Abstract
Background
Ovarian cancer is the primary cause of death in women diagnosed with gynecological malignancies worldwide. Absence of early symptoms prevents prompt diagnosis or successful therapeutic intervention. P16INK4a is a well-known tumor suppressor gene (TSG). Aberrant methylation of TSG promoter is an important epigenetic silencing mechanism leading to ovarian cancer progression. Studies have reported differences in methylation frequencies of the p16INK4a promoter between ovarian cancer and the corresponding control group. However, the association between p16INK4a promoter methylation and ovarian cancer remains unclear and controversial. Therefore, a meta-analysis was conducted to clarify the relationship between p16INK4a promoter methylation and ovarian cancer.
Methods
PubMed, Web of Science, EMBASE and CNKI were searched to identify eligible studies for the evaluation of the association between p16INK4a promoter methylation and ovarian cancer. Odds ratio (ORs) and 95% confidence intervals (95%CI) were calculated to determine the strength of association between p16INK4a promoter methylation and ovarian cancer.
Results
A total of 612 ovarian cancer patients and 289 controls from 12 eligible studies were included in the meta-analysis. Overall, a significant association was observed between p16INK4a methylation status and ovarian cancer risk using a fixed-effects model (OR = 2.02, 95% CI = 1.39–2.94).
Conclusion
The results of our meta-analysis show that aberrant methylation of p16INK4a promoter was significantly associated with ovarian cancer. It may represent a promising molecular marker to monitor the disease and provides new insights into the treatment of human ovarian cancer.
Introduction
Ovarian cancer is the primary cause of death in women with gynecological malignancies. Based on GLOBOCAN estimates, more than 238, 700 new cancer cases were diagnosed and nearly 151,900 died from ovarian cancer worldwide in 2012 [1]. Absence of early symptoms prevents prompt detection or therapy of ovarian cancer. Approximately 75% of ovarian cancers are diagnosed at an advanced stage [2,3]. Therefore, early diagnosis and prevention depend on the ability to identify genetic and epigenetic events underlying the initiation and progression of the disease. Recent advances in molecular oncology have facilitated the identification and understanding of several genetic and epigenetic events that contribute to ovarian carcinogenesis [4–7].
Molecular genetic alterations, including activation of proto-oncogenes and inactivation of tumor suppressor genes (TSG), may play a key role in tumorigenesis. Epigenetic inactivation of genes following methylation of CpG islands in promoters is one of the most frequent events encountered in human tumors. The tumor suppressor gene p16INK4a is a major target for carcinogenesis in various human tumors [8–11]. It is a negative regulator of cell cycle. P16INK4a prevents the inactivation of retinoblastoma (Rb) protein by inhibiting cyclin-dependent kinases (CDks). Retinoblastoma (Rb) protein has diverse tumor-suppressor functions and plays an important role in apoptosis and cell cycle regulation [12]. Studies have shown that methylation of p16INK4a promoter may play a critical role in the development of ovarian cancer.
Until now, a few studies reported the differences in methylation frequencies of p16INK4a promoter between ovarian cancer and non-cancerous tissues. However, these findings are inconsistent. Therefore, the objectives of this meta-analysis are to consolidate the available data and to clarify the association between p16INK4a promoter methylation and human ovarian cancer.
Materials and Methods
The meta-analysis was performed according to the latest Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA).
Search Strategy
Relevant studies were identified from online electronic databases (PubMed, Web of Science, EMBASE and CNKI) using the following key words: (ovarian OR ovary) AND (cancer OR carcinoma OR tumor) AND (p16 methylation). Articles were retrieved up to May 3, 2016.
Study Selection
Three independent reviewers (Xiyue Xiao, Yi Zhong, and Fucheng Cai) screened the titles and abstracts retrieved in the electronic search to identify relevant studies. The inclusion criteria were: (1) case-control study design, (2) data necessary for calculating odds ratios (ORs), (3) studies primarily evaluating the association between p16INK4a methylation and ovarian cancer, (4) incidence of p16INK4a methylation in both case and control groups, and (5) sample types limited to tissues. According to the inclusion criteria, the title and abstracts from the preliminary search were evaluated. All potentially relevant articles were evaluated in full. Abstracts, letters to the editor and case reports were not included. Finally, a total of 12 articles [13–24] were included in our meta-analysis involving 612 cases and 289 controls.
Data Extraction and Quality Assessment
The selected studies were reviewed by three independent reviewers (Xiyue Xiao, Yi Zhong, and Fucheng Cai). The following information was extracted from the eligible studies: first author’s name, year of publication, study population, the number of people with p16INK4a methylation in the case and control groups, the number of case and control groups, the measurement methods of methylation and control types. All the data in the included studies were checked by two reviewers (Xun Niu and Hao Shi) as described in the Cochrane Handbook for systematic reviews.
Statistical Analysis
The pooled odds ratios (ORs) and their 95% confidence intervals (CIs) were calculated to assess the strength of the association between p16INK4a methylation and ovarian cancer. Statistical heterogeneity was analyzed on the basis of I-square (I2) value. An I2 value above 75% indicated high heterogeneity, an I2 value between 50% and 75% suggested moderate heterogeneity, and an I2 value between 25% and 50% indicated low heterogeneity. A result was homogeneous when the I2 value was less than 25%. If I2< 50%, the studies were considered homogeneous or low in heterogeneity. A fixed-effects model was used to combine the effect size. If I2> 50%, the studies were believed to be moderately or highly heterogeneous, and the random-effects model was used to combine the effect size [25,26]. A subgroup analysis was conducted to assess the impact of race (Asia and Caucasus), method (BSP and MSP), and control type (blood and tissues). Potential publication bias was assessed by Funnel plot [27], Begg’s test, and Egger’s test [27,28]. According to the sensitivity analysis, the contribution of each study to the final results of the meta-analysis was evaluated. All the p values were two sided with a significant level at 0.05. All statistical analyses were performed with the Meta package (version 2.2–1) in R (version 3.00).
Results
Search Results
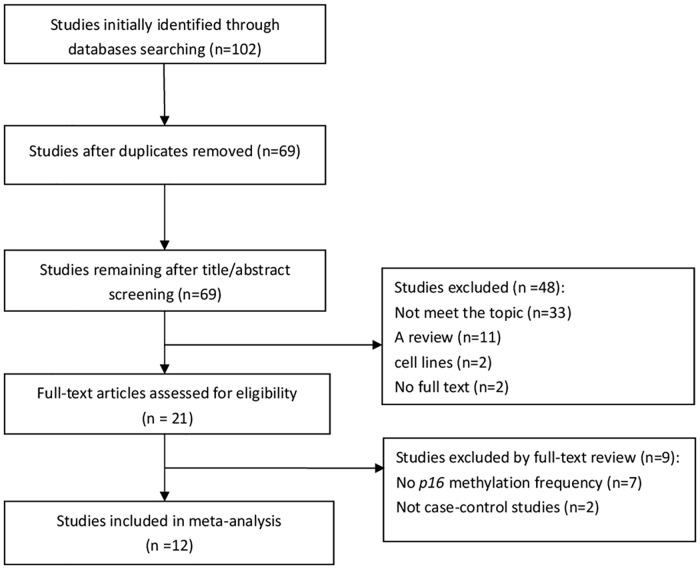
The initial search was independently executed by three reviewers (Xiyue Xiao, Yi Zhong, and Fucheng Cai), and 69 articles were initially selected. The 69 articles were quickly screened by abstract and title based on inclusion/exclusion criteria. After careful review by the three experts, 21 articles were found to be related toour meta-analysis. These articles underwent a second review. Finally, a total of 12 studies were included in the meta-analysis. The detailed steps involved in the literature search are shown in Fig 1.
Fig 1. Flowchart outlining study selection in the meta-analysis.
Study Characteristics
Twelve studies comprising data from a total of 612 cases and 289 controls were included in this review. Eight of these studies involved Asian subjects, and four studies investigated Caucasians. Among the included studies investigating p16INK4a methylation in ovarian cancer and controls, three utilized bisulfite sequencing PCR (BSP) and nine employed methylation-specific polymerase chain reaction (MSP). The control group included peripheral blood and tissues (benign ovarian tissues, normal ovarian tissues of cancer-free patients and adjacent tissues). Characteristics of the 12 studies are summarized in Table 1.
Table 1. Characteristics of included studies.
| Author | Year | Country | Age(y) | Case | Control | Method | Case type | Control type | ||
|---|---|---|---|---|---|---|---|---|---|---|
| M | U | M | U | |||||||
| Bammidi LS | 2012 | India | 20–75 | 28 | 22 | 18 | 32 | BSP | tissues | blood |
| Bhagat R | 2014 | India | 20–79 | 58 | 76 | 11 | 30 | MSP | tissues | tissues |
| Li M | 2006 | China | NA | 6 | 12 | 0 | 10 | MSP | tissues | tissues |
| Liu Z | 2005 | USA | 27–81 | 13 | 39 | 15 | 25 | MSP | tissues | tissues |
| Makarla P | 2005 | USA | 51.5(20–86) | 7 | 16 | 3 | 36 | MSP | tissues | tissues |
| Niederacher D | 1999 | Germany | NA | 6 | 17 | 0 | 10 | MSP | tissues | tissues |
| Shih YC | 1997 | Australia | NA | 0 | 45 | 0 | 2 | MSP | tissues | tissues |
| Tam KF | 2007 | China | 53.1±1.4 | 17 | 72 | 5 | 30 | MSP | tissues | tissues |
| Wong YF | 1999 | China | NA | 2 | 47 | 0 | 10 | MSP | tissues | blood |
| Shen W | 2008 | China | 52.2, 53.2 | 13 | 50 | 0 | 30 | BSP | tissues | tissues |
| Wei W | 1999 | China | 46.5±12.47 | 5 | 21 | 0 | 2 | BSP | tissues | tissues |
| Xu B | 2003 | China | NA | 4 | 36 | 0 | 20 | MSP | tissues | tissues |
Blood: peripheral blood samples from each patient with ovarian cancer; tissues: benign ovarian tissues, normal ovarian tissues of cancer-free patients and adjacent tissues; MSP: methylation-specific polymerase chain reaction; BSP: bisulfite sequencing PCR; NA: not available; M: methylation; U: unmethylation.
Combined Analysis of Included Studies
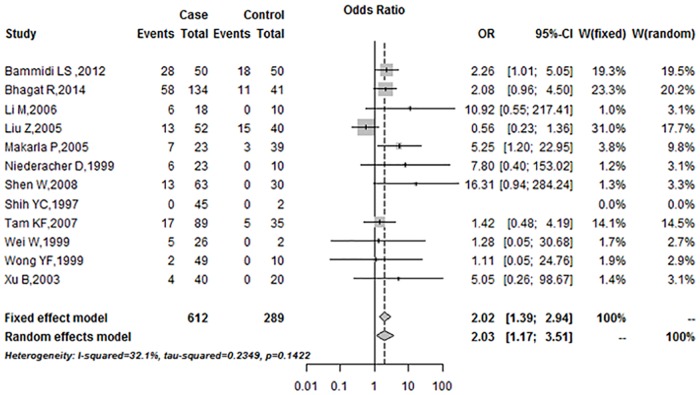
The combined analysis showed the relationship between p16INK4a promoter methylation and ovarian cancer risk (Fig 2). A fixed-effects model was employed due to low heterogeneity among the included studies (I2 = 32.1%). In the overall meta-analysis, p16INK4a promoter methylation frequency was significantly associated with ovarian cancer (Summary OR was 2.02, 95%CI = 1.39–2.94).
Fig 2. Summary estimates for p16INK4a promoter methylation frequency associated with ovarian cancer.
Sensitivity Analysis
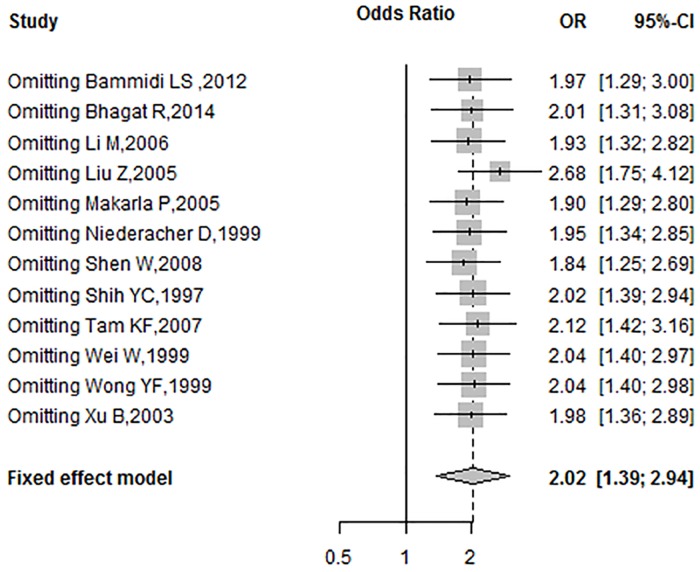
A sensitivity analysis was performed by omitting a single study and calculating the pooled OR for the remaining studies under the fixed-effects model, to determine the effects of each individual study. The results of sensitivity analysis are summarized in Fig 3. According to sensitivity analysis, the OR ranged from 1.84 (95%CI = 1.25–2.69) to 2.68 (95%CI = 1.75–4.12) by omitting a single study in the fixed-effects model. The pooled OR values between p16INK4a promoter methylation and ovarian cancer were reliable and stable.
Fig 3. Sensitivity analysis of pooled OR for p16INK4a methylation and ovarian cancer under the fixed-effects model.
Subgroup Analysis
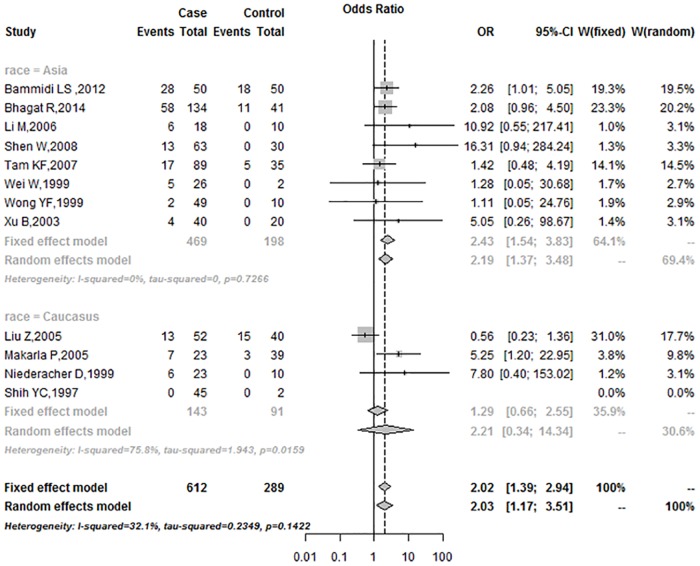
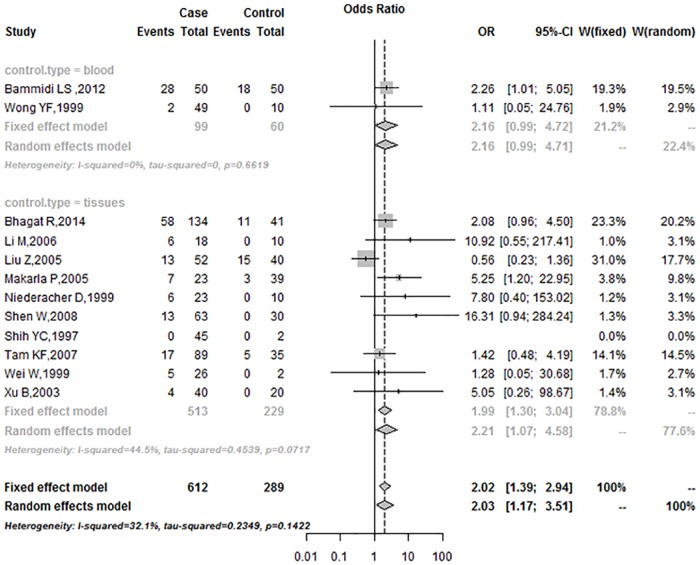
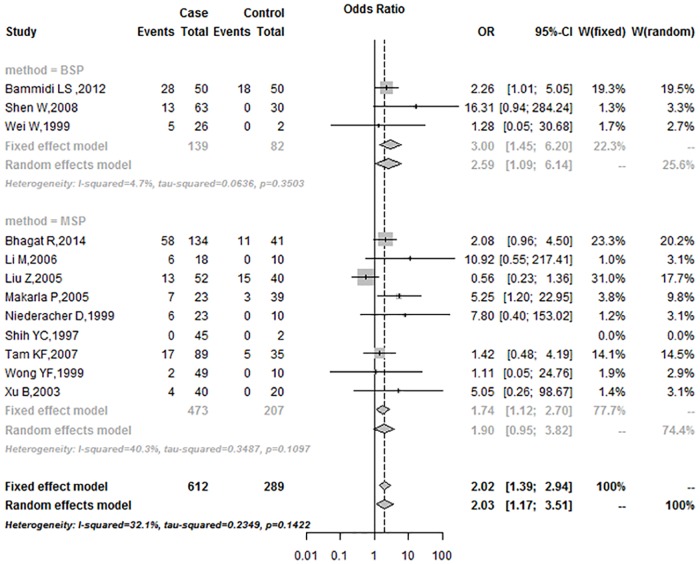
In the subgroup analysis based on race, the OR was 2.43 (95%CI = 1.54–3.83) in Asians, and 2.21 (95%CI = 0.34–14.34) in Caucasians (Fig 4). Subgroup analysis of the control sample showed that the frequencies of p16INK4a promoter methylation in the blood group were higher than in the tissue group under the fixed-effects model (Blood: 2.16, 95%CI = 0.99–4.72; Tissues: 1.99, 95%CI = 1.30–3.04; respectively) (Fig 5). According to the mode of methylation detection for p16INK4a promoter, the OR was 3.00 (95%CI = 1.45–6.20) in the BSP group and 1.74 (95%CI = 1.12–2.70) in the MSP group, under the fixed-effects model (Fig 6).
Fig 4. Subgroup analysis based on race.
Fig 5. Subgroup analysis based on control sample type.
Fig 6. Subgroup analysis based on methylation detection method.
Publication Bias
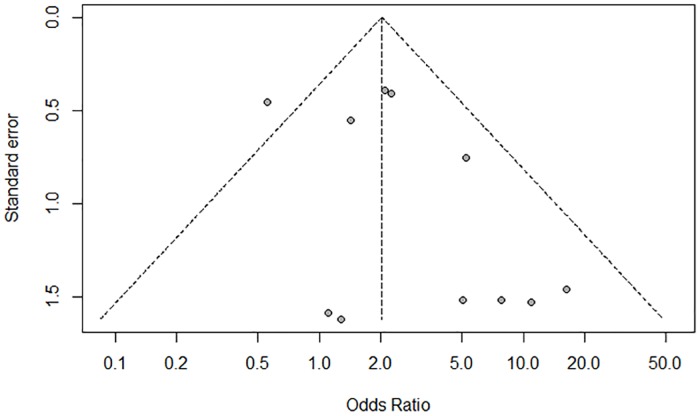
Funnel plot, Begg’s test, and Egger’s test were used to evaluate the publication bias of the studies. The funnel plot was not perfectly symmetrical (Fig 7), suggesting a slight publication bias. However, the Begg’s(P = 0.59) and Egger’s tests (P = 0.15) showed no evidence of publication bias in our meta-analysis. In addition, the ‘trim and fill’ method showed that no study required statistical correction for funnel plot asymmetry.
Fig 7. Funnel plot showing minor publication bias.
Discussion
The pathogenesis of human ovarian cancer involves complex, multistep and multi-factorial mechanisms including a variety of genetic and epigenetic abnormalities, signal transduction pathways, apoptosis, angiogenesis, and cell cycle regulation. Epigenetic inactivation of the tumor suppressor gene (TSG) following promoter methylation of CpG islands is one of the most frequent events in human tumors. Several studies suggest that inactivation of p16INK4a induced by aberrant hypermethylation may play an important role in the carcinogenesis of lung, liver, stomach, breast, and uterus [29–33]. In a recent meta-analysis from six eligible studies, including 261 patients, Hu et al found that p16INK4a promoter hypermethylation is correlated with an increased risk of endometrial carcinoma [33]. Similarly, our earlier meta-analysis reported a significant hypermethylation of p16INK4a promoter in head and neck squamous cell carcinoma (HNSCC) [34].
The current meta-analysis investigated the association between p16INK4a promoter methylation and human ovarian cancer. It included 12 studies comprising 612 cases and 289 controls. The pooled OR under a fixed-effects model was 2.02 (95%CI = 1.39–2.94) in the cancer cases compared with the controls. The result showed that methylation of p16INK4a promoter led to a 2.02-fold increased risk of human ovarian cancer compared with the control group. Begg’s tests (P = 0.59) and Egger’s tests (P = 0.15) revealed no publication bias in this study. The sensitivity analysis showed that exclusion of any single study did not affect the overall results or conclusions. Therefore, the results of our meta-analysis are reliable and show relatively strong statistical power.
In the subgroup analysis, the OR for was 2.43 (95%CI = 1.54–3.83) in Asians under the fixed-effects model, and 2.21 (95%CI = 0.34–14.34) in Caucasians under the random-effects model. The association between p16INK4a promoter methylation and human ovarian cancer in Asians was stronger than in Caucasians. The findings may be attributed in large part to a combination of differences in allele frequencies and complex epistasis or gene-environment interactions [35]. In subgroup analysis, the OR was 3.00 (95%CI = 1.45–6.20) in the BSP group under the fixed-effects model, and 1.74 (95%CI = 1.12–2.70) in the MSP group under the fixed-effects model, respectively. In the past few years, several methods were developed to detect aberrant gene methylation (e.g., BSP, MSP, QMSP, and Pyro). MSP is a simple, sensitive, and specific method for detection of methylation status in CpG-rich regions [36]. However, MSP requires specific gene sequence data for the design of PCR primers, and different primers may influence the results of methylation analyses. In addition, MSP (a non-quantitative non-fluorometric PCR method) failed to detect low levels. BSP provides a more direct and quantitative analysis of most CPG sites within a defined region than MSP [37]. In our study, the data from both BSP and MSP methods showed that the frequency of promoter methylation in p16INK4a was higher in ovarian cancer than in control. Subgroup analysis of the control sample type showed that the OR was 2.16 (95%CI = 0.99–4.72) in the blood group, and 1.99 (95%CI = 1.30–3.04) in the tissues, respectively, under the fixed-effects model. Subgroup analysis revealed that aberrant methylation of p16INK4a promoter was significantly associated with ovarian cancer, regardless of race, control sample type and detection method.
However, potential limitations need to be considered when interpreting the results of our meta-analysis. First, since the 12 included studies were retrospective, a potential unidentified confounding, information and selection bias may exist. Second, the population size of the studies included was relatively small. Further investigations with large sample sizes are required. Additionally, we did not explore the association between p16INK4a promoter methylation and disease characteristics (stage, metastasis, relapse and so on) in human ovarian cancer. The association between p16INK4a promoter methylation and disease characteristics may highlight the unique role of p16INK4a promoter methylation in human ovarian cancer.
In conclusion, the results demonstrate that aberrant methylation of p16INK4a promoter was associated with human ovarian cancer, suggesting that promoter methylation of p16INK4a plays a crucial role in human ovarian cancer. It may serve as a potential biomarker for early detection and diagnosis of human ovarian cancer.
Supporting Information
(DOC)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Nature Science Foundation of China (No.81200745 and No.81501297). The sponsors had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65(2):87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Landis SH, Murray T, Bolden S, Wingo PA. Cancer statistics, 1998. CA Cancer J Clin. 1998; 48(1):6–29. [DOI] [PubMed] [Google Scholar]
- 3.Hennessy BT, Coleman RL, Markman M. Ovarian cancer. Lancet. 2009; 374(9698):1371–1382. 10.1016/S0140-6736(09)61338-6 [DOI] [PubMed] [Google Scholar]
- 4.Wei SH, Balch C, Paik HH, Kim YS, Baldwin RL, Liyanarachchi S, et al. Prognostic DNA methylation biomarkers in ovarian cancer. Clin Cancer Res. 2006; 12(9):2788–2794. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007; 128(4):683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong R, Yu J, Pu H, Zhang Z, Xu X. Frequent SLIT2 promoter methylation in the serum of patients with ovarian cancer. J Int Med Res. 2012; 40(2):681–686. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Wang B, Zhang YM, Wang W. The association between CDH1 promoter methylation and patients with ovarian cancer: a systematic meta-analysis. J Ovarian Res. 2016; 9:23 10.1186/s13048-016-0231-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994; 368(6473):753–756. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto A, Demetrick DJ, Spillare EA, Hagiwara K, Hussain SP, Bennett WP, et al. Mutations and altered expression of p16INK4 in human cancer. Proc Natl Acad Sci U S A. 1994; 91(23):11045–11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim BN, Yamamoto H, Ikeda K, Damdinsuren B, Sugita Y, Ngan CY, et al. Methylation and expression of p16INK4 tumor suppressor gene in primary colorectal cancer tissues. Int J Oncol. 2005; 26(5):1217–1226. [PubMed] [Google Scholar]
- 11.Ghazizadeh M, Jin E, Shimizu H, Fujiwara M, Arai S, Ohaki Y, et al. Role of cdk4, p16INK4, and Rb expression in the prognosis of bronchioloalveolar carcinomas. Respiration. 2005; 72(1):68–73. [DOI] [PubMed] [Google Scholar]
- 12.de Assis LV, Locatelli J, Isoldi MC. The role of key genes and pathways involved in the tumorigenesis of Malignant Mesothelioma. Biochim Biophys Acta. 2014; 1845(2):232–247. 10.1016/j.bbcan.2014.01.008 [DOI] [PubMed] [Google Scholar]
- 13.Bammidi LS, Neerukonda GN, Murthy S, Kanapuram RD. p16 gene alterations in human ovarian cancers: comparison between tissue and blood samples. Int J Gynecol Cancer. 2012; 22(4):553–560. 10.1097/IGC.0b013e31823fa90c [DOI] [PubMed] [Google Scholar]
- 14.Bhagat R, Kumar SS, Vaderhobli S, Premalata CS, Pallavi VR, Ramesh G, et al. Epigenetic alteration of p16 and retinoic acid receptor beta genes in the development of epithelial ovarian carcinoma. Tumour Biol. 2014; 35(9):9069–9078. 10.1007/s13277-014-2136-1 [DOI] [PubMed] [Google Scholar]
- 15.Li M, Huang ZJ, Dong WH, Li XY, Wang XY, He XH, et al. [Disfigurement of p16INK4A gene expression in development of ovarian cancer and the mechanism]. Zhonghua Fu Chan Ke Za Zhi. 2006; 41(6):408–412. [PubMed] [Google Scholar]
- 16.Liu Z, Wang LE, Wang L, Lu KH, Mills GB, Bondy ML, et al. Methylation and messenger RNA expression of p15INK4b but not p16INK4a are independent risk factors for ovarian cancer. Clin Cancer Res. 2005; 11(13):4968–4976. [DOI] [PubMed] [Google Scholar]
- 17.Makarla PB, Saboorian MH, Ashfaq R, Toyooka KO, Toyooka S, Minna JD, et al. Promoter hypermethylation profile of ovarian epithelial neoplasms. Clin Cancer Res. 2005; 11(15):5365–5369. [DOI] [PubMed] [Google Scholar]
- 18.Niederacher D, Yan HY, An HX, Bender HG, Beckmann MW. CDKN2A gene inactivation in epithelial sporadic ovarian cancer. Br J Cancer. 1999; 80(12):1920–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen WJ, Dai DQ, Guo KJ, Li XM. [Promoter hypermethyIation of RASSFlA, BRCAl and p16 gene in epithelial ovarian cancer and its clinical significance]. Chin J Cancer Prev Treat.2007;15(7): 530–533. [Google Scholar]
- 20.Shih YC, Kerr J, Liu J, Hurst T, Khoo SK, Ward B, et al. Rare mutations and no hypermethylation at the CDKN2A locus in epithelial ovarian tumours. Int J Cancer. 1997; 70(5):508–511. [DOI] [PubMed] [Google Scholar]
- 21.Tam KF, Liu VW, Liu SS, Tsang PC, Cheung AN, Yip AM, et al. Methylation profile in benign, borderline and malignant ovarian tumors. J Cancer Res Clin Oncol. 2007; 133(5):331–341. [DOI] [PubMed] [Google Scholar]
- 22.Wei W, Meng ZZ, Cheng JX, Wang Y, Wang JP. [Methylation of the P16 promotor in ovarian carcinoma tissues and its clinical significance]. Current Adv Obstetr Gynecol. 1999;8(2):109–111. [Google Scholar]
- 23.Wong YF, Chung TK, Cheung TH, Nobori T, Yu AL, Yu J, et al. Methylation of p16INK4A in primary gynecologic malignancy. Cancer Lett. 1999; 136(2):231–235. [DOI] [PubMed] [Google Scholar]
- 24.Xu B, Dai SZ. [Hypermethylation and its clinical significance of 5’CpG island and in p16 gene in gynecologic malignancies]. MedJQilu. 2003;18(3):227–229. [Google Scholar]
- 25.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986; 7(3):177–188. [DOI] [PubMed] [Google Scholar]
- 26.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003; 327(7414):557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997; 315(7109):629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994; 50(4):1088–1101. [PubMed] [Google Scholar]
- 29.Belinsky SA, Klinge DM, Dekker JD, Smith MW, Bocklage TJ, Gilliland FD, et al. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res. 2005; 11(18):6505–6511. [DOI] [PubMed] [Google Scholar]
- 30.Hsu LS, Lee HC, Chau GY, Yin PH, Chi CW, Lui WY. Aberrant methylation of EDNRB and p16 genes in hepatocellular carcinoma (HCC) in Taiwan. Oncol Rep. 2006; 15(2):507–511. [PubMed] [Google Scholar]
- 31.Mino A, Onoda N, Yashiro M, Aya M, Fujiwara I, Kubo N, et al. Frequent p16 CpG island hypermethylation in primary remnant gastric cancer suggesting an independent carcinogenic pathway. Oncol Rep. 2006; 15(3):615–620. [PubMed] [Google Scholar]
- 32.Askari M, Sobti RC, Nikbakht M, Sharma SC. Promoter hypermethylation of tumour suppressor genes (p14/ARF and p16/INK4a): case-control study in North Indian population. Mol Biol Rep. 2013; 40(8):4921–4928. 10.1007/s11033-013-2592-5 [DOI] [PubMed] [Google Scholar]
- 33.Hu ZY, Tang LD, Zhou Q, Xiao L, Cao Y. Aberrant promoter hypermethylation of p16 gene in endometrial carcinoma. Tumour Biol. 2015; 36(3):1487–1491. 10.1007/s13277-014-2632-3 [DOI] [PubMed] [Google Scholar]
- 34.Shi H, Chen X, Lu C, Gu C, Jiang H, Meng R, et al. Association between P16INK4a promoter methylation and HNSCC: a meta-analysis of 21 published studies. PLoS One. 2015; 10(4):e0122302 10.1371/journal.pone.0122302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser HB, Lam LL, Neumann SM, Kobor MS. Population-specificity of human DNA methylation. Genome Biol. 2012; 13(2):R8 10.1186/gb-2012-13-2-r8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuck AE, Rubenstein LZ, Wieland D. Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. BMJ. 1998; 316(7129):469; author reply 470–461. [PMC free article] [PubMed] [Google Scholar]
- 37.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996; 93(18):9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.