Abstract
As a large, non-membrane bound organelle, the centrosome must rely heavily on protein-protein interactions to assemble itself in the cytoplasm and perform its functions as a microtubule-organizing center. Therefore, to understand how this organelle is built and functions, one must understand the protein-protein interactions made by each centrosome protein. Unfortunately, the highly interconnected nature of the centrosome, combined with its predicted unstructured, coil-rich proteins, has made the use of many standard approaches to studying protein-protein interactions very challenging. The yeast-two hybrid (Y2H) system is well suited for studying the centrosome and is an important complement to other biochemical approaches. In this chapter we describe how to carry out a directed Y2H screen to identify the direct interactions between a given centrosome protein and a library of others. Specifically, we detail using a bioinformatics based approach (structure prediction programs) to subdivide proteins and screen for interactions using an array-based Y2H approach. We also describe how to use the interaction information garnered from this screen to generate mutations to disrupt specific interactions using mutagenic-PCR and a “reverse” Y2H screen. Finally, we discuss how information from such a screen can be integrated into existing models of centrosome assembly and how it can initiate and guide extensive in vitro and in vivo experimentation to test these models.
Keywords: Yeast Two-Hybrid screen, Centrosome, Centriole
1. Introduction
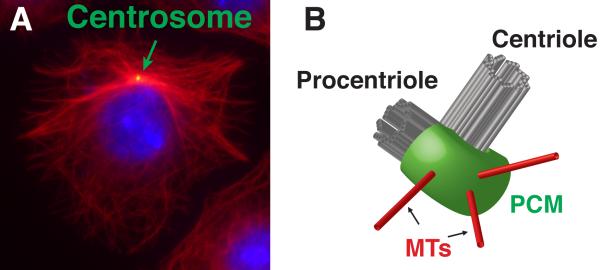
The centrosome is the major microtubule-organizing center (MTOC) of many cells (Figure 1A). As an MTOC, the centrosome functions in a variety of cellular processes including cell migration, neuronal path finding, axon selection, immune cell function, cell polarity and mitosis (reviewed in Bornens, 2012; Angus and Griffiths, 2013; Sakakibara et al., 2013; Elric and Etienne-Manneville, 2014; Bettencourt-Dias et al., 2011). Centrosomes consist of two major components, the core centriole pair and the pericentriolar material (PCM), both identifiable by light and electron microscopy. Centrioles are composed of 9 triplet microtubules (MTs) and many associated proteins arranged as a barrel (Figure 1B). Centrioles play many critical roles, such as ensuring proper centrosome duplication, serving as an anchor and organizer of PCM, and functioning as the basal body responsible for nucleating cilia and flagella. The PCM surrounds the centrioles and is the region from which MTs are nucleated and anchored (Figure 1A, B). While the PCM appears relatively unstructured by EM, super-resolution microscopy has revealed some amount of spatial order to several PCM proteins (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012).
Figure 1. The centrosome is the major MTOC of many cells.
A) Immnuofluorescent micrograph of an interphase Drosophila S2 cell showing the centrosome (green) organizing the minus ends of microtubules (red). B) Schematic of a centrosome, showing the centriole, procentriole, PCM and MTs.
The protein composition of centrosomes is increasingly well understood. Multiple groups have succeeded in purifying the entire organelle from cells and have revealed several hundred proteins associated with the centrosome (Andersen et al., 2003; Jakobsen et al., 2011; Muller et al., 2010). As is the case with other large molecular assemblies, a detailed understanding of the function of any centrosome protein must include an understanding of how it physically interacts with others. Only with this level of understanding can one begin to probe the true consequences of perturbing protein function by mutagenesis. In this chapter, we will discuss some of the challenges to studying the protein-protein interactions of the centrosome. We will then discuss the benefits of studying these interactions by Y2H and describe how to carry out an array-based screen to identify the interactions within a collection of centrosome proteins. Finally, we will discuss how the information gained from this method of Y2H screening can guide further experimentation, including the generation of highly specific, separation of function mutants using a reverse-Y2H approach. In combination with other powerful genetic, biochemical and cell biological approaches, these Y2H techniques should help propel our understanding of the biology of the centrosome.
1.1 Identifying direct protein interaction have had a profound impact on understanding centrosome function
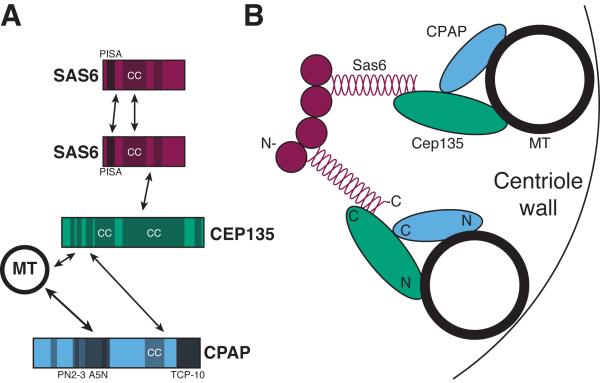
A few direct interactions among centrosome proteins have been successfully identified and the conclusions drawn from these studies have truly advanced our understanding of centrosome biogenesis. A particularly insightful set of interactions are those identified among the core centriole proteins, Sas6, STIL/Ana2/Sas5, Cep135/Bld10 and CPAP/Sas4 (Figure 2A). For this set of proteins the addition of direct protein-protein interaction data to the genetic and structural information has begun to crystalize a view of the centriole architecture. The interaction between Sas6 and STIL/Ana2/Sas5, which in some systems is regulated by the master centriole duplication kinase Plk4, is likely one of the earliest events in the construction of a new centriole, termed a procentriole (Leidel et al., 2005; Dzhindzhev et al., 2014; Ohta et al., 2014). The interactions that Sas6, and its Chlamydomonas reinhardtii ortholog Bld12, can make with itself seem likely to help establish the stereotypic centriole symmetry. Sas6 homodimerizes via its C-terminal tails and oligomerizes via its globular heads. Together, these interactions drive the formation of higher order structures that likely help establish the 9-fold radial symmetry of the procentriole's cartwheel (van Breugel et al., 2011; Kitagawa et al., 2011). In this higher order structure, the C-termini of 9 Sas6 dimers radiate out from a central hub (Figure 2B, two of nine Sas6 dimers are shown). The C-terminal end of Sas6 can interact with Cep135, which in turn, interacts with CPAP/Sas4. Since both Cep135 and CPAP/Sas4 can interact with MTs, an attractive model is that these interactions link the spokes of the Sas6 cartwheel to the MTs of the centriole wall, thus connecting the 9-fold symmetry of Sas6 tails to the triplet MTs (Lin et al., 2013; Hiraki et al., 2007; Roque et al., 2012). Therefore, the identification of direct interactions, in combination with other approaches, has helped shape this fundamental model of the centriole core.
Figure 2. The interactions among core centriole proteins dictate the organization of the structure.
A) Known direct protein-protein interactions among core centriole proteins. Sas6 is included twice to illustrate its homotypic interactions. Shaded regions denote known or predicted protein motifs, PISA (present in Sas6), CC (coiled-coil), PN2-3 (a MT destabilizing motif), A5N (a MT binding motif) and TCP-10 (T complex protein 10). B) Cartoon of the proteins that compose 2 of the 9 arms of the cartwheel of a centriole and how they might interact to connect the symmetry of Sas6 to the MTs of the centriole. Approximate locations of the amino-terminal (N) and carboxy-terminal (C) ends of the proteins are indicated. See section 1.2 for details and references.
Interactions between centrosome proteins have offered insight into other centrosomal processes, including regulation of centriole duplication (Dzhindzhev et al., 2014; Hatch et al., 2010; Ohta et al., 2014; Kim et al., 2013; Sonnen et al., 2013) and centriole length control (Spektor et al., 2007). Insight provided from these interactions bodes extremely well for the success of future endeavors to define more interactions among centrosome proteins.
1.2 Challenges to understanding protein-protein interactions in the centrosome
As illustrated by the examples above, understanding how centrosomes are assembled, regulated and perform their cellular functions will require a detailed understanding of how its proteins physically relate to each other. Loss-of-function and other genetic studies in vivo have been extremely fruitful in identifying proteins critical for major aspects of centrosome biology, such as centriole duplication and MTOC activity. In fact, much of our understanding of the initial steps of centriole duplication stems from pioneering genetic work in Caenorhabditis elegans (Dammermann et al., 2004; Delattre et al., 2004; Kemp et al., 2004; Leidel and Gonczy, 2003; Leidel et al., 2005; O'Connell et al., 2001; Pelletier et al., 2006) and later from RNAi based screens in cultured cells (Balestra et al., 2013; Dobbelaere et al., 2008; Goshima et al., 2007). However, these genetic studies have only provided limited insight into the interconnectedness of the centrosome. Some of this stems from the fact that disrupting many of the most important proteins leads to loss of the entire organelle, making it difficult to assess how the absence of one protein impacts the behaviors of others.
Understanding the nature of the protein-protein interactions within the centrosome, which are critical for its function, is challenging for a number of reasons. One major obstacle is the centrosome's size – it is a micron-scale, supra-molecular machine. It consists of hundreds of proteins, 50–100 of which are important for its centriole and MTOC functions. This predicts a very large number of possible interactions. Investigating each in a pairwise fashion using biochemical methods such as in vitro binding assays of purified components would be prohibitively laborious. Furthermore, it is becoming more evident that centrosome proteins are an unusually challenging class of proteins to study in vitro. More than half of the residues in human centrosome proteins are predicted to be in disordered structures and many are predicted to be part of coiled-coils (Dos Santos et al., 2013). It is possible that many proteins only adopt a stable state in the context of their binding partners and/or the local environment of the centrosome. In a high-throughput screen for soluble centrosome proteins, only 32.5% of centrosome proteins were soluble in lysates of E. coli expressing recombinant forms of the proteins. This success rate was not improved when only using putative globular domains (Dos Santos et al., 2013). These unusual characteristics of centrosome protein may contribute to the reason why only a few centrosome proteins have been purified for crystallography to date, all of which are relatively small portions of the protein. Furthermore, several have required complex, or unconventional procedures to ensure protein behavior (Cottee et al., 2013; Qiao et al., 2012; van Breugel et al., 2011; Zheng et al., 2014). The need for such protein specific treatment, although fruitful as in the case of the core centriole components, is a low-throughput approach and is not conducive to elucidating the large numbers of interaction that occur in the centrosome. Furthermore, many techniques for studying interactions in vitro are biased towards high-affinity interactions (Bruckner et al., 2009) and could miss lower-affinity interactions that might be critical for centrosome protein dynamics, especially within the PCM. As an alternative to using purified component, one can probe centrosome protein interactions in vivo using a co-immunoprecipitation (Co-IP) approach. However, Co-IP experiments of any centrosome protein leave open the possibility that the detected interaction is not direct. Thus, such an approach should be paired up with other methods such as direct in vitro binding assays (if possible) or a Y2H analysis as discussed below.
2. Dissection of complex, multi-component protein machines using Y2H
Given how little we know about the protein connections within the centrosome, there is a great need for a method that would uncover all the potential interactions in a quick and high-throughput manner. Here, we suggest that a Y2H approach is part of the solution – serving as a critical first step to solving the complex interactions within the centriole and PCM.
2.1 General principle of the Y2H
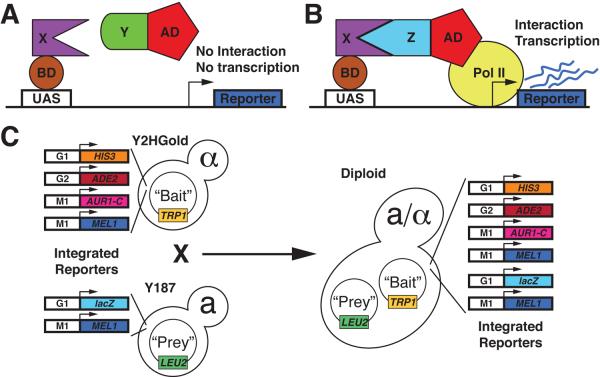
The Y2H system was originally described by Fields and Song (Fields and Song, 1989). The original system relied on the fact that the DNA-binding domain (BD) and transcriptional activation domain (AD) of the S. cerevisiae transcription factor GAL4 can be separated. When separated neither can drive transcription from Gal4 responsive promoters on their own. However, a functional transcription factor can be reconstituted and drive transcription when a protein fused to a BD interacts with a protein fused to an AD. In turn, reporter gene activity allows one to infer a direct interaction between the proteins fused to the AD and BD (Figure 3A, B). Since its introduction, many modifications and variations of the Y2H system have been developed to make it easier to use, improve its efficacy, etc., but the general principle of Y2H systems remains the same.
Figure 3. Schematic of the Y2H system.
A) Protein X is fused to the Gal4 BD. Protein Y is fused to the Gal4 AD. Since protein X does not interact with protein Y, no transcription is driven from the reporter gene. B) Protein X is fused to the Gal4 BD Protein Z is fused to the Gal4 AD. Since protein X and Z interact, a functional transcription factor is generated and reporter gene transcription occurs. C) Diagram of the mating based Matchmaker Gold Y2H system (Clonetech) showing the integrated reporter genes in the indicated yeast strains.
2.2 Advantages of the Y2H system for complex, multi-component protein machines
Testing interactions by the Y2H system has several advantages. Y2H requires little specialized equipment. Any lab capable of growing yeast cultures can perform it in its simplest forms. It does not require proteins to be purified to perform the assay, an advantage for studying challenging proteins, like those of the centrosome. The test for interaction by Y2H occurs in the nucleus of the yeast cells, so unless one is studying a protein normally native to the yeast nucleus, there is a reduced likelihood of detecting indirect interactions. Finally, with some modifications, Y2H is amenable to being used in a high-throughput setting, allowing a protein of interest to be tested for interactions with many proteins.
The Y2H system is well suited for studying complex multi-protein systems, because of its modularity and ease of use. In fact, several groups have used the Y2H to elucidate the interactions among proteins of whole organisms from viruses to yeast (reviewed in Roberts et al., 2012). In Drosophila melanogaster, our model system of choice, three large scale Y2H-based interactomes have been generated (Giot et al., 2003; Stanyon et al., 2004; Formstecher et al., 2005). However, a surprisingly small number of interactions between centrosomal proteins have been identified. Below, we discuss a possible explanation for this low number.
In addition to genome-scale interaction screens, targeted Y2H screens have been critical for understanding the interactions that occur within multi-protein assemblies involved in a variety of cellular processes. Of significant interest to those who study the centrosome are the interactions determined among proteins of complex structures related in form or function to the centrosome. For example, a large proportion of centrosome proteins are predicted to form coiled-coils (Dos Santos et al., 2013). Therefore, studies that used a Y2H system to map coiled-coil protein interactions in Saccharomyces cerevisiae could be very insightful (Newman et al., 2000; Wang et al., 2012). Interestingly these screens identified a number of interactions among the proteins of the kinetochore, responsible for connecting MTs to the chromosomes during mitosis, and also among the proteins of spindle pole body (SPB), the functional equivalent of the centrosome in yeast. Like the centrosome, both the kinetochore and the SPB are coiled-coil rich organelles involved in regulating MT attachments. The protein-protein interactions of the kinetochore (Shang et al., 2003; Ikeuchi et al., 2003; Ikeuchi et al., 2010; Wong et al., 2007) and the SPB (reviewed in Schramm et al., 2001; Adams and Kilmartin, 1999; Elliott et al., 1999; Schramm et al., 2000) have been extensively probed by Y2H, leading to important discoveries about protein function, as well as providing critical information for understanding the role of the larger protein assembly. In Drosophila, the Y2H system has been successfully used to uncover extensive direct interactions between the PCM proteins Cnn, Asl and Spd2 (Conduit et al., 2014). Extrapolating from this work, we predicts a massive number of centrosome interactions have yet to be discovered; investigating the importance of the Cnn-Asl-Spd2 and other interactions will be a challenging, yet exciting endeavor.
2.3 Limitations of the Y2H system
Like all approaches, there are several limitations to the Y2H method. The system generates false negatives that arise for a variety of reasons. In some cases the interaction between test proteins might place the BD and AD in a spatial configuration where they cannot form a functional unit, for example due to steric hindrances. False negatives also arise when one or both of the test proteins are not present in the yeast nucleus due to protein instability or its failure to enter the nucleus. Finally, many protein-protein interactions are regulated by post-translational modification. These modifications will likely be absent in an Y2H assay and interactions that require them will be missed (reviewed in Rajagopala and Uetz, 2009; Bruckner et al., 2009)).
The system can also produce false positives. Some proteins, when fused to either the AD or BD can activate transcription in the absence of an interaction partner. Fortunately, as discussed below, these proteins can be easily identified and the results discarded. Several approaches have been utilized to further reduce the number of false positive interactions that rely on increasing the stringency of the interaction. A widely used method is the addition of 3-AT, a competitive inhibitor of HIS3, to the yeast growth media. Increasing concentrations of 3-AT increases the amount of HIS3 that must be produced to support growth, thus reducing the background of spurious false positives (Durfee et al., 1993; Fields, 1993). Another method to reduce false positives is to use multiple reporters under the control of different promoters. The ability of the interaction to initiate transcription on multiple promoters increases the confidence in the detected interaction (Rajagopala and Uetz, 2009). As discussed below, we have found the use of multiple reporters greatly reduces the positive interaction hit rate. However, the inability to activate transcription on one promoter does not mean the interaction is necessarily false. Therefore, in the protocol described below, we suggest screening for interactions using different combinations of reporters. Testing an interaction using multiple stringencies can provide several layers of information to help eliminate false positives, while not missing weak, but significant interactions. We also recommend screening against a selection of non-centrosomal proteins, especially coiled-coil proteins, to serve as negative controls; this can help eliminate false positives due to “sticky” proteins. We stress here that the Y2H should be viewed as an initial guide to further secondary experimentation that could support, or refute a direct interaction. In the case of the centrosome, we strongly believe that the Y2H is a necessary first step because of the obstacles mentioned above. Once a Y2H interaction is identified, the proteins of interest should be tested for interaction in vitro (if possible) using purified components, or in vivo using Co-IPs. These studies can then guide genetic studies in vivo to truly show functional relevance of interactions. High standards and experimental rigor should naturally eliminate false positives.
3. Performing an array based screen for centrosome protein interactions
In this chapter we will describe how to generate an arrayed library of centrosome proteins using protein sub-fragments, and how to then screen a single protein or fragment against the entire library. However, the principles described can easily be modified to scale down to testing single interactions, or scaled up with automation to test a larger set of proteins. While we use the proteins of the centrosome as an example, it is by no means limited to this organelle. Our approach should be useful to examine the interactions among any interconnected protein complex. To allow for the greatest degree of flexibility, we assume the reader has already generated their own list of proteins from which they wish to construct their library.
3.1 Selecting a Y2H system
Since its original development, many variations on the principle of the Y2H system have been developed, utilizing different fusion proteins, different reporters and different host cell types. We direct the reader to a comprehensive review of Y2H systems that includes detailed discussion of their advantages and disadvantages (Bruckner et al., 2009). Some thought should be given to the system selected prior to beginning the screen. Of particular note is the finding that different systems used to probe the same set of proteins did not yield a completely overlapping set of interactions (Rajagopala et al., 2009; Stanyon et al., 2004). This work also highlights that an interaction might only be detected when protein X is used as a bait and protein Y is used as a prey, but not vice versa. This is consistent with our experience as well. We therefore test as many protein-protein pairs as possible in both directions. We describe a procedure to test interactions using an approach centered on the Matchmaker Gold system (Figure 3C, Clonetech, Mountain View, CA), but this protocol could be easily modified to accommodate other Y2H systems.
Since we are specifically interested in identifying how one centrosome protein can interact with other centrosome proteins, and not how it can interact with any protein per se, screening using an array-based Y2H system is perfectly suited for this application. In this type of screen, a collection of clones of known sequence is constructed in Y2H vectors. Bait and prey plasmids are separately transformed into yeast of opposite mating types. A collection of yeast strains of one mating type, each carrying a plasmid encoding a unique centrosome protein, are systematically arrayed in a manageable format, like that of a multiwell plate. Then, yeast of the opposite mating type, carrying the test Y2H vector, are mixed with the entire array. Yeast mating then brings the bait and prey plasmids together in the same diploid yeast cell. As a result each colony on the array contains a unique pair of proteins, which can be tested for interaction using reporter genes carried by the parental strains (reviewed in Uetz, 2002; Rajagopala and Uetz, 2009; Cagney and Uetz, 2001). This method accommodates testing a large number of combinations of potential interacting proteins in a systematic fashion and, as such, is ideally suited to understanding the interactions among proteins of a complex, an organelle and even a whole genome.
There are several advantages to performing a Y2H screen in an array-based format. Arrays allow for the immediate identification of the interacting proteins based on their position in the array. Unlike random library screening, plasmids from positive interactions do not need to be recovered and sequenced. This system can also be automated, allowing large sets of proteins to be systematically assayed. Finally, the array based format reduces the occurrence of certain types of false positives and makes the elimination of others easier due to the ease of retesting the interaction via a simple yeast mating (Reviewed in Uetz, 2002).
3.2 Dividing proteins into smaller fragments
Several large scale Y2H studies have indicated that it is advantageous to examine sub-regions of proteins in addition to full-length proteins (Flajolet et al., 2000; Fromont-Racine et al., 2000; Boxem et al., 2008; Flores et al., 1999; Formstecher et al., 2005). There are several advantages to including smaller protein fragments in a Y2H screen. One is that it can separate regions of proteins problematic for use in Y2H from the remainder of the protein. This might include regions that contain elements toxic to the yeast or that can activate transcription of the Y2H reporter genes in the absence of an interaction (auto-activation). When separated, regions of the protein that do not have these issues can still be used to test for interactions. Another advantage is that sub-fragments might uncover interactions not revealed by full-length proteins. The studies referenced above indicate that some full-length proteins, while expressed, do not yield interactions, while sub-fragments of the same proteins do. Our experience confirms this observation. A fragment might outperform full-length proteins for a number of reasons. For example, full-length proteins might not fold correctly in yeast, precluding proper interaction with its partner. It is also possible that the BD or AD may be placed in a position causing steric hindrance to protein-protein interaction or to transcriptional activation. The failure of full-length proteins to interact may also be reflective of their biology. For example, many proteins are regulated via intra-molecular auto-inhibition mechanisms that restrict access to portions of themselves until a specific cellular event occurs. Thus, dividing a protein into smaller fragments might alleviate this auto-inhibition and expose a necessary binding domain or surface. The use of full-length proteins might be a factor contributing to the limited number of centrosomal protein interactions identified in genome wide Y2H screens mentioned above. We therefore recommend that proteins be assayed for interaction as both full-length and as small protein fragments, if possible.
We suggest a rational, structure-based (existing or predicted) approach to subdividing proteins prior to use in Y2H screens. For each centrosome protein we first determined if any structures of the protein has been solved. In the absence of existing structural information, we perform secondary and tertiary protein structure predictions utilizing two available structure prediction servers, Jpred3 and Phyre2, (Cole et al., 2008; Kelley and Sternberg, 2009). We then screen the protein for known structural or functional motifs using the SMART web server (Letunic et al., 2014). Finally, since centrosome proteins are rich in sequences predicted to participate in the formation of coiled-coils, we use the COILS web server to predict such regions (Lupas et al., 1991). With this information in hand we divide these proteins into smaller fragments with the least disruption to the above features. As an alternative, several groups referenced above describe screening protocols where a protein of interest is screened against a collection of protein fragments that have been randomly generated prior to screening.
3.3 Generating the Y2H library
Commercial Y2H systems provide vectors that contain multiple cloning sites allowing for restriction enzyme based cloning. To reduce the labor in producing an array of protein fragments, bait and prey vectors modified to accommodate cloning techniques more conducive for use in high throughput circumstances can be used. One such modification was to make the Y2H vectors pGBKT7 and pGADT7 compatible with the Gateway cloning system (Rossignol et al., 2007); Life Technologies, Grand Island, NY). Our lab has further modified the Gateway compatible pGBKT7 vector by replacing the kanamycin resistance cassette with one providing resistance against ampicillin so that it could be used with Gateway Entry clones (Galletta et al., 2014). Sequences encoding the fragments should be generated by PCR and then cloned into Entry vectors. After verification by DNA sequencing, Gateway recombination reactions are performed to transfer these sequences into bait (pGBKT7) and prey (pGADT7) vectors. Other cloning systems can also be used, such as plasmid construction by homologous recombination in yeast.
As discussed above, bait and prey plasmids carried in yeast of opposite mating type are used to introduce pairs of proteins into the same yeast by mating. For this procedure, bait plasmids (pGBKT7) are transformed into the Y2HGold yeast strain, a MATα strain, and prey (pGADT7) into the Y187 yeast strain, a MATa strain. Single colonies of each are selected, propagated and stocks of each bait in Y2HGold and each prey in Y187 are generated.
3.4 Autoactivation and false positive rate identification
A common limitation to testing protein interactions by Y2H is that some protein fragments, when introduced into the system, can activate the Y2H reporters in the absence of any binding partner. While this is more commonly a problem with fragments fused to the GAL4-BD (bait), this can occur in GAL4-AD (prey) fusions as well (Serebriiskii and Golemis, 2001). Prior to use in testing interactions, all strains carrying Y2H vectors should be tested for autoactivation by first generating “empty strains” (Prey-empty = Y187 containing empty pGADT7; Bait-empty = Y2HGold containing empty pGBKT7). Then, all newly generated stocks should be crossed to these `empty stains' and tested for reporter activity as described below. If the process is performed in a more high-throughput fashion, these `empty stains' should be placed into the array. Plates showing a positive signal in these control wells can be eliminated from the analysis. Protein fragments that cause autoactivation cannot be used in interaction testing. It is unlikely, however, that a protein will autoactivate as both prey and bait, and it is also unlikely that all subfragments of the same proteins cause autoactivation. Therefore, in a properly designed screen, little information is lost or missed due to autoactivation events. If this proves to be a major problem, we suggest repeating the process by designing, generating and testing new protein subfragments. Another useful control is to test for interactions against random, non-centrosomal proteins, to serve as control for particularly sticky proteins. Since so many proteins in the centrosome contain coiled-coils and disordered regions, proteins with these types of regions should be included.
3.5 Pre-screen planning
Depending on the number of interactions that one will test, screening using an array Y2H based system will involve growing a large number of yeast strains, moving and mixing a large number of cultures in a very specific order, dealing with many yeast plates and keeping track of a large dataset on the backend. Prescreen planning to ensure the availability of the correct amount of media, number of tubes and plates, etc. can help ensure the fewest missteps on the day of the screen. Tables 1 and 2 list the yeast media and equipment required for the screen we describe.
Table 1.
Yeast media recipes
| SD media base | 5 g ammonium sulfate, 3.4 g yeast nitrogen base without amino acids and ammonium sulfate, 20g D-glucose, appropriate DO mix, water to 1 L. Add 20 g agar / L for plates. |
| Complete amino acid (CAA) mix | 800 mg adenine, 2 g arginine, 800 mg histidine, 2 g isoleucine, 4 g leucine, 2 g lysine, 800 mg methionine, 2 g phenylalanine, 8 g threonine, 2 g tryptophan, 2 g tyrosine, 800 mg uracil, 6 g valine |
| -leu drop out (DO) mix | CAA, omit leucine – use 730 mg / L |
| -trp DO mix | CAA, omit tryptophan – use 780 mg / L |
| -leu -trp DO mix | CAA, omit leucine and tryptophan – use 680 mg / L |
| -ade - leu -trp –ura DO mix | CAA, omit adenine, leucine, tryptophan and uracil – use 640 mg / ml |
| SD –leu | SD media base, –leu DO mix |
| SD –trp | SD media base, –trp DO mix |
| DDO | SD media base, –leu –trp DO mix |
| QDO | SD media base, –ade –leu -trp –ura DO mix |
| DDOXA | SD –leu –trp DO mix, Aureobasidin A (Clonetech, Mountain View, CA), X-α-Gal Clonetech, Mountain View, CA and Gold Biotechnology, St. Louis, MO) |
| QDOXA | SD –ade –leu -trp –ura DO mix, Aureobasidin A, X-α-Gal |
| 2X YPAD | 20 g tryptone, 10 g yeast extract, 80 mg adenine, 20 g D-glucose, water to 1L |
Table 2.
Materials needed
| Item | Commercial Example (Company – Catolog #) |
|---|---|
| Culture tubes | Falcon - 352059 |
| 96 well flat bottom dishes | Olympus – 25–104 |
| 96 deep well plate | Axygen – P-2ML-SQ-C-S |
| Reagent reservoirs | Corning – 4870 |
| Multichannel pipettor | Gilson - F14404 |
| 48 pin Multi-Blot Replicator | V&P Scientific – VP 407AH |
| Petri dishes | Fisher Scientific – FB0875712 |
| Replica-plating tool | Bel Art Products- 378480000 |
| Sterile velvet squares | Bel Art Products- 378480002 |
Some thought should also be given to the design of the array. We recommend including blank wells on each half of the 96 well array, in different positions, to aid in plate orientation (see below). We also recommend including a well for each of the `empty stains' to function as negative controls on each plate; they will serve as a readout of any unexpected autoactivation that might occur on the day of the experiment. Finally, this type of screen will yield a large data set that must be tracked and organized. We recommend building a database, using software such as Filemaker Pro (FileMaker, Inc., Santa Clara, CA) or Access (Microsoft, Redmond, WA), to track all the data.
3.6 Screening for interactions
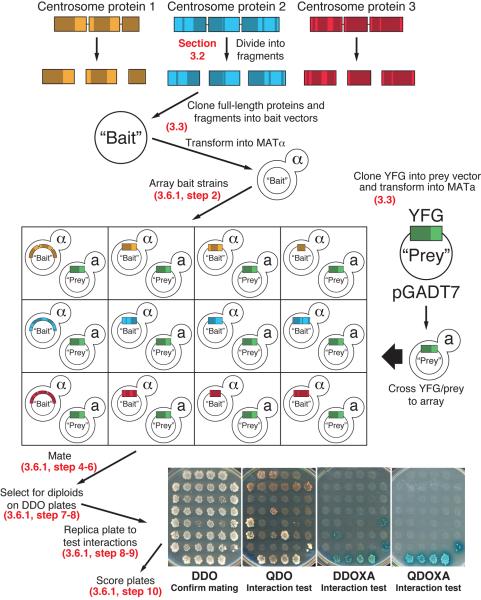
Here we describe the details for how to screen a single protein or protein fragment, against the collection of centrosome proteins generated above. As discussed, some protein-protein interactions only occur when protein X serves as the bait and protein Y as the prey, and not vice versa. Therefore, one must test any protein of interest against a bait array and a prey array to maximize the number of interactions identified. For simplicity, we will describe testing the centrosome protein encoded by Your Favorite Gene in a prey plasmid (YFG-prey) against a collection of strains carrying bait plasmids encoding a variety of centrosome proteins in an array format (ARRAY-bait). However, we strongly suggest that the tests also be done in the opposite direction (YFG-bait against ARRAY-prey). Figure 4 diagrams the steps in the screening procedure.
Figure 4. Workflow of an array based Y2H screen for centrosome protein interactions.
Refer to sections indicated on the figure for details describing each step.
3.6.1 Protocol
-
1)
Grow fresh cultures of all yeast strains to be tested. Inoculate liquid cultures of yeast carrying Y2H plasmids for the array (ARRAY-bait), as well as for the protein or fragment to be tested (YFG-prey), at 30°C with shaking in SD –leu media or SD -trp media, as appropriate to maintain plasmid selection. This can be done in individual culture tubes or directly in a 96 well format using a deep well plate, although the latter may not be optimal for yeast growth. Grow to OD600 ≈ 0.5. Some strains may grow faster than others. Generally this takes 1 – 3 days. It may be usefully to estimate that growth rate of the strains prior to starting. Then the time of growth for individual strains can be adjusted so that all strains reach the desired OD600 at approximately the same time.
-
2)
Array the ARRAY-bait cultures by transferring 20 μl of each into a single well of a 96-well, flat bottom plate. If more than one YFG-prey strain is to be tested against the array, it is useful to set up the ARRAY-bait in a master plate (using a deep well, 96-well plate if necessary) and then use a multi-channel pipette to transfer the array to multiple, identical ARRAY-bait plates.
-
3)
In a sterile reagent reservoir, mix 2 ml of YFG-prey culture with 10 ml of 2X YPAD media.
-
4)
Using a multichannel pipette, transfer 120 μl of the YFG-prey / 2X YPAD mixture into each well of the 96-well ARRAY-bait plate. Mix by pipetting up and down a few times. This is now referred to as the Mating-plate.
-
5)
Repeat steps 3 – 4 until all YFG-prey samples have been crossed with the ARRAY-bait.
-
6)
Grow Mating-plates for 20 – 24 hours at 30°C with shaking to allow the yeast to mate. The success of the mating reaction can be assayed by examining a small sample of the culture for the presence of zygotes by phase contrast microscopy, although this is usually not necessary.
-
7)
Transfer approximately 3 μl of each mating culture from the Mating-plate onto DDO plates. This can be facilitated using a 48 pin Multi-Blot Replicator (VP 407AH, V&P Scientific, San Diego, CA). In this case, the cultures from one 96 well Mating-plate are transferred as two 48-sample halves to each of two DDO plates. These plates will select for growth of diploids that have received both the bait and prey plasmids from their parents. Parental haploids that have failed to mate will not grow on this media.
Sterilize the replicator before each use by immersing the pins into a dish of ethanol or isopropanol. Gently shake off excess and place the pins in the flame of a Bunsen burner. Allow the pins to cool. Introduce the replicator into one half of the 96 well Mating-plate and swirl it in the media to ensure the yeast is evenly suspended. Remove the replicator from the Mating-plate, taking care not to touch the sides of the wells. Gently set the replicator down onto the surface of a DDO plate, taking care to not let the replicator slide laterally. Lift the replicator off the plate, leaving ~ 3 μl of culture behind. Place the replicator back in the dish with alcohol. Repeat for the other half of the 96 well Mating-plate. Mark each DDO plate so that the orientation relative to the array can be determined. These plates will be referred to as Diploid-plates. Repeat for all Mating-plates.
-
8)
Allow the yeast on Diploid-plates to grow for 3 – 5 days at 30°C until robust patches of yeast are seen on the plate, as in Figure 4.
-
9)
Replica each Diploid-plate onto DDO, QDO, DDOXA and QDOXA plates, all labeled to match the orientation of the Diploid-plate. To replica, place a sterile velvet cloth onto the replica plating tool and secure with the ring. Press the surface of the Diploid-plate onto the velvet, with the top of the array facing away from you. Remove the Diploid-plate. Press each of the fresh plates onto the velvet and remove to make a copy. These new DDO, QDO, DDOXA and QDOXA plates will be referred to as Test-plates. Repeat for all Diploid-plates.
-
9)
Grow Test-plates for 5 days at 30°C.
-
10)
Test-plates can now be scored to determine if any of the proteins in the array interact with YFG. Score each patch independently for its growth on each of the Test-plates. We have found it useful to score the result of protein pair on each test plate on a scale of 0 – 3, where 0 = no growth, 1 = minimal growth/color, 2 = moderate growth/color, and 3 = robust growth/color. The plates are scored as follows.
DDO – Media lacks leucine and tryptophan, which selects diploids carrying both bait and prey plasmids. Ensures that replica plating was successful at all positions.
QDO (2 growth interaction reporters) – Scored for growth. Media lacks leucine and tryptophan, which maintains selection for the bait and prey plasmids. Growth on this media, which lacks histidine and adenine indicates activation of the HIS3 and ADE2 Y2H reporters respectively and indicates a bait/prey interaction.
DDOXA (2 drug interaction reporters) – Scored for growth and development of blue colony color. Media lacks leucine and tryptophan, which maintains selection for the bait and prey plasmids. Growth on this media, which contains the antibiotic agent Aureobasidin A indicates activation of the AUR1-C Y2H reporter. Development of a blue color on this media, which contains X-α-Gal indicates activation of the MEL1 Y2H reporter. Activation of both these reporters indicates a bait/prey interaction.
QDOXA (2 growth interaction reporters, 2 drug reporters) - Scored for growth and development of blue colony color. Media lacks leucine and tryptophan, which maintains selection for the bait and prey plasmids. This media lacks histidine and adenine, and contains Aureobasidin A and X-α-Gal. Growth and development of the blue color requires activation of the ADE2, HIS3, AUR1-C and MEL1 Y2H reporters and indicates an interaction under the most stringent conditions.
3.7 Interpreting screening results
As discussed above, the yeast strains used in this Y2H system carry multiple reporters driven by different promoters. Each of these reporters should have subtle differences in the false positives they yield and when used in combination they should reduce the incidence of false positives. The plates used in the protocol test for activity of these reporters in different combinations. QDO plates are similar to the plates used historically in many yeast two hybrids screens. We have found that these plates show a much greater number of interactions than the other plates. In our experience, of the centrosomal protein pairs that show an interaction on QDO, only about 60% of these pairs show growth on DDOXA and only 50% show growth on QDOXA (Galletta and Rusan, unpublished observation). This is consistent with an increased stringency with additional promoters and likely a significant elimination of false positives.
Once the initial screen is scored, all pairs showing an interaction should be retested by taking the original yeast stocks and preforming small scale mating assays to validate positive interactions. This simple retesting will eliminate a significant number of false positives (Rajagopala and Uetz, 2009; Uetz, 2002). The interactions identified can then be used in combination with biochemical, cellular biological and other approaches to truly determine protein function. One particularly powerful use of the information gained in this type of screen is to guide a genetic approach to identify mutations to disrupt specific protein-protein interactions.
4. Generating specific, separation of function mutations by reverse Y2H
Mutations are powerful tools for elucidating protein function. Even more powerful are mutations that specifically disrupt the interaction between a protein and only one of its binding partners. It is critical to note that any mutation, even a single point mutation, has the potential to disrupt more than one interaction. This is especially a concern in a complex, multi-protein structure like the centrosome, which is highly interconnected. However, with the knowledge obtained from the interaction studies described in the previous section it is possible to generate mutations that disrupt specific subsets of interactions, and possibly exclusively a single interaction. In this section we describe how to generate such a mutant by a reverse-Y2H approach.
4.1 Rationale
The reverse two-hybrid approach used here is based on the method described by Bennett et al. (Bennett et al., 2004) with significant modifications. This method utilizes low-fidelity PCR to introduce random mutations into DNA encoding a protein of interest. The mutagenized DNA is then cloned into the Y2H vectors directly in the Y2H strains by homologous recombination mediated repair. These mutant alleles can then be screened to identify ones that disrupt a known interactor.
The major modification we have made is to adapt the procedure for use in a mating-based, arrayed format. Similar to Bennett et al. (Bennett et al., 2004), we generate random mutations in the sequence encoding YFG by low-fidelity PCR and use homologous recombination mediated repair to clone the mutated YFG fragments. However, instead of cotransforming the mutated-YFG with a plasmid encoding the interaction partner being tested against, we perform the recombination in a haploid Y2H strain without its interaction counterpart. The YFG mutants are then clonally collected and put into an array. Once the YFG mutant array is generated, it can be tested for the loss of interactions by mating the array to Y2H strains carrying plasmids encoding the interacting protein of interest to identify mutations that abolish the interaction.
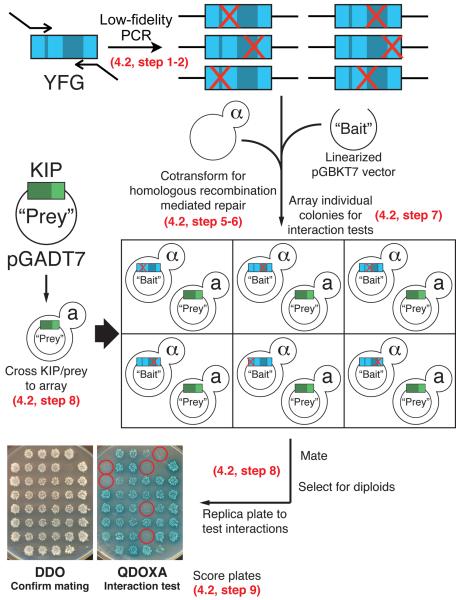
Performing the screen in the fashion described below has several advantages over cotransforming random mutants with their interaction partner. Most significantly, to ensure that the generated mutation only disrupts a specific protein-protein interaction of interest, a candidate clone can easily be pulled from the master array and tested for its ability to interact with all interaction partners. There is no need to first isolate the mutant plasmid from yeast, a labor-intensive process. This secondary screen allows the investigator to eliminate non-specific mutants simply by performing additional yeast matings. The investigator would only recover the few mutants that fit the desired criteria. This method saves a significant amount of time and effort. A workflow diagram of the mutagenesis and screen is found in Figure 5.
Figure 5. Workflow of an array based reverse Y2H screen to generate and identify mutations that disrupt protein-protein interactions.
Refer to sections indicated on the figure for details describing each step. Red X's represent random point mutations introduced by mutagenic PCR. Yeast colonies carrying mutations that disrupt the protein-protein interaction are circled in red.
4.2 Generating mutant library and screening for loss of interaction
To facilitate the use of this system with any protein or fragment of interest we have designed universal primers that allow amplification from the Y2H vectors (pGADT7 and pGBKT7) generated in section 3.3 above (Table 3). PCR products of putative YFG mutants are cloned by co-transforming them into the Y2H yeast strains with linearized Y2H vectors and then selecting for the plasmid. For simplicity, we describe mutagenizing Your Favorite Gene (YFG) and cloning it into the bait vector (pGBKT7) in the bait Y2H strain (Y2HGold). An array of YFG mutants is then mated to a Known Interacting Protein (KIP) in a prey vector (pGADT7) in the prey strain (Y187) and screened to identify mutations that disrupt the YFG/KIP interaction. While we describe mutagenizing a bait and testing it against a prey, this process works equally well when mutagenizing the prey. Simply replace the primer “pGBKT7 Mut” with “pGADT7 Mut” listed in Table 3 for amplification and switch to the appropriate plasmids and yeast hosts.
Table 3.
Primers for low fidelity PCR
| Primer name | Sequence |
|---|---|
| T7 Sequencing | 5'- TAATACGACTCACTATAGGGCG-3' |
| pGBKT7 Mut | 5'- CGGAATTAGCTTGGCTGC - 3' |
| pGADT7 Mut | 5'- ATGGTGCACGATGCACAG - 3' |
The mutagenic PCR we describe generates a mutation approximately every 250 base pair. If mutations are desired more or less frequently, we direct the reader to studies focusing on low-fidelity PCR (Cadwell and Joyce, 1992; Wilson and Keefe, 2001).
4.2.1 Protocol
- Mutagenic PCR mix:
- Taq polymerase
- 1X Taq polymerase buffer (supplied buffer by the manufacturer)
- 0.05 mM MnCl2
- 0.06 mM dATP
- 0.25 mM dCTP
- 0.25 mM dGTP
- 0.25 mM dTTP
- YFG in pGBKT7 (from section 3.3) – PCR template
- T7 Sequencing Primer
- pGBKT7 Mut Primer
- The following conditions for PCR were used for the pGBKT7 primers to amplify a product of ~1 kb. Adjust conditions as necessary.
-
1)95°C 2 minutes
-
2)95°C 30 seconds
-
3)54°C 30 seconds
-
4)72°C 1 minute
-
5)Repeat 2 – 4 for 30 cycles.
-
1)
Gel purify mutant YFG PCR product.
Linearize pGBKT7 by restriction co-digestion with EcoRI and PstI. (If mutagenizing prey clones, pGADT7 can be linearized by co-digesting with EcoRI and XhoI.) Gel purify linearized vector to ensure there is no uncut plasmid present, as any will increase the background of clones that appear to lose interaction.
Co-transform equimolar amounts of the mutant YFG PCR product with the linearized pGBKT7 vector, for a total of 0.5 – 1 μg DNA, into Y2HGold. The exact amount of DNA needed will have to be determined empirically to yield optimal results. The goal is to find amounts that yield a plate full of colonies with adequate separation to allow individual colonies to be picked.
Plate on SD –trp plates to select for repaired plasmids containing mutant versions of YFG.
-
Load a 96 well plate with 100 μl / well SD – trp liquid media. Inoculate individual colonies from the plate in step 6 into each well. Each well now contains a unique mutant version of YFG in pGBKT7 in Y2HGold. Grow at 30°C with shaking for 1–2 days until OD600 = ~0.5. Pin a copy of the array onto solid SD – trp media, as in section 3.6.1, Step 7; this will serve as a duplicate of the array.
Note: The number of mutant colonies that must be screened to identify a desired mutant cannot be known a priori and must be determined empirically. We have found that the number can vary dramatically. In some cases we have identified the desired mutation after screening only a few hundred mutants, others have taken several thousand, and others we have never been able to generate.
Transfer the YFG mutants from step 7 into an array as described in section 3.6.1 above. Follow steps 2 – 9 above and mate the array with KIP in pGADT7.
Score plates. Ensure the presence of both the YFG mutant plasmid and the KIP plasmid by growth on DDO. Score colonies for interactions using QDO, DDOXA and QDOXA plates. In this application, the experimenter is looking for colonies that grow on DDO, but show reduced or no growth on QDO, DDOXA and QDOXA plates as compared to the results of unmutaginzed YFG and KIP.
Recover all clones that displayed a loss of the YFG / KIP interaction from the YFG mutant array. Retest these against KIP. This should help eliminate certain types of false hits.
Screen the selected mutant YFG clones from step 10 for their ability to interact with all other proteins known to interact with YFG. This is accomplished by crossing the YFG mutant clones to the known interactors in pGADT7 in Y187 generated in the original screen and testing them as described above.
Once a clone harboring the desired mutation is found the insert contained in the clone can be recovered by performing colony PCR (Sambrook and Russell, 2006; Sathe et al., 1991) using the same primers used for mutagenesis. This PCR product can be sequenced to identify the mutations it harbors. These mutations can be engineered back into the sequence encoding the fragment to confirm they are causative of the loss of interaction.
5. Summary
Many important cellular functions rely on large, multi-protein assemblies. In order to truly understand the function of these complexes, and the functions of their constituent parts, an understanding of the connections among these proteins is critical. This is especially true for the centrosome, which as a non-membrane bound organelle is, in many respects, a truly enormous and highly interconnected protein complex. As discussed above, there are numerous challenges to understanding of the protein-protein interactions within a complex like the centrosome. The Y2H system is a powerful tool for probing direct protein-protein interactions within complexes. It allows the experimenter to identify interactions within the structure that might not be accessible using other techniques, such as low-affinity and transient interactions.
On the simplest level, interaction information can provide an understanding of how the proteins of the complex fit together. But beyond this, interaction information can be critical to direct experiments to probe function. Mutagenesis is one of the most powerful tools used to understand protein function in the cell. However, multi-protein complexes present special challenges to interpreting the results of these studies. The potential interconnectedness means that complete loss-of-function mutations might alter many protein-protein interactions within the complex. However, once the interactions within a complex are known, the consequence of a given mutation on all of the interactions can be assessed. This can allow increased confidence that the consequence of a mutation is the result of disruption of a specific interaction. Interaction data can also guide reconstitution and structure determination experiments. As discussed above, the proteins of the centrosome are frequently challenging to working with in vitro. Proteins functioning in complexes can sometimes not fold correctly in the absence of their partners. The Y2H data from a screen like the one described will provide an excellent resource to identify pairs or groups of proteins with which to begin co-purifying proteins together with their partners. The data from this screen could also help identify interactions critical for building a structure, but not necessarily present in the final structure. In addition, Y2H of fragments can also reveal regulated interactions, for example interactions involving domains that are masked in the context of the full-length protein until some event uncovers them. Finally, Y2H screens can reveal interactions that are important in certain contexts that are not important in others. For example it might identify interactions that take place in mitotic centrosomes, but not in interphase centrosomes; or in basal bodies, but not in centrioles.
Y2H screens like this have the potential to yield large sets of interaction data. This leaves the task of understanding how the interactions we identify contribute to how the centrosome assembles and performs its functions. This will certainly involve integrating Y2H interaction data with data generated by other techniques. For example, combining Y2H interaction data with proximity-mapping techniques, like the BirA biotin ligase system (Firat-Karalar et al., 2014), and crosslinking studies, like those done by S-CROSS (Lukinavicius et al., 2013), will be a powerful approach to probe the landscape of protein assemblies within the centrosome. We also envision the Y2H method playing an important role in understanding the growing number of human congenital disease linked to mutations in centrosome and cilia proteins. Testing these mutants against arrays of centrosomal proteins might reveal the interactions altered by these mutations. This type of detailed interaction information allows for the generation of separation of function mutations that only affect a single protein-protein interaction. This combination has the promise of becoming a powerful reverse genetic approach to understand the molecular details of protein function in the centrosome.
Acknowledgements
We thank Alexander Kelly and Colleen Skau for critical reading of the manuscript. NMR is supported by the division of intramural research at the National Institutes of Health / NHLBI (1ZIAHL006104).
References
- Adams IR, Kilmartin JV. Localization of core spindle pole body (spb) components during spb duplication in saccharomyces cerevisiae. J Cell Biol. 1999;145:809–23. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–4. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Angus KL, Griffiths GM. Cell polarisation and the immunological synapse. Curr Opin Cell Biol. 2013;25:85–91. doi: 10.1016/j.ceb.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestra FR, Strnad P, Fluckiger I, Gonczy P. Discovering regulators of centriole biogenesis through sirna-based functional genomics in human cells. Dev Cell. 2013;25:555–71. doi: 10.1016/j.devcel.2013.05.016. [DOI] [PubMed] [Google Scholar]
- Bennett MA, Shern JF, Kahn RA. Reverse two-hybrid techniques in the yeast saccharomyces cerevisiae. Methods Mol Biol. 2004;261:313–26. doi: 10.1385/1-59259-762-9:313. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–15. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. The centrosome in cells and organisms. Science. 2012;335:422–6. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul JD, van de Peut D, Devos M, Simonis N, Yildirim MA, Cokol M, Kao HL, de Smet AS, Wang H, Schlaitz AL, Hao T, Milstein S, Fan C, Tipsword M, Drew K, Galli M, Rhrissorrakrai K, Drechsel D, Koller D, Roth FP, Iakoucheva LM, Dunker AK, Bonneau R, Gunsalus KC, Hill DE, Piano F, Tavernier J, van den Heuvel S, Hyman AA, Vidal M. A protein domain-based interactome network for c. Elegans early embryogenesis. Cell. 2008;134:534–45. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner A, Polge C, Lentze N, Auerbach D, Schlattner U. Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci. 2009;10:2763–88. doi: 10.3390/ijms10062763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell RC, Joyce GF. Randomization of genes by pcr mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Cagney G, Uetz P. High-throughput screening for protein-protein interactions using yeast two-hybrid arrays. Current protocols in protein science. 2001;Chapter 19(Unit 19.6) doi: 10.1002/0471140864.ps1906s24. [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Richens JH, Wainman A, Holder J, Vicente CC, Pratt MB, Dix CI, Novak ZA, Dobbie IM, Schermelleh L, Raff JW. A molecular mechanism of mitotic centrosome assembly in drosophila. Elife. 2014;3:e03399. doi: 10.7554/eLife.03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee MA, Muschalik N, Wong YL, Johnson CM, Johnson S, Andreeva A, Oegema K, Lea SM, Raff JW, van Breugel M. Crystal structures of the cpap/stil complex reveal its role in centriole assembly and human microcephaly. Elife. 2013;2:e01071. doi: 10.7554/eLife.01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–29. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gonczy P. Centriolar sas-5 is required for centrosome duplication in c. Elegans. Nat Cell Biol. 2004;6:656–64. doi: 10.1038/ncb1146. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Josue F, Suijkerbuijk S, Baum B, Tapon N, Raff J. A genome-wide rnai screen to dissect centriole duplication and centrosome maturation in drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos HG, Abia D, Janowski R, Mortuza G, Bertero MG, Boutin M, Guarin N, Mendez-Giraldez R, Nunez A, Pedrero JG, Redondo P, Sanz M, Speroni S, Teichert F, Bruix M, Carazo JM, Gonzalez C, Reina J, Valpuesta JM, Vernos I, Zabala JC, Montoya G, Coll M, Bastolla U, Serrano L. Structure and non-structure of centrosomal proteins. PLoS One. 2013;8:e62633. doi: 10.1371/journal.pone.0062633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–69. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Tzolovsky G, Lipinszki Z, Schneider S, Lattao R, Fu J, Debski J, Dadlez M, Glover DM. Plk4 phosphorylates ana2 to trigger sas6 recruitment and procentriole formation. Curr Biol. 2014 doi: 10.1016/j.cub.2014.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S, Knop M, Schlenstedt G, Schiebel E. Spc29P is a component of the spc110p subcomplex and is essential for spindle pole body duplication. Proc Natl Acad Sci U S A. 1999;96:6205–10. doi: 10.1073/pnas.96.11.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elric J, Etienne-Manneville S. Centrosome positioning in polarized cells: Common themes and variations. Exp Cell Res. 2014 doi: 10.1016/j.yexcr.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–6. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fields S. The two-hybrid system to detect protein-protein interactions. Methods. 1993;5:116–24. [Google Scholar]
- Firat-Karalar EN, Rauniyar N, Yates J.R.r., Stearns T. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol. 2014 doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajolet M, Rotondo G, Daviet L, Bergametti F, Inchauspe G, Tiollais P, Transy C, Legrain P. A genomic approach of the hepatitis c virus generates a protein interaction map. Gene. 2000;242:369–79. doi: 10.1016/s0378-1119(99)00511-9. [DOI] [PubMed] [Google Scholar]
- Flores A, Briand JF, Gadal O, Andrau JC, Rubbi L, Van Mullem V, Boschiero C, Goussot M, Marck C, Carles C, Thuriaux P, Sentenac A, Werner M. A protein-protein interaction map of yeast RNA polymerase iii. Proc Natl Acad Sci U S A. 1999;96:7815–20. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, Jacq B, Arpin M, Bellaiche Y, Bellusci S, Benaroch P, Bornens M, Chanet R, Chavrier P, Delattre O, Doye V, Fehon R, Faye G, Galli T, Girault JA, Goud B, de Gunzburg J, Johannes L, Junier MP, Mirouse V, Mukherjee A, Papadopoulo D, Perez F, Plessis A, Rosse C, Saule S, Stoppa-Lyonnet D, Vincent A, White M, Legrain P, Wojcik J, Camonis J, Daviet L. Protein interaction mapping: A drosophila case study. Genome Res. 2005;15:376–84. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Mayes AE, Brunet-Simon A, Rain JC, Colley A, Dix I, Decourty L, Joly N, Ricard F, Beggs JD, Legrain P. Genome-wide protein interaction screens reveal functional networks involving sm-like proteins. Yeast. 2000;17:95–110. doi: 10.1002/1097-0061(20000630)17:2<95::AID-YEA16>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open biology. 2012;2:120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta BJ, Guillen RX, Fagerstrom CJ, Brownlee CW, Lerit DA, Megraw TL, Rogers GC, Rusan NM. Drosophila pericentrin requires interaction with calmodulin for its function at centrosomes and neuronal basal bodies but not at sperm basal bodies. Mol Biol Cell. 2014;25:2682–94. doi: 10.1091/mbc.E13-10-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RLJ, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of drosophila melanogaster. Science. 2003;302:1727–36. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in drosophila s2 cells. Science. 2007;316:417–21. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–9. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10P constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:1778–83. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Ikeuchi A, Nakano H, Kamiya T, Yamane T, Kawarasaki Y. A method for reverse interactome analysis: High-resolution mapping of interdomain interaction network in dam1 complex and its specific disorganization based on the interaction domain expression. Biotechnol Prog. 2010;26:945–53. doi: 10.1002/btpr.403. [DOI] [PubMed] [Google Scholar]
- Ikeuchi A, Sasaki Y, Kawarasaki Y, Yamane T. Exhaustive identification of interaction domains using a high-throughput method based on two-hybrid screening and pcr-convergence: Molecular dissection of a kinetochore subunit spc34p. Nucleic Acids Res. 2003;31:6953–62. doi: 10.1093/nar/gkg888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, Uhlen M, Hyman AA, Andersen JS. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. The EMBO journal. 2011;30:1520–35. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the web: A case study using the phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF. Centrosome maturation and duplication in c. Elegans require the coiled-coil protein spd-2. Dev Cell. 2004;6:511–23. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- Kim TS, Park JE, Shukla A, Choi S, Murugan RN, Lee JH, Ahn M, Rhee K, Bang JK, Kim BY, Loncarek J, Erikson RL, Lee KS. Hierarchical recruitment of plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, cep192 and cep152. Proc Natl Acad Sci U S A. 2013;110:E4849–57. doi: 10.1073/pnas.1319656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Fluckiger I, Gonczy P, Steinmetz MO. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–75. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol. 2012;14:1148–58. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. Sas-6 defines a protein family required for centrosome duplication in c. Elegans and in human cells. Nat Cell Biol. 2005;7:115–25. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Leidel S, Gonczy P. Sas-4 is essential for centrosome duplication in c elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4:431–9. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. Smart: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Chang CW, Hsu WB, Tang CJ, Lin YN, Chou EJ, Wu CT, Tang TK. Human microcephaly protein cep135 binds to hsas-6 and cpap, and is required for centriole assembly. EMBO J. 2013;32:1141–54. doi: 10.1038/emboj.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavicius G, Lavogina D, Orpinell M, Umezawa K, Reymond L, Garin N, Gonczy P, Johnsson K. Selective chemical crosslinking reveals a cep57-cep63-cep152 centrosomal complex. Curr Biol. 2013;23:265–70. doi: 10.1016/j.cub.2012.12.030. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–4. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol. 2012;14:1159–68. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Schmidt D, Steinbrink S, Mirgorodskaya E, Lehmann V, Habermann K, Dreher F, Gustavsson N, Kessler T, Lehrach H, Herwig R, Gobom J, Ploubidou A, Boutros M, Lange BM. Proteomic and functional analysis of the mitotic drosophila centrosome. The EMBO journal. 2010;29:3344–57. doi: 10.1038/emboj.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Wolf E, Kim PS. A computationally directed screen identifying interacting coiled coils from saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:13203–8. doi: 10.1073/pnas.97.24.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The c. Elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–58. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, Inagaki M, Oyama M, Kitagawa D. Direct interaction of plk4 with stil ensures formation of a single procentriole per parental centriole. Nat Commun. 2014;5:5267. doi: 10.1038/ncomms6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O'Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in caenorhabditis elegans. Nature. 2006;444:619–23. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Qiao R, Cabral G, Lettman MM, Dammermann A, Dong G. Sas-6 coiled-coil structure and interaction with sas-5 suggest a regulatory mechanism in c. Elegans centriole assembly. EMBO J. 2012;31:4334–47. doi: 10.1038/emboj.2012.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopala SV, Hughes KT, Uetz P. Benchmarking yeast two-hybrid systems using the interactions of bacterial motility proteins. Proteomics. 2009;9:5296–302. doi: 10.1002/pmic.200900282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopala SV, Uetz P. Analysis of protein-protein interactions using array-based yeast two-hybrid screens. Methods in molecular biology. 2009;548:223–45. doi: 10.1007/978-1-59745-540-4_13. [DOI] [PubMed] [Google Scholar]
- Roberts GG, 3rd., Parrish JR, Mangiola BA, Finley RL., Jr. High-throughput yeast two-hybrid screening. Methods Mol Biol. 2012;812:39–61. doi: 10.1007/978-1-61779-455-1_3. [DOI] [PubMed] [Google Scholar]
- Roque H, Wainman A, Richens J, Kozyrska K, Franz A, Raff JW. Drosophila cep135/bld10 maintains proper centriole structure but is dispensable for cartwheel formation. J Cell Sci. 2012;125:5881–6. doi: 10.1242/jcs.113506. [DOI] [PubMed] [Google Scholar]
- Rossignol P, Collier S, Bush M, Shaw P, Doonan JH. Arabidopsis pot1a interacts with tert-v(i8), an n-terminal splicing variant of telomerase. Journal of cell science. 2007;120:3678–87. doi: 10.1242/jcs.004119. [DOI] [PubMed] [Google Scholar]
- Sakakibara A, Ando R, Sapir T, Tanaka T. Microtubule dynamics in neuronal morphogenesis. Open Biol. 2013;3:130061. doi: 10.1098/rsob.130061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Analyzing yeast colonies by pcr. CSH Protoc. 2006;2006 doi: 10.1101/pdb.prot4015. [DOI] [PubMed] [Google Scholar]
- Sathe GM, O'Brien S, McLaughlin MM, Watson F, Livi GP. Use of polymerase chain reaction for rapid detection of gene insertions in whole yeast cells. Nucleic Acids Res. 1991;19:4775. doi: 10.1093/nar/19.17.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm C, Elliott S, Shevchenko A, Schiebel E. The bbp1p-mps2p complex connects the spb to the nuclear envelope and is essential for spb duplication. EMBO J. 2000;19:421–33. doi: 10.1093/emboj/19.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm C, Janke C, Schiebel E. Molecular dissection of yeast spindle pole bodies by two hybrid, in vitro binding, and co-purification. Methods Cell Biol. 2001;67:71–94. doi: 10.1016/s0091-679x(01)67006-7. [DOI] [PubMed] [Google Scholar]
- Serebriiskii IG, Golemis EA. Two-hybrid system and false positives. Approaches to detection and elimination. Methods Mol Biol. 2001;177:123–34. doi: 10.1385/1-59259-210-4:123. [DOI] [PubMed] [Google Scholar]
- Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, Barnes G. Kinetochore protein interactions and their regulation by the aurora kinase ipl1p. Mol Biol Cell. 2003;14:3342–55. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Gabryjonczyk AM, Anselm E, Stierhof YD, Nigg EA. Human cep192 and cep152 cooperate in plk4 recruitment and centriole duplication. J Cell Sci. 2013;126:3223–33. doi: 10.1242/jcs.129502. [DOI] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3d-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1:965–76. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and cp110 suppress a cilia assembly program. Cell. 2007;130:678–90. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Stanyon CA, Liu G, Mangiola BA, Patel N, Giot L, Kuang B, Zhang H, Zhong J, Finley RLJ. A drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P. Two-hybrid arrays. Current opinion in chemical biology. 2002;6:57–62. doi: 10.1016/s1367-5931(01)00288-5. [DOI] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa H-, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong I-O, Robinson CV, Johnson CM, Veprintsev D, Zuber B. Structures of sas-6 suggest its organization in centrioles. Science (New York, NY) 2011;331:1196–9. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang X, Zhang H, Lu Y, Huang H, Dong X, Chen J, Dong J, Yang X, Hang H, Jiang T. Coiled-coil networking shapes cell molecular machinery. Mol Biol Cell. 2012;23:3911–22. doi: 10.1091/mbc.E12-05-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DS, Keefe AD. Random mutagenesis by pcr. Curr Protoc Mol Biol. 2001;Chapter 8(Unit8.3) doi: 10.1002/0471142727.mb0803s51. [DOI] [PubMed] [Google Scholar]
- Wong J, Nakajima Y, Westermann S, Shang C, Kang JS, Goodner C, Houshmand P, Fields S, Chan CS, Drubin D, Barnes G, Hazbun T. A protein interaction map of the mitotic spindle. Mol Biol Cell. 2007;18:3800–9. doi: 10.1091/mbc.E07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Gooi LM, Wason A, Gabriel E, Mehrjardi NZ, Yang Q, Zhang X, Debec A, Basiri ML, Avidor-Reiss T, Pozniakovsky A, Poser I, Saric T, Hyman AA, Li H, Gopalakrishnan J. Conserved tcp domain of sas-4/cpap is essential for pericentriolar material tethering during centrosome biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E354–63. doi: 10.1073/pnas.1317535111. [DOI] [PMC free article] [PubMed] [Google Scholar]