Abstract
Phosphorus is an essential nutrient for all plants, but also one of the least mobile, and consequently least available, in the soil. Plants have evolved a series of molecular, metabolic and developmental adaptations to increase the acquisition of phosphorus and to maximize the efficiency of use within the plant. In Arabidopsis (Arabidopsis thaliana), the AtPHO1 protein regulates and facilitates the distribution of phosphorus. To investigate the role of PHO1 proteins in maize (Zea mays), the B73 reference genome was searched for homologous sequences, and four genes identified that were designated ZmPho1;1, ZmPho1;2a, ZmPho1;2b and ZmPho1;3. ZmPho1;2a and ZmPho1;2b are the most similar to AtPHO1, and represent candidate co-orthologs that we hypothesize to have been retained following whole genome duplication. Evidence was obtained for the production of natural anti-sense transcripts associated with both ZmPho1;2a and ZmPho1;2b, suggesting the possibility of regulatory crosstalk between paralogs. To characterize functional divergence between ZmPho1;2a and ZmPho1;2b, a program of transposon mutagenesis was initiated using the Ac/Ds system, and, here, we report the generation of novel alleles of ZmPho1;2a and ZmPho1;2b.
Introduction
Phosphorus (P) is an essential nutrient for all plants and a limitation on productivity in many agricultural systems [1]. Current levels of agricultural phosphorus inputs are recognized to be both unsustainable and environmentally undesirable [2]. Rational strategies to improve P efficiency in agricultural systems demand a greater understanding of P relations in crops, both in terms of P uptake from the soil and P translocation and use within the plant.
The protein PHO1 has been characterized in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) to play a key role both in the export of inorganic P (Pi) to the xylem apoplast for translocation [3] and in the modulation of long-distance signals underlying the P-deficiency response [4]. The Arabidopsis Atpho1 mutant hypo-accumulates P in the shoots and displays associated symptoms of phosphate deficiency, including reduced growth rate, thinner stalks, smaller leaves, very few secondary inflorescences, delayed flowering and elevated levels of anthocyanin accumulation [3]. In rice, disruption of OsPHO1;2, the ortholog of AtPHO1, results in a phenotype similar to that of the Atpho1 mutant [5], suggesting that the two genes are functionally equivalent. Indeed, expression of OsPHO1;2 in the Atpho1 background was found to partially complement the mutant phenotype [6]. A feature distinguishing the rice OsPHO1;2 gene from its Arabidopsis ortholog is the P-regulated production of a cis-Natural Antisense Transcript (cis-NATOsPHO1;2) [5] [7] which acts as a translational enhancer [8].
PHO1 proteins contain two conserved domains: the N-terminal hydrophilic SPX domain (named for the yeast proteins Syg1 and Pho81, and the human Xpr1) and the C-terminal hydrophobic EXS domain (named for the yeast proteins ERD1 and Syg1 and the mammalian Xpr1) [9]. The SPX domain is subdivided into three well-conserved sub-domains, separated from each other by regions of low conservation [7]. SPX domain containing proteins are key players in a number of processes involved in P homeostasis, including fine tuning of Pi transport and signaling by physical interactions with other proteins [7]. E Following the SPX domain there are a series of putative membrane-spanning α-helices that extend into the C-terminal EXS domain [9]. In AtPHO1, the EXS domain is crucial for protein localization to the Golgi/trans-Golgi network and for Pi export activity, as well as playing a role in the modulation of long-distance root-to-shoot signaling under P limitation [4].
Despite the importance of maize as a staple crop and the dependence of maize production on large-scale input of phosphate fertilizers, the molecular components of maize P uptake and translocation remain poorly characterized [10]. Although it has been possible to identify maize sequences homologous to known P-related genes from other species, functional assignment has been based largely on patterns of transcript accumulation. With the development of accessible public-sector resources, it is now feasible to conduct reverse genetic analyses in maize. Here, we extend the molecular characterization of maize P response by generating mutant alleles of maize Pho1 genes using endogenous Activator/Dissociation (Ac/Ds) transposable elements. The (Ac/Ds) system consists of autonomous Ac elements that encode a transposase (TPase) and non-autonomous Ds elements that are typically derived from Ac elements by mutations within the TPase gene. Lacking TPase, Ds elements are stable, unless mobilized by TPase supplied in trans by an Ac. Ac/Ds elements move via a cut-and-paste mechanism [11], with a preference for transposition to linked sites [12] that makes the system ideal for local mutagenesis [13]. To exploit the system for reverse genetics, Ac and Ds elements have been distributed throughout the genome and placed on the maize physical map, providing potential “launch pads” or mutagenesis of nearby genes [14] [15].
In this study, we identify four maize Pho1 genes in the maize (var. B73) genome, including two (ZmPho1;2a and ZmPho1;2b) that we consider co-orthologs of AtPHO1. Structure of the ZmPho1;2a and ZmPho1;2b genes was confirmed experimentally, and accumulation of transcripts characterized in the roots and shoots of seedlings grown in P-replete or P-limiting conditions. Novel insertional alleles of ZmPho1;2a and ZmPho1;2b are reported, generated using the Ac/Ds transposon system. Availability of mutant alleles will be central in determining the functional role of ZmPho1;2a and ZmPho1;2b in maize.
Materials and Methods
Identification of maize Pho1 genes
The AtPHO1 cDNA sequence (GenBank ID: AF474076.1) was used to search the maize working gene set peptide database (www.maizesequence.org) in a BLASTX search performed under the default parameters. Identified maize sequences were in turn used to reciprocally search Arabidopsis thaliana (www.phytozome.net). Four sequences were identified encoding proteins with a high level of similarity to AtPHO1: GRMZM5G891944 (chr 3:28,919,073-28,923,871); GRMZM2G466545 (chr 4:171,946,555-171,952,268); a sequence split into the four adjacent gene models GRMZM5G801969,GRMZM5G851655, GRMZM5G815128 and GRMZM5G872499 (chr 5:215,110,603-215,115,635) and GRMZM2G064657 (chr 6:122,577,593-122,582,074). The putative protein sequences were confirmed to contain canonical PHO1 domain structure by PFam analysis (pfam.sanger.ac.uk) and NCBI conserved domains search (www.ncbi.nlm.nih.gov). Additional gene models in the region of Pho1;2 and their putative orthology relationships were obtained from Ensembl plants (www.plants.ensembl.org).
Amplification of full length Pho1;2 cDNAs
Total RNA was extracted using Trizol-chloroform from the roots of 10-day-old B73 seedlings grown under phosphate limiting conditions (sand substrate; fertilized with modified P-free Hoagland solution [16]). 1μg of RNA was used to synthesize cDNA with oligo(dT) primer and SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) in a reaction volume of 20μl. PCR amplification of full length Pho1;2 cDNAs was performed using using Platinum Taq High Fidelity DNA Polymerase (Invitrogen) under the following cycling conditions: initial incubation at 95°C for 3min, followed by 35 cycles of 94°C for 30sec, 61°C for 30sec and 68°C for 30sec, final extension at 68°C for 5min. Primers used are shown in Table 1. Confirmed gene models were submitted to www.maizegdb.org.
Table 1. Primers used in this study for CDS amplification and transcript accumulation.
| Target | ID | Primer 5’-3’ | Description |
|---|---|---|---|
| Pho1;2a | A | MS103- TCGCGGAGGATGGTGAAGTTCT | Full CDS (2463bp) |
| H | MS062- GTGTGTCCATTCCTGGAACTCT | ||
| F | MS002- AGGTGGCCATGAAGTACCTG | Exon 6,7,8,9 and 10 (734bp/1130bp) | |
| G | MS056- CCTGCATTGCTCTCCAGTAGTAA | ||
| Pho1;2b | I | MS013- ATCCCACGATGGTGAAGTTCT | Full CDS (2263bp) |
| N | MS077- ACAATTCTCAATCGACCACTAGC | ||
| L | RS138- AACTCCGTCTCGGTATGGTGGAGTCT | Exon 13, 14 and 3’UTR (383bp/475bp) | |
| M | RS139- TGACCAGAACGCCTCATGTTATACCC | ||
| SbPho1;2 | Q | MS105- CATGGACTGGGGCTTCTTAAAC | Exon 13, 14 and 15(293bp/475bp) |
| R | MS106- GTAATGGGACAGTCTTCACTGCT | ||
| cis-NATPho1;2a | B | MS121- GAGCATCCTGATTCCATATCTACC | Flanking putative exon (478bp) |
| E | MS120- CCCGTAATGGAAGCTTTTACTG | ||
| cis-NATPho1;2b | J | MS131- CGTACTGCTGATCGCATGCATA | Flanking putative exon (544bp) |
| K | MS132- AGTACGTGATCAGTGATCTACACTC | ||
| cis-NATSbPho1;2 | O | MS162- CTGATCGCTGACAGATGGCCATA | Flanking putative exon (475bp) |
| P | MS158- ACCAGCATCCAGCACCCAAAC | ||
| Nested cis-NAT | C | MS155- GGAGGTGATGGCGGCGCTGGA | Putative exon (263bp(Pho1;2a/b), |
| D | MS163- TCTCGGCGTGCTGGACCTTCTT | 266bp(SbPho1;2)) | |
| ZmUBQ/SbUBQ | FWD- CTACAACATTCAGAAGGAGAGCAC | ||
| REV- TCTGCAAGGGTACGGCCATCC | |||
| ZmCDK | FWD- GGAAGGTATGCACAGGACAGAT | ||
| REV- TTCAGCACAATCTTGGCAAAAC |
Analysis of Pho1;2 transcript accumulation
Total RNA was extracted using Trizol-Chloroform from the roots of 10-day-old B73 seedlings grown under P replete (sand substrate; plants fertilized with complete Hoagland solution containing 1mM concentration of PO4) or P limiting (sand substrate; fertilized with modified P-free Hoagland solution). cDNA was synthesized as described above. PCR amplification of sense genes was performed using the primers MS002-MS056 (Pho1;2a), RS138-RS139 (Pho1;2b) and MS105-MS106 (SbPho1;2) under the following cycling conditions: initial incubation at 95°C for 5min, followed by 32 cycles of 95°C for 30sec, 63°C for 30sec and 72°C for 1min, final extension 72°C for 5min. PCR amplification of cis-NATs was performed with the primers MS121-MS120 (cis-NATPho1;2a), MS131-MS132 (cis-NATPho1;2b) and MS162-MS158 (cis-NATSbPho1;2) under the following cycling conditions: initial incubation at 95°C for 5min, followed by 38 cycles of 95°C for 30sec, 59°C for 30sec and 72°C for 30sec, final extension 72°C for 5min. Products from the primary PCR reaction were diluted 1:100,000 and used as template for nested PCR with MS155-MS163 primers under the following conditions: initial incubation at 95°C for 5min, followed by 25 cycles of 95°C for 30sec, 58°C for 30sec and 72°C for 15sec, final extension 72°C for 5min. Primers are shown in Table 1. Maize and sorghum poly-ubiquitin (GRMZM2G419891/Sb04g004260) or the maize CDK (GRMZM2G149286) were used as a control, along with amplification from genomic DNA (gDNA) template, using 50ng gDNA in 20μl for 32-cycle reactions and 10ng gDNA in 20μl for 38-cycle reactions. Products were analyzed on 1.5% agarose gels. RNA sequence data for Fig B in S1 File as reported previously [17]
Transposon mutagenesis
The strategy for Ac/Ds mutagenesis was as previously described [18] [14]; [15]. Genetic stocks were maintained in the T43 background, a color-converted W22 stock carrying r1-sc::m3, a Ds6-like insertion in the r1 locus that controls anthocyanin accumulation in aleurone and scutellar tissues [19]. The frequency of purple spotting in the aluerone resulting from somatic reversion of r1-sc::m3 was used to monitor Ac activity [20]. Donor Ac and Ds stocks were selected from existing collections [14] [15]: the element bti31094::Ac is placed on the B73 physical map 650.8Kb from ZmPho1;2a; the element I.S06.1616::Ds is inserted in intron 13 of ZmPho1;2b, and was subsequently designated ZmPho1;2b-m1::Ds. To generate a testcross population for mutagenesis of ZmPho1;2a, 207 individuals homozygous for bti31094::Ac were crossed as females by T43, and rare, finely spotted progeny kernels were selected for screening. To re-mobilize the Ds element I.S06.1616::Ds within ZmPho1;2b, homozygous ZmPho1;2b-1::Ds individuals carrying the unlinked stable transposase source Ac-Immobilized (Ac-im) [21], were used as males to pollinate T43, and coarsely spotted progeny kernels were selected for screening.
To identify novel Ac/Ds insertions in ZmPho1;2a and ZmPho1;2b, selected kernels were germinated in the greenhouse and DNA was isolated from pools of 18 seedlings. The candidate gene space was explored by PCR using a range of gene-specific primers in combination with “outward-facing” Ac/Ds-end primers. All primers are listed in Table 2. PCR reactions contained 400ng gDNA and.25μM of each primer. Reactions for ZmPho1;2a were performed using Platinum Taq High Fidelity DNA Polymerase (Invitrogen) under the following cycling conditions: denaturation at 94°c for 4 min; 30 cycles of 94°c for 30 sec, 58°c for 30 sec, 68°c for 3min 30 sec; final extension at 68°c for 10 min. Reactions for ZmPho1;2b were performed using Kapa Taq DNA polymerase (Kapa Biosystems, Wilmington, Massachusetts, USA) under the following cycling conditions: denaturation at 95°c for 5 min; 35 cycles of 95°c for 30 sec, 58°c for 30 sec, 72°c for 3min 30 secs; final extension at 72°c for 5 min. Positive pools were re-analyzed as individuals, following the same cycling conditions. PCR reactions were analyzed on 1.5% agarose gels. Products from positive individuals were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany), ligated into pGEM T-easy vector (Promega, Fitchburg, Wisconsin, USA) and sequenced. Genotyping of Zmpho1;2a-m1::Ac was performed by amplification of fragments spanning the insertion with specific-Ac primers (MS124-MS052, MS124-JRS01 and MS052-JGp3; see Table 2), using Kapa Taq DNA polymerase (Kapa Biosystems), under the following cycling conditions: denaturation at 95°c for 5 min; 35 cycles of 95°c for 30 sec, 58°c for 30 sec, 72°c for 3min 30 secs; final extension at 72°c for 5 min. Somatic excision events were identified by BseYI (New England BioLabs, Ipswich, Massachusetts, USA) digest of MS124-MS052 PCR products, according to the manufacturer’s protocol.
Table 2. Primers used in this study for genotyping.
| Target | ID | Primer 5’-3’ | Description |
|---|---|---|---|
| ZmPho1;2a | MS069- ACCTTTCTACACTGCCTGTACC | 1st fragment (1244bp) | |
| MS044- GTTCGACCTACCTAACATGGACT | |||
| MS043- GTTTGTACGTACCCATGCCGTAT | 2nd fragment (1577bp) | ||
| MS048- GGAAGGGAAGTACCTTGTCAGAG | |||
| MS047- CTCCCTCAATGTGAAGGCTTT | 3rd fragment (2835bp) | ||
| MS062- GTGTGTCCATTCCTGGAACTCT | |||
| ZmPho1;2b | MS085-CTCATTTGTTTCCAGTTTCTCTCC | 1st fragment (Promoter—2981bp) | |
| MS028- AGCTAGCTACCTGACATGGACT | |||
| MS027- CAGGAGCAAGAGTTTGTGGAGA | 2nd fragment (2003bp) | ||
| MS016- GTGCTGGAGAAGTCGAAGATG | |||
| MS033- GTTCACAGGCACATTTGTGTC | 3rd fragment (2088bp) | ||
| MS077- ACAATTCTCAATCGACCACTAGC | |||
| Zmpho1;2a’-m1.1 | S | MS124- CACACTCATCATCTGAACAAAGCAAG | flanking Ac excision (421bp) |
| T | MS052- GCATCCTAATAAAGCCTGGAAGA | ||
| Ac/Ds | JGp3-ACCCGACCGGATCGTATCGG | Transposon primers | |
| JRS01-GTTCGAAATCGATCGGGATA |
To generate footprint alleles, individuals homozygous for Zmpho1;2a-m1::Ac were crossed as males to T43 females. Rare, non-spotted progeny kernels were selected and screened for excision by PCR amplification across the site of Ac insertion using primers MS124-MS052 under the following cycling conditions: denaturation at 95°c for 5 min; 35 cycles of 95°c for 30 sec, 58°c for 30 sec, 72°c for 3min 30 secs; final extension at 72°c for 5 min. PCR products from each individual were purified using the QIAquick PCR Purification Kit (Qiagen), ligated into pGEM T-easy vector (Promega) and sequenced. Ac excision was confirmed by BseYI digestion as described above.
Results
The maize genome contains four PHO1 homologs
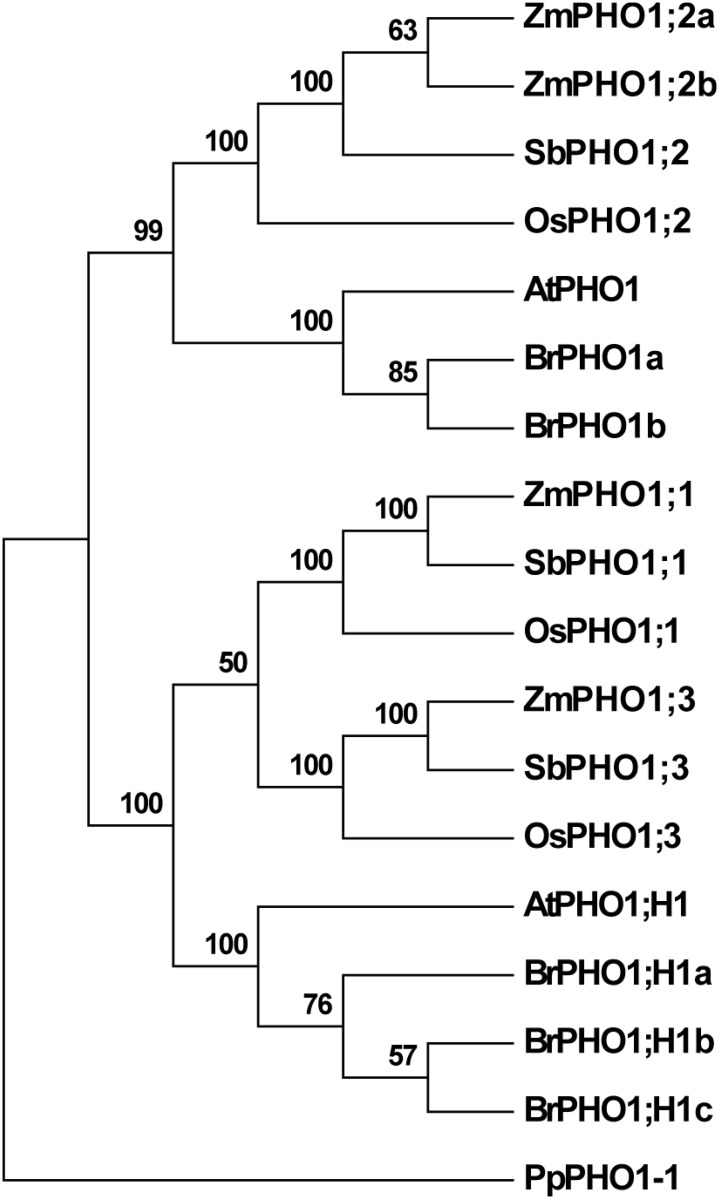
To identify maize PHO1 genes, the B73 reference genome (B73 RefGen_v3; www.maizegdb.org) was searched to identify gene models whose putative protein products exhibited a high degree of similarity to the Arabidopsis protein AtPHO1 (Table 3). Four such maize gene models were identified, and, on the basis of similarity to previously annotated rice genes [5], designated ZmPho1;1 (GRMZM5G891944), ZmPho1;2a (GRMZM2G466545), ZmPho1;2b (split across the gene models GRMZM5G801969, GRMZM5G851655, GRMZM5G815128 and GRMZM5G872499) and ZmPho1;3 (GRMZM2G064657). To investigate orthology among Arabidopsis thaliana and grass PHO1 genes, additional sequences were identified from sorghum (Sorghum bicolor) and canola (Brassica rapa), and used to generate a multiple alignment and distance tree (Fig 1).
Table 3. MaizePho1 genes and correspondingArabidopsis and rice orthologs.
| At Gene | Os Gene | Zm Gene | Zm Gene Model | Zm Position |
|---|---|---|---|---|
| AtPHO1 | OsPho1;2 | ZmPho1;2a | GRMZM2G466545 | chr 4:171,946,555-171,952,268 |
| AtPHO1 | OsPho1;2 | ZmPho1;2b | GRMZM5G801969 | chr 5:215,110,603-215,115,635 |
| GRMZM5G851655 | ||||
| GRMZM5G815128 | ||||
| GRMZM5G872499 | ||||
| AtPHO1;H1 | OsPho1;1 | ZmPho1;1 | GRMZM5G891944 | chr 3:28,919,073-28,923,871 |
| AtPHO1;H1 | OsPho1;3 | ZmPho1;3 | GRMZM2G064657 | chr 6:122,577,593-122,582,074 |
The gene ZmPho1;2b is split across four adjacent annotated gene models
Fig 1. Maximum likelihood reconstruction of the complete set of maize (Zm), sorghum (Sb) and rice (Os) PHO1 proteins along with selected PHO1 proteins of Arabidopsis (At) and canola (Br).
The moss (Physcomitrella patens) protein PpPHO1-1 is shown as an outgroup. Bootstrap support shown as a percentage at the nodes.
From Arabidopsis, only the proteins AtPHO1 and AtPHO1;H1 were included in the analysis, leaving aside a large clade of divergent functionally distinct PHO1 proteins that are specific to dicotyledonous plants [22]. The analysis supported the previously reported divergence of PHO1 and PHO1;H1 clades, dating from before the divergence of monocotyledonous and dicotyledonous plants [5, 22]. Within the PHO1;H1 clade, a duplication event was observed specific to the grasses in the analysis. As a result, the three grass species each contain two co-orthologs of AtPHO1;H1—encoded by the genes annotated Pho1;1 and Pho1;3. We observed also an expansion of the PHO1;H1 clade in canola, although this expansion is lineage specific, and there is no indication that PHO1;H1 was not encoded by a single gene at the base of this clade. The PHO1 clade itself contains the products of single-copy PHO1/PHO1;2sequences in all species in our analysis, with the exception of a lineage-specific duplication in maize. As a consequence, the paralogous maize genes ZmPho1;2a and ZmPho1;2b are considered to be co-orthologos to AtPHO1.
ZmPho1;2a and ZmPho1;2b show features of syntenic paralogs retained following to genome duplication
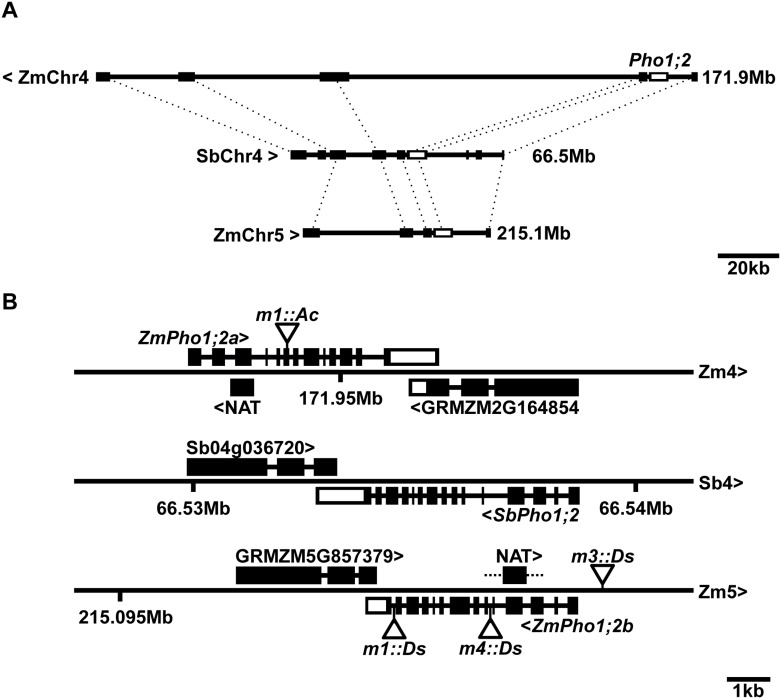
The high degree of sequence similarity (85% protein identity) between ZmPho1;2a and ZmPho1;2b suggests that they result from a recent gene duplication event. It has been hypothesized that the last whole genome duplication (WGD) event in maize occurred between 5 and 12 million-years-ago, sometime after divergence from the sorghum lineage, as the result of polyploidization [23]. The observation that Pho1;2 is a single copy sequence in both rice and sorghum is consistent with the maize duplication arising during this most recent WGD. Further inspection revealed that the two genomic regions carrying maize Pho1;2 genes are both syntenic to the sorghum region carrying SbPho1;2, and that the two maize regions have been assigned previously to distinct pre-tetraploid ancestral genomes (Chr4:168,085,162…179,711,318 to sub-genome 2; Chr5:208,925,180…217,680,842 to sub-genome 1; [23]). The return of the maize genome to a diploid organization following WGD has been accompanied by the loss of the majority of duplicate genes through a process known as fractionation [23]. In certain cases, however, pairs of syntenic paralogs have been retained. The genomic region surrounding the Pho1;2 genes exhibits a number of such candidate pairs in addition to Pho1;2a and Pho1;2b (Fig 2A), providing ample evidence of micro-synteny between the regions. In both sorghum and maize, Pho1;2 genes are adjacent to a putative WD40 protein encoding gene, present on the opposite strand and partially overlapping the annotated 3’ UTR region of the Pho1;2 sequence (Fig 2B), a feature not observed in the other maize or sorghum Pho1 paralogs.
Fig 2.
Microsynteny among Pho1;2 loci A) Annotated genes (filled boxes) in the region ofSbPho1;2 on Sorghum chromosome 4 (SbChr4) and candidate orthologs on maize (B73) chromosomes 4 (ZmChr4) and 5 (ZmChr5). Orthologous genes are connected by dashed lines. Pho1;2 genes on the three chromosomes shown in unfilled boxes. Regions shown to scale, the right hand position indicated. SbChr4 and ZmChr5 run left to right, ZmChr4 right to left. B) Pho1;2 gene models on ZmChr4, SbChr4 and ZmChr5. Exons are shown as boxes, coding regions filled, UTR unfilled. Putative anti-sense transcripts associated with ZmPho1;2a and ZmPho1;2b shown as filled boxes (NAT). Angle brackets indicate the direction of transcription. The maize syntenic paralog pair GRMZM2G164854/GRMZM5G853379 and their sorghum ortholog are also shown. Triangles indicate the position of Activator and Dissociation insertion. Refer to Table 4 for full description alleles. Shown to scale.
Transcripts encoded by ZmPho1;2a and ZmPho1;2b accumulate preferentially in the roots
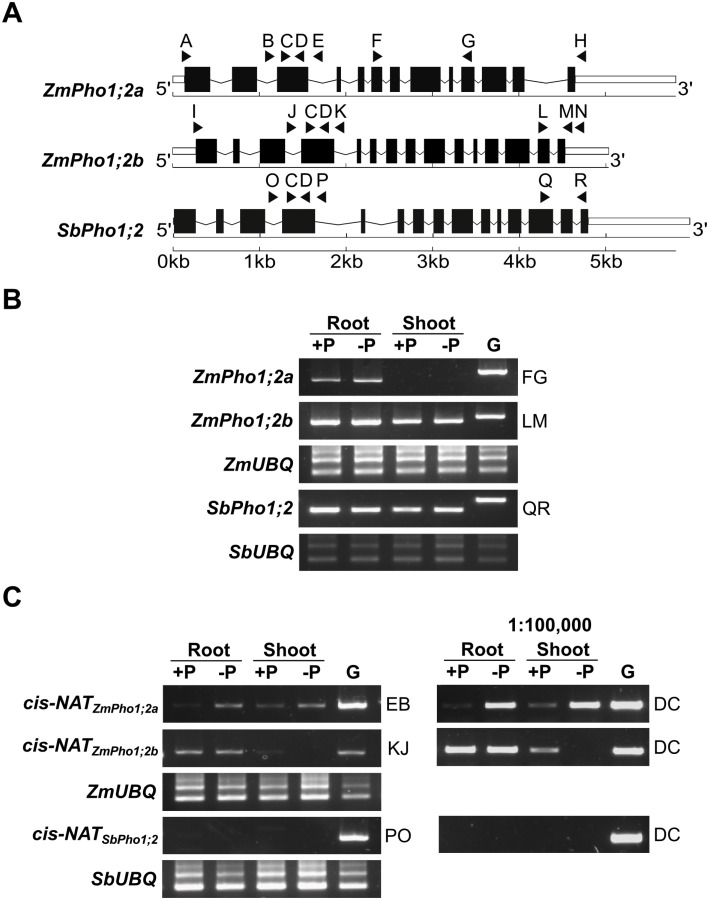
To determine the pattern of accumulation of transcripts encoded by ZmPho1;2a and ZmPho1;2b, RT-PCR was used to amplify gene-specific fragments of the two genes from cDNA generated from roots or leaves of 10-day-old seedlings (B73) grown in sand, watered with either complete (+P) or a modified P-free Hoagland solution (-P) (Fig 3A and 3B). The pattern of transcript accumulation was similar for ZmPho1;2a and ZmPho1;2b: stronger amplification in roots than shoots; no difference between P replete and P limiting conditions. The strength of amplification, however, was greater for ZmPho1;2b than ZmPho1;2a, the latter being detectable in shoots only with a high number of PCR cycles (Fig A in S1 File). The accumulation of SbPho1;2 transcripts in sorghum (BTx623) seedlings was also examined, under the same growth conditions. Transcripts of SbPho1;2 accumulated in a pattern similar to that observed for the maize genes, suggesting this pattern to be the ancestral, pre-WGD, state. To investigate further the possibility of divergence at the level of transcript abundance between ZmPho1;2a and ZmPho1;2b, we examined a previously published transcriptome data set [17]. Transcriptome data confirmed both the pattern of transcript accumulation with respect to tissue type and P availability and the difference in the level of transcript accumulation between the two maize ZmPho1;2 paralogs (Fig B in S1 File). In addition, transcriptome data indicated ZmPho1;1 transcripts to accumulate constitutively to low levels and ZmPho1;3 transcripts to accumulate preferentially in leaves rather than roots. Further gene-specific PCR primers were designed and used to amplify the complete ZmPho1;2a and ZmPho1;2b cDNAs that were sequenced to confirm the gene-model structure.
Fig 3. Accumulation of Pho1;2 sense and antisense transcripts under contrasting phosphate conditions.
(A) Schematic of Pho1;2 genes indicating the position of primers (solid triangles; see Table 1). (B) Amplification of fragments corresponding to the mature sense-RNAs of ZmPho1;2a, ZmPho1;2b and SbPho1;2 from oligo-dT primed cDNA prepared from roots and shoot of 10-day-old seedlings, fertilized with Hoagland solution adjusted to 1mM (+P) or 0mM (-P) inorganic phosphate. Primer pairs used for amplification indicated by letter codes on the right-hand side of panel. (C) Amplification of fragments corresponding to putative anti-sense transcripts encoded by ZmPho1;2a, ZmPho1;2b from cDNA as Panel A. Primary PCR (left) and nested PCR performed from 1:100,000 dilution of primary reaction (right). Amplification of ZmUBQ and SbUBQ fragments and amplification from genomic DNA template (G) were used as controls. Primer pairs indicated as in (B).
ZmPho1;2a and ZmPho1;2b are associated with phosphate-regulated putative cis-Natural Anti-sense Transcripts
Although we did not observe differential accumulation of ZmPho1;2 transcripts with respect to P availability, it has been reported that the Pho1;2 in rice is largely regulated at the post-transcriptional level by a P-regulated cis-Natural Anti-sense Transcript cis-NATOsPho1;2 [5]; [8]. The cis-NATOsPho1;2 transcript has been shown to act as a translational enhancer, and has been proposed to act by direct interaction with the sense transcript [8]. The rice cis-NATOsPho1;2 initiates in Intron 4 of OsPho1;2 and extends into the 5’ UTR region [8]. A putative ZmPho1;2a anti-sense sequence is annotated in the maize reference genome in a homologous position to the rice transcript (www.maizegdb.org), although, on the basis of cDNA evidence, the transcript is considerably shorter than cis-NATOsPho1;2, being trunctated at the 3’ end and extending only as far as Intron 2 of ZmPho1;2a (Fig 2B). No paralogous sequence has been annotated associated with ZmPho1;2b.
To investigate the presence of cis-NAT transcripts associated with ZmPho1;2a and explore the possibility that a paralogous cis-NAT transcript might be generated from ZmPho1;2b, gene-specific primers were designed to the introns flanking the homologous Exons 4 and 3 of ZmPho1;2a and ZmPho1;2b, respectively. These primers were used to attempt to amplify products from cDNA prepared from seedling root and leaves as described above. Products of the predicted size were successfully amplified using both ZmPho1;2a and ZmPho1;2b primer sets, consistent with the accumulation of cis-NATs (Fig 3B). No products were amplified from no-RT control samples (data not shown). Putative cis-NAT products were sequenced and confirmed to originate from the ZmPho1;2a and ZmPho1;2b genes. There was no evidence of the accumulation of alternatively or partially spliced transcripts during the previous amplification of full length Pho1;2 cDNAs.
The accumulation of the putative cis-NATZmPho1;2a was observed to be induced under -P conditions in both roots and leaves. In contrast, the putative cis-NATZmPho1;2b transcript showed no response to P availability in roots and reduced accumulation in shoots under -P (Fig 3B), providing evidence of functional divergence between paralogs at the level of anti-sense production. Interestingly, using the approach we employed in maize, we found no evidence of an equivalent cis-NAT associated with SbPho1;2 (Fig 3B), although additional experiments will be required to rule out the possibility that anti-sense transcripts might initiate from other regions of the sorghum gene.
Transposon mutagenesis of ZmPho1;2a and ZmPho1;2b
To investigate functional divergence between ZmPho1;2a and ZmPho1;2b, we initiated a program to mutagenize both loci using the endogenous Activator/Dissociation (Ac/Ds) transposon system. Ac and Ds elements show a strong preference for linked transposition, allowing a given element to be used for mutagenesis of nearby candidate genes. Once established, it becomes possible to generate multiple alleles from a single test-cross population.
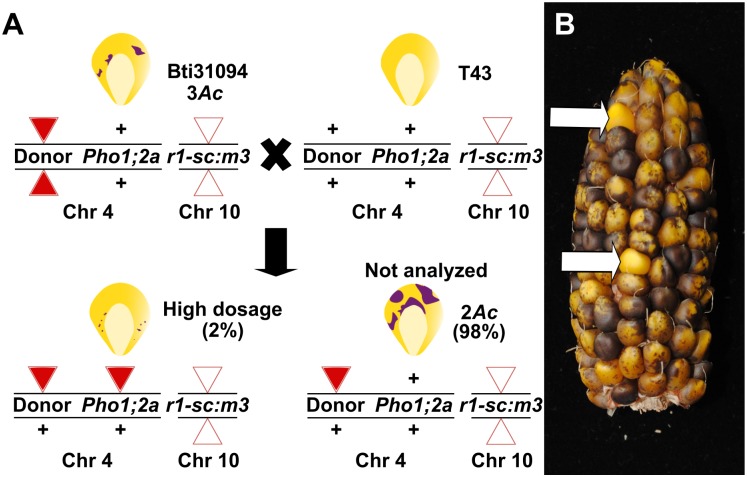
To mutagenize ZmPho1;2a, we recovered 1082 novel transposition events from an Acelement (Bti31094::Ac) located 650.8kb upstream, selecting rare high Ac dosage kernels from a testcross population (Fig 4). A PCR-based strategy was designed to screen for reinsertion of Ac into ZmPho1;2a: the gene was divided into three overlapping fragments, and, allowing for both possible orientations of Ac insertion, we performed a total of 12 reactions to cover the gene space, screening first pools of 18 seedlings, and subsequently the individuals constituting positive pools. Putative insertions were re-amplified using DNA extracted from a second seedling leaf to reduce the probability of selecting somatic events. Using this strategy, we recovered a novel germinal Ac insertion in Exon 6 of ZmPho1;2a (Zmpho1;2a-m1::Ac) (Fig 5). Left and right flanking border fragments were amplified and sequenced, confirming the exact location of the element and identifying an 8bp target site duplication (TSD; AGCCCAGG) typical of Ac insertion. Analysis of progeny derived from the original positive plant revealed a high-frequency of somatic excision from Zmpho1;2a-m1::Ac: apparently wild-type fragments were routinely amplified spanning the insertion site from individuals selected on the basis of kernel spotting pattern to be genotypically homozygous for Zmpho1;2a-m1::Ac. Sequencing of these products revealed them to be the result of somatic Ac excision (data not shown), as indicated by a short, typically 8bp, insertion adjacent to the excision site. Excision of Ac from Zmpho1;2a-m1::Ac coupled with a TSD is predicted to generate a BseYI restriction site (CCCAGC), providing a means to confirm events by digestion of PCR products (Fig 6A, 6B and 6D).
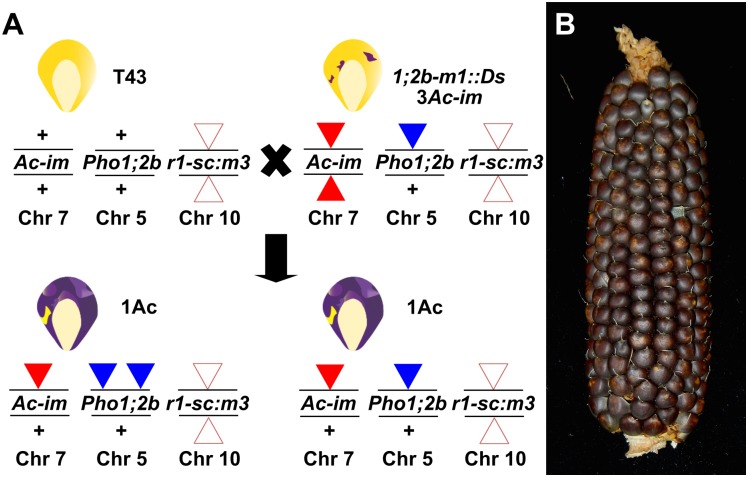
Fig 4. Ac mutagenesis of ZmPho1;2a.
(A) Genetic strategy to mobilize the donor element bti31094::Ac. Individuals homozygous for the donor element, displaying the characteristic 3 Ac dosage pattern of excision from r1-sc:m3 in the triploid aleurone, were crossed as females by T43. A small proportion (~2%) of the progeny kernels were finely spotted, indicating high Ac dose as a result of replicative transposition, and were screened for potential re-insertion into the target gene. (B) Two high Ac dose kernels (white arrows) highlighted among the largely 2 Ac dose progeny of a typical test cross ear.
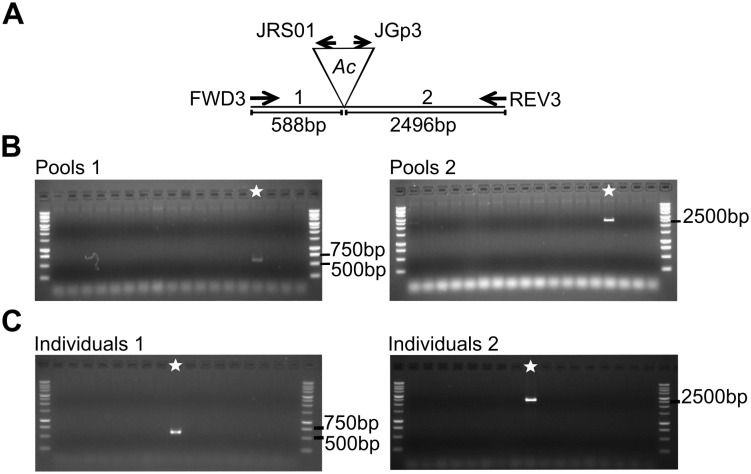
Fig 5. PCR screening for novel Ac insertion in ZmPho1;2a.
(A) Schematic of Zmpho1;2a-m1 allele. Reaction 1 (588bp expected product) and reaction 2 (2496bp) are indicated. (B) PCR reactions using gene and Ac specific primers to amplify potential products from DNA pools. Pool with a star shows amplification in complementary reactions 1 and 2. (C) PCR reactions using individual DNA. Amplification from a positive individual in complementary reactions 1 and 2 indicated with a star.
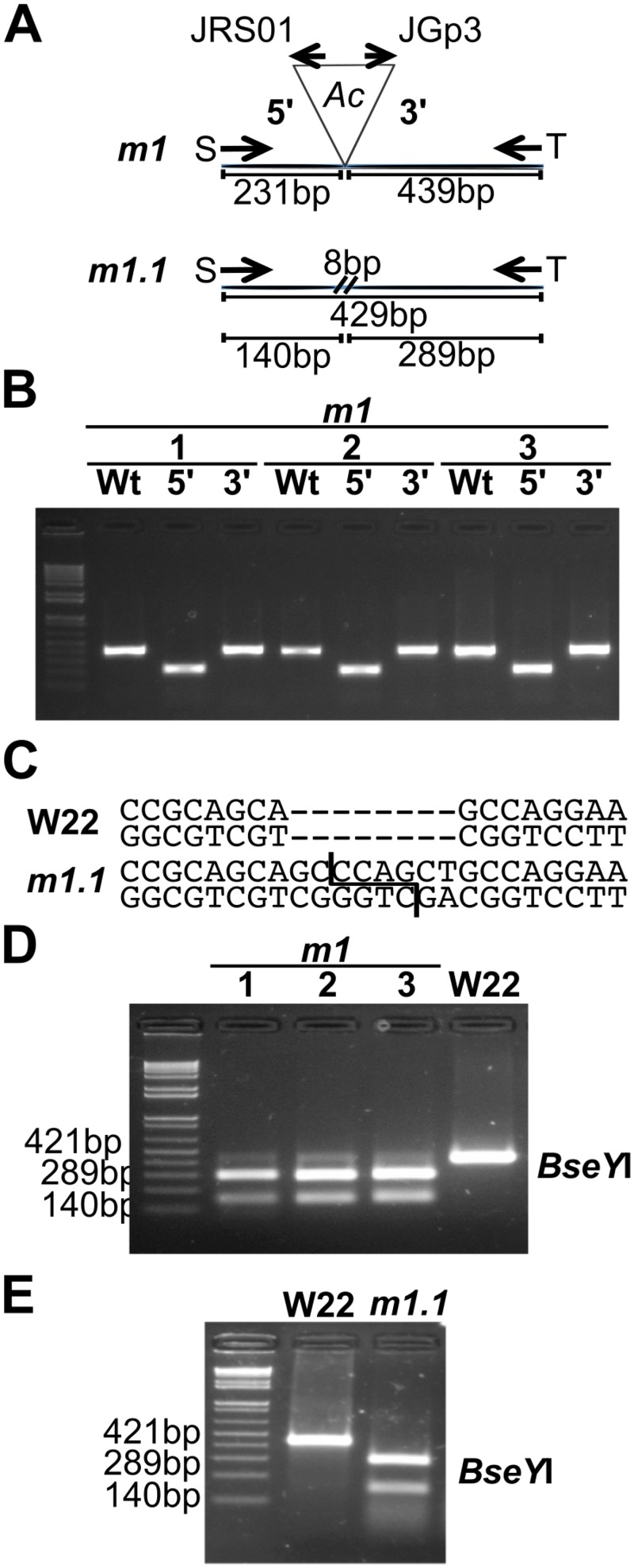
Fig 6.
Recovery of Ac excision events from Zmpho1;2a-m1. (A) Schematic of primer position and fragment lengths corresponding to Zmpho1;2a-m1 (m1) and derived excision events (m1.1). (B) Amplification of apparent wild-type (Wt) and flanking products (5’, 3’) from three different individuals genotypically homozygous Zmpho1;2a-m1 on the basis of kernel phenotype and pedigree. (C) Generation of a BseYI cutting site, not present in wild-type (W22), as the result of an insertion of 8bp following excision of Ac from Zmpho1;2a-m1. (D) BseYI digestion of products amplified in (B) from three genotypically Zmpho1;2a-m1 homozygous individuals and a wild-type (W22) individual. (E) BseYI digestion of products amplified with the insertion spanning primers MS124/MS052 from a wild-type individual (W22) and an individual homozygous for the stable germinal excision allele pho1;2a’m1.1. 1Kb plus DNA ladder (Invitrogen) is loaded on the first lane in B,C and E.
To identify novel Ds insertions in ZmPho1;2b, I.S06.1616::Ds (designated Zmpho1;2b-m1::Ds), inserted in intron 13 of the target gene, was re-mobilized. Plants homozygous for the Zmpho1;2b-m1::Ds allele did not present any observable phenotype and RT-PCR analysis of transcript accumulation indicated such plants to accumulate correctly-spliced transcript to normal levels (data not shown). To derive further alleles, individuals homozygous for Zmpho1;2b-m1::Ds carrying the unlinked stable transposase source Ac-immobilized (Ac-im; [21]) were crossed as males to T43 (Fig 7), and test-cross progeny screened using a strategy comparable to that employed in the mutagensis of ZmPho1;2a. Two novel Ds insertions were identified, one in the promoter region, 591bp upstream of the ATG (Zmpho1;2b-m3::Ds) and the second in intron 5 (Zmpho1;2b-m4::Ds). Sequencing of the region flanking each novel insertion identified the expected 8bp TSD (Table 4).
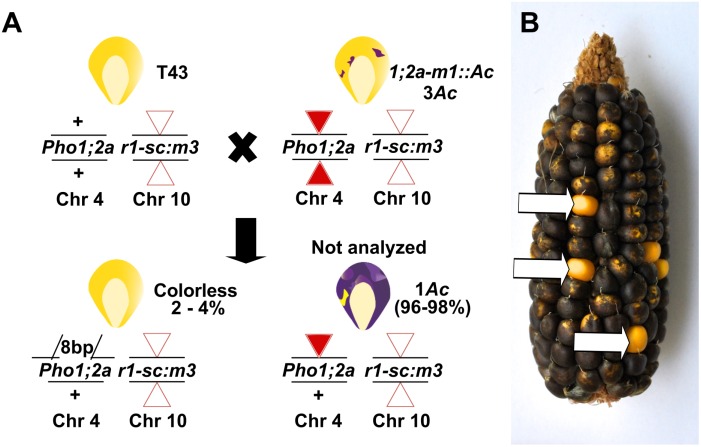
Fig 7. Ds mutagenesis of ZmPho1;2b.
(A) Genetic strategy to re-mobilize the Ds element ZmPho1;2b-m1::Ds. Individuals carrying ZmPho1;2b-m1::Ds and the stable transposase source Ac-im were crossed as males to T43. All progeny were screened as rare kernels carrying transposed Ds were indistinguishable from other progeny on the basis of aluerone phenotype. (B) Ear carrying kernels with one copy of Ac-im in the chr7 expressed in the triploid aleurone.
Table 4. Transposon insertion alleles of maize Pho1;2 genes.
| Alleles | Position (bp) | Description |
|---|---|---|
| pho1;2a-m1::Ac | 2261 | Ac in Exon 6; TSD: AGCCCAGG |
| pho1;2a’-m1.1 | 2260 | Footprint (CTGCCCAG)from pho1;2a-m1::Ac in Exon 6 |
| pho1;2a’-m1.2 | 2260 | Footprint (CCCAG) from pho1;2a-m1::Ac in Exon 6 |
| pho1;2b-m1::Ds | 4104 | Ds in intron 13; TSD: GGTGGGAG |
| pho1;2b-m3::Ds | -591 | Ds in promoter; TSD: ATCACTAT |
| pho1;2b-m4::Ds | 1943 | Ds Intron 5; TSD: CACACGCT |
Position relative to ATG. B73 genomic sequence (RefGen v3) as reference. TSD—target site duplication.
Derivation of stable derivatives of ZmPho1;2a
To generate stable “footprint” alleles by Ac excision from Zmpho1;2a-m1::Ac, a homozygous Zmpho1;2a-m1::Ac individual was crossed as a male to multiple T43 females, and resulting colorless progeny screened by PCR amplification across the Zmpho1;2a-m1::Ac insertion site (Fig 8). Products of a size consistent with Ac excision were cloned and sequenced. Two footprint alleles were identified, one with an 8bp insertion (CTGCCCAG) (Zmpho1;2a’-m1.1) and the second with a 5bp insertion (CCCAG) (Zmpho1;2a’-m1.2). The region spanning the excision site was re-amplified and digested with the enzyme BseYI, confirming the presence of the footprint (Fig 6A, 6C and 6E). As a result of non-triplet insertion, both ZmPho1;2a’-m1.1 and ZmPho1;2a’-m1.2 alleles disrupt the DNA reading frame and are predicted to result in a premature termination of translation (Table 4).
Fig 8. Generation of stable footprint alleles by Ac excision.
(A) Genetic strategy to re-mobilize to recover stable germinal excision of Ac from ZmPho1;2a-m1::Ac. An individual homozygous for ZmPho1;2a-m1::Ac was crossed as male to T43. Rare colorless kernels were selected for PCR screening. (B) Colorless kernels (white arrows) highlighted among the largely 1 Ac dose progeny of a typical test cross ear.
Discussion
Maize is the most widely grown cereal in the world (ref http://faostat3.fao.org). Much of this cultivated area is P limited. And yet, the molecular basis of P uptake and translocation in maize remains poorly characterized (reviewed in [10]). In this study, we have described the maize Pho1 gene family and generated novel mutant insertion alleles of the two ZmPho1;2 genes using the endogenous maize Ac/Ds transposon system. The genetic material described here initiates the functional analysis of P homeostasis in maize.
The maize PHO1 family consists of four genes, corresponding to the three gene (PHO1;1, PHO1;2, PHO1;3) structure reported previously in rice (Oryza sativa) [5], with the elaboration of a duplication of the maize ZmPho1;2. The sorghum PHO1 family was found also to consist of three genes. The restricted PHO1 family present in these cereals is in contrast to larger 11-member family of Arabidopsis [9]. Specifically, the cereals lack a large clade of PHO1 related sequences present in Arabidopsis that has been implicated in a range of biological functions extending beyond P homeostasis [9, 22, 24]. Indeed, in experiments to complement the Atpho1 phenotype by expression of other Arabidopsis PHO1 family members, it was only AtPHO1;H1 that could rescue the mutant [22]. Although phylogenetic analysis and experimental data from Arabidopsis and rice suggest all four maize PHO1 to be directly involved in P homeostasis, further work in heterologous systems, and ultimately the analysis of the mutants described here, will be required to determine functional equivalence across species and the biological role in maize.
The lineage leading to maize experienced a tetraploidy event resulting in whole genome duplication (WGD) sometime after the split with the sorghum lineage, 5-12 million years ago [23]. Taking contemporary sorghum to represent the pre-duplication state, immediately following the tetraploid event maize would have carried six Pho1 genes, represented by three pairs of syntenic paralogs (homeologs). Subsequently, the maize genome has returned to a diploid state through a process of reorganization that has been coupled with extensive fractionation—the loss of one of a pair of syntenic paralogs [25]. Large scale gene loss following WGD appears to be a general trend observed across taxa and across timescales [26]. Gene loss is presumed to be buffered by the presence of functionally equivalent paralogs. Where both paralogs of a syntenic pair are retained, it may indicate either selection or simply incomplete fractionation. The former case would imply functional divergence or a selective advantage of increased dosage. In maize, it is estimated that 3228 pairs of syntenic paralogs have been retained, representing ~20% of the total complement of ~32,000 total genes, or closer to ~10% of the pre-duplication gene set [23] [27]. While gene loss is the more likely outcome following genome duplication, it is difficult to determine the balance of selective gene-by-gene reduction and the largely random loss of larger sections of DNA. Similarly, where a pair of syntenic paralogs are retained, as is the case with Pho1;2, it may indicate selection directly on the gene pair or a genomic context that insulates the gene pair from larger scale DNA loss events. It is noticeable that a number of syntenic paralog pairs have been retained close to the Pho1;2 locus, potentially “hitchhiking” on direct selection to maintain one or more of the adjacent pairs. In the case of the pair GRMZM2G164854/GRMZM5G853379, the two paralogs overlap directly with Pho1;2 sequence on the opposite DNA strand. Consequently, selection to maintain either the Pho1;2 or GRMZM2G164854/GRMZM5G853379 pair would protect the adjacent genes from silencing or deletion.
Transcripts of ZmPho1;2a and ZmPho1;2b were found to accumulate similarly with respect to tissue-specificity and P availability. The absolute level of transcript accumulation, however, was observed to differ between paralogs, consistent with a model of ongoing fractionation in which one member of a paralog pair becomes functionally less significant prior to loss. We did, however, observe regulatory divergence at the level of putative cis-NAT transcripts: accumulation of cis-NATZmPho1;2a was induced by P limitation, in a manner similar to that observed forcis-NATOsPho1;2, while cis-NATZmPho1;2b accumulation mirrored that of the ZmPho1;2b sense transcript, suggesting that, with respect to anti-sense transcript production, it is ZmPho1;2a that is maintaining the ancestral, and functionally significant, role. Although, as their name implies, characterized cis-NATs act on adjacent protein coding genes, a translational enhancer function, such as that observed in rice for the PHO1;2 NAT, may allow for trans action when, as in maize, two highly similar ZmPho1;2 paralogs are present. Indeed, one intriguing hypothesis, suggested by our transcript accumulation data, is that in maize there has been sub-functionalization of PHO1;2, with the primary production of sense transcripts from ZmPho1;2b and the primary production of anti-sense transcripts from ZmPho1;2a.
Characterization of the insertional alleles described here will be central in determining the function of ZmPho1;2a and ZmPho1;2b. We are continuing to mobilize Ac and Ds elements at the maize Pho1;2 loci, taking full advantage of the capacity of the system to generate allelic series, impacting variously sense and anti-sense transcripts. Such material will be invaluable in the fine-scale evaluation of regulatory crosstalk and functional redundancy between between ZmPho1;2 paralogs and, ultimately, the biological role of PHO1 proteins in maize.
Supporting Information
(A) Accumulation of Pho1;2 sense transcripts under contrasting phosphate conditions. Amplification of fragments corresponding to the mature sense-RNAs of ZmPho1;2a and ZmPho1;2b from oligo-dT primed cDNA prepared from roots and shoot of 10-day-old B73 seedlings, fertilized with Hoagland solution adjusted to 1mM (+P) or 0mM (-P) inorganic phosphate. PCR was run for 27 (27X), 32 (32X) or 37 (37X) cycles. Amplification of ZmCDK and amplification from genomic DNA template (G) were used as controls. (B) Accumulation of Pho1;2 sense transcripts detected by RNA sequencing. RNA was extracted from roots and leaves of 21-day old B73 seedlings fertilized with 1000μM (P+) or 10μM (P-) phosphate, and used to generate libraries for RNA sequencing. Data presented as Log2 counts per million (CPM).
(PDF)
Acknowledgments
We thank Juan M. Estévez-Palmas for valuable comments on the manuscript. This work was supported by the National Science Foundation grant IOS-0922701 to TB and the Mexican National Council of Science and Technology (CONACYT) grant CB2012-151947 to RS.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Science Foundation (www.nsf.gov) award IOS-0922701 to TB and Consejo Nacional de Ciencia y Tecnologia (Mexico; www.conacyt.mx) award CB2012-151947 to RS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Veneklaas EJ, Lambers H, Bragg J, Finnegan PM, Lovelock CE, Plaxton WC, et al. Opportunities for improving phosphorus-use efficiency in crop plants. New Phytologist. 2012;195:306–320. 10.1111/j.1469-8137.2012.04190.x [DOI] [PubMed] [Google Scholar]
- 2. Vance CP. Update on the State of Nitrogen and Phosphorus Nutrition Symbiotic Nitrogen Fixation and Phosphorus Acquisition. Plant Nutrition in a World of Declining Renewable Resources. 2014; [PMC free article] [PubMed] [Google Scholar]
- 3. Poirier Y, Thoma S, Somerville C, Schiefelbein J. Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant physiology. 1991;97:1087–1093. 10.1104/pp.97.3.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wege S, Khan GA, Jung JY, Vogiatzaki E, Pradervand S, Aller I, et al. The EXS domain of PHO1 participates in the response of shoots to phosphate deficiency via a root-to-shoot signal. Plant physiology. 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Secco D, Baumann A, Yves P. Characterization of the rice PHO1 gene family reveals a key role for OsPHO1; 2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant physiology. 2010;152(March):1693–1704. 10.1104/pp.109.149872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rouached H, Secco D, Arpat B, Poirier Y. The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC plant biology. 2011;11(1):19 10.1186/1471-2229-11-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Secco D, Wang C, Arpat Ba, Wang Z, Poirier Y, Tyerman SD, et al. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytologist. 2012;193(4):842–851. 10.1111/j.1469-8137.2011.04002.x [DOI] [PubMed] [Google Scholar]
- 8. Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y. A Rice cis-Natural Antisense RNA Acts as a Translational Enhancer for Its Cognate mRNA and Contributes to Phosphate Homeostasis and Plant Fitness. The Plant Cell. 2013;25:4166–4182. 10.1105/tpc.113.116251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y, Ribot C, Rezzonico E, Poirier Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant physiology. 2004;135(May):400–11. 10.1104/pp.103.037945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calderon-Vazquez C, Sawers RJ, Herrera-Estrella L. Phosphate deprivation in maize: genetics and genomics. Plant Physiol. 2011;156(3):1067–1077. 10.1104/pp.111.174987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bai L, Singh M, Pitt L, Sweeney M, Brutnell TP. Generating novel allelic variation through Activator insertional mutagenesis in maize. Genetics. 2007;175(3):981–92. 10.1534/genetics.106.066837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dooner HK, Belachew A. Transposition Pattern of the Maize Element Ac from the Bz-M2(ac) Allele. Genetics. 1989;122(2):447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brutnell TP, Conrad LJ. Transposon tagging using Activator (Ac) in maize. Methods in molecular biology (Clifton, NJ). 2003;236:157–76.. [DOI] [PubMed] [Google Scholar]
- 14. Kolkman JM, Conrad LJ, Farmer PR, Hardeman K, Ahern KR, Lewis PE, et al. Distribution of Activator (Ac) throughout the maize genome for use in regional mutagenesis. Genetics. 2005;169(2):981–995. 10.1534/genetics.104.033738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vollbrecht E, Duvick J, Schares JP, Ahern KR, Deewatthanawong P, Xu L, et al. Genome-wide distribution of transposed Dissociation elements in maize. Plant Cell. 2010;22(6):1667–1685. 10.1105/tpc.109.073452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoagland DR, Broyer TC. General nature of the process of salt accumulation by roots with description of experimental methods. Plant physiology. 1936;11(3):471–507. 10.1104/pp.11.3.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. González-Muñoz E, Avendaño-Vázquez AO, Montes RaC, de Folter S, Andrés-Hernández L, Abreu-Goodger C, et al. The maize (Zea mays ssp. mays var. B73) genome encodes 33 members of the purple acid phosphatase family. Frontiers in Plant Science. 2015;6(May):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahern KR, Deewatthanawong P, Schares J, Muszynski M, Weeks R, Vollbrecht E, et al. Regional mutagenesis using Dissociation in maize. Methods (San Diego, Calif). 2009;49:248–54. 10.1016/j.ymeth.2009.04.009 [DOI] [PubMed] [Google Scholar]
- 19. Alleman M, Kermicle JL. Somatic variegation and germinal mutability reflect the position of transposable element Dissociation within the maize R gene. Genetics. 1993;135(1):189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McClintock B. Chromosome organization and genic expression. Cold Spring Harbor symposia on quantitative biology. 1951;16:13–47. 10.1101/SQB.1951.016.01.004 [DOI] [PubMed] [Google Scholar]
- 21. Conrad LJ, Brutnell TP. Ac-immobilized, a stable source of Activator transposase that mediates sporophytic and gametophytic excision of Dissociation elements in maize. Genetics. 2005;171(4):1999–2012. 10.1534/genetics.105.046623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stefanovic A, Ribot C, Rouached H, Wang Y, Chong J, Belbahri L, et al. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. The Plant journal: for cell and molecular biology. 2007;50(6):982–94. 10.1111/j.1365-313X.2007.03108.x [DOI] [PubMed] [Google Scholar]
- 23. Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci U S A. 2011;108(10):4069–4074. 10.1073/pnas.1101368108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ribot C, Zimmerli C, Farmer EE, Reymond P, Poirier Y. Induction of the Arabidopsis PHO1;H10 gene by 12-oxo-phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1-dependent pathway. Plant physiology. 2008;147(June):696–706. 10.1104/pp.108.119321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woodhouse MR, Schnable JC, Pedersen BS, Lyons E, Lisch D, Subramaniam S, et al. Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS biology. 2010;8(6):e1000409 10.1371/journal.pbio.1000409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sankoff D, Zheng C, Zhu Q. The collapse of gene complement following whole genome duplication. BMC genomics. 2010;11:313 10.1186/1471-2164-11-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hughes TE, Langdale JA, Kelly S. The impact of widespread regulatory neofunctionalization on homeolog gene evolution following whole-genome duplication in maize. Genome research. 2014;24(8):1348–55. 10.1101/gr.172684.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Accumulation of Pho1;2 sense transcripts under contrasting phosphate conditions. Amplification of fragments corresponding to the mature sense-RNAs of ZmPho1;2a and ZmPho1;2b from oligo-dT primed cDNA prepared from roots and shoot of 10-day-old B73 seedlings, fertilized with Hoagland solution adjusted to 1mM (+P) or 0mM (-P) inorganic phosphate. PCR was run for 27 (27X), 32 (32X) or 37 (37X) cycles. Amplification of ZmCDK and amplification from genomic DNA template (G) were used as controls. (B) Accumulation of Pho1;2 sense transcripts detected by RNA sequencing. RNA was extracted from roots and leaves of 21-day old B73 seedlings fertilized with 1000μM (P+) or 10μM (P-) phosphate, and used to generate libraries for RNA sequencing. Data presented as Log2 counts per million (CPM).
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.