Abstract
Mammalian p53 is a super tumor suppressor and plays a key role in guarding genome from DNA damage. However, p53 has not been found in plants which do not bear cancer although they constantly expose to ionizing radiation of ultraviolet light. Here we introduced p53 into the model plant Arabidopsis and examined p53-conferred phenotype in plant. Most strikingly, p53 caused early senescence and fasciation. In plants, fasciation has been shown as a result of the elevated homologous DNA recombination. Consistently, a reporter with overlapping segments of the GUS gene (1445) showed that the frequency of homologous recombination was highly induced in p53-transgenic plants. In contrast to p53, SUPPRESSOR OF NPR1-1 INDUCIBLE 1 (SNI1), as a negative regulator of homologous recombination in plants, is not present in mammals. Comet assay and clonogenic survival assay demonstrated that SNI1 inhibited DNA damage repair caused by either ionizing radiation or hydroxyurea in human osteosarcoma U2OS cancer cells. RAD51D is a recombinase in homologous recombination and functions downstream of SNI1 in plants. Interestingly, p53 rendered the sni1 mutants madly branching of inflorescence, a phenotype of fasciation, whereas rad51d mutant fully suppressed the p53-induced phenotype, indicating that human p53 action in plant is mediated by the SNI1-RAD51D signaling pathway. The reciprocal species-swap tests of p53 and SNI1 in human and Arabidopsis manifest that these species-specific proteins play a common role in homologous recombination across kingdoms of animals and plants.
Introduction
Mammalian p53 plays a pivotal role in suppression of tumor. More than half of human cancers bear mutations in the p53 gene [1]. p53 functions as a node in coordinating the cellular responses to a broad range of stresses with apoptosis, cell cycle arrest, senescence, DNA repair, cell metabolism, or autophagy [2]. p53 not just functions as a transcriptional factor to activate genes during stress responses but also acts independent of transcription especially during apoptosis and DNA damage repair (DDR) [1, 3–6]. p53 has been called “the guardian of the genome” because of its central role in DNA damage repair [2]. In response to DNA damage, the two phosphatidyl inositol 3-kinase-like kinases (PIKKs), ATAXIA TELANGIECTASIA MUTATED (ATM) and ATM-RAD3-RELATED (ATR), activate p53 proteins through phosphorylation on its serine-15 [7–9]. The activated p53 physically interacts with DNA repair proteins including REPLICATION PROTEIN A (RPA), RAD51, RAD54, BREAST CANCER 1 (BRCA1), and BRCA2 [10–18] to non-transcriptionally regulate homologous DNA recombination. p53 is a double-edged sword in regulation of homologous recombination: on the one edge, p53 complexes with RAD51 to inhibit homologous recombination [16, 17, 19, 20]; on the other edge, p53 activates homologous recombination though topoisomerase I [21].
In addition to immunity, SUPPRESSOR OF NPR1-1 INDUCIBLE 1 (SNI1) is a negative regulator of homologous recombination in plant as the frequency of homologous recombination is highly elevated in sni1 mutants [22, 23]. The sni1 mutant was first identified as a suppressor of nonexpressor of pr1 genes 1 (npr1) mutant [22]. NPR1 is a master regulator of plant immunity. The npr1 mutants were notably susceptible to pathogens [24]. To dissect the SNI1 signaling pathway, a genetic approach was employed to isolate suppressor of sni1 (ssn) mutants which restored the wild type phenotype of sni1 mutant. So far, the characterized SSN proteins are all involved in homologous recombination including RAD51D (SSN1), SSN2 (SWI2/SNF2 and MuDR with SWIM domain), BREAST CANCER 2 (BRCA2, SSN3), RAD51, RAD17 (SSN4), and ATR1 [23, 25–27], suggesting that these positive regulators of homologous recombination function downstream of SNI1 and SNI1 may serve as a brake to attenuate DNA damage response in a proper manner.
The p53 family proteins, including p53, P63 and P73, have been found in Choanozoa and animals. However, they are absent in yeast and plants [28]. In contrast to p53, SNI1 is absent in animals [22, 29]. Previous study showed that overexpression of human p53 inhibited cell growth of the fission yeast Schizosaccharomyces pombe [30] and induced cell death of the budding yeast Saccharomyces cerevisiae [31]. In this study, we introduced human p53 into the model plant Arabidopsis and examined p53-conferred plant phenotype. Since both species-specific proteins p53 and SNI1 function in homologous recombination, we further investigated whether p53 action in plant is mediated by the SNI1 signaling pathway.
Materials and Methods
Plant materials
The wild-type background of the mutants used in this study is Columbia (Col-0). Mutants of sni1 and rad51d are as described [22, 23].
Constructs
To generate 35S:p53 for expression of human p53 in Arabidopsis, the 35S promoter from pBI121 [32] was amplified by PCR and inserted between BstEII and HindIII of the binary vector pCAMBIA1301 (CAMBIA, Canberra, Australia). Subsequently, the NOS terminator was released from pBI121 by SacI and EcoRI digestion and inserted between the corresponding sites of the above 35S promoter-integrated pCAMBIA1301. Finally, the coding DNA sequence (CDS) of p53, obtained from OriGene (Cat. No.: RC200003), was amplified by PCR and inserted between the 35S promoter and the NOS terminator (35S:p53).
Measurement of homologous DNA recombination
The frequency of somatic homologous DNA recombination was analyzed using a reporter containing overlapping segments of the GUS gene in inverted orientation (line 1445) as described previously [33].
Microarray analysis
Total RNA of ten-day-old wild type and p53-transgenic seedlings was extracted using the RNeasy Plant Mini Kit (Qiagen). The MessageAmp Premier RNA Amplification Kit (Ambion) was used for RNA labeling. Hybridization with the GeneChip Arabidopsis ATH1 Genome Array (Affymetrix), washing, and scanning were performed at the Duke Microarray Facility. Experiments were performed in triplicate. Statistical analysis was performed using GeneSpring GX 11.5 (Agilent).
Quantitative PCR (qPCR)
Arabidopsis RNA was extracted using TRIzol Reagent (Invitrogen) and measured by NanoDrop 2000 Spectrophotometer (Thermo Fisher). Five μg of RNA was treated with DNase (Ambion TURBO DNA-free Kit, Thermo Fisher). Two μg of DNase-treated RNA was used to synthesize cDNA using the SuperScript III cDNA Synthesis (Invitrogen). The synthesized cDNA, diluted 5 times, was used as templates of qPCR. qPCR was performed using SYBR Green PCR Kit (Roche Applied Science) in Mastercycler ep realplex (Eppendorf). The primers of RAD51D used for qPCR are RAD51D-qPCR-F, TTTCGCTATCACGTGACCAT and RAD51D-qPCR-R, TGAAGGCAAGGATGTGTGTT. UBIQUITIN 5 (UBQ5) was used as an internal control which was amplified using primers of UBQ5-qPCR-F, GTAAACGTAGGTGAGTCCA and UBQ5-qPCR-R, GACGCTTCATCTCGTCC.
Transfection of human U2OS cells
Human osteosarcoma U2OS cancer cells were obtained from the Duke Cell Culture Facility and maintained in McCoy’s 5A medium with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The hemagglutinin (HA)-tagged Arabidopsis SNI1 coding DNA sequence (CDS) was integrated into the retroviral vector of pQCXIP (Clontech) for transfection of U2OS cells.
Immunoblots
Plant tissues were homogenized in liquid nitrogen. Total protein was extracted using a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, 0.2% Nonidet P-40, and 1% Protease Inhibitor Cocktail (Sigma, P9599). U2OS cancer cells were lysed in RIPA buffer (Sigma, R0278) supplemented with Protease Inhibitor Cocktail (Sigma, P8340). Cell lysates were resolved on sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS/PAGE). Western blot analysis was performed as described previously [26]. Anti-α-tubulin antibody (Sigma, T5168) was used as an internal loading control.
Comet assay
The repair kinetics of ionizing radiation (IR)-induced DNA damage was evaluated by the Alkaline Comet Assay according to the manufacturer's protocol (Trevigen). Briefly, cells were exposed to the indicated doses of IR and harvested at various recovery time points for comet assay. Nuclei were stained with SYBR green and comets were visualized by epifluorescence on a Zeiss microscope.
Clonogenic survival assay
U2OS cells were plated at 500 cells per well in 6-well plates and pulse-treated with the indicated doses of hydroxyurea (HU, Sigma, H8627) for 24 hours to introduce DNA damage. After treatment, cells were rinsed twice with PBS and allowed to recover in drug-free medium. The cultures were then incubated for 14 days with the medium being changed every 3 days. Colonies were stained with crystal violet (Sigma, C6158), counted, and normalized to untreated control.
Statistical Analysis
Statistical analysis was performed by one-way ANOVA with Bonferroni post hoc test. The letter above the bar indicates a statistically significant difference between groups at p value < 0.01.
Accession numbers
The Gene Expression Omnibus (GEO) accession number for the microarray data used in this study is GSE79678 which is available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79678.
Results
Human p53 induces early senescence and fasciation in Arabidopsis
Ectopic expression of human p53 under a constitutive promoter, cauliflower mosaic virus (CaMV) 35S (35S:p53) [34], in Arabidopsis Columbia (Col-0) resulted in pleiotropic developmental phenotype (Fig 1). We carried out transformation of the 35S:p53 construct three times and got 19, 27, and 21 transgenic lines, respectively. p53 proteins were detected by immunoblot in the p53-transgenic plants (S1 Fig). All of the p53-transgenic lines displayed similar phenotype, except for those died with severe phenotype.
Fig 1. Human p53-conferred phenotype in plant.
(A) Leaves of 3-week-old wild type (WT) and p53-transgenic plants (p53). Arrows indicate cotyledons. (B) Four-week-old WT and p53 plants. Arrows indicate the first pair of true leaves. (C) Bolting WT and p53 plants. (D) Left panel: inflorescences of WT and p53. Insets show enlarged stems (in yellow box). Right panel: number (#) of secondary inflorescences. Error bars represent standard errors (SEs). ***, p value < 0.001, compared to WT by binomial test. Experiments were carried out in triplicate (n > 30) with similar results. (E) Siliques of WT and p53. Arrow indicates clustered (fascinated) siliques.
Firstly, p53 induced early senescence which appeared in the cotyledons of three-week-old plants (Fig 1A), in the first pair of true leaves of four-week-old plants (Fig 1B), and most of the leaves of the flowering plants (Fig 1C). In mammals, p53 plays a critical role in senescence which is an irreversible cell-cycle arrest [35]. Overexpression of p53 in human tumor cells triggers senescence [36, 37]. Two cyclin-dependent kinase inhibitors (CKIs) that are often expressed in senescent cells are p21 and p16 [38]. p21 is a crucial transcriptional target of p53 and mediator of p53-dependent senescence [39, 40]. Both p16 and p21 inhibit pRB phosphorylation to activate senescence [38]. Mammalian CKIs are classified into two families: the CDK INTERACTING PROTEIN/KINASE INHIBITORY PROTEIN (Cip/Kip) family, including p21, p27, p57, and the INHIBITORS OF CDK4 (INK4) family, including p16, p15, p18, p19 [41]. Unlike mammals, the involvement of CKIs in plant senescence has not been well understood. CONSTITUTIVE EXPRESSER OF PATHOGENESIS-RELATED GENES 5 (CPR5) is a negative regulator of plant senescence [42]. Recently, SIAMESE (SIM) and SIAMESE-RELATED 1 (SMR1), homologs of mammalian Cip/Kip CKI family proteins, were discovered to function redundantly downstream of CPR5 to activate plant senescence [43]. These data suggest that p53 as well as its mammalian target p21 could activate senescence in both mammals and plants.
Secondly, the p53-transgenic plants exhibited fascinated phenotype including thick stems, increased secondary inflorescences, and clustered siliques (Fig 1D and 1E). Fascination is the term used to describe fused or distorted organs along a plant stem [44]. The bolting stems of p53-transgenic plants were obviously thicker than those of wild type plants (Fig 1D). The number of secondary inflorescences (lateral branches) of p53-transgenic plants was doubled as the average numbers of those of wild type and p53-transgenic plants were 4.2 and 8.3, respectively (Fig 1D). Moreover, most of the p53-transgenic plants produced clustered (fasciated) siliques (Fig 1E). In Arabidopsis, fasciation has been found in mutants of fasciata1 (fas1), fasciata2 (fas2) and clavata1 (clv1) [44, 45]. CHROMATIN ASSEMBLY FACTOR 1 (CAF-1) is a heterotrimeric complex [46]. Homologs of CAF-1 have been found in yeast, insects, plants, and vertebrates [47]. The Arabidopsis CAF-1 subunits corresponding to the human subunits p150, p60, and p48 are encoded by FAS1, FAS2, and MSI1, respectively [48, 49]. CAF-1 functions in depositing H3/H4 histones onto replicating DNA during S-phase and DNA repair [50–53]. The yeast CHROMATIN ASSEMBLY COMPLEX 1 (CAC1) is the homolog of a subunit of human CAF-1. The frequency of homologous recombination considerably increased in yeast cac1 mutants [54]. Accordingly, a dominant-negative mutation of a subunit of human CAF-1 resulted in double-strand breaks and activating DNA repair proteins ATM and ATR [55]. ATM was activated by double-strand breaks, whereas ATR was activated by single-strand breaks or stalled replication forks [56]. Like caf-1 mutants in both yeast and human, the frequency of homologous recombination also dramatically increased (~40-fold) in Arabidopsis fas1 mutant [47, 57]. Epistatic analysis showed that FAS1 functioned upstream of ATM in homologous recombination as the phenotype of fas1 mutant was suppressed by atm mutant [58]. These data suggest that p53-induced fasciation in plant likely results from the elevated homologous recombination.
The species-specific p53 and SNI1 play a common role in DNA repair across plants and mammals
In mammals, p53 is a double-edged sword which could either inhibit or activate homologous recombination [20, 21]. The p53-induced fasciation indicates that p53 may activate homologous recombination in plant. To quantify the homologous recombination, we introduced the overlapping segments of the GUS gene (1445) [33] through crosses into three independent p53-transgenic lines. The GUS reporter (1445) demonstrated that the frequency of homologous recombination was highly induced by p53 as the sectors per plant of wild type and p53-transgenic plants are 1.6 and 6.2, respectively (Fig 2A and 2B and S1 Table). Previously, genetic study revealed that SNI1 was a negative regulator of plant homologous recombination [23]. Intriguingly, comet assay and clonogenic survival assay showed that SNI1 could inhibit DDR caused by either ionizing radiation (IR) or hydroxyurea (HU) in human osteosarcoma U2OS cancer cells (Fig 2C–2G). Immunoblot showed that the SNI1 proteins remained stable with these treatments (S2 Fig). These data demonstrate that both p53 and SNI1 could play a common role in DNA repair across Arabidopsis and human, although they are species-specific proteins.
Fig 2. The reciprocal species-swap test of p53 and SNI1 between Arabidopsis and human.
(A) Somatic recombination in wild type (WT) and p53-transgenic (p53) plants is shown in blue sectors by a reporter with overlapping segments of the GUS gene (1445). (B) Quantitative result of panel A. Experiments were performed in three p53-transgenic lines (n = 50 ~ 100) (S1 Table). The result of line 1 is shown. Error bars represent SEs. ***, p value < 0.001 compared to WT by binomial test. (C) Human osteosarcoma U2OS cancer cells transfected with empty vector (EV) or hemagglutinin (HA)-tagged SNI1 (SNI1). Proteins extracted from the transfected U2OS cancer cells were blotted with anti-HA antibody (abcam, ab1265). Anti-α-tubulin was used as an internal loading control. (D) The comet assay was carried out on the transfected U2OS cancer cells which were treated with 10 Gy of ionizing radiation (IR) and recovered with indicated time. The level of DNA break repair was visualized with the length of comet tail. (E) Images in panel B were analyzed using CometScore software (Tritek) to quantify the comet tail moment of at least 75 cells for each sample. Error bars represent SEs. ***, p value < 0.001, compared to EV by binomial test. Experiments were performed three times with similar results. (F) The transfected U2OS cancer cells were pulse-treated with hydroxyurea (HU) for 24 hours to introduce DNA damage and recovered in drug-free medium. (G) Quantitative results of panel D. After 14 days of culture, colonies were counted and normalized to untreated control. Error bars represent SEs. Experiments were carried out in triplicate.
Human p53 action in plant is mediated by the SNI1-RAD51D signaling pathway
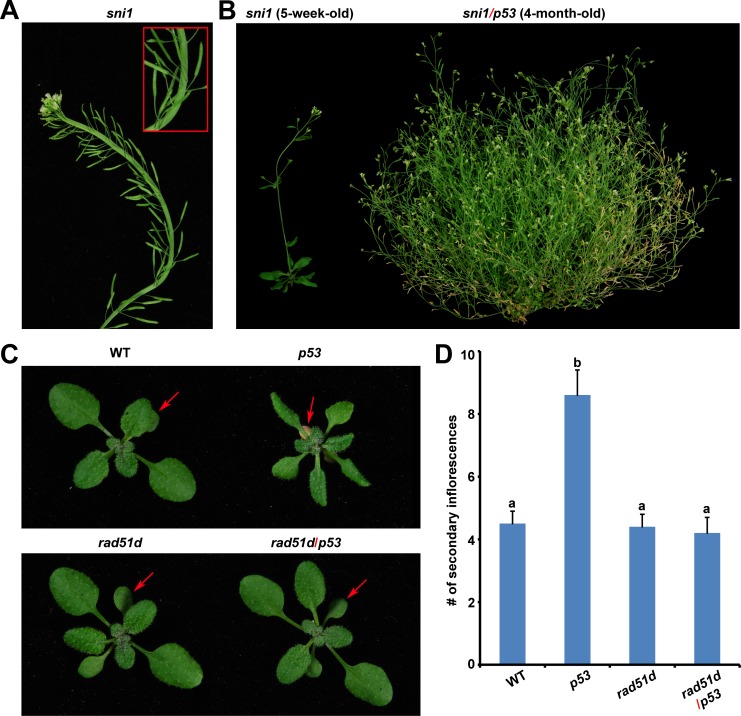
Fasciation was also occasionally observed in sni1 mutants (Fig 3A), which is consistent with its heightened homologous recombination. Since both p53-transgenic plants and sni1 mutants exhibit fasciation which is presumably associated with homologous recombination, we expect that p53 acts in Arabidopsis through the SNI1 signaling pathway. To test this hypothesis, we introduced 35S:p53 construct into sni1 mutant through crosses with three independent transgenic lines. Surprisingly, p53-transgenic sni1 mutants madly and long lastly (over 4 months) developed secondary inflorescences (Fig 3B), which was a phenotype of fasciation in fas1and fas2 mutants and p53-transgenic plants. Since both fas1 and sni1 mutants are suppressed by disruptions of DNA repair proteins, we further expect that p53 functions in plant through the SNI1-RAD51D signaling pathway. To testify this possibility, we introduced 35S:p53 construct into rad51d mutant through crosses with three independent transgenic lines. RAD51D is a paralog of RAD51 which is a key player of homologous recombination. The rad51d mutant is the first identified suppressor of sni1 (ssn1) mutant. As expected, rad51d fully suppressed p53-induced phenotype including early senescence and fasciation (Fig 3C and 3D), demonstrating that ectopic p53 proteins act in plant through DNA repair protein RAD51D. These data suggest that p53 action is mediated by the SNI1-RAD51D signaling pathway of homologous recombination in plant.
Fig 3. Human p53 acts through the SNI1-RAD51D signaling pathway in plant.
(A) Fascinated inflorescence of sni1 mutant. Inset (box in red): part of the fascinated inflorescence is enlarged. (B) A single plant of five-week-old sni1 mutant and four-month-old p53-transgenic sni1 mutant (sni1/p53). (C) Three-week-old WT, p53-transgenic (p53), rad51d and p53-transgenic rad51d (rad51d/p53) plants. Arrows indicate cotyledons. (D) Number (#) of secondary inflorescences of WT, p53, rad51d and rad51d/p53 plants was plotted. The letter above the bar indicates a statistically significant difference between groups at p value < 0.01. Experiments were conducted in triplicate (n > 30) with similar results.
Regulation of homologous DNA recombination by p53 is independent of transcription in plant
It has been shown that the actions of p53 in mammalian homologous recombination are independent of its transcriptional activity [59, 60]. We adopted a genomic approach to explore how p53 acts in plants. The quality control of microarray is summarized in S2 Table. Gene Ontology (GO) analysis of the differential expressed genes (p value < 0.05 and fold change > 2) induced by ectopic p53 proteins revealed that stress was the most significantly enriched biological process (9.2%) (Fig 4A). However, the expression of SNI1, 7 SSNs and 3 fascination-associated genes (FAS1, FAS2 and CLV1) was not significantly altered (fold change < 1.5) by introduction of p53 in plant (Fig 4B). We adopted quantitative PCR (qPCR) to validate the microarray data. The quality assessments showed that the qPCR assays for UBQ5 and RAD51D were comparable as indicated by the key parameters of the qPCR reaction including slope, precision, amplification and efficiency (S3 Table and S3B–S3D Fig). qPCR analysis validated that p53 did not affect the expression of RAD51D (Fig 4C and S3 Table). These data suggest that the influence of human p53 on plant homologous recombination may also be independent of its transactivation as it does in mammals.
Fig 4. Influence of p53 on plant transcriptome.
(A) Gene Ontology (GO) analysis of microarray data. Ten-day-old wild type (WT) and p53-transgenic (p53) seedlings were used for microarray analysis (GEO accession number: GSE79678). The differential expressed genes (t test, p value < 0.05 and fold change > 2) were analyzed for enriched biological processes by Gene Ontology (GO: https://www.arabidopsis.org/tools/bulk/go/index.jsp). Experiments were performed in triplicate. (B) The expressions of SNI1, 7 SSNs (SUPPRESSORS OF SNI1s) and 3 fascination-associated genes in p53-transgenic plants were compared to those in WT plants. The red line indicates the expression with no change. (C) Ten-day-old wild type (WT) and p53-transgenic (p53) seedlings were used for RNA extraction. RAD51D transcripts were quantified by qPCR. UBQ5 was used as an internal control. Error bars represent SEs. Experiments were conducted in triplicate.
Discussion
Homologous recombination (HR) and nonhomologous end joining (NHEJ) are two main pathways to repair double strand break (DSB) of DNA. The machinery of homologous recombination is highly conserved between animals and plants [61]. As shown in S4A Fig, plants possess most of the key players in homologous recombination [62]. In mammals, the MRE11/RAD50/NBS1 (MRN) complex functions as DSB sensors. RAD17 recruits the MRN complex to the DSB site. In response to DSBs, the MRN complex activates two PIKK kinases: ATM and ATR. ATM is recruited to DSBs by the MRN complex, whereas ATR is recruited by ATR-interacting protein (ATRIP) to RPA-coated single-stranded DNA (ssDNA) [63–66]. ATM and ATR phosphorylate multiple substrates, including p53, and the checkpoint kinases, CHECKPOINT KINASE 1 (CHEK1) and CHECKPOINT KINASE 2 (CHEK2), which inhibits CYCLIN-DEPENDENT KINASE (CDK) activity to allow cell cycle arrest and DNA repair [67]. p53 activates the expression of p21 which is an inhibitor of CDK [1]. Subsequently, de-phosphorylation of BREAST CANCER 2 (BRCA2), which is a substrate of CDK, allows RAD51 to bind DNA and to form nucleofilament that invades a homologous sequence and activates strand exchange to further carry out homologous recombination repair of DSBs [68–70]. Although how plant-specific SNI1 is regulated remains elusive, epistatic analysis suggests that SNI1 functions upstream of the key players of homologous recombination including RAD17, ATR1, BRCA2, RAD51, and RAD51D in plant [23, 26, 27]. Therefore, both p53 and SNI1 play important role in the signaling pathway of homologous recombination.
Our data demonstrate that human p53-conferred plant phenotype including senescence and fasciation is coupled with the elevated homologous recombination which is mediated by the SNI1-RAD51D signaling pathway (Figs 1–3 and S4B Fig). There are at least four possibilities that ectopic p53 proteins could activate homologous recombination. Firstly, p53 is a transcription factor and its excessive accumulation, under the constitutive 35S promoter, in nucleus could directly cause DNA damage. Secondly, the transcriptional profiling shows that the most significantly enriched biological process is stress which often produces reactive oxygen species (ROS) and indirectly causes DNA damage [71]. Thirdly, p53 may function as a transcription factor to transcriptionally regulate homologous recombination. Fourthly, p53 may complex with protein to non-transcriptionally regulate homologous recombination. It has been discovered that RADIATION (RAD) genes, including RAD17 and RAD51, are highly upregulated in response to DNA damage in plants [26, 72–75]. As shown in Fig 4B, the expression of RAD17 and RAD51 was not significantly affected by the alien p53, ruling out the possibility that human p53-induced homologous recombination in Arabidopsis could be an artifact as a result of the excessive p53-caused DNA damage. Although we cannot exclude with certainty the possibility that p53 transcriptionally regulates homologous recombination through genes other than SNI1, 7 SSNs and 3 fascination-associated genes, it is likely that p53 acts in plant through interaction with player in the signaling pathway of homologous recombination as it does in mammals [12, 21].
The underlying mechanism of how the SNI1-RAD51D signaling pathway is activated by p53 remains to be investigated. In mammals, p53 is activated through posttranscriptional modifications in response to DNA damage and stress. The activated p53 physically interacts with DNA repair proteins to non-transcriptionally regulate homologous recombination [23, 26, 27]. There is an array of posttranslational modifications including phosphorylation, ubiquitination, acetylation, methylation, sumoylation, neddylation, glycosylation, and ribosylation [76]. It would be fascinating to explore the modifications of p53 protein in response to DNA damage and stress in plants. In this study, our reciprocal species-swap test showed that human p53 caused fasciation through activation of homologous recombination in plants and plant SNI1 inhibited homologous recombination in mammalian cells (Fig 2). It has long been a mystery that plants do not get cancer although they constantly expose to ionizing radiation of ultraviolet light [77, 78]. Our results will stimulate future studies of these species-specific homologous recombination players in a bigger picture which may lead us to better understand why cancers do not bother plants but animals and may help us to better handle human cancers.
Supporting Information
(A) Proteins were extracted from 10-day-old seedlings of wild type (WT) and three lines of p53-transgenic plants (p53, L1-L3), resolved on sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS/PAGE), and immunoblotted with anti-p53 antibody. Anti-α-tubulin was used as an internal loading control. (B) Proteins were extracted from 10-day-old seedlings of wild type (WT) and p53-transgenic (L1: line1) WT plants, sni1mutant, rad51d mutant, and GUS (1445) reporter. Immunoblot was performed as panel A.
(TIF)
The transfected U2OS cancer cells were treated with 10 Gy of ionizing radiation (IR) or hydroxyurea (HU). Proteins extracted from the transfected transfected U2OS cancer cells were blotted with anti-HA antibody (abcam, ab1265). Anti-α-tubulin was used as an internal loading control.
(TIF)
(A) Total RNA was extracted from ten-day-old wild type and p53-transgenic seedlings. The A260/A280 ratio of the total RNA was about 2.0 as measured on NanoDrop 2000 Spectrophotometer. The quality of RNA was further assessed by agarose gel electrophoresis. The 28S/18S ratio was about 2.0, indicating that the isolated RNAs were not degraded. (B) The qPCR products of UBQ5 and RAD51D were viewed by agarose gel electrophoresis. The qPCR product size of UBQ5 and RAD51D is about 250 bp and 100 bp, respectively. M, DNA marker. (C) The melting curves from qPCR analysis of UBQ5 and RAD51D. There was only one peak appeared in the melting curves of qPCR analysis in both UBQ5 and RAD51D, which was consistent with the agarose gel electrophoresis of qPCR product in panel B. (D) Serial dilutions of reverse transcription product (cDNA) were used for the qPCR quality assay of UBQ5 and RAD51D. Ct, cycle threshold.
(TIF)
(A) The signaling pathway of DNA damage repair through homologous recombination in mammals and plants. In mammals, the MRE11/RAD50/NBS1 (MRN) complex functions as double strand brake (DSB) sensors. RAD17 recruits the MRN complex to the DSB site. In response to DNA damage, two phosphoinositide 3-kinase-like kinases, ATM and ATR, are activated by the MRN complex to phosphorylate the transcription factor p53. p21, a target of p53, is an inhibitor of cyclin-dependent kinase (CDK). KIP-RELATED PROTEIN (KRP) is the homolog of p21 in plant. CDK phosphorylates BREAST CANCER 2 (BRCA2) which is a mediator of the recombinase RAD51. BRCA2 first loads RAD51 on the DSB site. After the DSB is repaired by RAD51, BRCA2 is then phosphorylated by CDK to remove RAD51 from DNA. Plants possess the homologs of these DNA repair proteins, except for p53. On the contrary, mammals do not bear SNI1. SNI1 is a negative regulator of homologous recombination in plants. Although genetic study reveals that SNI1 functions upstream of ATR, RAD17, BRCA2, and RAD51, how SNI1 is regulated remains unknown. (B) A proposed model of p53 action in plant. p53 induces homologous recombination through the SNI1-RAD51D signaling pathway in plant, leading to senescence and fasciation.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors would like to thank Drs. X Dong and X Wang for their support and discussion of this study and Drs. J Feng, TP Wakeman and Q Wang for their help in experiments at Duke University.
Data Availability
Microarray data are available from the Gene Expression Omnibus (GEO) database (accession number: GSE79678) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79678). All other relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grant 31571254 from National Natural Science Foundation of China (http://www.nsfc.gov.cn).
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000; 408(6810): 307–10. 10.1038/35042675 . [DOI] [PubMed] [Google Scholar]
- 2.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992; 358(6381): 15–6. 10.1038/358015a0 . [DOI] [PubMed] [Google Scholar]
- 3.Marchenko ND, Moll UM. The role of ubiquitination in the direct mitochondrial death program of p53. Cell cycle. 2007; 6(14): 1718–23. . [DOI] [PubMed] [Google Scholar]
- 4.Vousden KH, Lane DP. p53 in health and disease. Nature reviews Molecular cell biology. 2007; 8(4): 275–83. 10.1038/nrm2147 . [DOI] [PubMed] [Google Scholar]
- 5.Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Current opinion in cell biology. 2005; 17(6): 631–6. 10.1016/j.ceb.2005.09.007 . [DOI] [PubMed] [Google Scholar]
- 6.Speidel D. Transcription-independent p53 apoptosis: an alternative route to death. Trends in cell biology. 2010; 20(1): 14–24. 10.1016/j.tcb.2009.10.002 . [DOI] [PubMed] [Google Scholar]
- 7.Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998; 281(5383): 1674–7. . [DOI] [PubMed] [Google Scholar]
- 8.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998; 281(5383): 1677–9. . [DOI] [PubMed] [Google Scholar]
- 9.Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, et al. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes & development. 1999; 13(2): 152–7. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturzbecher HW, Donzelmann B, Henning W, Knippschild U, Buchhop S. p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. The EMBO journal. 1996; 15(8): 1992–2002. . [PMC free article] [PubMed] [Google Scholar]
- 11.Marmorstein LY, Ouchi T, Aaronson SA. The BRCA2 gene product functionally interacts with p53 and RAD51. Proceedings of the National Academy of Sciences of the United States of America. 1998; 95(23): 13869–74. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verma S, Rao BJ. p53 suppresses BRCA2-stimulated ATPase and strand exchange functions of human RAD51. Journal of biochemistry. 2013; 154(3): 237–48. 10.1093/jb/mvt040 . [DOI] [PubMed] [Google Scholar]
- 13.Mizuta R, LaSalle JM, Cheng HL, Shinohara A, Ogawa H, Copeland N, et al. RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proceedings of the National Academy of Sciences of the United States of America. 1997; 94(13): 6927–32. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen RB, Ozes A, Kim T, Estep A, Kowalczykowski SC. BRCA2 is epistatic to the RAD51 paralogs in response to DNA damage. DNA repair. 2013; 12(4): 306–11. 10.1016/j.dnarep.2012.12.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchhop S, Gibson MK, Wang XW, Wagner P, Sturzbecher HW, Harris CC. Interaction of p53 with the human Rad51 protein. Nucleic acids research. 1997; 25(19): 3868–74. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romanova LY, Willers H, Blagosklonny MV, Powell SN. The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene. 2004; 23(56): 9025–33. 10.1038/sj.onc.1207982 . [DOI] [PubMed] [Google Scholar]
- 17.Linke SP, Sengupta S, Khabie N, Jeffries BA, Buchhop S, Miska S, et al. p53 interacts with hRAD51 and hRAD54, and directly modulates homologous recombination. Cancer research. 2003; 63(10): 2596–605. . [PubMed] [Google Scholar]
- 18.Zhang H, Somasundaram K, Peng Y, Tian H, Zhang H, Bi D, et al. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene. 1998; 16(13): 1713–21. 10.1038/sj.onc.1201932 . [DOI] [PubMed] [Google Scholar]
- 19.Yoon D, Wang Y, Stapleford K, Wiesmuller L, Chen J. P53 inhibits strand exchange and replication fork regression promoted by human Rad51. Journal of molecular biology. 2004; 336(3): 639–54. 10.1016/j.jmb.2003.12.050 . [DOI] [PubMed] [Google Scholar]
- 20.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nature reviews Molecular cell biology. 2005; 6(1): 44–55. 10.1038/nrm1546 . [DOI] [PubMed] [Google Scholar]
- 21.Baumann C, Boehden GS, Burkle A, Wiesmuller L. Poly(ADP-RIBOSE) polymerase-1 (Parp-1) antagonizes topoisomerase I-dependent recombination stimulation by P53. Nucleic acids research. 2006; 34(3): 1036–49. 10.1093/nar/gkj509 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Zhang Y, Clarke JD, Li Y, Dong X. Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell. 1999; 98(3): 329–39. . [DOI] [PubMed] [Google Scholar]
- 23.Durrant WE, Wang S, Dong X. Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proceedings of the National Academy of Sciences of the United States of America. 2007; 104(10): 4223–7. 10.1073/pnas.0609357104 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997; 88(1): 57–63. . [DOI] [PubMed] [Google Scholar]
- 25.Song J, Durrant WE, Wang S, Yan S, Tan EH, Dong X. DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell host & microbe. 2011; 9(2): 115–24. 10.1016/j.chom.2011.01.011 . [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Durrant WE, Song J, Spivey NW, Dong X. Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2010; 107(52): 22716–21. Epub 2010/12/15. 10.1073/pnas.1005978107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan S, Wang W, Marques J, Mohan R, Saleh A, Durrant WE, et al. Salicylic acid activates DNA damage responses to potentiate plant immunity. Molecular cell. 2013; 52(4): 602–10. 10.1016/j.molcel.2013.09.019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutkowski R, Hofmann K, Gartner A. Phylogeny and function of the invertebrate p53 superfamily. Cold Spring Harbor perspectives in biology. 2010; 2(7): a001131 10.1101/cshperspect.a001131 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosher RA, Durrant WE, Wang D, Song J, Dong X. A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. The Plant cell. 2006; 18(7): 1750–65. 10.1105/tpc.105.039677 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bischoff JR, Casso D, Beach D. Human p53 inhibits growth in Schizosaccharomyces pombe. Molecular and cellular biology. 1992; 12(4): 1405–11. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadj Amor IY, Smaoui K, Chaabene I, Mabrouk I, Djemal L, Elleuch H, et al. Human p53 induces cell death and downregulates thioredoxin expression in Saccharomyces cerevisiae. FEMS yeast research. 2008; 8(8): 1254–62. 10.1111/j.1567-1364.2008.00445.x . [DOI] [PubMed] [Google Scholar]
- 32.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO journal. 1987; 6(13): 3901–7. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucht JM, Mauch-Mani B, Steiner HY, Metraux JP, Ryals J, Hohn B. Pathogen stress increases somatic recombination frequency in Arabidopsis. Nature genetics. 2002; 30(3): 311–4. 10.1038/ng846 . [DOI] [PubMed] [Google Scholar]
- 34.Odell JT, Nagy F, Chua NH. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 1985; 313(6005): 810–2. . [DOI] [PubMed] [Google Scholar]
- 35.Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: the critical roles of p53. Oncogene. 2013; 32(43): 5129–43. 10.1038/onc.2012.640 . [DOI] [PubMed] [Google Scholar]
- 36.Sugrue MM, Shin DY, Lee SW, Aaronson SA. Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53. Proceedings of the National Academy of Sciences of the United States of America. 1997; 94(18): 9648–53. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu D, Ou L, Clemenson GD Jr., Chao C, Lutske ME, Zambetti GP, et al. Puma is required for p53-induced depletion of adult stem cells. Nature cell biology. 2010; 12(10): 993–8. 10.1038/ncb2100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nature reviews Molecular cell biology. 2007; 8(9): 729–40. 10.1038/nrm2233 . [DOI] [PubMed] [Google Scholar]
- 39.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997; 277(5327): 831–4. . [DOI] [PubMed] [Google Scholar]
- 40.Jackson JG, Pereira-Smith OM. p53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer research. 2006; 66(17): 8356–60. 10.1158/0008-5472.CAN-06-1752 . [DOI] [PubMed] [Google Scholar]
- 41.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Developmental cell. 2008; 14(2): 159–69. Epub 2008/02/13. 10.1016/j.devcel.2008.01.013 . [DOI] [PubMed] [Google Scholar]
- 42.Bowling SA, Clarke JD, Liu YD, Klessig DF, Dong XN. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. The Plant cell. 1997; 9(9): 1573–84. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R, et al. A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell host & microbe. 2014; 16(6): 787–94. 10.1016/j.chom.2014.10.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leyser HMO, Furner LI. Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development. 1992; 116: 397–403. [Google Scholar]
- 45.Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993; 119(2): 397–418. . [DOI] [PubMed] [Google Scholar]
- 46.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989; 58(1): 15–25. . [DOI] [PubMed] [Google Scholar]
- 47.Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, et al. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. The EMBO journal. 2006; 25(23): 5579–90. 10.1038/sj.emboj.7601434 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell. 2001; 104(1): 131–42. . [DOI] [PubMed] [Google Scholar]
- 49.Hennig L, Taranto P, Walser M, Schonrock N, Gruissem W. Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development. 2003; 130(12): 2555–65. . [DOI] [PubMed] [Google Scholar]
- 50.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004; 116(1): 51–61. . [DOI] [PubMed] [Google Scholar]
- 51.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999; 96(4): 575–85. . [DOI] [PubMed] [Google Scholar]
- 52.Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, et al. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Molecular and cellular biology. 2000; 20(4): 1206–18. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridgway P, Almouzni G. CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. Journal of cell science. 2000; 113 (Pt 15): 2647–58. . [DOI] [PubMed] [Google Scholar]
- 54.Prado F, Cortes-Ledesma F, Aguilera A. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO reports. 2004; 5(5): 497–502. 10.1038/sj.embor.7400128 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Molecular cell. 2003; 11(2): 341–51. . [DOI] [PubMed] [Google Scholar]
- 56.Harper JW, Elledge SJ. The DNA damage response: Ten years after. Molecular cell. 2007; 28(5): 739–45. 10.1016/j.molcel.2007.11.015 . [DOI] [PubMed] [Google Scholar]
- 57.Kirik A, Pecinka A, Wendeler E, Reiss B. The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. The Plant cell. 2006; 18(10): 2431–42. 10.1105/tpc.106.045088 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hisanaga T, Ferjani A, Horiguchi G, Ishikawa N, Fujikura U, Kubo M, et al. The ATM-Dependent DNA Damage Response Acts as an Upstream Trigger for Compensation in the fas1 Mutation during Arabidopsis Leaf Development. Plant physiology. 2013; 162(2): 831–41. 10.1104/pp.113.216796 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dudenhoffer C, Kurth M, Janus F, Deppert W, Wiesmuller L. Dissociation of the recombination control and the sequence-specific transactivation function of P53. Oncogene. 1999; 18(42): 5773–84. 10.1038/sj.onc.1202964 . [DOI] [PubMed] [Google Scholar]
- 60.Willers H, McCarthy EE, Wu B, Wunsch H, Tang W, Taghian DG, et al. Dissociation of p53-mediated suppression of homologous recombination from G1/S cell cycle checkpoint control. Oncogene. 2000; 19(5): 632–9. 10.1038/sj.onc.1203142 . [DOI] [PubMed] [Google Scholar]
- 61.Hartung F, Puchta H. What Comparative Genomics Tells Us About the Evolution of Eukaryotic Genes Involved in Recombination. Current Genomics. 2004; 5(2): 109–21. [Google Scholar]
- 62.Manova V, Gruszka D. DNA damage and repair in plants—from models to crops. Frontiers in plant science. 2015; 6: 885 10.3389/fpls.2015.00885 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001; 294(5547): 1713–6. 10.1126/science.1065521 . [DOI] [PubMed] [Google Scholar]
- 64.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003; 300(5625): 1542–8. 10.1126/science.1083430 . [DOI] [PubMed] [Google Scholar]
- 65.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005; 434(7033): 605–11. 10.1038/nature03442 . [DOI] [PubMed] [Google Scholar]
- 66.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005; 308(5721): 551–4. 10.1126/science.1108297 . [DOI] [PubMed] [Google Scholar]
- 67.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nature cell biology. 2006; 8(1): 37–45. 10.1038/ncb1337 . [DOI] [PubMed] [Google Scholar]
- 68.Carreira A, Kowalczykowski SC. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2011; 108(26): 10448–53. 10.1073/pnas.1106971108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nature structural & molecular biology. 2007; 14(6): 475–83. 10.1038/nsmb1251 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nature structural & molecular biology. 2007; 14(6): 468–74. 10.1038/nsmb1245 . [DOI] [PubMed] [Google Scholar]
- 71.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual review of plant biology. 2004; 55: 373–99. 10.1146/annurev.arplant.55.031903.141701 . [DOI] [PubMed] [Google Scholar]
- 72.Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A. AtATM is essential for meiosis and the somatic response to DNA damage in plants. The Plant cell. 2003; 15(1): 119–32. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Missirian V, Conklin PA, Culligan KM, Huefner ND, Britt AB. High atomic weight, high-energy radiation (HZE) induces transcriptional responses shared with conventional stresses in addition to a core "DSB" response specific to clastogenic treatments. Frontiers in plant science. 2014; 5: 364 10.3389/fpls.2014.00364 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen IP, Haehnel U, Altschmied L, Schubert I, Puchta H. The transcriptional response of Arabidopsis to genotoxic stress—a high-density colony array study (HDCA). The Plant journal: for cell and molecular biology. 2003; 35(6): 771–86. . [DOI] [PubMed] [Google Scholar]
- 75.Heitzeberg F, Chen IP, Hartung F, Orel N, Angelis KJ, Puchta H. The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. The Plant journal: for cell and molecular biology. 2004; 38(6): 954–68. 10.1111/j.1365-313X.2004.02097.x . [DOI] [PubMed] [Google Scholar]
- 76.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009; 137(4): 609–22. 10.1016/j.cell.2009.04.050 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Doonan J, Hunt T. Cell cycle. Why don't plants get cancer? Nature. 1996; 380(6574): 481–2. 10.1038/380481a0 . [DOI] [PubMed] [Google Scholar]
- 78.Doonan JH, Sablowski R. Walls around tumours—why plants do not develop cancer. Nature reviews Cancer. 2010; 10(11): 794–802. 10.1038/nrc2942 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Proteins were extracted from 10-day-old seedlings of wild type (WT) and three lines of p53-transgenic plants (p53, L1-L3), resolved on sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS/PAGE), and immunoblotted with anti-p53 antibody. Anti-α-tubulin was used as an internal loading control. (B) Proteins were extracted from 10-day-old seedlings of wild type (WT) and p53-transgenic (L1: line1) WT plants, sni1mutant, rad51d mutant, and GUS (1445) reporter. Immunoblot was performed as panel A.
(TIF)
The transfected U2OS cancer cells were treated with 10 Gy of ionizing radiation (IR) or hydroxyurea (HU). Proteins extracted from the transfected transfected U2OS cancer cells were blotted with anti-HA antibody (abcam, ab1265). Anti-α-tubulin was used as an internal loading control.
(TIF)
(A) Total RNA was extracted from ten-day-old wild type and p53-transgenic seedlings. The A260/A280 ratio of the total RNA was about 2.0 as measured on NanoDrop 2000 Spectrophotometer. The quality of RNA was further assessed by agarose gel electrophoresis. The 28S/18S ratio was about 2.0, indicating that the isolated RNAs were not degraded. (B) The qPCR products of UBQ5 and RAD51D were viewed by agarose gel electrophoresis. The qPCR product size of UBQ5 and RAD51D is about 250 bp and 100 bp, respectively. M, DNA marker. (C) The melting curves from qPCR analysis of UBQ5 and RAD51D. There was only one peak appeared in the melting curves of qPCR analysis in both UBQ5 and RAD51D, which was consistent with the agarose gel electrophoresis of qPCR product in panel B. (D) Serial dilutions of reverse transcription product (cDNA) were used for the qPCR quality assay of UBQ5 and RAD51D. Ct, cycle threshold.
(TIF)
(A) The signaling pathway of DNA damage repair through homologous recombination in mammals and plants. In mammals, the MRE11/RAD50/NBS1 (MRN) complex functions as double strand brake (DSB) sensors. RAD17 recruits the MRN complex to the DSB site. In response to DNA damage, two phosphoinositide 3-kinase-like kinases, ATM and ATR, are activated by the MRN complex to phosphorylate the transcription factor p53. p21, a target of p53, is an inhibitor of cyclin-dependent kinase (CDK). KIP-RELATED PROTEIN (KRP) is the homolog of p21 in plant. CDK phosphorylates BREAST CANCER 2 (BRCA2) which is a mediator of the recombinase RAD51. BRCA2 first loads RAD51 on the DSB site. After the DSB is repaired by RAD51, BRCA2 is then phosphorylated by CDK to remove RAD51 from DNA. Plants possess the homologs of these DNA repair proteins, except for p53. On the contrary, mammals do not bear SNI1. SNI1 is a negative regulator of homologous recombination in plants. Although genetic study reveals that SNI1 functions upstream of ATR, RAD17, BRCA2, and RAD51, how SNI1 is regulated remains unknown. (B) A proposed model of p53 action in plant. p53 induces homologous recombination through the SNI1-RAD51D signaling pathway in plant, leading to senescence and fasciation.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Microarray data are available from the Gene Expression Omnibus (GEO) database (accession number: GSE79678) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79678). All other relevant data are within the paper and its Supporting Information files.