SUMMARY
RNA has the intrinsic property to base pair, forming complex structures fundamental to its diverse functions. Here we develop PARIS, a method based on reversible psoralen-crosslinking for global mapping of RNA duplexes with near base-pair resolution in living cells. PARIS analysis in three human and mouse cell types reveals frequent long-range structures, higher order architectures, and RNA:RNA interactions in trans across the transcriptome. PARIS determines base-pairing interactions on an individual-molecule level, revealing pervasive alternative conformations. We used PARIS-determined helices to guide phylogenetic analysis of RNA structures, and discovered conserved long-range and alternative structures. XIST, a lncRNA essential for X chromosome inactivation, folds into evolutionarily conserved RNA structural domains that span many kilobases. XIST A-repeat forms complex inter-repeat duplexes that nucleate higher order assembly of the key epigenetic silencing protein SPEN. PARIS is a generally applicable and versatile method that provides novel insights into the RNA structurome and interactome.
Graphical abstract
INTRODUCTION
RNA structure and intermolecular interactions are essential in nearly every step of the gene expression program. Structured RNAs are critical components of key molecular machines in the cell, such as the spliceosome, ribosome, and telomerase, and RNA structures play important roles in the control of messenger and noncoding RNA functions (Cech and Steitz, 2014). Base pairing dominates the energetics of both RNA folding and RNA-RNA interactions. Despite recent advances in measuring RNA structures in living cells (Ding et al., 2014; Rouskin et al., 2014; Smola et al., 2015; Spitale et al., 2015), current methods largely provide one-dimensional information. That is, these methods identify which bases are single- or double-stranded, but do not directly reveal the counter-parties in each base pair (Lu and Chang, 2016). Inferring transcriptome structure in living cells is especially challenging, due to the presence of long-range structures, pseudoknots, alternative structures, repetitive sequences and RNA:RNA interactions. One example illustrating these difficulties is XIST, a long noncoding RNA (lncRNA) required for X chromosome inactivation in female cells of eutherian animals (Penny et al., 1996). The key region for XIST-mediated epigenetic silencing, termed the A-repeat, is comprised of 7.5 or 8.5 near-identical copies of a sequence, and multiple structural models have been proposed (Fang et al., 2015; Maenner et al., 2010; Wutz et al., 2002). The structural basis for XIST interaction with key protein partners like SPEN is also not known (Chu et al., 2015; McHugh et al., 2015; Moindrot et al., 2015; Monfort et al., 2015). These challenges highlight the need for further advances to address the structures of the vast majority of coding and noncoding RNAs in the cell.
RNA affinity capture and proximity ligation may offer the next generation of solutions (Engreitz et al., 2014; Helwak et al., 2013; Ramani et al., 2015; Sugimoto et al., 2015). While these methods can identify RNA base pairs, current methods are limited by specific protein or RNA baits which are performed one at a time, and have limited resolution especially for longer RNAs (Engreitz et al., 2014). Here we describe a general method that directly identifies base-pairing interactions in living cells and by doing so, determines both RNA structures and RNA-RNA interactions. PARIS (Psoralen Analysis of RNA Interactions and Structures) combines several critical steps (in vivo crosslinking, 2D purification of RNA duplexes and proximity ligation) that yield excellent sensitivity and specificity, as validated by numerous known structures and evolutionary conservation. We discovered a large number of long-range and alternative structures. PARIS-determined structures contain many targets of double-stranded RNA binding proteins (STAU1, DICER1, DGCR8). Furthermore, the high confidence structures guide two new approaches for phylogenetic analysis of RNA structures, revealing conserved architectures in housekeeping gene mRNAs. The combination of PARIS, icSHAPE (in vivo click SHAPE), phylogenetic analysis and iCLIP reveals the overall architecture of the XIST lncRNA and the mechanism of SPEN binding to XIST A-repeat.
RESULTS
The PARIS method and validation
Current methods for in vivo probing generate averaged reactivity profiles and fail to capture the complexity of RNA structures that include long-range structures, pseudoknots and alternative conformations. To address these challenges, we developed PARIS to directly identify base-paired helices, the most basic elements in RNA structures and RNA-RNA interactions (Figure 1, Figure S1, Table S1 and Experimental Procedures). The PARIS method employs the highly specific and reversible nucleic acid crosslinker psoralen-derivative 4’-aminomethyltrioxsalen (AMT) to fix base pairs in living cells (Calvet and Pederson, 1979). AMT intercalates in RNA helices and, upon photo-activation, crosslinks the two strands, with a preference for staggered uridines (Cimino et al., 1985). Partial RNase and complete proteinase digestion during RNA purification ensures that the identified crosslinks are limited to small and directly base-paired RNA fragments (Figure S1A-C). Two-dimension electrophoresis of the RNase-digested fragments enables purification of only crosslinked fragments (above the main diagonal, Figure 1A, 1B and S1D,E). The 2D purification consistently recovers 0.2%-0.5% of input RNA as double-stranded (above the diagonal), demonstrating that 2D purification is essential for enriching dsRNA fragments. Proximity ligation of duplex RNA fragments, photo-reversal of crosslinks and high throughput sequencing reveal the direct base pairing between fragments. Each PARIS read is an individual-molecule evidence of a duplex between two RNA fragments (arms). The multiplicity of PARIS reads can thus reveal a single common structure, multiple alternative structures, or interactions between two RNAs in trans (Figure 1D-G). The combination of these important features allows us to model RNA structures and interactions with high specificity and sensitivity.
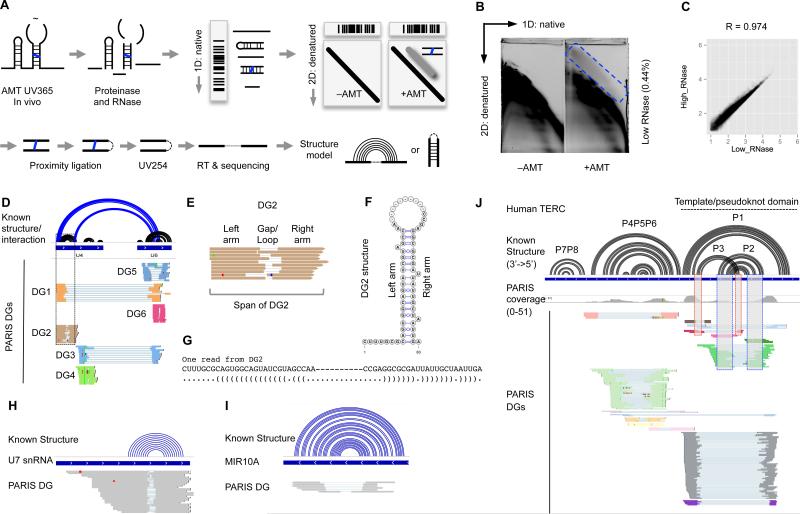
Figure 1. PARIS identifies RNA helices and interactions in living cells.
(A) Schematic diagram of PARIS with three critical steps: in vivo AMT crosslinking, 2D gel purification and proximity library. The blue line is AMT. The dashed lines indicate ligations. Note that the ligation could happen on either ends, resulting in normal gapped or chiastic reads.
(B) 2D purification of the crosslinked RNA. The blue box indicates the region that contain crosslinked RNA. Percentage of recovery of crosslinked RNA from total RNA is indicated in parentheses. See Figure S1 for the high RNase digestion 2D gel.
(C) PARIS sequenced reads are highly reproducible between the high RNase and low RNase conditions in HeLa cells.
(D) Comparison of known structures (black arcs) and interactions (blue arcs) of the U4 and U6 snRNAs to PARIS DGs. Ten reads are shown for each DG. Dashed box highlights DG2 (see E-G). DG2 and DG4: U4 stem-loops . DG5 and DG6: U6 stem-loops. DG1 and DG3: U4:U6 interaction.
(E) An example duplex group (DG2) in U4 snRNA and the definition of terms (DG, arm, gap/loop and span) used in this paper. Note that the staggered termini for the two arms indicate that these reads come from distinct RNase cleavage sites from individual RNA molecules, i.e. each gapped read is an individual molecule measurement of a stem-loop or an RNA-RNA interaction duplex.
(F-G) The structure model of the duplex group (DG2) is consistent with known base pairs from the crystal structure of U4. Dashes are the gaps.
(H-I) PARIS identifies the stem-loop structure in the low-abundance snRNA U7 (H) and MIR10A precursor (I).
(J) PARIS identifies known structures in telomerase RNA (TERC). The boxes indicate interlocking DGs corresponding to the P2/P3 pseudoknot.
We performed PARIS on human HeLa, HEK293T and mouse embryonic stem (mES) cells, generated a total of 350 million reads after removing duplicates. The gapped reads, arising from RNase digestion of single-stranded loops in RNA structure, constitute 2.5%-6% of all mappable reads (Figure 1B, Figure S1, Table S1). Given the absence of any background above the diagonal in the –AMT controls (Figure 1B), the non-gapped reads come from failed ligations of duplexes due to steric hindrance (Sugimoto et al., 2015). PARIS is highly reproducible across biological replicates in each of the three cell types (R= 0.94-0.98 between replicates, Figure 1C, Figure S1F,G).
We assembled gapped reads into duplex groups (DG), each corresponding to an RNA stem-loop, with the two arms from the stem and the gap from the RNase-cleaved loop, or an RNA-RNA interaction, with the two arms from the two interacting RNAs (Figure 1E-G). DGs are filtered to retain only the ones with high confidence supported by multiple reads. To visualize this new type of RNA structurome data and associated structure models, we developed new features in the Integrative Genome Viewer (Robinson et al., 2011) (Figure 1D). Gapped reads are displayed in groups by DG, and structure models are visualized as arcs connecting the two arms of each DG (see Supplemental Experimental Procedures for the detailed analysis methods, directions and links to visualization of PARIS data).
We validated the sensitivity and specificity of PARIS using a number of well-studied RNAs, such as ribosomal RNA, snRNAs and microRNAs (Figure 1H-I, Figure S2). Complex RNA structures are currently difficult to detect using one-dimensional chemical probing or computational prediction. Among the most difficult structures to predict are pseudoknots, comprised of interlocked helices, where the loop of one stem-loop participates in base-pairing with an outside region. We were able to detect well-known pseudoknots in telomerase RNA (TERC, Figure 1J), RMRP and RPPH1 (the RNA components of RNase MRP and RNase P, data not shown) in both human and mouse PARIS data.
Global properties of the RNA structurome revealed by PARIS
Having established the PARIS method, we investigated the global properties of the RNA structurome. Most previous experimental and computational methods can only identify short-range structures (i.e. the span from the beginning of the left arm of the duplex to the end of the right arm), typically focusing on <200 nt windows. We found that a large number of RNA duplexes (29-40%) span greater than 200 nt in the three cell types and 4-11% of duplexes span greater than 1000 nt (Figure 2A).
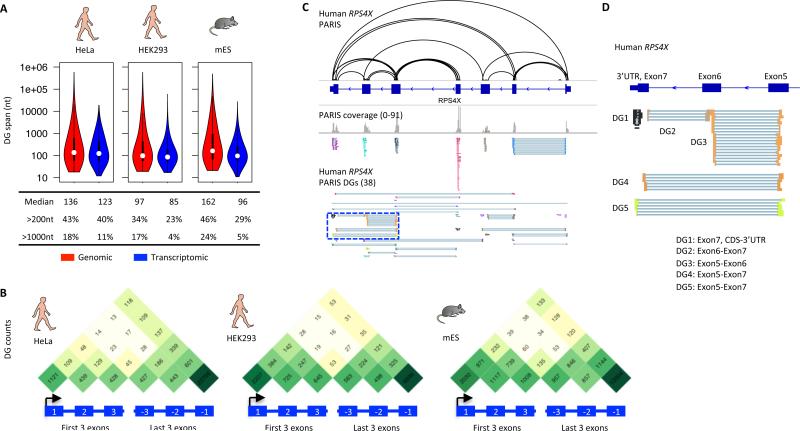
Figure 2. Global properties of RNA structures in living cells.
(A) Size distribution of RNA structures. One replicate from each cell type is shown here. Genomic span is the distance between the ends of gapped reads in the genome, while the transcriptomic span excludes introns.
(B) Metagene distribution of PARIS determined helices among exons. Only the first three and last three exons were plotted. One biological replicate is plotted for each cell type. The gradation of green color correlates to number of DGs in log scale.
(C,D) Example higher order architecture of human RPS4X mRNA (C). The blue boxed region is zoomed in to highlight DGs connecting different parts of the mRNA (D).
We next investigated the extent to which RNA duplexes are organized into higher-level architectures. Many genomic studies categorize messenger RNAs into 5’ untranslated region (UTR), coding sequence (CDS), and 3’ UTR, and perform analyses on these units assuming they are separate entities. We observed extensive RNA duplexes that cross these artificial boundaries. To illustrate the long-range structures, we plotted the number of DGs connecting among the first three and last three exons (Figure 2B). Even though most structures are local, as shown in Figure 2A, we observed many structures that span multiple exons (Figure 2B). For example in the RPS4X mRNA and other mRNAs, we observe multiple independent loops between the 5’ UTR and CDS, CDS and 3’ UTR, and between 5’ and 3’ UTRs (Figure 2C,D, Figure S3A, B), and structures that cover the start and stop codons (Figure S3C, D). In addition, we also identified structures formed by repetitive elements like Alu elements (Figure S3E,F). The RNA structural features that dictate the specific recognition of double-stranded RNA binding proteins (dsRBPs) to their cognate targets are not known, and PARIS identified the RNA structures associated with the dsRBP binding sites (Figure S4).
PARIS-guided analysis of RNA structure conservation and covariation
The large number and diversity of RNA duplexes identified by PARIS poses a challenge to distinguish the subset of structures with important biological functions. Evolutionary conservation of RNA secondary structure across several species is a strong indicator of function (Smith et al., 2013). Conserved RNA duplexes are supported by conservation of base pairs between the two arms, or more convincingly, by covariation in evolution (e.g. swapping Watson-Crick base pairs across the helix, i.e. less conservation). Genomic screens of conserved structures usually employ sliding window analysis of in multiple sequence alignments and therefore are limited by the window size, sliding step and generally lack experimental validation. Whereas typical covariation analysis uses sliding windows of 200 nucleotides to achieve reasonable runtimes (Smith et al., 2013), PARIS data reveal that a substantial fraction of the RNA duplexes span more than 200 nt. This observation suggests that a large number of the structures (at least 23%-46%, Figure 2A) have been missed and are in fact incorrectly assigned to nearby neighbors by current methods. We reasoned that PARIS data can focus evolutionary analyses to the biologically relevant helix arms, overcome length limitation imposed by current methods and evolutionary conservation can globally validate and highlight functional RNA duplexes.
RNA duplex determination by PARIS in human and mouse cells enables direct analysis of global structure conservation in two ways. First, direct determination of RNA duplexes by PARIS enabled us to precisely position the two arms of RNA helices in whole-genome alignments and guide covariation analyses regardless of their linear distance (Figure 3A). We measured the significance of base-pair covariation and structure conservation by shuffling sequences within each duplex, and calculating a Z-score based on the distribution of structure energies in 100 shuffled alignments for each DG (Gesell and von Haeseler, 2006). This guided analysis revealed 25% of the well-aligned helices in amniotes genomes are highly conserved (Z-score < −2.326, corresponding to p-value 0.01). Many of these conserved structures also show strong covariation (46% conserved DGs with less than −10kcal/mol covariation energy contribution, Figure 3B, Table S2). Among these conserved structures, we found that 43% of them span long distances (200nt) (Figure 3C). This analysis further validates the PARIS method by showing that a significant fraction of the experimentally derived structures are potentially functional (examples in Figure S3D and S5).
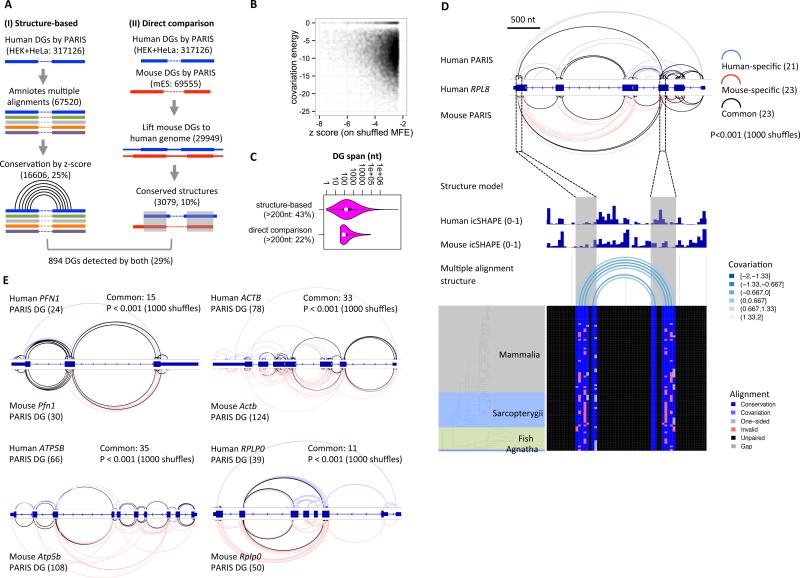
Figure 3. PARIS guides global phylogenetic analysis of RNA structures.
(A) Two approaches of PARIS-guided phylogenetic analysis of RNA structures. The numbers of structures are in parentheses.
(B) Scatterplot of z-scores and covariation energies for the structure-based analysis of conservation in amniotes. All 16606 structures with Z-score < −2.326 (p<0.01) were plotted.
(C) Distribution of the linear span of the conserved structures identified by the two methods.
(D) Evolutionarily conserved structures in RPL8 mRNA using direct comparison of human (HEK293) and mouse (ES cells) PARIS data. An example conserved long-range structure, connecting the third and sixth exons in human and mouse is supported by both icSHAPE and phylogenetic analysis in multiz100 multiple genome alignments. Significance of the overlap was tested by random shuffling of DGs in the exons. In this structure, 6.5% of all potential base pairs are one- or two- sided covariants (E). Four more examples of conserved mRNA architectures between human and mouse.
Prior computational genomic screens have identified large numbers of conserved elements, yet little is known about their function. Bejerano et al. reported the identification of a 481 ultra-conserved elements (UCEs) in human, mouse and rat, and 95 of them are located in mature RNA transcripts (Bejerano et al., 2004). We intersected the 95 UCEs with the PARIS-defined structures and found 14 overlapping with mES cell PARIS DGs and 34 overlapping with human PARIS DGs, and 12 of them overlap with both human and mouse PARIS DGs (Figure S5B-D, Table S3). This analysis suggests that at least some of the UCEs encode structural elements.
Second, the PARIS-determined structures in two distantly related species--human and mouse--allowed us to directly compare the structures on homologous sequences. We lifted the coordinates of mouse RNA structures to the human genome based on human-mouse pairwise genome alignments and intersected the helices between the two species (Figure 3A, Table S4). Despite the limited coverage of homologous RNAs between the two cell types, different cell type origins, and the dramatic difference of noncoding regions, we identified 10% of the structures to be shared between human and mouse. Among these ~3000 structures shared between human and mouse, 22% of them span regions longer than 200nt (Figure 3C). In addition, 29% of the direct-comparison-discovered (approach II) conserved helices are also found by structure-based phylogenetic analysis (approach I) (Figure 3A).
Direct comparison of PARIS data in human and mouse validated conserved long-range structures in mRNAs and lncRNAs (Figure 3D,E, Figure S5A). In the RPL8 mRNA, 23 of the 44 DGs identified in human cells and of 46 in mouse cells are shared (Figure 3D). Many of these conserved structures span different exons, revealing conserved architecture of the RPL8 mRNA (P<0.001 with 1000 shuffles). The conserved long-range structures that connect exon3 to exon6 are also supported by icSHAPE data (low SHAPE reactivity in the base paired region) in both species and phylogenetic analysis of vertebrates (Figure 3D). In addition, analysis of five mRNAs and the well-known lncRNA MALAT1 with similar numbers of PARIS-detected DGs in human and mouse showed that architectures are conserved for all of them (Figure 3E, Figure S5E-H).
PARIS reveals pervasive alternative RNA structures
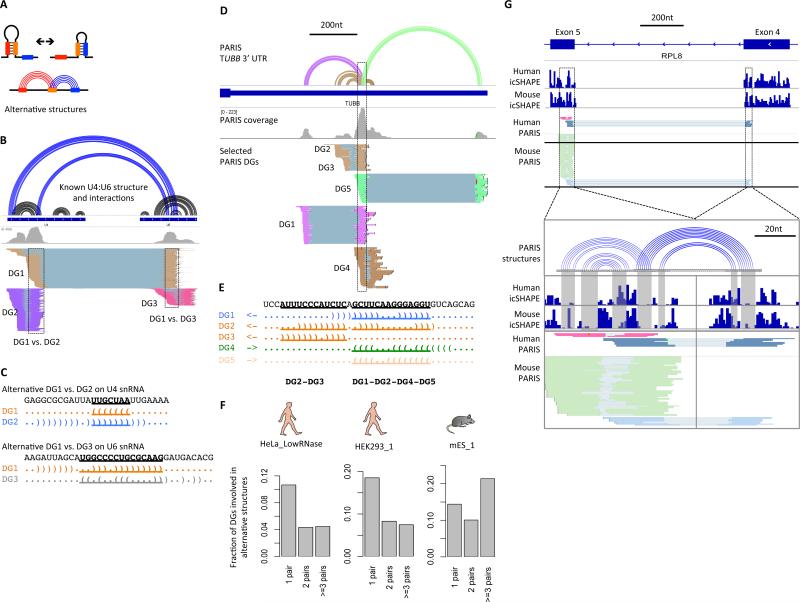
Dynamic RNA structures play important roles in regulating gene expression and catalyzing enzymatic reactions (Dethoff et al., 2012). Previous methods for identifying dynamic or alternative structures typically use McCaskill's partition functions, with or without flexibility measurements as soft constraints (McCaskill, 1990; Ritz et al., 2013). These methods are often limited by sequence length and lack experimental validation. Since PARIS detects individual RNA duplexes in cells, alternative structures are directly detected as conflicting duplexes (Figure 4A). As a positive control, we detected the important U4:U6 alternative structures in the U4:U6 dimer in addition to their individual structures (Figure 4B, C).
Figure 4. PARIS reveals pervasive alternative structures.
(A) Diagram of alternative structures.
(B-C) PARIS identifies alternative structure/interactions in the U4:U6 snRNA heterodimer (B). Two alternative structures are shown here: DG1 vs. DG2 and DG1 vs. DG3 (C).
(D) An example of extensive alternative structures in the 3’UTR of TUBB mRNA from HeLa PARIS data. Only DGs involved in this cluster of alternative structures are shown. First track: PARIS-based structure models. The corresponding structure models and DGs are color-coded.
(E) The hub of the alternative structures. The five alternative structures are displayed in dot-bracket format and color-matched to panel D. Nucleotides involved in conflicts are highlighted and underlined.
(F) Fraction of DGs involved in alternative structures that comprise 1, 2, or at least three pairs of alternative structures are plotted as a fraction of all DGs. Top 50 mRNAs were used for each of the 3 panels. One replicate was plotted for each cell type. HeLa_LowRNase: 744 out of 3801 DGs (20%) are involved in alternative structures (711 pairs) and 31 pairs of alternative structures (4.4%) are supported by conservation/covariation (both structures in each pair). HEK293_1: 459 out of 1338 DGs (34%) involved in alternative structures (448 pairs) and 7 pairs of alternative structures (1.6%) are supported by conservation/covariation. mES_1: 592 out of 1291 DGs (46%) involved in alternative structures.
(G) An example alternative structure in RPL8 mRNA supported by both human and mouse icSHAPE and PARIS data. The structure models show the perfect correspondence between the icSHAPE data and base pairs (gray shaded area).
We also identified new alternative structures, for example in the 3’UTR of TUBB mRNA (Figure 4D,E) and lncRNAs MALAT1 and XIST (Figure S5E, S7B). The TUBB cluster of alternative structures consists of 5 helices (DG1-DG5,. Among these structures, DG1, 2, 4 and 5 appear to be mutually exclusive (Figure 4E). DG2 and 3 also have strong conflicts with each other, and thus cannot simultaneous take place on the same molecule. We analyzed the top 50 mRNAs with the highest numbers of detected helices in the three cell types and found that about 20% to 50% of them are involved in at least one pair of alternative structures, suggesting that alternative structures are pervasive (Figure 4F, Table S5). Interestingly, a substantial amount of the helices are involved in more than 3 pairs of alternative structures, suggesting highly complex networks of structures in living cells. These results are consistent with recent in vitro studies showing mRNAs sampling multiple structures (Kutchko et al., 2015).
Alternative RNA structures could be simply a result of the degeneracy of base pairing, or in contrast, be important for the RNA's function. The latter scenario predicts that some of the alternative structures should be evolutionarily conserved. To test this, we integrated PARIS, icSHAPE, and phylogenetic analysis to examine both high-level architecture and high-resolution structures in a functional context. The matched PARIS and icSHAPE datasets in HEK293 and mES cells showed that the alternative structures are evolutionarily conserved. Out of the 44 DGs for human RPL8, 32 of them form 42 alternative structure pairs; 19 of the 42 pairs of alternative structures are conserved between human and mouse. An example alternative structure is shown in the coding region of RPL8 mRNA (Figure 4G). Both human and mouse PARIS and icSHAPE in the same cell types support this pair of alternative structures. Approximately 5% of the alternative structures examined have both structures supported by sequence conservation or covariation in evolution. Thus, some alternative structures in mRNAs are evolutionarily conserved and therefore likely functional.
PARIS identifies RNA-RNA interactions in trans with high precision
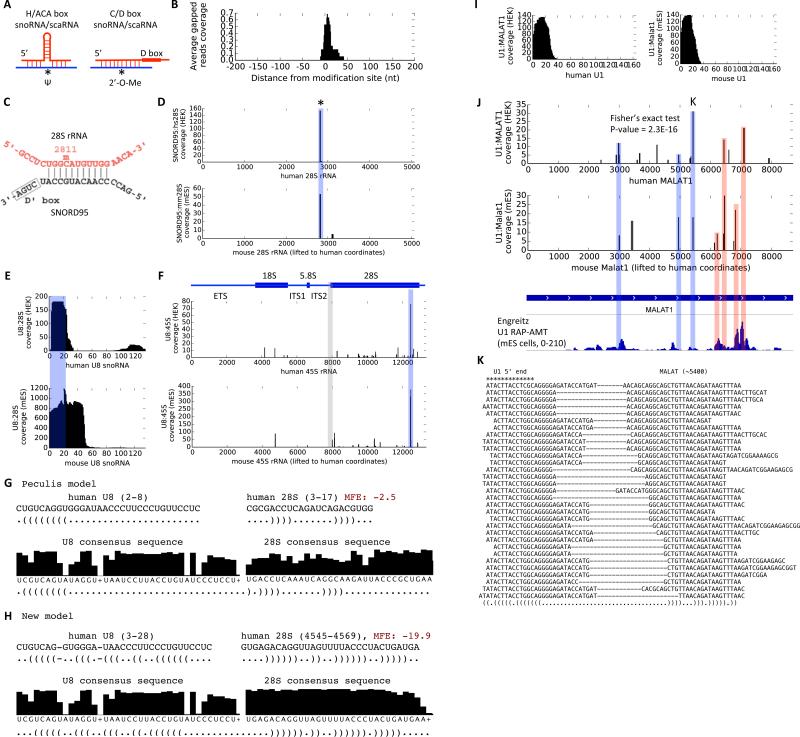
RNA-RNA interactions are used by many ncRNAs to build macromolecular complexes and regulate gene expression (Lee et al., 2015). Current methods to identify RNA-RNA interactions require a “bait” protein or RNA; thus can be limited in scope (Helwak et al., 2013; Sugimoto et al., 2015). In contrast, PARIS is a general method that can detect RNA-RNA interactions in a protein/RNA-agnostic fashion. SnoRNAs and scaRNAs guide modification and processing of rRNAs and snRNAs (Kiss, 2001) (Figure 5A). We compared all known snoRNA:rRNA interactions with the PARIS data from HEK293 cells. All the arms mapped to the rRNAs are centered on the modification sites, with a very narrow distribution (~20nt at half height, Figure 5B, D and Figure S6). Given that snoRNA:rRNA interactions are around 10-20 base pairs, PARIS determines the interaction with near base pair resolution. Furthermore, because rRNA and snoRNAs are among the most abundant RNAs, the precise mapping of their interaction sites confirms the high specificity of PARIS. The availability of both human and mouse PARIS data and the identical location of the interaction sites provide even stronger evidence to the authenticity of the interactions (Figure 5C,D and Figure S6).
Figure 5. PARIS determines new RNA:RNA interactions with high resolution.
(A) Models of H/ACA box sno/scaRNA guided RNA pseudouridylation and C/D box sno/scaRNA guided 2’-O-methylation. Ψ: pseudouridine. 2’-O-Me: 2’-O-methyl.
(B) Specificity and resolution of the snoRNA-guided modification of human ribosomal RNAs. For each known snoRNA:rRNA interaction, the number of reads were normalized so that the maximum is 1. All identified snoRNA:rRNA interactions from HEK293 cells were averaged.
(C-D) Base pairing model from snoRNABase (C) and PARIS data (D) were shown for the SNORD95:28S interaction. The asterisk indicates the known modification site.
(E-F) PARIS in human and mouse cells reveals the interaction site on U8 snoRNA (E) and 28S rRNA (F). PARIS-determined interaction sites were marked by the blue box, while the previously reported binding site is shaded gray (Peculis 1997).
(G-H) The original U8:rRNA interaction was not supported by phylogenetic conservation and hybridization energy (G), whereas the newly identified U8:rRNA interaction is (H). The consensus sequences were from Rfam.
(I) Meta analysis of the U1 target site. The U1:MALAT1 interactions use the 5’ end of the U1 snRNA in both human and mouse cells.
(J) U1 snRNA interacts with MALAT1 RNA in human and mouse cells. PARIS achieves higher resolution than RAP (Engreitz 2014). The blue shaded peaks are shared between human/mouse PARIS and RAP data. The red shaded peaks are shared between one of the PARIS datasets and RAP data. Fisher's exact test was used to show the significant overlap between human and mouse PARIS-determined U1 sites.
(K) Example gapped reads for a conserved U1:MALAT1 interaction. The 5’ end of the U1 snRNA interacts with MALAT1 (at nt position ~5400)
See also Figure S6.
We highlight two applications of PARIS to understand RNA:RNA interactions. First, PARIS can identify new RNA interactions, such as between snoRNAs and rRNAs. U8 snoRNA is essential for the processing of 5.8S and 28S rRNAs (Peculis and Steitz, 1993). U8 depletion leads to accumulation of pre-rRNA intermediates in Xenopus. Previous studies suggested that the 5’ end of U8 snoRNA base pairs with the 5’ end of 28S rRNA based on accessibility measurement (Peculis, 1997). Phylogenetic analysis revealed high conservation of ~15nt at U8 snoRNA 5’ end (Peculis, 1997), suggesting that this region is essential. We found that in both human and mouse cells, the primary U8 snoRNA interaction sequence is located on the 5’ end, consistent with previous studies (Figure 5E). However, the 28S interaction site is near the 3’ end in both human and mouse cells (Figure 5F, blue shaded area); no crosslinking is observed on the previously proposed binding site, even though uridine crosslinking sites are present (Figure 5F, gray shaded area). Phylogenetic analysis using the Rfam database provided independent support and showed that the highly conserved nucleotides correspond to the base-paired nucleotides in the new model (Figure 5G,H). The new model is more energetically favorable, with a minimum free energy of −19.9 kcal/mol vs. −2.5 kcal/mol for the current model (Peculis model). Thus, PARIS can nominate new RNA interactions that derive further support from comparisons of human and mouse PARIS data, evolutionary conservation, and computational modeling.
Second, PARIS can refine the resolution of RNA:RNA interaction sites. U1 snRNA has been shown to bind 5’ splice sites and other cognate sequences throughout the transcriptome (Almada et al., 2013; Lu et al., 2014; Ntini et al., 2013). Engreitz et al. used RAP-RNA to enrich for U1-associated RNAs and identify U1 binding sites across transcripts (Engreitz et al., 2014). However, this purification approach recovers broad regions (Figure 5J). In contrast, PARIS determines high resolution binding sites for U1. In both human and mouse, the first ~20 nucleotides of U1 are involved in trans interactions, consistent with the accessibility of the first 12nt (Figure 5I), and the interaction with target RNAs is focal (Figure 5J). For instance, Engreitz et al. reported strong interactions between U1 and Malat1 in mES cells. We find precise PARIS interactions between U1 and MALAT1 within the broad RAP peaks. The U1:MALAT1 PARIS interactions are conserved between human and mouse (p= 2.3 × 10−16, Fisher's exact test, Figure 5J). These results are consistent with strong predictive power of complementary U1 motifs in target RNAs for U1-dependent RNA stabilization (Almada et al., 2013), indicative of precise sequence-dependent interactions.
XIST structure informs higher order assembly of XIST-Spen complex
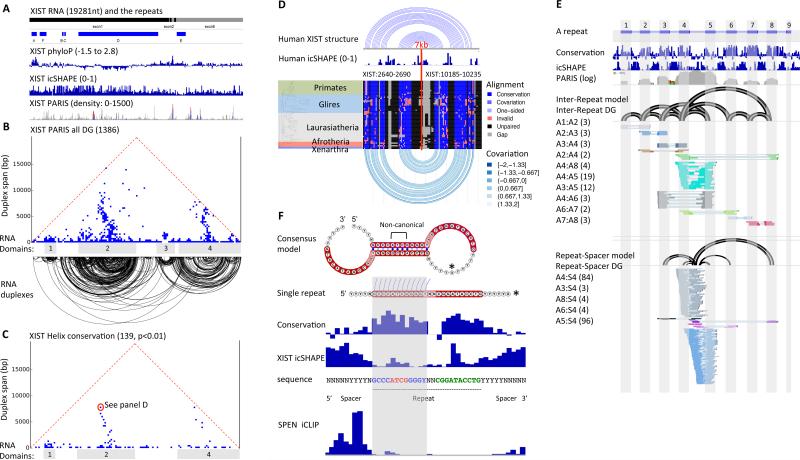
XIST is a 19kb lncRNA essential for X chromosome inactivation in placental mammals (Brown et al., 1991; Penny et al., 1996). However, one-dimensional methods have produced conflicting models of its structure. We used a combination of three orthogonal methods -- PARIS, icSHAPE, and phylogenetic conservation – to determine the structure of the XIST lncRNA in living cells (Figure 6A, B). Global analysis of the PARIS data reveal both local helices and multiple long-range structures that span up to 7kb (Figure 6B). The long-range helices organize regions of the RNA into four major domains. To determine if the identified secondary structures are biologically meaningful, we used the PARIS-determined helices to guide phylogenetic analysis. Our analysis reveals that 10% of the PARIS determined helices are conserved; and the domain structures for domains 1, 2, and 4 are conserved (Figure 6C and S7A, Table S6). A conserved long-range structure over 7kb that anchors domain 2 is shown in Figure 6D. A large number of the helices in XIST are involved in alternative structures suggesting that this lncRNA is highly dynamic (Figure S7B,C). Interestingly, another lncRNA MALAT1 also contains many long-range structures, yet NEAT1 does not (Figure S5B, S7D).
Figure 6. Integrated structure analysis of the human XIST RNA.
(A) Overview of XIST lncRNA. Xist exons and repeat, phylogenetic conservation (PhyloP), icSHAPE, and PARIS data in HEK293 cells are shown.
(B) Architecture of the XIST RNA. Each point in the triangular heatmap shows the PARIS connection between the two regions indicated by the feet of the triangle. Data are plotted in 100nt × 100nt bins. Each RNA duplex detected by PARIS are plotted below. The duplex loops are clustered into four major RNA structure domains. The repeat A region is a small domain before the domain 1.
(C) Conservation of RNA duplexes determined by phylogenetic analysis of eutherian XIST homologs. Conserved helices (p-value<0.01) are plotted.
(D) An example long range (~7kb gap) structure with PARIS, icSHAPE, and phylogenetic support (9.4% of all base pairs are one- or two-sided covariants).
(E) Integrated structure analysis of the conserved repeats in the A-repeat region. Conservation track: phyloP score for the eutherian alignments. PARIS coverage is shown in log scale. All detected inter-repeat and are illustrated in the arcs of structure models. A1-A8, repeats. Numbers in parentheses are the numbers of reads in each DG. The non-conserved repeat-spacer DGs (lower part) were shown separately from the conserved ones (upper part).
(F) Consensus model of the A-repeat inter-repeat structure. The consensus model depicts two repeats base-paired to each other. The red highlighted regions indicate the conserved repeats, while the non-highlighted regions indicate the spacers. Non-canonical: non-Watson-Crick base pairs with intermediate icSHAPE reactivity (constrained by the surrounding base pairs). Conservation and icSHAPE: average for all 8 repeats. Mouse Xist in vitro SHAPE is similar to the HEK293 XIST icSHAPE. SPEN is crosslinked to 3-5nt upstream of the inter-repeat duplex (see Figure 7 for SPEN iCLIP).
The A-repeat, located at the 5’ end of the XIST RNA, contains up to 8.5 copies of a highly conserved sequence separated by uridine-rich variable spacers (8.5 repeats in human and 7.5 in mouse, ~400nt; Figure 6E). A mouse Xist mutant lacking the A-repeat is unable to silence genes, but still capable of coating the X chromosome (Wutz et al., 2002). The A-repeat is thus a critical link in RNA-mediated epigenetic silencing. The A-repeat was recently found to be required for Xist to interact with a small number of proteins (Chu et al., 2015), and among these, Spen emerged as a factor linking Xist to histone deacetylase complexes and gene silencing (Chu et al., 2015; McHugh et al., 2015; Moindrot et al., 2015; Monfort et al., 2015). Despite its importance, the repetitive nature of the A-repeat has complicated structural studies. Indeed, several contradictory models have been proposed, suggesting that each repeat base pairs within itself (“intra-repeat”, (Wutz et al., 2002)), base pairs with other repeats (“inter-repeat” (Maenner et al., 2010)), or a combination of both (Fang et al., 2015). Prior studies were limited by the use of one-dimensional RNA structure data and computational models that arbitrarily precluded long-range RNA interactions (e.g.(Fang et al., 2015; Maenner et al., 2010; Wutz et al., 2002)).
PARIS highlighted several key structural features of the ~400nt A-repeat region in vivo. First, the A-repeat does not form duplexes with any sequence far from the region, suggesting that this region mostly folds as an isolated domain (Figure 6B). Second, the repeats form extensive duplexes. All the detected RNA duplexes are between repeats (Figure 6E). While we cannot rule out the possibility that intra-repeat structures can form, our data suggest inter-repeat structures are more likely to occur in vivo, consistent with the higher stability of inter-repeat helices (ΔG= −15.2 kcal/mol for inter-repeat vs. −5.8 kcal/mol for intra-repeat duplex). Each repeat tends to contact the closest repeats, but long-range contacts (bigger arcs) are also observed, suggesting 3D folding of the A-repeat region. In addition to the inter-repeat structures, we also observed structures between spacer 4 and several repeats. Repeat 4 and spacer 4 are not conserved rodents (Elisaphenko et al., 2008); these spacer-repeat structures may have species-specific function. Notably, the inter-repeat helices form between the first halves of the two repeats, flanked by single-stranded U-rich sequences on the 5’ and the second half of the repeat on the 3’ side (Figure 6F). Each inter-repeat unit has nearly identical structure, which is also supported by icSHAPE data that delineate precisely the complementarity (Figure 6F). Since each instance of the A-repeat can contact one of several other repeats, our data imply that the A-repeat exists as a family of multiple complex structures in living cells.
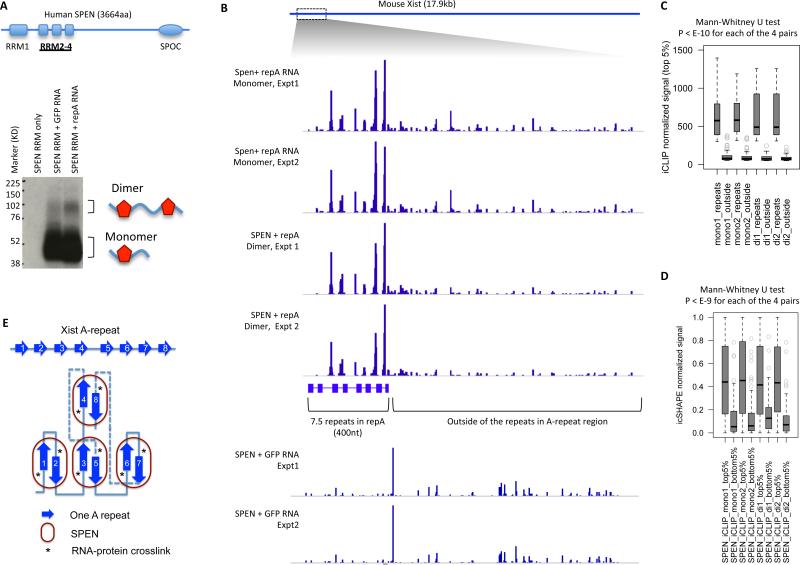
The presence of at least 7.5 copies of repeats in XIST and the unique structural unit raised the hypothesis that its higher order structure may be important for the interaction with the key silencing factor SPEN. Previous studies of SPEN RRM domains suggest that they bind many RNA species, without preference for single copies of the A-repeat motif (Monfort et al., 2015). To address this issue, we performed individual nucleotide crosslinking and IP (iCLIP) with recombinant SPEN RRM domains (RRM2-4) and a ~1.6 kb region of mouse Xist RNA containing the A-repeat in vitro. We used a GFP mRNA matched in length as a negative control (Figure 7A, S7E). iCLIP on both GFP mRNA and A-repeat RNA generated a radioactive SPEN band, but A-repeat RNA also generated a higher molecular weight band the size of a dimer SPEN RRM2-4 crosslinked to RNA (Figure 7A, Figure S7E).
Figure 7. The A-repeat structure promotes SPEN binding and higher order RNP formation.
(A) In vitro iCLIP with human SPEN RRM2-4 and IRES-GFP or mouse repA RNA. The diagram shows the domain organization of SPEN. The autoradiograph shows one iCLIP experiment. The entire A-repeat region is 1630nt. The IRES-GFP RNA is 1533nt. The dimer band relative intensity is 1 for the repA RNA and 0.61 for the GFP RNA control. See Figure S7 for another replicate of the iCLIP experiment.
(B) All the 6 iCLIP tracks are normalized by total read count and scaled to 0-2300.
(C) For each of the four SPEN+repA iCLIP tracks, the crosslinking frequency for top 5% of crosslinked nucleotides were extracted from the repeats region and the outside region. This analysis shows that SPEN binds the repeats region more than the outside region.
(D) Nucleotides with the top 5% and bottom 5% of iCLIP signal were extracted from each of the 4 tracks, and then the icSHAPE signals were compared. This analysis shows that SPEN RRM2-4 are preferentially crosslinked to single-stranded regions (high icSHAPE signal).
(E) Model of SPEN-repA association. The base pairing among the repeats are stochastic and only one specific conformation is shown here. SPEN binding requires both single-stranded and double-stranded regions, but is only crosslinked to the single stranded nucleotides 3-5nt upstream of the inter-repeat duplex.
See also Figure S7.
We sequenced RNA from the monomer and dimer bands separately, and found that SPEN interacted nearly exclusively with the A-repeat region (Figure 7C). SPEN is crosslinked to the single-stranded spacers immediately upstream of the inter-repeat duplexes (summarized in Figure 6F). SPEN binds single stranded nucleotides as determined by icSHAPE even in the non-specific regions (Figure 7D). The dimer SPEN complex is even more enriched for the A-repeats and depleted of the rest of the RNA (Figure 7B, quantified in Figure 7C,D, Figure S7F). iCLIP experiments with GFP mRNA showed that SPEN-RRM can interact with other RNAs, but did not show the clustered interaction in A-repeat. These results suggest that the secondary structure of the A-repeats facilitates the binding and clustering of SPEN into a higher order structure (Figure 7E). Consistent with this model, quantitative binding experiments showed that RRM2-4 binds the A-repeat with high cooperativity, switching from all unbound to nearly all bound within a two-fold concentration range; no such cooperativity is observed in SPEN interaction with size-matched control RNA (Figure S7G). Collectively, these results and the PARIS-determined inter-repeat helices that span multiple repeats revealed the high level architecture and the precise structure of a key lncRNA-protein interface.
DISCUSSION
PARIS reveals the RNA structurome and interactome
Here we introduced PARIS as a method to map RNA helices and RNA:RNA interactions in living cells across the transcriptome. This work represents a culmination of pioneering efforts since the 1970s to map nucleic acid duplexes in living cells with psoralen (Calvet and Pederson, 1979; Cech and Pardue, 1976; Shen and Hearst, 1976). The major advantage of PARIS is that the nucleotides forming RNA helices are directly identified on a global scale. The strategy described here achieves high precision and specificity. PARIS has many advantages over other methods recently developed to determine RNA structures in vivo (Lu and Chang, 2016). Compared with icSHAPE and DMS-seq, which measure nucleotide flexibility (Ding et al., 2014; Rouskin et al., 2014; Spitale et al., 2015), PARIS directly determines the locations of long-range duplexes and can resolve complex structures such as pseudoknots and alternative structures. Compared with protein-directed methods such as hiCLIP or CLASH (Helwak et al., 2013; Sugimoto et al., 2015), PARIS can address higher order transcriptome structure without the limit of a bait protein. The relationship of PARIS to HiCLIP and CLIP is a comparison of ‘all-to-all’ vs. ‘targeted’, analogous to Hi-C vs. ChIA-PET (Fullwood et al., 2009). PARIS also has better transcriptome coverage than RPL (Ramani et al., 2015). Compared with RNA-RAP (Engreitz et al., 2014), PARIS provides near base-pair resolution, independent of the RNA of interest. Conversely, these targeted approaches are more appropriate if an experiment is focused on a specific RNP. Psoralen crosslinking also has sequence bias; although the preferred UpA dinucleotide should occur frequently (once every 16 base pairs) in a random duplex. Use of multiple orthogonal approaches will continue to be most powerful in future studies, as demonstrated by the integration of multiple methods in determining alternative structures and conserved architectures for mRNAs and lncRNAs.
We found that RNA duplexes can form across long distances, and they pervasively occur with alternative structures where one sequence can base pair with two or more different partners. By using experimentally determined RNA duplex data to guide phylogenetic analysis of evolutionary conservation, this approach can evaluate the potential biological significance of any RNA duplex. We show that many long-range, trans-acting, and alternative RNA structures are evolutionarily conserved, highlighting novel dimensions of transcriptome organization. RNA binding proteins and microRNAs interact with target RNAs and function in a structure-dependent manner (Kedde et al., 2010; Sugimoto et al., 2015). The precise determination of RNA helices set these important interactions in a structural context, which will greatly facilitate the discovery of novel regulatory events and mechanistic studies.
XIST: Higher order lncRNA structure guides epigenetic silencing
Our analysis of XIST RNA illustrates the potential utility of a structural approach to guide the discovery of lncRNA functions. LncRNAs are distinguished from mRNAs by the former's limited conservation at the primary sequence level and the frequent presence of repeats. Using XIST as a model, we found that long-range structures that organize the lncRNA into four major modular domains; each domain is quite compact due to extensive duplex formation. This model of lncRNA organization is consistent with recent super-resolution imaging studies of Xist in situ (Sunwoo et al., 2015). PARIS data were particularly useful in deciphering the structure formed by the 8.5 repeat units in the XIST A-repeat. Our studies suggest that SPEN scans RNAs in a sequence-independent manner, but will nucleate a higher-order, nuclease-resistant RNP structure with the proper structural context of the A-repeat. The long-range, inter-repeat helices should cause the A-repeat to fold up, and create multiple copies of a uniform duplex structure, flanked by U-rich sequence motif recognized by SPEN RRM. This arrangement of both single- and double- stranded RNAs for interaction is consistent with recent crystallographic studies of Spen RRM domains in vitro (Arieti et al., 2014). This model of A-repeat architecture is also quite analogous to the structural organization of Drosophila roX RNA (Ilik et al., 2013). The advent of PARIS and related methods should catalyze discoveries of higher order lncRNA structures in the future.
EXPERIMENTAL PROCEDURES
PARIS experimental method
HeLa, HEK293T and mES cells were treated with or without AMT and crosslinked with 365nm UV. Cell lysates were digested with S1 nuclease and RNA purified using TRIzol. Purified RNA was further digested with ShortCut RNase III to smaller fragments. RNA was separated by 12% native polyacrylamide gel and then the first dimension gel slices were further electrophoresed in a second dimension 20% urea-denatured gel. Crosslinked RNA above the main diagonal was eluted, proximity ligated with T4 RNA ligase I and photo-reversed with 254nm UV. The proximity ligated RNA molecules were then ligated to barcoded adapters, and converted to libraries for Illumina sequencing. See Extended Experimental Procedures for details.
Determination of RNA structure and interactions
Sequencing reads were mapped to the human, mouse or artificial genomes (such as the rDNA unit, or the snRNAs) using STAR (Dobin et al., 2013), allowing chimeric mapping (in a chiastic manner). Mapped reads were filtered to retain only gapped reads and the gapped reads were assembled into duplex groups (DGs) and visualized in together with the predicted or known secondary structures using newly implemented features in IGV. To analyze RNA:RNA interactions, reads were mapped to the Rfam database and chimeric reads mapped to two RNA molecules were assembled into DGs. See Extended Experimental Procedures for details.
Analysis of structure conservation/covariation and alternative structures
For structure-based analysis (approach I), DG coordinates in hg38 were used to extract alignment blocks from the amniote23 or other multiple genome alignments. The extracted alignments were scored for structure conservation and covariation. For direct comparison (approach II), DGs in mm10 were lifted to hg38 coordinates and conserved structures were defined as human and mouse DGs with both arms overlapped. Alternative structures were extracted such that for each pair of DGs, one arm should overlap while the other not. See Extended Experimental Procedures for details.
In vitro SPEN iCLIP
SPEN RRM2-4 was mixed with the repA RNA or control GFP mRNA, crosslinked with 254nm UV, digested with RNase and labeled with radioactivity. The monomer and dimer bands were purified separately for iCLIP library construction.
Supplementary Material
Highlights.
PARIS yields in vivo RNA duplex maps of human and mouse cells.
In vivo maps discover extensive long-range and alternative RNA structures.
PARIS guides evolution analysis and validation of duplex function.
Unique duplex fold of XIST A-repeat nucleates XIST-SPEN lncRNP.
ACKNOWLEDGEMENT
We thank Anton Petrov (Georgia Tech) for the compiled ribosomal RNA structures, Alexey Amunts (Stockholm University) for the mitochondrial rRNA structure, Sebastian Will (MIT) for help with LocARNA software. We also want to thank Yoon-Jae Cho and Sekyung Oh and Y. Grace Chen for reagents. Supported by NIH R01-HG004361 and P50-HG007735 (H.Y.C). Z.L. is a Layton Family Fellow of the Damon Runyon-Sohn Foundation Pediatric Cancer Fellowship Award (DRSG-14-15). M.A.S and J.S.M. are supported by a Cancer Council NSW project grant (RG 14-18).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Z.L. conceived this project and designed the PARIS method and overall analysis strategy. Q.C.Z. and Z.L. implemented the PARIS analysis programs. Z.L. and B.L. performed all the PARIS experiments. R.A.F. performed the icSHAPE experiments. Z.L. and Q.C.Z. performed the analysis on most of the data. Z.L., B.L., C.D., A.R.G., K.J.G. and T.R.C. performed the in vitro studies on XIST and SPEN. Z.L., M.A.S. and J.S.M. performed conservation and covariation analysis. Z.L., J.T.R. and J.P.M. implemented the new features for structure visualization in IGV. H.Y.C. supervised the project. Z.L. and H.Y.C. wrote the manuscript with input from all authors.
ACCESSION NUMBERS
The accession number for the sequencing data reported in this paper is GEO: GSE74353
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures and six tables and can be found with this article online at xxxx.
REFERENCES
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arieti F, Gabus C, Tambalo M, Huet T, Round A, Thore S. The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs. Nucleic acids research. 2014;42:6742–6752. doi: 10.1093/nar/gku277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Calvet JP, Pederson T. Heterogeneous nuclear RNA double-stranded regions probed in living HeLa cells by crosslinking with the psoralen derivative aminomethyltrioxsalen. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:755–759. doi: 10.1073/pnas.76.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Pardue ML. Electron microscopy of DNA crosslinked with trimethylpsoralen: test of the secondary structure of eukaryotic inverted repeat sequences. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:2644–2648. doi: 10.1073/pnas.73.8.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annual review of biochemistry. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elisaphenko EA, Kolesnikov NN, Shevchenko AI, Rogozin IB, Nesterova TB, Brockdorff N, Zakian SM. A dual origin of the Xist gene from a protein-coding gene and a set of transposable elements. PloS one. 2008;3:e2521. doi: 10.1371/journal.pone.0002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Moss WN, Rutenberg-Schoenberg M, Simon MD. Probing Xist RNA Structure in Cells Using Targeted Structure-Seq. PLoS genetics. 2015;11:e1005668. doi: 10.1371/journal.pgen.1005668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesell T, von Haeseler A. In silico sequence evolution with site-specific interactions along phylogenetic trees. Bioinformatics. 2006;22:716–722. doi: 10.1093/bioinformatics/bti812. [DOI] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, Toscano S, Wan Y, Spitale RC, Luscombe N, Backofen R, et al. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Molecular cell. 2013;51:156–173. doi: 10.1016/j.molcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nature cell biology. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. The EMBO journal. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchko KM, Sanders W, Ziehr B, Phillips G, Solem A, Halvorsen M, Weeks KM, Moorman N, Laederach A. Multiple conformations are a conserved and regulatory feature of the RB1 5′ UTR. Rna. 2015;21:1274–1285. doi: 10.1261/rna.049221.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Moss WN, Yario TA, Steitz JA. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015;160:607–618. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Chang HY. Decoding the RNA structurome. Current opinion in structural biology. 2016;36:142–148. doi: 10.1016/j.sbi.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Guan X, Schmidt CA, Matera AG. RIP-seq analysis of eukaryotic Sm proteins identifies three major categories of Sm-containing ribonucleoproteins. Genome biology. 2014;15:R7. doi: 10.1186/gb-2014-15-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS biology. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G, Schermelleh L, Nesterova TB, Brockdorff N. A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell reports. 2015;12:562–572. doi: 10.1016/j.celrep.2015.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, Wutz A. Identification of Spen as a Crucial Factor for Xist Function through Forward Genetic Screening in Haploid Embryonic Stem Cells. Cell reports. 2015;12:554–561. doi: 10.1016/j.celrep.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntini E, Jarvelin AI, Bornholdt J, Chen Y, Boyd M, Jorgensen M, Andersson R, Hoof I, Schein A, Andersen PR, et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nature structural & molecular biology. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- Peculis BA. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Molecular and cellular biology. 1997;17:3702–3713. doi: 10.1128/mcb.17.7.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Ramani V, Qiu R, Shendure J. High-throughput determination of RNA structure by proximity ligation. Nature biotechnology. 2015;33:980–984. doi: 10.1038/nbt.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz J, Martin JS, Laederach A. Evolutionary evidence for alternative structure in RNA sequence co-variation. PLoS computational biology. 2013;9:e1003152. doi: 10.1371/journal.pcbi.1003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CK, Hearst JE. Psoralen-crosslinked secondary structure map of single-stranded virus DNA. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:2649–2653. doi: 10.1073/pnas.73.8.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Gesell T, Stadler PF, Mattick JS. Widespread purifying selection on RNA structure in mammals. Nucleic acids research. 2013;41:8220–8236. doi: 10.1093/nar/gkt596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Calabrese JM, Weeks KM. Detection of RNA-Protein Interactions in Living Cells with SHAPE. Biochemistry. 2015;54:6867–6875. doi: 10.1021/acs.biochem.5b00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D'Ambrogio A, Luscombe NM, Ule J. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519:491–494. doi: 10.1038/nature14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Wu JY, Lee JT. The Xist RNA-PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E4216–4225. doi: 10.1073/pnas.1503690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nature genetics. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.