Abstract
GP63 or leishmanolysin is the major surface protease of Leishmania spp. involved in parasite virulence and host cell interaction. As such, GP63 is a potential target of eventual vaccines against these protozoa. In the current study we evaluate the polymorphism of gp63 in Leishmania (Viannia) braziliensis isolated from two sets of American tegumentary leishmaniasis (ATL) cases from Corte de Pedra, Brazil, including 35 cases diagnosed between 1994 and 2001 and 6 cases diagnosed between 2008 and 2011. Parasites were obtained from lesions by needle aspiration and cultivation. Genomic DNA was extracted, and 405 bp fragments, including sequences encoding the putative macrophage interacting sites, were amplified from gp63 genes of all isolates. DNA amplicons were cloned into plasmid vectors and ten clones per L. (V.) braziliensis isolate were sequenced. Alignment of cloned sequences showed extensive polymorphism among gp63 genes within, and between parasite isolates. Overall, 45 different polymorphic alleles were detected in all samples, which could be segregated into two clusters. Cluster one included 25, and cluster two included 20 such genotypes. The predicted peptides showed overall conservation below 50%. In marked contrast, the conservation at segments with putative functional domains approached 90% (Fisher’s exact test p<0.0001). These findings show that gp63 is very polymorphic even among parasites from a same endemic focus, but the functional domains interacting with the mammalian host environment are conserved.
Introduction
The Leishmania species parasite cause a variety of clinical syndromes in individuals living in tropical and subtropical areas of the globe [1]. An estimated 1.5 to 2 million people develop symptomatic leishmaniasis per year, and approximately 12 million people are infected worldwide [2]. American Tegumentary Leishmaniasis (ATL) refers to cutaneous forms of leishmaniasis in the New World, caused by species of the subgenera Leishmania Viannia and Leishmania Leishmania [1, 3]. L. (V.) braziliensis is the main cause of ATL in South America, including Brazil [4, 5].
The standard treatment for ATL in Brazil is pentavalent antimony [Sb(V)] at a daily dose of 15–20 mg/Kg for 20 to 30 days [5]. However, up to 50% of patients fail Sb(V) therapy [6–9]. This scenario is even worse in areas of high L. (V.) braziliensis endemicity such as Corte de Pedra in the state of Bahia, Brazil. The most common form of disease due to L. (V.) braziliensis, and the most responsive to therapy, is cutaneous leishmaniasis (CL). More difficult to treat forms of ATL in the region include the classically recognized mucosal leishmaniasis (ML), and emerging clinical forms such as disseminated leishmaniasis (DL) and atypical cutaneous leishmaniasis (ACL). Patients with these unusual forms tend to respond poorly to Sb(V), with failure occurring in up to 60–90% of these individuals [10–16]. Such experience has led investigators to realize the need for new treatment modalities for ATL [9, 16, 17].
Leishmanolysin, or glycoprotein 63 (GP63), is a major surface protease (MSP) of Leishmania [18–21], capable of hydrolyzing a variety of substrates in the parasite’s immediate environment within the host [18]. GP63 genes are expressed in promastigotes and amastigotes, and their products are involved in the adhesion to and internalization of the parasite by the host macrophages [19, 22–25]. Furthermore, GP63 is in part responsible for Leishmania spp. to migrate through extracellular matrix, to avoid lysis by inactivating components of the complement system, and to hydrolyze intracellular macrophage targets [26, 27]. Its increased expression has been correlated with increased virulence of L. (V.) braziliensis [28].
Given its role in pathogenesis, GP63 might prove a good target for treatment of or prophylactic immunization against leishmaniasis [21, 29]. Its use might be complicated by the variability in genes encoding these molecules [30, 31]. However, peptides synthesized according to short regions in GP63, conserved across different species of Leishmania, have been shown to inhibit internalization of the parasite by macrophages [32]. Effective inhibitory oligopeptides surround the sequences SRYD, involved in binding macrophage surface receptors, and HExxH, an essential region for metalloprotease activity [32]. The goal of the current study was to determine whether these short regions are conserved in gp63 genes from a panel of L. (V.) braziliensis isolates from a variety of patients whose diagnosis is temporally distributed over time. The study utilized parasites isolated from ATL patients of Corte de Pedra, Brazil. The results provide the basis for considering these peptides for therapeutic use in management of leishmaniasis. Furthermore, polymorphic gp63 alleles could potentially serve as molecular markers of functionally distinct L. (V.) braziliensis isolates.
Materials and Methods
Study area
Corte de Pedra is composed of 20 municipalities in a rural area located in the southeastern region of the state of Bahia, in the northeast of Brazil within geographic coordinates (latitude/longitude) 14°/39°, 13°/39°, 14°/40°, 13°/40°. Lutzomyia (Nyssomyia) whitmany and L. (N.) intermedia sandflies are the most important vectors of L. (V.) braziliensis in this endemic area [33]. The residents work mostly in agriculture, a vocation that often takes them into primary or secondary forests. There is little population migration in or out of this region. Study participants’ mean time of residence at their addresses at the time of diagnosis and parasite sampling was 17 years. 90% of the study participants lived on farms.
ATL patients’ disease definitions
CL consists of an ulcerated skin lesion at a single body site with no more than two secondary or satellite lesions, without clinical evidence of mucosal involvement. ML was defined as the presence of an inflamed or ulcerated mucosal lesion(s) at a site that is noncontiguous with any cutaneous lesion. Subjects with ML might have concomitant lesions of CL, but not always. DL was defined as 10 or more skin lesions of mixed types (acneiform, papular, nodular, and/or ulcerated) located on two or more body parts (head, trunk, arms and legs).
Parasites
The L. (V.) braziliensisis isolates analyzed in the present study were obtained by culture of aspirates from the borders of skin or mucosal lesions. Aspirate material was immediately suspended in biphasic liver infusion tryptose-Novy, McNeal and Nicolle medium (LIT/NNN) and incubated at 26°C for 1 to 2 weeks. The suspension was transferred to Schneider’s medium complemented with 10% heat-inactivated fetal calf serum and 2 mM L-glutamine, and incubated at 26°C for up to an additional 2 weeks. Parasites were frozen without further subculture in 10% dimethyl sulfoxide (DMSO) 90% growth medium and maintained in liquid nitrogen until used.
Thirty-five L. (V.) braziliensis isolates were obtained from 17 individuals with CL, 9 with ML and 9 with DL, between 1992 and 2001. Six L. (V.) braziliensis isolates derived from 2 individuals with CL, 2 with ML and 2 with DL diagnosed between 2008 and 2011. All patients were diagnosed at the outpatient clinic in the health post of Corte de Pedra.
L. (V.) braziliensis genomic DNA extraction and parasite species determination by PCR
Genomic DNA was extracted from approximately 106 promastigotes of each isolate. Briefly, parasites were pelleted and suspended in 150μL of TELT buffer (50mM Tris-HCl pH 8.0, 62.5mM EDTA pH 9.0, 2.5M LiCl, 4% v/v Triton x 100) for 5 min at room temperature, followed by phenol-chloroform extraction (150μL) to remove protein and lipids. Nucleic acids were precipitated with ethanol (300μL), followed by an ethanol rinse (1,000μL). The pellets were suspended in 100μL of TE buffer (Tris-HCl 10mM, EDTA 1mM pH 8.0). Samples were stored at either –20°C or -70°C until used. The determination of the infecting Leishmania species was performed by real-time qPCR assay, using primers based on sequences of KDNA1, KDNA3 and MAG1 as previously described [34].
PCR amplification and cloning of selected targets
The gp63 used as reference for the alignment encodes a 557 amino acids peptide: "Leishmania braziliensis MHOM/BR/75/M2904 GP63 leishmanolysin (LBRM_10_0540)", accessed at www.ncbi.nlm.nih.gov/nuccore/XM_001562773.2. We focused the regions that encode the putative functional sites (macrophage binding, protease activity) of GP63 within the host [32]. A fragment of gp63 genes containing these sites was amplified with the forward 5:ATGTCCCGCGACCGCAGCAG and reverse 5:TCACACCGCCGCTGTGTCGG primers in a 50μL reaction volume, in a 96-well thermal cycler Veriti® from Applied Biosystems.
Amplified products were separated in 1.3% agarose gels, stained with ethidium bromide and visualized with a UV trans-illuminator—digital imager (UVP Labworks Laboratory Imaging and Analysis System Inc., CA, EUA) in order to confirm the amplicons were the expected 405 bp size.
Amplified gp63 fragments were cloned using the Original TA Cloning Kit pCR 2.1 VECTOR (Invitrogen), according to manufacturer’s instructions. The recombinant PCR 2.1 plasmids were transformed into competent DH5α Escherichia coli [35]. Ten colonies with inserts were selected per studied L. (V.) braziliensis isolate.
Sequence analysis
Plasmid inserts were sequenced by the Sanger method at Macrogen Inc. (Seoul, South Korea), with M13 sequencing primers flanking PCR 2.1 cloning sites. Sequencing was bidirectional and only sequences with 100% identity among overlapping insert strands were recorded. Insert sequences were aligned to the Leishmania braziliensis MHOM/BR/75/M2904 GP63 leishmanolysin (LBRM_10_0540) gene sequence with MEGA 5.0 software. ClustaW algorithm was used in the process and no manual adjustments to maximize sequence alignments were necessary.
Upon alignment we searched for events of SNP and indels among the cloned 405 bp gp63 fragments. Polymorphism among sequences of gp63 fragments and their predicted peptides was evaluated manually and confirmed using the Dna Sequence Polymorphism software, version 5.10.01 [36, 37]. We defined a polymorphism as a single bp difference between isolates; a polymorphic allele as a linear gp63 DNA sequence detected in more than one clone per parasite isolate, and in more than one isolate of L. (V.) braziliensis in our study sample.
Classification of gp63 alleles employed Neighbor Joining algorithm. Consistency of nodes within dendrogram was tested by bootstrap.
Statistical Analysis
Differences in the distribution frequencies of each polymorphism detected among gp63 fragments were analyzed by Fisher’s exact test. Results with p ≤ 0.05 were considered statistically significant.
Ethics statement
This study was approved by the institutional review board of the Federal University of Bahia Medical College, under document number CAAE: 37297614.0.0000.5577.
Results
Identification of polymorphisms in the vicinity of the putative macrophage binding region of gp63 among human isolates of L. (V.) braziliensis
Forty-five alleles of gp63 could be distinguished among 410 clones of the gene fragment evaluated in thirty-five isolates of L. (V.) braziliensis collected between 1992 and 2001, and six isolates of the parasite drawn between 2008 and 2011 from ATL patients of Corte de Pedra. The median number of alleles per parasite isolate was 5, ranging from 2 to 9. One hundred eighty-six polymorphic positions were found in the 405 bp fragment, considering all forty-five alleles of the gene (Fig 1). This results in an average of one polymorphic position every 2.2 base-pairs. The frequency of polymorphic positions varied from eight in allele 28 to one hundred one in allele 8 (Fig 1), with a median of 93 polymorphisms per allele (Table 1). 169 polymorphisms consisted of nucleotide substitutions. Many variations occurred in the first two nucleotides of codons, predicting amino acid replacements in 73 (54%) of the translated proteins. Silent changes were also observed in only 52 (31%) nucleotide positions. Nucleotide positions 579, 594, 795 and 906 had either amino acid replacement or silent polymorphisms in different gp63 alleles (Fig 1).
Fig 1. Alignment of polymorphic nucleotide positions of 45 polymorphic alleles found for the gp63 gene fragment studied.
Alleles 1 to 28 were detected among 35 different isolates of L. (V.) braziliensis from ATL patients diagnosed between 1992 and 2001 in Corte de Pedra, Brazil. Alleles 29 to 45 were detected among isolates of the parasite drawn from six different patients of the same area between 2008 and 2011. Alignment was generated by MEGA 5.0 software. Numbers boxed in top three rows correspond to nucleotide position in the gp63 gene used as reference (see methods). Top sequence in bold corresponds to the nucleotides found in each displayed position in the reference gp63 sequence. Rows numbered 1 to 45 at left correspond to the 45 alleles of gp63 detected in Corte de Pedra. Dots indicate same nucleotide as reference sequence, while letters indicate the substituting nucleotides in the study fragments. (*) Positions with silent polymorphisms in study gp63 fragments. (#) Positions with polymorphisms that may be silent or lead to predicted amino acid substitution (highlighted in gray), depending on the study gp63 allele. All other positions resulted in predicted amino acid substitution in study gp63 alleles.
Table 1. Counts of nucleotide/amino acid polymorphisms in nucleic acid/predicted amino acid sequences from 405 base-pairs study fragments of L. (V.) braziliensis gp63.
Columns show data for all forty-five gp63 alleles (total) identified and alleles stratified according to clustering analysis depicted in Fig 2 (clades A to D).
| TOTAL | CLADE A | CLADE B | CLADE C | CLADE D | |
|---|---|---|---|---|---|
| (nt/aa) | (nt/aa) | (nt/aa) | (nt/aa) | (nt/aa) | |
| Number of alleles | 45 | 25 | 8 | 5 | 7 |
| Lowest count of polymorphisms per allele | 8/7 | 86/39 | 89/47 | 50/29 | 8/7 |
| Highest count of polymorphisms per allele | 101/52 | 101/45 | 93/52 | 54/35 | 49/30 |
| Median count of polymorphisms per allele | 93/43 | 94/43 | 92,5/51 | 53/34 | 42/27 |
* nt/aa = nucleotides/amino acids.
Classification of gp63 alleles
Neighbor-Joining classification revealed two distinct clusters of alleles, which could be further subdivided into four smaller clades (Fig 2; clades A to D). We defined as clade each discrete group of gp63 alleles immediately lower hierarchically to the cluster nodes in the dendrogram. Cluster 1 included 25 gp63 alleles, all belonging to clade A. Cluster 2 included 20 alleles distributed across clades B, C and D.
Fig 2. Neighbor-Joining classification of 45 alleles of the gp63 study fragment found in 41 isolates of L. (V.) braziliensis from Corte de Pedra, Brazil.
Nucleotide sequence alignments and gp63 allele classification employed MEGA 5.0 software. Branch tips correspond to each gp63 fragment allele. Clusters 1 and 2 correspond to the major aggregates, while clades A to D correspond to the secondary aggregates of alleles. Black dots indicate alleles detected among 35 different isolates of L. (V.) braziliensis obtained from ATL patients in Corte de Pedra between 1992 and 2001. White dots indicate alleles in parasites drawn from six different patients of the same area between 2008 and 2011. Percentages at the nodes of the dendrogram consist in bootstrap values.
The predicted peptides reveal that segments of gp63 encoding functional peptides are highly conserved in the natural population of L. (V.) braziliensis
The predicted peptide encoded by the gp63 fragment is 135 amino acids long. The number of polymorphic amino acid positions varied from seven in allele 28 to fifty-two in alleles 38 and 40, with a median of 43 amino acid changes per allele (Fig 3, Table 1). In 54% of these polymorphic positions there were changes in classes of predicted amino acids between alleles (Fig 3). The 45 gp63 translated alleles presented an overall conservation of 46%.
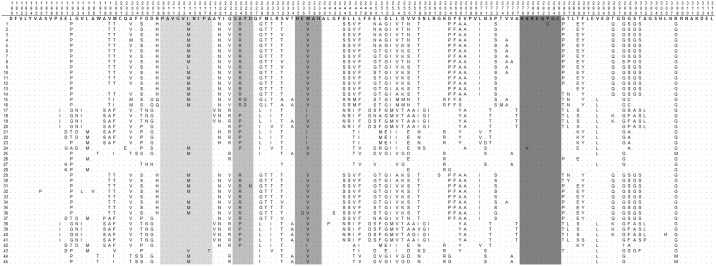
Fig 3. Alignment of amino acid positions among peptides predicted from 45 alleles found for the gp63 gene fragment studied.
The 45 translated alleles were detected among 41 different isolates of L. (V.) braziliensis from ATL patients of Corte de Pedra. Alleles 1 to 28 derived from 35 parasite isolates obtained from ATL patients between 1992 and 2001. Alleles 29 to 45 derived from 6 parasite isolates obtained from ATL patients between 2008 and 2011. Alignment was generated by MEGA 5.0 software. Numbers boxed in top three rows correspond to amino acid positions in the gp63 gene used a reference (see methods). Top sequence in bold corresponds to the amino acids found for each displayed position in the reference gp63 sequence. Rows numbered 1 to 45 at left correspond to the 45 alleles of gp63 detected in Corte de Pedra. Dots indicate same amino acids as in reference sequence, while letters indicate the substituting amino acids in the study fragments. Regular letters indicate hydrophilic, italicized letters indicate intermediary, and bold letters indicate hydrophobic amino acids. Shaded positions correspond to the segments PAVGVINIPA, SRYD, HEVAH and KAREQYGC referred to in text.
Despite the overall low conservation of the predicted GP63 fragment studied, the two previously described functional regions, defined by the primary sequences SRYD (macrophage receptor binding) and HEVAH (metalloprotease activity) [32] exhibited approximately 70% identity across the 45 gp63 alleles. There were two other highly conserved short regions that flanked the SRYD and HEVAH sequences (PAVGNIPA and KAREQYGC). Altogether, these four stretches of amino acids encompassed 27 of the 135 evaluated residues (Fig 3, shaded positions). The data showed approximately 89% identity between these peptides across the predicted polypeptides of all gp63 alleles sequenced. This is in marked contrast with the overall identity of 46% of predicted protein residues in the full peptide fragments (Fisher’s exact p <0.0001; Fig 3).
A BLAST search of Genbank deposited sequences using PAVGNIPA, HEVAH, SRYD and KAREQYGC further displayed that these segments were also conserved among L. (V.) braziliensis [38], L. (V.) guyanensis [39], L. (V.) panamensis [40], L. (L.) Mexicana [38], L. major [41], L.donovani [42] and L. infantum [38].
Novel gp63 alleles arise in natural L. (V.) braziliensis populations overtime
Twenty-eight alleles of gp63 could be distinguished among 350 clones of the gene fragment evaluated in thirty-five isolates of L. (V.) braziliensis, collected between 1992 and 2001 from ATL patients of Corte de Pedra. The median number of alleles per parasite isolate was 5, ranging from 2 to 9. In the sample of parasites obtained from six ATL patients of Corte de Pedra between 2008 and 2011, twenty-five alleles could be discriminated among 60 clones of the 405 bp gp63 fragment evaluated. The median number of alleles per parasite isolate was 4.5, ranging from 1 to 8. Remarkably, seventeen novel alleles could be identified in this latter sample (Fig 1, gp63 29 to 45). These alleles distributed throughout all defined clades of gp63 in Corte de Pedra (Fig 2, white dots).
Discussion
Polymorphic alleles of mammalian hosts have been a focus of investigations into the diverse outcome of infection with different Leishmania species. Our prior work has underscored the fact that polymorphic isolates of L. (V.) braziliensis are independently associated with different clinical outcomes of infection [43, 44]. These analyses have focused on anonymous markers that are not associated with any functional significance, in part due to a lack of knowledge of genomic markers in genes of functional significance. In the current study, we investigated the variability in the coding regions of gp63 genes, focusing on peptides with known functional significance in the host. We were able to analyze the variability between gp63 genes among clinical isolates from a population of individuals naturally infected with L. (V.) braziliensis. We found that gp63 genes are highly polymorphic, but that sequences encoding the functional peptides involved in macrophage binding, or in protease activity were remarkably conserved among parasites from one of the regions with highest endemicity of ATL, in northeast Brazil.
GP63 is encoded by tandemly repeated gene clusters in all species in which the gene organization has been investigated [28]. Study of gp63 gene cluster in L. (V.) braziliensis revealed approximately 37 genes in the cluster, with 8 distinct classes [31]. Our experimental approach, in which we selected 10 clones from each isolate for sequence analysis, could not distinguish the full spectrum of polymorphic gp63 genes in single isolates. Nonetheless the polymorphism of the total population was much higher than expected from a single parasite clone, and the number of polymorphic alleles greatly exceeded the expected 8 reported. As such, this analysis leads to a reasonable conclusion that the polymorphism of gp63 genes in the entire population exceeds that expected in individual L. (V.) braziliensis isolates. Further documentation of isolates containing different gp63 genes will require the generation of allele-specific markers from our sequence information, and re-examination of these specific polymorphisms in the collection of clinical isolates.
Polymorphism concentrated in stretches of the gene that encode segments of GP63 that do not directly participate in the interaction between parasite and host cell. We speculate that population-wise the extensive variability of GP63 may be in part driven by selective pressure caused by the host immune responses, during infection with Leishmania spp. Studies suggest that GP63 polymorphisms are more abundant in segments of the protein that serve as epitopes for T and B cells [45, 46]. The existence of several dozen, and potentially hundreds of gp63 alleles within a single L. (V.) braziliensis population might perhaps allow the parasite overcome host herd immunity, and maintain endemicity at each successive transmission season of ATL.
We detected four different clades of alleles, and a median of approximately five distinct alleles per parasite isolate. This suggests that gp63 polymorphism in the natural population of L. (V.) braziliensis from Corte de Pedra may be warranted by multiple loci and / or gene copies distributed in different chromosomes. This hypothesis is consistent with observations in previous studies of both physical mapping and whole genome sequencing of L. (V.) braziliensis strains [47, 48]. It is important to note that we analyzed gp63 amplified from the genomes of non-cloned parasites by specific PCR primers. As such, the findings reported herein may consist in an underestimation of the actual complexity of these loci, in as much as they may also be influenced by isolates comprised of multiple strains of L. (V.) braziliensis. Future studies should employ deep sequencing approaches to address these limitations. Besides, since we did not evaluate gene expression then we cannot conclude on the functionality of detected alleles.
GP63 and the complement receptor CR3 on host-cells seem to interact in part via the leishmanolysin segment containing SRYD amino acids [49]. SRYD is highly conserved among Leishmania spp., and monoclonal antibodies against this oligo-peptide inhibit internalization of L. infantum by macropages [32]. Four short stretches of the putative macrophage interacting segment of GP63 proved highly conserved among L. (V.) braziliensis from Corte de Pedra. These include SRYD and HExxH. The active site at the N-terminal proteinase domain of leishmanolysin contains the HExxH sequence conserved in all species of Leishmania [32].
The other two conserved amino acid stretches were PAVGNIPA and KAREQYGC, which flank SRYD and HExxH. The consistent conservation of PAVGNIPA and KAREQYGC among parasites of Corte de Pedra suggests that these segments may also play a role in L. (V.) braziliensis host cell interaction. Reinforcing this hypothesis, the review of sequences for other species of Leishmania, deposited in Genbank or published in different papers, reveal the presence for PAVGNIPA and KAREQYGC in all of them [[32], GeneBank accession numbers: CBZ24358.1, AIN96110.1, AAC39120.1, XP_001463700.1, AAA29240.1,XP_001562820.1 and AAA53688.1]. Nevertheless biological testing with inhibition assays is necessary to ascertain that these segments are really functional for the interplay between host and parasite cells.
Proteases fulfill important roles during host infection, microbe survival and pathogenicity in several protozoa [50, 51]. GP63 (also called leishmanolysin or MSP) is one such metallo-enzyme produced by Leishmania spp. [52–54]. GP63 has been described to participate in complement resistance, migration through extracellular matrix, establishment of intracellular parasitism and interference with intracellular microbicidal mechanisms of infected cells [55–59]. Finally, several studies in experimental cutaneous and visceral leishmaniasis (VL) have shown efficacy of various formulations of GP63 administered through different routes in protective immunity in mouse models [21, 60–68].
Experimental studies do not take into account the complexity of natural pathogen populations. As in the reports cited above, immunization and challenge are usually carried out with a single pathogenic strain and its components. This approach is sound, but does not take into account the variability that some antigens used as immunogens may present within and / or between human disease transmission foci.
As we demonstrate in this molecular epidemiology study, GP63 is one such example of a highly variable molecule. Thus experimental results may not easily translate into its successful use as an immune prophylaxis reagent in affected regions. Its ample variability reported for Corte de Pedra likely reflects the realm found in other affected regions as well.
The polymorphisms in gp63 genes reported herein might prove particularly interesting as molecular markers of different parasite isolates. As we previously showed, distinct L. (V.) braziliensis clades are associated with different clinical outcomes of infection [43, 44]. If future investigations are able to discover that polymorphisms in this highly important functional protein associate with distinct outcomes of infection, this could have implications for GP63 function in different disease forms. Focusing on polymorphic markers avoids the need to define all gp63 genes present in isolates, but rather highlights only those genes that are different between isolates. This approach might be expanded to study polymorphisms in other known Leishmania spp. proteins important in pathogenesis.
Acknowledgments
We would like to deeply thank all personnel in the health post of Corte de Pedra, Bahia / Brazil, for their careful help with patient management, and Ms. Kátia Salgado for laboratory support with parasite isolation and management.
Data Availability
All relevant files are available at https://figshare.com under the following DOIs: https://dx.doi.org/10.6084/m9.figshare.3507908.v2; https://dx.doi.org/10.6084/m9.figshare.3509066.v2; https://dx.doi.org/10.6084/m9.figshare.3509429.v2; https://dx.doi.org/10.6084/m9.figshare.3509432.v2; https://dx.doi.org/10.6084/m9.figshare.3509435.v2; https://dx.doi.org/10.6084/m9.figshare.3509438.v2; https://dx.doi.org/10.6084/m9.figshare.3509441.v2; https://dx.doi.org/10.6084/m9.figshare.3509447.v2; https://dx.doi.org/10.6084/m9.figshare.3509453.v2; https://dx.doi.org/10.6084/m9.figshare.3509456.v2; https://dx.doi.org/10.6084/m9.figshare.3509459.v2; https://dx.doi.org/10.6084/m9.figshare.3509462.v2; https://dx.doi.org/10.6084/m9.figshare.3509465.v2; https://dx.doi.org/10.6084/m9.figshare.3509468.v2; https://dx.doi.org/10.6084/m9.figshare.3509471.v2; https://dx.doi.org/10.6084/m9.figshare.3509474.v2; https://dx.doi.org/10.6084/m9.figshare.3509483.v2; https://dx.doi.org/10.6084/m9.figshare.3509495.v2; https://dx.doi.org/10.6084/m9.figshare.3509498.v2; https://dx.doi.org/10.6084/m9.figshare.3509525.v2; https://dx.doi.org/10.6084/m9.figshare.3509531.v2; https://dx.doi.org/10.6084/m9.figshare.3509537.v2; https://dx.doi.org/10.6084/m9.figshare.3509540.v2; https://dx.doi.org/10.6084/m9.figshare.3509555.v2; https://dx.doi.org/10.6084/m9.figshare.3509561.v2; https://dx.doi.org/10.6084/m9.figshare.3509573.v2; https://dx.doi.org/10.6084/m9.figshare.3509579.v2; https://dx.doi.org/10.6084/m9.figshare.3509582.v2; https://dx.doi.org/10.6084/m9.figshare.3509585.v2; https://dx.doi.org/10.6084/m9.figshare.3509591.v2; https://dx.doi.org/10.6084/m9.figshare.3509594.v2; https://dx.doi.org/10.6084/m9.figshare.3509600.v2; https://dx.doi.org/10.6084/m9.figshare.3509603.v2; https://dx.doi.org/10.6084/m9.figshare.3509609.v2; https://dx.doi.org/10.6084/m9.figshare.3509615.v2; https://dx.doi.org/10.6084/m9.figshare.3509621.v2; https://dx.doi.org/10.6084/m9.figshare.3509627.v2; https://dx.doi.org/10.6084/m9.figshare.3509639.v2; https://dx.doi.org/10.6084/m9.figshare.3509642.v2; https://dx.doi.org/10.6084/m9.figshare.3509645.v2; https://dx.doi.org/10.6084/m9.figshare.3509648.v2; https://dx.doi.org/10.6084/m9.figshare.3509651.v2; https://dx.doi.org/10.6084/m9.figshare.3509654.v2; https://dx.doi.org/10.6084/m9.figshare.3509657.v2; https://dx.doi.org/10.6084/m9.figshare.3509660.v2.
Funding Statement
This work was supported by the National Institutes of Health (NIH), USA, through grants NIH P50-AI30639 and NIH R03 A167663. AS and MW were recipients of NIH R03 A167663 and EMC, AS and MW were recipients of NIH P50-AI30639. LSM, AQ and LHG were recipients of CAPES MS and PhD scholarships. BAS was the recipient of a CNPq Scientific Initiation Fellowship scholarship.
References
- 1.Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366(9496):1561–77. 10.1016/S0140-6736(05)67629-5 . [DOI] [PubMed] [Google Scholar]
- 2.(WHO) WHO. Leishmaniasis 2011 [cited 2015 16/12]. Available: http://www.who.int/mediacentre/factsheets/fs375/en/
- 3.Azulay RD, Azulay DR Junior. Immune-clinical-pathologic spectrum of leishmaniasis. Int J Dermatol. 1995;34(5):303–7. Epub 1995/05/01. . [DOI] [PubMed] [Google Scholar]
- 4.Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, Cano J, et al. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7(5):e35671 Epub 2012/06/14. 10.1371/journal.pone.0035671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ministério da Saúde SdVe, Saúde. Manual de Vigilância da Leishmaniose Tegumentar Americana. 2 ed ed. Brasília: 2007. [Google Scholar]
- 6.Romero GA, Guerra MV, Paes MG, Cupolillo E, Bentin Toaldo C, Macedo VO, et al. Sensitivity of the polymerase chain reaction for the diagnosis of cutaneous leishmaniasis due to Leishmania (Viannia) guyanensis. Acta Trop. 2001;79(3):225–9. Epub 2001/06/20. S0001706X01001401 [pii]. . [DOI] [PubMed] [Google Scholar]
- 7.Santos JB, de Jesus AR, Machado PR, Magalhaes A, Salgado K, Carvalho EM, et al. Antimony plus recombinant human granulocyte-macrophage colony-stimulating factor applied topically in low doses enhances healing of cutaneous Leishmaniasis ulcers: a randomized, double-blind, placebo-controlled study. J Infect Dis. 2004;190(10):1793–6. Epub 2004/10/23. JID32461 [pii] 10.1086/424848 . [DOI] [PubMed] [Google Scholar]
- 8.Newlove T, Guimaraes LH, Morgan DJ, Alcantara L, Glesby MJ, Carvalho EM, et al. Antihelminthic therapy and antimony in cutaneous leishmaniasis: a randomized, double-blind, placebo-controlled trial in patients co-infected with helminths and Leishmania braziliensis. Am J Trop Med Hyg. 2011;84(4):551–5. Epub 2011/04/05. 84/4/551 [pii] 10.4269/ajtmh.2011.10-0423 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brito G, Dourado M, Polari L, Celestino D, Carvalho LP, Queiroz A, et al. Clinical and immunological outcome in cutaneous leishmaniasis patients treated with pentoxifylline. Am J Trop Med Hyg. 2014;90(4):617–20. 10.4269/ajtmh.12-0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho EM, Barral A, Costa JM, Bittencourt A, Marsden P. Clinical and immunopathological aspects of disseminated cutaneous leishmaniasis. Acta Trop. 1994;56(4):315–25. Epub 1994/04/01. . [DOI] [PubMed] [Google Scholar]
- 11.Costa JM, Marsden PD, Llanos-Cuentas EA, Netto EM, Carvalho EM, Barral A, et al. Disseminated cutaneous leishmaniasis in a field clinic in Bahia, Brazil: a report of eight cases. J Trop Med Hyg. 1986;89(6):319–23. Epub 1986/12/01. . [PubMed] [Google Scholar]
- 12.Turetz ML, Machado PR, Ko AI, Alves F, Bittencourt A, Almeida RP, et al. Disseminated leishmaniasis: a new and emerging form of leishmaniasis observed in northeastern Brazil. J Infect Dis. 2002;186(12):1829–34. Epub 2002/11/26. JID020615 [pii] 10.1086/345772 . [DOI] [PubMed] [Google Scholar]
- 13.Unger A, O'Neal S, Machado PR, Guimaraes LH, Morgan DJ, Schriefer A, et al. Association of treatment of American cutaneous leishmaniasis prior to ulcer development with high rate of failure in northeastern Brazil. Am J Trop Med Hyg. 2009;80(4):574–9. Epub 2009/04/07. 80/4/574 [pii]. . [PMC free article] [PubMed] [Google Scholar]
- 14.Guimaraes LH, Machado PR, Lago EL, Morgan DJ, Schriefer A, Bacellar O, et al. Atypical manifestations of tegumentary leishmaniasis in a transmission area of Leishmania braziliensis in the state of Bahia, Brazil. Trans R Soc Trop Med Hyg. 2009;103(7):712–5. Epub 2009/06/02. S0035-9203(09)00162-X [pii] 10.1016/j.trstmh.2009.04.019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machado PR, Rosa ME, Costa D, Mignac M, Silva JS, Schriefer A, et al. Reappraisal of the immunopathogenesis of disseminated leishmaniasis: in situ and systemic immune response. Trans R Soc Trop Med Hyg. 2011;105(8):438–44. Epub 2011/07/05. S0035-9203(11)00092-7 [pii] 10.1016/j.trstmh.2011.05.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machado PR, Rosa ME, Guimaraes LH, Prates FV, Queiroz A, Schriefer A, et al. Treatment of Disseminated Leishmaniasis With Liposomal Amphotericin B. Clin Infect Dis. 2015;61(6):945–9. Epub 2015/06/07. civ416 [pii] 10.1093/cid/civ416 . [DOI] [PubMed] [Google Scholar]
- 17.Schriefer A, Guimaraes LH, Machado PR, Lessa M, Lessa HA, Lago E, et al. Geographic clustering of leishmaniasis in northeastern Brazil. Emerg Infect Dis. 2009;15(6):871–6. Epub 2009/06/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bianchini G, Bocedi A, Ascenzi P, Gavuzzo E, Mazza F, Aschi M. Molecular dynamics simulation of Leishmania major surface metalloprotease GP63 (leishmanolysin). Proteins. 2006;64(2):385–90. Epub 2006/05/19. 10.1002/prot.21009 . [DOI] [PubMed] [Google Scholar]
- 19.Sadlova J, Volf P, Victoir K, Dujardin JC, Votypka J. Virulent and attenuated lines of Leishmania major: DNA karyotypes and differences in metalloproteinase GP63. Folia Parasitol (Praha). 2006;53(2):81–90. Epub 2006/08/11. . [PubMed] [Google Scholar]
- 20.Hsiao CH, Yao C, Storlie P, Donelson JE, Wilson ME. The major surface protease (MSP or GP63) in the intracellular amastigote stage of Leishmania chagasi. Mol Biochem Parasitol. 2008;157(2):148–59. Epub 2007/12/11. S0166-6851(07)00300-3 [pii] 10.1016/j.molbiopara.2007.10.008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaur T, Sobti RC, Kaur S. Cocktail of gp63 and Hsp70 induces protection against Leishmania donovani in BALB/c mice. Parasite Immunol. 2011;33(2):95–103. Epub 2011/01/14. 10.1111/j.1365-3024.2010.01253.x . [DOI] [PubMed] [Google Scholar]
- 22.Pandey S, Chakraborti P, Sharma R, Bandyopadhyay S, Sarkar D, Adhya S. Involvement of Leishmania donovani major surface glycoprotein gp63 in promastigote multiplication. J Biosci. 2004;29(1):15–22. Epub 2004/08/03. . [DOI] [PubMed] [Google Scholar]
- 23.Thiakaki M, Kolli B, Chang KP, Soteriadou K. Down-regulation of gp63 level in Leishmania amazonensis promastigotes reduces their infectivity in BALB/c mice. Microbes Infect. 2006;8(6):1455–63. Epub 2006/05/16. S1286-4579(06)00074-8 [pii] 10.1016/j.micinf.2006.01.006 . [DOI] [PubMed] [Google Scholar]
- 24.Mauricio IL, Gaunt MW, Stothard JR, Miles MA. Glycoprotein 63 (gp63) genes show gene conversion and reveal the evolution of Old World Leishmania. Int J Parasitol. 2007;37(5):565–76. Epub 2007/02/07. S0020-7519(06)00443-7 [pii] 10.1016/j.ijpara.2006.11.020 . [DOI] [PubMed] [Google Scholar]
- 25.Cuervo P, Santos AL, Alves CR, Menezes GC, Silva BA, Britto C, et al. Cellular localization and expression of gp63 homologous metalloproteases in Leishmania (Viannia) braziliensis strains. Acta Trop. 2008;106(3):143–8. Epub 2008/04/22. S0001-706X(08)00068-5 [pii] 10.1016/j.actatropica.2008.03.005 . [DOI] [PubMed] [Google Scholar]
- 26.Gomez MA, Contreras I, Halle M, Tremblay ML, McMaster RW, Olivier M. Leishmania GP63 alters host signaling through cleavage-activated protein tyrosine phosphatases. Sci Signal. 2009;2(90):ra58 Epub 2009/10/03. 2/90/ra58 [pii] 10.1126/scisignal.2000213 . [DOI] [PubMed] [Google Scholar]
- 27.Contreras I, Gomez MA, Nguyen O, Shio MT, McMaster RW, Olivier M. Leishmania-induced inactivation of the macrophage transcription factor AP-1 is mediated by the parasite metalloprotease GP63. PLoS Pathog. 2010;6(10):e1001148 Epub 2010/10/27. 10.1371/journal.ppat.1001148 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao C, Donelson JE, Wilson ME. The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Mol Biochem Parasitol. 2003;132(1):1–16. Epub 2003/10/18. S0166685103002111 [pii]. . [DOI] [PubMed] [Google Scholar]
- 29.Lieke T, Nylen S, Eidsmo L, McMaster WR, Mohammadi AM, Khamesipour A, et al. Leishmania surface protein gp63 binds directly to human natural killer cells and inhibits proliferation. Clin Exp Immunol. 2008;153(2):221–30. Epub 2008/08/21. CEI3687 [pii] 10.1111/j.1365-2249.2008.03687.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quispe Tintaya KW, Ying X, Dedet JP, Rijal S, De Bolle X, Dujardin JC. Antigen genes for molecular epidemiology of leishmaniasis: polymorphism of cysteine proteinase B and surface metalloprotease glycoprotein 63 in the Leishmania donovani complex. J Infect Dis. 2004;189(6):1035–43. Epub 2004/03/05. 10.1086/382049 JID31299 [pii]. . [DOI] [PubMed] [Google Scholar]
- 31.Victoir K, Arevalo J, De Doncker S, Barker DC, Laurent T, Godfroid E, et al. Complexity of the major surface protease (msp) gene organization in Leishmania (Viannia) braziliensis: evolutionary and functional implications. Parasitology. 2005;131(Pt 2):207–14. Epub 2005/09/09. . [DOI] [PubMed] [Google Scholar]
- 32.Puentes F, Guzman F, Marin V, Alonso C, Patarroyo ME, Moreno A. Leishmania: fine mapping of the Leishmanolysin molecule's conserved core domains involved in binding and internalization. Exp Parasitol. 1999;93(1):7–22. Epub 1999/08/28. 10.1006/expr.1999.4427 S0014-4894(99)94427-2 [pii]. . [DOI] [PubMed] [Google Scholar]
- 33.Miranda JC, Reis E, Schriefer A, Goncalves M, Reis MG, Carvalho L, et al. Frequency of infection of Lutzomyia phlebotomines with Leishmania braziliensis in a Brazilian endemic area as assessed by pinpoint capture and polymerase chain reaction. Mem Inst Oswaldo Cruz. 2002;97(2):185–8. . [DOI] [PubMed] [Google Scholar]
- 34.Weirather JL, Jeronimo SM, Gautam S, Sundar S, Kang M, Kurtz MA, et al. Serial quantitative PCR assay for detection, species discrimination, and quantification of Leishmania spp. in human samples. J Clin Microbiol. 2011;49(11):3892–904. Epub 2011/11/02. 49/11/3892 [pii] 10.1128/JCM.r00764-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Rozas J. DNA sequence polymorphism analysis using DnaSP. Methods Mol Biol. 2009;537:337–50. Epub 2009/04/21. 10.1007/978-1-59745-251-9_17 . [DOI] [PubMed] [Google Scholar]
- 37.Rozas J, Rozas R. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput Appl Biosci. 1995;11(6):621–5. Epub 1995/12/01. . [DOI] [PubMed] [Google Scholar]
- 38.Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, Depledge DP, et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21(12):2129–42. Epub 2011/11/01. gr.122945.111 [pii] 10.1101/gr.122945.111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinkraus HB, Langer PJ. The protein sequence predicted from a Leishmania guyanensis gp63 major surface glycoprotein gene is divergent as compared with other Leishmania species. Mol Biochem Parasitol. 1992;52(1):141–4. Epub 1992/05/01. 0166-6851(92)90045-L [pii]. . [DOI] [PubMed] [Google Scholar]
- 40.Llanes A, Restrepo CM, Del Vecchio G, Anguizola FJ, Lleonart R. The genome of Leishmania panamensis: insights into genomics of the L. (Viannia) subgenus. Sci Rep. 2015;5:8550 Epub 2015/02/25. srep08550 [pii] 10.1038/srep08550 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voth BR, Kelly BL, Joshi PB, Ivens AC, McMaster WR. Differentially expressed Leishmania major gp63 genes encode cell surface leishmanolysin with distinct signals for glycosylphosphatidylinositol attachment. Mol Biochem Parasitol. 1998;93(1):31–41. Epub 1998/07/14. S0166685198000139 [pii]. . [DOI] [PubMed] [Google Scholar]
- 42.Roberts SC, Swihart KG, Agey MW, Ramamoorthy R, Wilson ME, Donelson JE. Sequence diversity and organization of the msp gene family encoding gp63 of Leishmania chagasi. Mol Biochem Parasitol. 1993;62(2):157–71. Epub 1993/12/01. . [DOI] [PubMed] [Google Scholar]
- 43.Queiroz A, Sousa R, Heine C, Cardoso M, Guimaraes LH, Machado PR, et al. Association between an emerging disseminated form of leishmaniasis and Leishmania (Viannia) braziliensis strain polymorphisms. J Clin Microbiol. 2012;50(12):4028–34. Epub 2012/10/05. 10.1128/jcm.02064-12. PubMed Central PMCID: PMCPmc3503016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schriefer A, Schriefer AL, Goes-Neto A, Guimaraes LH, Carvalho LP, Almeida RP, et al. Multiclonal Leishmania braziliensis population structure and its clinical implication in a region of endemicity for American tegumentary leishmaniasis. Infect Immun. 2004;72(1):508–14. Epub 2003/12/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guerbouj S, Victoir K, Guizani I, Seridi N, Nuwayri-Salti N, Belkaid M, et al. Gp63 gene polymorphism and population structure of Leishmania donovani complex: influence of the host selection pressure? Parasitology. 2001;122 Pt 1:25–35. Epub 2001/02/24. . [DOI] [PubMed] [Google Scholar]
- 46.Victoir K, Dujardin JC. How to succeed in parasitic life without sex? Asking Leishmania. Trends Parasitol. 2002;18(2):81–5. Epub 2002/02/08. S1471492201021997 [pii]. . [DOI] [PubMed] [Google Scholar]
- 47.Victoir K, Dujardin JC, de Doncker S, Barker DC, Arevalo J, Hamers R, et al. Plasticity of gp63 gene organization in Leishmania (Viannia) braziliensis and Leishmania (Viannia) peruviana. Parasitology. 1995;111 (Pt 3):265–73. Epub 1995/09/01. . [DOI] [PubMed] [Google Scholar]
- 48.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39(7):839–47. Epub 2007/06/19. ng2053 [pii] 10.1038/ng2053 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soteriadou KP, Remoundos MS, Katsikas MC, Tzinia AK, Tsikaris V, Sakarellos C, et al. The Ser-Arg-Tyr-Asp region of the major surface glycoprotein of Leishmania mimics the Arg-Gly-Asp-Ser cell attachment region of fibronectin. J Biol Chem. 1992;267(20):13980–5. Epub 1992/07/15. . [PubMed] [Google Scholar]
- 50.Rosenthal PJ. Proteases of protozoan parasites. Adv Parasitol. 1999;43:105–59. Epub 1999/04/24. . [DOI] [PubMed] [Google Scholar]
- 51.Sajid M, McKerrow JH. Cysteine proteases of parasitic organisms. Mol Biochem Parasitol. 2002;120(1):1–21. Epub 2002/02/19. S0166685101004388 [pii]. . [DOI] [PubMed] [Google Scholar]
- 52.North MJ, Coombs GH. Proteinases of Leishmania mexicana amastigotes and promastigotes: analysis by gel electrophoresis. Mol Biochem Parasitol. 1981;3(5):293–300. Epub 1981/09/01. 0166-6851(81)90003-7 [pii]. . [DOI] [PubMed] [Google Scholar]
- 53.Coombs GH. Proteinases of Leishmania mexicana and other flagellate protozoa. Parasitology. 1982;84(1):149–55. Epub 1982/02/01. . [DOI] [PubMed] [Google Scholar]
- 54.Pupkis MF, Coombs GH. Purification and characterization of proteolytic enzymes of Leishmania mexicana mexicana amastigotes and promastigotes. J Gen Microbiol. 1984;130(9):2375–83. Epub 1984/09/01. 10.1099/00221287-130-9-2375 . [DOI] [PubMed] [Google Scholar]
- 55.Chang CS, Chang KP. Monoclonal antibody affinity purification of a Leishmania membrane glycoprotein and its inhibition of leishmania-macrophage binding. Proc Natl Acad Sci U S A. 1986;83(1):100–4. Epub 1986/01/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russell DG, Wilhelm H. The involvement of the major surface glycoprotein (gp63) of Leishmania promastigotes in attachment to macrophages. J Immunol. 1986;136(7):2613–20. Epub 1986/04/01. . [PubMed] [Google Scholar]
- 57.Liu X, Chang KP. Extrachromosomal genetic complementation of surface metalloproteinase (gp63)-deficient Leishmania increases their binding to macrophages. Proc Natl Acad Sci U S A. 1992;89(11):4991–5. Epub 1992/06/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mottram JC, Souza AE, Hutchison JE, Carter R, Frame MJ, Coombs GH. Evidence from disruption of the lmcpb gene array of Leishmania mexicana that cysteine proteinases are virulence factors. Proc Natl Acad Sci U S A. 1996;93(12):6008–13. Epub 1996/06/11. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexander J, Coombs GH, Mottram JC. Leishmania mexicana cysteine proteinase-deficient mutants have attenuated virulence for mice and potentiate a Th1 response. J Immunol. 1998;161(12):6794–801. Epub 1998/12/23. . [PubMed] [Google Scholar]
- 60.Kaur T, Thakur A, Kaur S. Protective immunity using MPL-A and autoclaved Leishmania donovani as adjuvants along with a cocktail vaccine in murine model of visceral leishmaniasis. J Parasit Dis. 2013;37(2):231–9. Epub 2014/01/17. 10.1007/s12639-012-0171-7 171 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mazumder S, Maji M, Ali N. Potentiating effects of MPL on DSPC bearing cationic liposomes promote recombinant GP63 vaccine efficacy: high immunogenicity and protection. PLoS Negl Trop Dis. 2011;5(12):e1429 Epub 2011/12/30. 10.1371/journal.pntd.0001429 PNTD-D-11-00184 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mazumder S, Maji M, Das A, Ali N. Potency, efficacy and durability of DNA/DNA, DNA/protein and protein/protein based vaccination using gp63 against Leishmania donovani in BALB/c mice. PLoS One. 2011;6(2):e14644 Epub 2011/02/12. 10.1371/journal.pone.0014644 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sachdeva R, Banerjea AC, Malla N, Dubey ML. Immunogenicity and efficacy of single antigen Gp63, polytope and polytopeHSP70 DNA vaccines against visceral Leishmaniasis in experimental mouse model. PLoS One. 2009;4(12):e7880 Epub 2009/12/04. 10.1371/journal.pone.0007880 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhowmick S, Ali N. Recent developments in leishmaniasis vaccine delivery systems. Expert Opin Drug Deliv. 2008;5(7):789–803. Epub 2008/07/02. 10.1517/17425247.5.7.789 . [DOI] [PubMed] [Google Scholar]
- 65.Jaafari MR, Ghafarian A, Farrokh-Gisour A, Samiei A, Kheiri MT, Mahboudi F, et al. Immune response and protection assay of recombinant major surface glycoprotein of Leishmania (rgp63) reconstituted with liposomes in BALB/c mice. Vaccine. 2006;24(29–30):5708–17. Epub 2006/06/03. S0264-410X(06)00485-3 [pii] 10.1016/j.vaccine.2006.04.062 . [DOI] [PubMed] [Google Scholar]
- 66.Dumonteil E, Maria Jesus RS, Javier EO, Maria del Rosario GM. DNA vaccines induce partial protection against Leishmania mexicana. Vaccine. 2003;21(17–18):2161–8. Epub 2003/04/23. S0264410X02007697 [pii]. . [DOI] [PubMed] [Google Scholar]
- 67.Papadopoulou G, Karagouni E, Dotsika E. ISCOMs vaccine against experimental leishmaniasis. Vaccine. 1998;16(9–10):885–92. Epub 1998/07/31. S0264-410X(97)00308-3 [pii]. . [DOI] [PubMed] [Google Scholar]
- 68.Xu D, Liew FY. Protection against leishmaniasis by injection of DNA encoding a major surface glycoprotein, gp63, of L. major. Immunology. 1995;84(2):173–6. Epub 1995/02/01. . [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant files are available at https://figshare.com under the following DOIs: https://dx.doi.org/10.6084/m9.figshare.3507908.v2; https://dx.doi.org/10.6084/m9.figshare.3509066.v2; https://dx.doi.org/10.6084/m9.figshare.3509429.v2; https://dx.doi.org/10.6084/m9.figshare.3509432.v2; https://dx.doi.org/10.6084/m9.figshare.3509435.v2; https://dx.doi.org/10.6084/m9.figshare.3509438.v2; https://dx.doi.org/10.6084/m9.figshare.3509441.v2; https://dx.doi.org/10.6084/m9.figshare.3509447.v2; https://dx.doi.org/10.6084/m9.figshare.3509453.v2; https://dx.doi.org/10.6084/m9.figshare.3509456.v2; https://dx.doi.org/10.6084/m9.figshare.3509459.v2; https://dx.doi.org/10.6084/m9.figshare.3509462.v2; https://dx.doi.org/10.6084/m9.figshare.3509465.v2; https://dx.doi.org/10.6084/m9.figshare.3509468.v2; https://dx.doi.org/10.6084/m9.figshare.3509471.v2; https://dx.doi.org/10.6084/m9.figshare.3509474.v2; https://dx.doi.org/10.6084/m9.figshare.3509483.v2; https://dx.doi.org/10.6084/m9.figshare.3509495.v2; https://dx.doi.org/10.6084/m9.figshare.3509498.v2; https://dx.doi.org/10.6084/m9.figshare.3509525.v2; https://dx.doi.org/10.6084/m9.figshare.3509531.v2; https://dx.doi.org/10.6084/m9.figshare.3509537.v2; https://dx.doi.org/10.6084/m9.figshare.3509540.v2; https://dx.doi.org/10.6084/m9.figshare.3509555.v2; https://dx.doi.org/10.6084/m9.figshare.3509561.v2; https://dx.doi.org/10.6084/m9.figshare.3509573.v2; https://dx.doi.org/10.6084/m9.figshare.3509579.v2; https://dx.doi.org/10.6084/m9.figshare.3509582.v2; https://dx.doi.org/10.6084/m9.figshare.3509585.v2; https://dx.doi.org/10.6084/m9.figshare.3509591.v2; https://dx.doi.org/10.6084/m9.figshare.3509594.v2; https://dx.doi.org/10.6084/m9.figshare.3509600.v2; https://dx.doi.org/10.6084/m9.figshare.3509603.v2; https://dx.doi.org/10.6084/m9.figshare.3509609.v2; https://dx.doi.org/10.6084/m9.figshare.3509615.v2; https://dx.doi.org/10.6084/m9.figshare.3509621.v2; https://dx.doi.org/10.6084/m9.figshare.3509627.v2; https://dx.doi.org/10.6084/m9.figshare.3509639.v2; https://dx.doi.org/10.6084/m9.figshare.3509642.v2; https://dx.doi.org/10.6084/m9.figshare.3509645.v2; https://dx.doi.org/10.6084/m9.figshare.3509648.v2; https://dx.doi.org/10.6084/m9.figshare.3509651.v2; https://dx.doi.org/10.6084/m9.figshare.3509654.v2; https://dx.doi.org/10.6084/m9.figshare.3509657.v2; https://dx.doi.org/10.6084/m9.figshare.3509660.v2.