations. According to another report, G12C is associated with smokers and G12D is associated with never smoking[7]. However, in our data, smoking was not an independent factor in multivariate analysis.
Generally, EGFR, KRAS, and ALK mutations are mutually exclusive. There are few reports of lung cancer with concurrent mutations of these genes[36–39]. In many of these, the secondary mutation was not detected at diagnosis, but after targeted therapy. These secondary mutations in other genes can promote resistance to targeted therapy. We identified 12 tumors (0.2%) having an EGFR mutation and an additional KRAS or ALK mutation. Only one patient had received prior targeted therapy. Of the 12 EGFR mutations, 7 were of a common type (exon 19 deletion and L858R point mutation), 4 were G719X point mutations, and 1 was a R803W point mutation. The proportion of rare mutations like the G719X point mutation was high in these tumors. The rare S768I point mutation was identified frequently in another study[39]. Intratumoral heterogeneity has been reported in lung cancer having both EGFR and ALK alterations[54]. Here, 9 of 12 cases were higher than stage III. It is likely that a second mutation occurred during tumor progression.
Twelve second or third primary tumors were included in this study. Among them, 10 had distinct genetic changes from the prior tumors. A second or third primary tumor is not uncommon in lung cancer. Distinguishing a second primary tumor from recurrence by clinical features or histologic features can be difficult, though genetic profiling can be helpful. If the genetic alteration differs from the prior tumor, this identifies the second as another primary tumor[55].
EGFR test results are influenced by several factors. When tissue was obtained from lymph nodes or bronchus, the EGFR mutation rate was lower (odds ratio: 0.56 and 0.67 each). It can be concluded that EGFR tests done with lymph node or bronchus specimens have a one-third false negative rate. Since the lymph node and bronchus usually are biopsied by bronchoscopy, the tissue sample is small. Dense lymphocytes in lymph nodes also dilute tumor DNA. These facters make the tests less sensitive. The EGFR mutation rate did not differ between tissues obtained from bone or body fluid. Tumor proportion was also important. When tumor proportion was below 20%, the EGFR mutation rate decreased. When tumor proportion was below 5%, the EGFR mutation was detected less than half as often. To make accurate tests, tumor proportion must be above the analytical sensitivity of the testing method. When the tumor proportion is low, a more sensitive method should be used[56].
Since our data were extracted from past medical records, some data were missing, and the data may contain inaccuracies. The number of cases was large enough to measure detailed trends of association between clinicopathologic features and genetic alterations of EGFR, KRAS, and ALK. EGFR exon 19 deletions and L858R point mutations tend to occur at different ages. The EGFR G719X point mutation differs from other subtypes in that age and sex are equal, and G719X commonly coexists with another gene mutation. The KRAS G12C point mutation was less frequently associated with mucinous type. However, more cases are required to characterize other rare subtypes of EGFR and KRAS mutations.
In this study, we analyzed the clinicopathologic features associated with three major driving mutations of lung cancer. Each subtype of driving mutation will occur by different mechanisms of mutagenesis in a different environment which is related to age, sex, and smoking history. The driving mutation and related risk factors are associated with morphology and behavior of the tumor. These data are valuable in understanding the characteristics of lung cancer.
MATERIALS AND METHODS
Study design
We conducted a retrospective study of all patients whose tumors were tested for EGFR, KRAS, and ALK mutation at the Samsung Medical Center (Seoul, Korea) from 2006 to 2014. The study was approved by the Institutional Review Board of the Samsung Medical Center. The requirement for informed consent was waived, as the study was based on existing data.
Data collection
Study data were automatically or manually extracted from electronic medical records. Clinical data included sex, age when testing was performed, smoking history, origin of cancer, and EGFR, KRAS and ALK mutation status. Data regarding EGFR testing methods included biopsy methods, organs biopsied, tumor proportion of material sampled, test methods, report date, ΔCT-1(the difference in CT value between the negative control and test sample[44]) and test results including the type of EGFR mutation. Pathologic data included tumor type, histologic pattern, tumor size, pathologic stage, and the presence of lymphatic, vascular, or pleural invasion. All pathologic data except type of tumor refers only to resected tumors. Histologic pattern was assessed only for adenocarcinoma. When an EGFR mutation was identified together with a KRAS or ALK mutation, the tissue slide and chromatogram of Sanger sequencing were reviewed.
Detection of alterations of EGFR, KRAS and ALK
EGFR gene alteration was detected by either real-time PCR with PNA-clamping methods, direct sequencing, or both methods. The PNA-Clamp™EGFR Mutation Detection kit (PANAGENE, Inc., Daejeon, Korea) was used for real-time PCR, performed as described[45]. When detection was done only with direct sequencing, exon 18, 19, 20, and 21 were sequenced as previously described[44]. When both methods were used, exons containing mutations detected by real-time PCR were sequenced, and exon 19 was sequenced if no mutation was detected by real-time PCR.
KRAS gene alteration was also detected by either real-time PCR with PNA-clamping methods, direct sequencing, or both methods. The PNA-Clamp™KRAS Mutation Detection kit (PANAGENE, Inc., Daejeon, Korea) was used for real-time PCR, performed as described[46]. KRAS exon 2, which contains codons 12 and 13, was sequenced by direct sequencing as previously described[44].
ALK gene alteration was detected by immunohistochemistry or fluorescence in situ hybridization(FISH)[47].
Statistical analysis
We used means and standard deviations to summarize continuous variables and counts and numbers with percentages to summarize categorical variables. Age was categorized into three groups: group 1, younger than 40 years; group 2, between 40 and 64 years; group 3, older than 64 years. Univariate and multivariate logistic regression analyses were used to determine the association between each variable and EGFR, KRAS and ALK mutations. Differences between subtypes of EGFR and KRAS mutations were tested using multinomial logistic regression. P-values of less than 0.01 were considered statistically significant.
SUPPLEMENTARY FIGURES AND TABLES
Acknowledgments
This work was supported by the R&D Program of the Society of the National Research Foundation funded by the Ministry of Science, ICT & Future Planning (NFR-2013M3C8A1078501 and NRF-2013R1A2A2A01068922).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest with regard to the present paper.
Authors' contributions
BL worked on experimental work, data acquisition, data analysis, data interpretation, and drafting of the manuscript. TL participated in data acquisition and data analysis. YLC and JH worked on study design, data interpretation, and manuscript writing. All authors read and approved the final manuscript.
REFERENCES
- 1.Cancer Genome Atlas Research N Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Choi YL, Sun JM, Cho J, Rampal S, Han J, Parasuraman B, Guallar E, Lee G, Lee J, Shim YM. EGFR mutation testing in patients with advanced non-small cell lung cancer: a comprehensive evaluation of real-world practice in an East Asian tertiary hospital. PLoS One. 2013;8:e56011. doi: 10.1371/journal.pone.0056011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee DH, Srimuninnimit V, Cheng R, Wang X, Orlando M. Epidermal Growth Factor Receptor Mutation Status in the Treatment of Non-small Cell Lung Cancer: Lessons Learned. Cancer Res Treat. 2015;47:549–554. doi: 10.4143/crt.2014.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, Jenkins RB, Kwiatkowski DJ, Saldivar JS, Squire J, Thunnissen E, Ladanyi M. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol. 2013;8:823–859. doi: 10.1097/JTO.0b013e318290868f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dogan S, Shen R, Ang DC, Johnson ML, D'Angelo SP, Paik PK, Brzostowski EB, Riely GJ, Kris MG, Zakowski MF, Ladanyi M. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bae NC, Chae MH, Lee MH, Kim KM, Lee EB, Kim CH, Park TI, Han SB, Jheon S, Jung TH, Park JY. EGFR, ERBB2, and KRAS mutations in Korean non-small cell lung cancer patients. Cancer Genet Cytogenet. 2007;173:107–113. doi: 10.1016/j.cancergencyto.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, Heeroma K, Itoh Y, Cornelio G, Yang PC. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9:154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girard N, Sima CS, Jackman DM, Sequist LV, Chen H, Yang JC, Ji H, Waltman B, Rosell R, Taron M, Zakowski MF, Ladanyi M, Riely G, Pao W. Nomogram to predict the presence of EGFR activating mutation in lung adenocarcinoma. Eur Respir J. 2012;39:366–372. doi: 10.1183/09031936.00010111. [DOI] [PubMed] [Google Scholar]
- 11.Kim HJ, Choi EY, Jin HJ, Shin KC. Relationship between EGFR mutations and clinicopathological features of lung adenocarcinomas diagnosed via small biopsies. Anticancer Res. 2014;34:3189–3195. [PubMed] [Google Scholar]
- 12.Motoi N, Szoke J, Riely GJ, Seshan VE, Kris MG, Rusch VW, Gerald WL, Travis WD. Lung adenocarcinoma: Modification of the 2004 WHO mixed subtype to include the major histologic subtype suggests correlations between papillary and micropapillary adenocarcinoma subtypes, EGFR mutations and gene expression analysis. American Journal of Surgical Pathology. 2008;32A:810–827. doi: 10.1097/PAS.0b013e31815cb162. [DOI] [PubMed] [Google Scholar]
- 13.Ninomiya H, Hiramatsu M, Inamura K, Nomura K, Okui M, Miyoshi T, Okumura S, Satoh Y, Nakagawa K, Nishio M, Horai T, Miyata S, Tsuchiya E, Fukayama M, Ishikawa Y. Correlation between morphology and EGFR mutations in lung adenocarcinomas: Significance of the micropapillary pattern and the hobnail cell type. Lung Cancer. 2009;63:235–240. doi: 10.1016/j.lungcan.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 14.Sun PL, Seol H, Lee HJ, Yoo SB, Kim H, Xu X, Jheon S, Lee CT, Lee JS, Chung JH. High incidence of EGFR mutations in Korean men smokers with no intratumoral heterogeneity of lung adenocarcinomas: correlation with histologic subtypes, EGFR/TTF-1 expressions, and clinical features. J Thorac Oncol. 2012;7:323–330. doi: 10.1097/JTO.0b013e3182381515. [DOI] [PubMed] [Google Scholar]
- 15.Blons H, Cote JF, Le Corre D, Riquet M, Fabre-Guilevin E, Laurent-Puig P, Danel C. Epidermal growth factor receptor mutation in lung cancer are linked to bronchioloalveolar differentiation. Am J Surg Pathol. 2006;30:1309–1315. doi: 10.1097/01.pas.0000213285.65907.31. [DOI] [PubMed] [Google Scholar]
- 16.Ha SY, Choi SJ, Cho JH, Choi HJ, Lee J, Jung K, Irwin D, Liu X, Lira ME, Mao M, Kim HK, Choi YS, Shim YM, et al. Lung cancer in never-smoker Asian females is driven by oncogenic mutations, most often involving EGFR. Oncotarget. 2015;6:5465–5474. doi: 10.18632/oncotarget.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finberg KE, Sequist LV, Joshi VA, Muzikansky A, Miller JM, Han M, Beheshti J, Chirieac LR, Mark EJ, Iafrate AJ. Mucinous differentiation correlates with absence of EGFR mutation and presence of KRAS mutation in lung adenocarcinomas with bronchioloalveolar features. J Mol Diagn. 2007;9:320–326. doi: 10.2353/jmoldx.2007.060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi YH, Lee JK, Kang HJ, Lee TS, Kim HR, Kim CH, Koh JS, Baek HJ, Lee JC, Na II. Association between age at diagnosis and the presence of EGFR mutations in female patients with resected non-small cell lung cancer. J Thorac Oncol. 2010;5:1949–1952. doi: 10.1097/jto.0b013e3181f38816. [DOI] [PubMed] [Google Scholar]
- 19.Ueno T, Toyooka S, Suda K, Soh J, Yatabe Y, Miyoshi S, Matsuo K, Mitsudomi T. Impact of age on epidermal growth factor receptor mutation in lung cancer. Lung Cancer. 2012;78:207–211. doi: 10.1016/j.lungcan.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Sun Y, Pan Y, Li C, Shen L, Li Y, Luo X, Ye T, Wang R, Hu H, Li H, Wang L, Pao W, Chen H. Frequency of Driver Mutations in Lung Adenocarcinoma from Female Never-Smokers Varies with Histologic Subtypes and Age at Diagnosis. Clinical Cancer Research. 2012;18:1947–1953. doi: 10.1158/1078-0432.CCR-11-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Pan Y, Li Y, Li C, Wang R, Hu H, Zhang Y, Ye T, Wang L, Shen L, Sun Y, Chen H. Frequency of well-identified oncogenic driver mutations in lung adenocarcinoma of smokers varies with histological subtypes and graduated smoking dose. Lung Cancer. 2013;79:8–13. doi: 10.1016/j.lungcan.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Tam IY, Chung LP, Suen WS, Wang E, Wong MC, Ho KK, Lam WK, Chiu SW, Girard L, Minna JD, Gazdar AF, Wong MP. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 23.Dacic S, Shuai Y, Yousem S, Ohori P, Nikiforova M. Clinicopathological predictors of EGFR/KRAS mutational status in primary lung adenocarcinomas. Mod Pathol. 2009;23:159–168. doi: 10.1038/modpathol.2009.154. [DOI] [PubMed] [Google Scholar]
- 24.Rekhtman N, Ang DC, Riely GJ, Ladanyi M, Moreira AL. KRAS mutations are associated with solid growth pattern and tumor-infiltrating leukocytes in lung adenocarcinoma. Mod Pathol. 2013;26:1307–1319. doi: 10.1038/modpathol.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inamura K, Takeuchi K, Togashi Y, Hatano S, Ninomiya H, Motoi N, Mun M-y, Sakao Y, Okumura S, Nakagawa K, Soda M, Lim Choi Y, Mano H, Ishikawa Y. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol. 2009;22:508–515. doi: 10.1038/modpathol.2009.2. [DOI] [PubMed] [Google Scholar]
- 26.Kim HR, Shim HS, Chung J-H, Lee YJ, Hong YK, Rha SY, Kim SH, Ha S-J, Kim SK, Chung KY, Soo R, Kim JH, Cho BC. Distinct clinical features and outcomes in never-smokers with nonsmall cell lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement. Cancer. 2012;118:729–739. doi: 10.1002/cncr.26311. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Li Y, Yang T, Wei S, Wang J, Wang M, Wang Y, Zhou Q, Liu H, Chen J. Clinical significance of EML4-ALK fusion gene and association with EGFR and KRAS gene mutations in 208 Chinese patients with non-small cell lung cancer. PLoS One. 2013;8:e52093. doi: 10.1371/journal.pone.0052093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodig SJ, Mino-Kenudson M, Dacic S, Yeap BY, Shaw A, Barletta JA, Stubbs H, Law K, Lindeman N, Mark E, Janne PA, Lynch T, Johnson BE, Iafrate AJ, Chirieac LR. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15:5216–5223. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC, Ho KK, Au JS, Chung LP, Pik Wong M. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 30.Nishino M, Klepeis VE, Yeap BY, Bergethon K, Morales-Oyarvide V, Dias-Santagata D, Yagi Y, Mark EJ, Iafrate AJ, Mino-Kenudson M. Histologic and cytomorphologic features of ALK-rearranged lung adenocarcinomas. Mod Pathol. 2012;25:1462–1472. doi: 10.1038/modpathol.2012.109. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Pan Y, Wang R, Sun Y, Hu H, Shen X, Lu Y, Shen L, Zhu X, Chen H. ALK-rearranged lung cancer in Chinese: a comprehensive assessment of clinicopathology, IHC, FISH and RT-PCR. PLoS One. 2013;8:e69016. doi: 10.1371/journal.pone.0069016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi T, Sonobe M, Kobayashi M, Yoshizawa A, Menju T, Nakayama E, Mino N, Iwakiri S, Sato K, Miyahara R, Okubo K, Manabe T, Date H. Clinicopathologic Features of Non-Small-Cell Lung Cancer with EML4–ALK Fusion Gene. Annals of Surgical Oncology. 2010;17:889–897. doi: 10.1245/s10434-009-0808-7. [DOI] [PubMed] [Google Scholar]
- 33.Paik JH, Choi CM, Kim H, Jang SJ, Choe G, Kim DK, Kim HJ, Yoon H, Lee CT, Jheon S, Choe JY, Chung JH. Clinicopathologic implication of ALK rearrangement in surgically resected lung cancer: a proposal of diagnostic algorithm for ALK-rearranged adenocarcinoma. Lung Cancer. 2012;76:403–409. doi: 10.1016/j.lungcan.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 34.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, Solomon B, Stubbs H, Admane S, McDermott U, Settleman J, Kobayashi S, Mark EJ, et al. Clinical Features and Outcome of Patients With Non–Small-Cell Lung Cancer Who Harbor EML4-ALK. Journal of Clinical Oncology. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gainor JF, Varghese AM, Ou SH, Kabraji S, Awad MM, Katayama R, Pawlak A, Mino-Kenudson M, Yeap BY, Riely GJ, Iafrate AJ, Arcila ME, Ladanyi M, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res. 2013;19:4273–4281. doi: 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, Choi HG, Kim J, Chiang D, Thomas R, Lee J, Richards WG, Sugarbaker DJ, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo YW, Wu SG, Ho CC, Shih JY. Good response to gefitinib in lung adenocarcinoma harboring coexisting EML4-ALK fusion gene and EGFR mutation. J Thorac Oncol. 2010;5:2039–2040. doi: 10.1097/JTO.0b013e3181f43274. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Zhang S, Yang X, Yang J, Zhou Q, Yin L, An S, Lin J, Chen S, Xie Z, Zhu M, Wu YL. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer. 2010;9:188. doi: 10.1186/1476-4598-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Li L, Zhu Y, Huang C, Qin Y, Liu H, Ren-Heidenreich L, Shi B, Ren H, Chu X, Kang J, Wang W, Xu J, et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer. 2014;110:2812–2820. doi: 10.1038/bjc.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popat S, Vieira de Araújo A, Min T, Swansbury J, Dainton M, Wotherspoon A, Lim E, Nicholson AG, O'Brien MER. Lung Adenocarcinoma with Concurrent Exon 19 EGFR Mutation and ALK Rearrangement Responding to Erlotinib. Journal of Thoracic Oncology. 2011;6:1962–1963. doi: 10.1097/JTO.0b013e31822eec5e. [DOI] [PubMed] [Google Scholar]
- 41.Rossing HH, Grauslund M, Urbanska EM, Melchior LC, Rask CK, Costa JC, Skov BG, Sorensen JB, Santoni-Rugiu E. Concomitant occurrence of EGFR (epidermal growth factor receptor) and KRAS (V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog) mutations in an ALK (anaplastic lymphoma kinase)-positive lung adenocarcinoma patient with acquired resistance to crizotinib: a case report. BMC Res Notes. 2013;6:489. doi: 10.1186/1756-0500-6-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeda M, Okamoto I, Fujita Y, Arao T, Ito H, Fukuoka M, Nishio K, Nakagawa K. De novo resistance to epidermal growth factor receptor-tyrosine kinase inhibitors in EGFR mutation-positive patients with non-small cell lung cancer. J Thorac Oncol. 2010;5:399–400. doi: 10.1097/JTO.0b013e3181cee47e. [DOI] [PubMed] [Google Scholar]
- 43.Tiseo M, Gelsomino F, Boggiani D, Bortesi B, Bartolotti M, Bozzetti C, Sammarelli G, Thai E, Ardizzoni A. EGFR and EML4-ALK gene mutations in NSCLC: A case report of erlotinib-resistant patient with both concomitant mutations. Lung Cancer. 2011;71:241–243. doi: 10.1016/j.lungcan.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 44.Lee B, Lee B, Han G, Kwon MJ, Han J, Choi YL. KRAS Mutation Detection in Non-small Cell Lung Cancer Using a Peptide Nucleic Acid-Mediated Polymerase Chain Reaction Clamping Method and Comparative Validation with Next-Generation Sequencing. Korean J Pathol. 2014;48:100–107. doi: 10.4132/KoreanJPathol.2014.48.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HJ, Lee KY, Kim Y-C, Kim K-S, Lee SY, Jang TW, Lee MK, Shin K-C, Lee GH, Lee JC, Lee JE, Kim SY. Detection and comparison of peptide nucleic acid-mediated real-time polymerase chain reaction clamping and direct gene sequencing for epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Lung Cancer. 2012;75:321–325. doi: 10.1016/j.lungcan.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 46.Ha SY, Han J, Lee JJ, Kim YE, Choi Y-L, Kim HK. Mucoepidermoid Carcinoma of Tracheobronchial Tree: Clinicopathological Study of 31 Cases. The Korean Journal of Pathology. 2011;45:175. [Google Scholar]
- 47.Sun JM, Lira M, Pandya K, Choi YL, Ahn JS, Mao M, Han J, Park K, Ahn MJ, Kim J. Clinical characteristics associated with ALK rearrangements in never-smokers with pulmonary adenocarcinoma. Lung Cancer. 2014;83:259–264. doi: 10.1016/j.lungcan.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Lee SH, Kim WS, Choi YD, Seo JW, Han JH, Kim MJ, Kim L, Lee GK, Lee CH, Oh MH, Kim GY, Sung SH, Lee KY, et al. Analysis of Mutations in Epidermal Growth Factor Receptor Gene in Korean Patients with Non-small Cell Lung Cancer: Summary of a Nationwide Survey. J Pathol Transl Med. 2015 doi: 10.4132/jptm.2015.09.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inukai M, Toyooka S, Ito S, Asano H, Ichihara S, Soh J, Suehisa H, Ouchida M, Aoe K, Aoe M, Kiura K, Shimizu N, Date H. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66:7854–7858. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 51.Lee Y, Lee GK, Hwang JA, Yun T, Kim HT, Lee JS. Clinical likelihood of sporadic primary EGFR T790M mutation in EGFR-mutant lung cancer. Clin Lung Cancer. 2015;16:46–50. doi: 10.1016/j.cllc.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Falini B, Bigerna B, Fizzotti M, Pulford K, Pileri SA, Delsol G, Carbone A, Paulli M, Magrini U, Menestrina F, Giardini R, Pilotti S, Mezzelani A, et al. ALK Expression Defines a Distinct Group of T/Null Lymphomas (“ALK Lymphomas”) with a Wide Morphological Spectrum. The American Journal of Pathology. 1998;153:875–886. doi: 10.1016/S0002-9440(10)65629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawrence B, Perez-Atayde A, Hibbard MK, Rubin BP, Dal Cin P, Pinkus JL, Pinkus GS, Xiao S, Yi ES, Fletcher CDM, Fletcher JA. TPM3-ALK and TPM4-ALK Oncogenes in Inflammatory Myofibroblastic Tumors. The American Journal of Pathology. 2000;157:377–384. doi: 10.1016/S0002-9440(10)64550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai W, Lin D, Wu C, Li X, Zhao C, Zheng L, Chuai S, Fei K, Zhou C, Hirsch FR. Intratumoral Heterogeneity of ALK-Rearranged and ALK/EGFR Coaltered Lung Adenocarcinoma. J Clin Oncol. 2015;33:3701–9. doi: 10.1200/JCO.2014.58.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Girard N, Deshpande C, Azzoli CG, Rusch VW, Travis WD, Ladanyi M, Pao W. Use of epidermal growth factor receptor/Kirsten rat sarcoma 2 viral oncogene homolog mutation testing to define clonal relationships among multiple lung adenocarcinomas: comparison with clinical guidelines. Chest. 2010;137:46–52. doi: 10.1378/chest.09-0325. [DOI] [PubMed] [Google Scholar]
- 56.Ellison G, Zhu G, Moulis A, Dearden S, Speake G, McCormack R. EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol. 2013;66:79–89. doi: 10.1136/jclinpath-2012-201194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


