Abstract
5-Azacytidine is a well-known anticancer drug that is clinically used in the treatment of breast cancer, melanoma and colon cancer. It has been reported that 5-azacytidine suppresses the biological behavior of esophageal cancer cells. However, corresponding mechanisms remain unclear. In this study, using Transwell invasion and cell proliferation assays, we demonstrated that 5-azacytidine significantly inhibited the metastasis and proliferation of EC9706 cells, and upregulated the expression of cadherin 1 (CDH1) and SRY-box containing gene 17 (SOX17). Moreover, the inhibition of the metastasis of the 5-azacytidine-treated EC9706 cells was impaired following transfection with siRNA targeting CDH1 (CDH1 siRNA), and the inhibition of cell proliferation was attenuated following the downregulation of SOX17 by siRNA targeting SOX17 (SOX17 siRNA). Furthermore, 5-azacytidine remarkably reduced the CDH1 and SOX17 promoter methylation levels, suggesting that 5-azacytidine upregulates the expression of SOX17 and CDH1 by inhibiting the methylation of the SOX17 and CDH1 promoter. The findings of our study confirm that 5-azacytidine suppresses the proliferation and metastasis of EC9706 esophageal cancer cells by upregulating the expression of CDH1 and SOX17. The expression levels of CDH1 and SOX17 negatively correlate with the promoter methylation levels. CDH1 and SOX17 are potential indicators of the clinical application of 5-azacytidine.
Keywords: 5-azacytidine, SRY-box containing gene 17, cadherin-1, proliferation, metastasis
Introduction
Esophageal cancer is the eighth most common cancer, and approximately 25-30 million new cases are confirmed each year. There are two major histological subtypes of esophageal cancer: esophageal adenocarcinoma and esophageal squamous cell carcinoma (1-3). Given the fact that the incidence of esophageal cancer is rising (4), it is imperative to assess the effectiveness of chemotherapeutic drugs for the treatment of this type of cancer, and to elucidate the mechanisms responsible for its development, in order to select effective chemotherapeutic regimens.
5-Azacytidine is a common anticancer drug. It is mainly used in chemotherapy for various types of cancer, including breast cancer, melanoma and acute myeloid leukemia (5-12). One of the most important mechanisms responsible for its antitumor activity previously proposed is that the drug can incorporate into DNA during DNA replication and inhibit DNA methylation (13). DNA methylation plays an important role in tumorigenesis and cancer progression (14-17). Aberrant DNA methylation leads to the inactivation of tumor suppressor genes in the development and progression of cancers, including esophageal cancer (18-20).
Cadherin 1 (CDH1) and SRY-box containing gene 17 (SOX17) are two members that are aberrantly methylated in esophageal cancer cells. CDH1 belongs to the cadherin superfamily, and is also known as epithelial cadherin (E-cadherin) or uvomorulin (21). CDH1, as a tumor suppressor gene, plays an essential role in maintaining cell adhesion and adherent junctions in normal tissues (22). The expression of CDH1 is frequently absent in a variety of epithelial tumors in which normal intercellular junctions are lost (23-27). SOX17 belongs to the high-mobility group (HMG)-box transcriptive factor superfamily. It is highly conserved during evolution and plays an important role in the formation of embryonic germ layers (28). Frequent methylation of SOX17 promoter was detected in colon, liver, breast and lung cancers (29-31). The hypermethylation of the SOX17 promoter and the lower expression of SOX17 has previously been shown in breast cancer, resulting in cancer cell proliferation (32). In lung cancer cells, the promoter of SOX17 is also hypermethylated, leading to the silencing or the inhibition of SOX17 expression. In addition, SOX17 promoter methylation is remitted by 5-azacytidine in lung cancer cells (31). The hypermethylation of the CDH1 and SOX17 promoters and the reduced expression of CDH1 and SOX17 have been reported to occur in esophageal cancer (33-35). These data suggest that these two genes are epigenetically involved in the development and progression of esophageal cancer.
Based on the facts that 5-azacytidine can affect DNA methylation and that the promoters of tumor suppressor genes are usually downregulated by high hypermethylation in esophageal cancer, in this study, we aimed to examine the effects of treatment with 5-azacytidine on esophageal cancer cells and to elucidate the mechanisms responsible for its antitumor activity in esophageal cancer. Our findings may help clinicians select an effective chemotherapeutic regimen for the treatment of esophageal cancer, and may also aid in the prognostic evaluation.
Materials and methods
Cell culture
The EC9706 esophageal cancer cell line was purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (both from Thermo Fisher Scientific, Waltham, MA, USA) and 100 U/ml penicillin/streptomycin (Invitrogen Life Technologies, Carlsbad, CA, USA), and were routinely incubated at 37˚C under a 5% CO2 atmosphere.
Treatment with 5-azacytidin
The EC9706 cells were seeded into 6-well culture plate (NEST Biotechnology, Jiangsu, China) at a density of 106 cells/well and cultured overnight. The cells were treated with 5-azacytidine (50 µM) (5-aza; Sigma-Aldrich, St. Louis, MO, USA) for 72 h and dimethyl sulfoxide (DMSO; 1:100) as the negative control.
Reverse transcription-quantitative PCR (RT-qPCR)
Following treatment with 5-azacytidine for 72 h, total RNA was isolated from the EC9706 cells using TRIzol reagent (Life Technologies) according to the manufacturer's instructions. RNA was reverse transcribed using the PrimeScript® First Strand cDNA Synthesis kit (Takara, Shiga, Japan) and then amplified with PCR primers on the iQ5 Real-time Quantitative PCR system (Bio-Rad, Hercules, CA, USA). The average Ct, from triplicate assays, was used for further calculations. Relative expression levels were normalized to the control and actin as the internal control. The reverse transcribed reaction included approximately 1 µg of cDNA, 0.5 µg of each primer, 20 nmoles of dNTPs, 4 µl of 5X PCR buffer, 12 units of RiboLock™ Ribonuclease inhibitor and 1 unit of ReverTra Ace polymerase in a final reaction volume of 20 µl. The PCR cycling conditions were as follows: 42˚C for 60 min; and 72˚C for 10 min. The primers used for PCR were as follows: SOX17 forward, 5'-GGGATACGCCAGTGACGAC-3' and reverse, 5'-CCTTA GCCCACACCATGAAA-3'; CDH1 forward, 5'- AATGCCG CCATCGCTTAC-3' and reverse, 5'-TCAGGCACCTGACC CTTGTA-3'; and actin forward, 5'-GCACCCAGCACAA TGAAGA-3' and reverse, 5'-AATAAAGCCATGCCAA TCTCA-3'.
Western blot analysis
Total proteins were mixed by loading buffer and boiled for 5 min. Equivalent quantities of protein were separated by 12% sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred onto polyvinylidene fluoride membranes. The membranes were then blocked with 10% defatted milk for 1 h at 4˚C. This was followed by the addition of primary antibodies specific to the SOX17 markers (1:1,000; species raised in, rabbit; specificity, rat, human and mouse; polyclonal antibody; Ab191699; Abcam, Cambridge, UK), anti-E-Cadherin antibody IgG (CDH1; 1:1,000; species raised in, rabbit; specificity, rat, human and mouse; monoclonal antibody; Ab133597; Abcam), anti-MMP2 antibody IgG (1:1000; species raised in, rabbit; specificity, rat, mouse, human; polyclonal antibody; 4022; Cell Signaling Technology, Danvers, MA, USA) and anti-MMP9 antibody IgG (1:1000; species raised in, rabbit; specificity, human; polyclonal antibody; 2270; Cell Signaling Technology). in phosphate-buffered saline (PBS) containing 5% BSA incubated with the membranes at 4˚C overnight. The membranes were then washed and incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000; SC-2004; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) for 1 h. The bound secondary antibody was visualized using an enhanced chemiluminescence (ECL) system (Pierce Biotechnology, Rockford, IL, USA).
siRNA transfection
siRNAs targeting SOX17 (SOX17 siRNA) and CDH1 (CDH1 siRNA) were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China) and transfection was performed using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer's instructions. The negative control siRNA was also synthesized by Shanghai GenePharma Co., Ltd. The EC9706 cells were seeded in a 6-well plate at a density of 106 cells/well and allowed to attach overnight (16 h). Subsequently, SOX17 siRNA, CDH1 siRNA or NC siRNA were transfected into the EC9706 cells at a final concentration of 50 nM using Lipofectamine RNAiMAX. The interference efficiency was detected by RT-qPCR at 48 h post-transfection.
Cell viability assay
The EC9706 cells were seeded into 96-well culture plates (5,000 cells/well). 5-Azacytidine was added to the wells at a concentration of 50 µM. Cell viability was assayed using the Cell Counting kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan) at 1-5 days following treatment with 5-azacytidine. The absorbance was measured at 450 nm using a microplate reader.
Cell colony formation assay
Soft agar assays were performed to detect the colony-forming ability of the EC9706 cells at 96 h following treatment with 5-azacytidine (50 µM). The cells were then resuspended and seeded into 6-well culture plates (1,000 cells/well). On day 7, the cells were washed with PBS twice and fixed with 75% ethanol for 30 min and stained with 0.2% crystal violet for visualization and photographing by microscope (Mi 8; Leica, Wetzlar, Germany).
Apoptosis assay
Cell apoptosis was determined by flow cytometry using the Annexin V-FITC Apoptosis Detection kit (Sungene, Tianjin, China). The EC9706 cells were seeded in 6-well culture plates. 5-Azacytidine was added to the wells at the concentration of 50 µM. The cells were harvested 96 h following treatment with 5-azacytidine, washed in PBS and incubated with Annexin V and propidium iodide (PI) in binding buffer in the dark at room temperature for 10 min. The stained cells were analyzed using the BD FASAria Cell Sorter (BD Biosciences, Franklin Lakes, NJ, USA).
Cell invasion assay
Transwell invasion chambers coated with Matrigel (50 µl/filter) (BD Biosciences) were performed to detect EC9706 cell invasion. The EC9706 cells were transferred on the top of the chambers (50,000 cells/chamber) in DMEM and the lower chambers were supplemented with DMEM containing 10% fetal calf serum 96 h following treatment with 5-azacytidine (50 µM). The cells were fixed and stained with GenMed crystal violet after 48 h of culture.
Detection of methylation by bisulfite-assisted genomic sequencing PCR (BSP)
EC9706 cell genomic DNA was extracted 72 h following treatment with 5-azacytidine (50 µM). Genomic DNA (1 µg) was modified and purified according to the instructions provided with the EZ DNA Methylation-Gold™ kit (D5005, Zymo Research, Shanghai, China). Modified and purified genomic DNA was amplified using the primers listed in Table I. The PCR products were cloned and sequenced. The methylation rates of each promoter were analyzed in the website: http://quma.cdb.riken.jp/.
Table I.
Primer used for bisulfite-assisted genomic sequencing PCR (BSP).
| Name | Forward primer (5'→3') | Reverse primer (5'→3') |
|---|---|---|
| CDH1-1 | TGATTTTAGGTTTTAGTGAG | CAAACTCACAAATACTTTAC |
| CDH1-2 | GAATTGTAAAGTATTTGTGAGTTTG | AATACCTACAACAACAACAACAAC |
| CDH1-3 | GTTGTTGTTGTTGTTGTAGGTATTT | CCACTCCCATCACTAAAAAATC |
| SOX17-1 | AGAGTGAAGGAATATTGGA | CAAAACTACACCTACCCC |
| SOX17-2 | GGGGTAGGTGTAGTTTTG | TACCCAAAACCCCCAACC |
| SOX17-3 | GGTTGGGGGTTTTGGGTA | CCCTTCACCTTCATATCC |
| SOX17-4 | GGATATGAAGGTGAAGGG | CTACACACCCCTAATTTT |
| SOX17-5 | GTTTAAAATTAGGGGTGTG | CTCCCCCCTCAAACTTTA |
CDH, cadherin; SOX, SRY-box containing gene.
Statistical analysis
Values are expressed as the means ± standard error of 3 independent experiments. The statistical significance of the differences was calculated using Prism software (version 4.0a; GraphPad Software, Inc., La Jolla, CA, USA) by one-way analysis of variance. Values of P<0.05 and P<0.01 were considered to indicate statistically significant differences.
Results
5-Azacytidine suppresses the proliferation and invasion of EC9706 cells
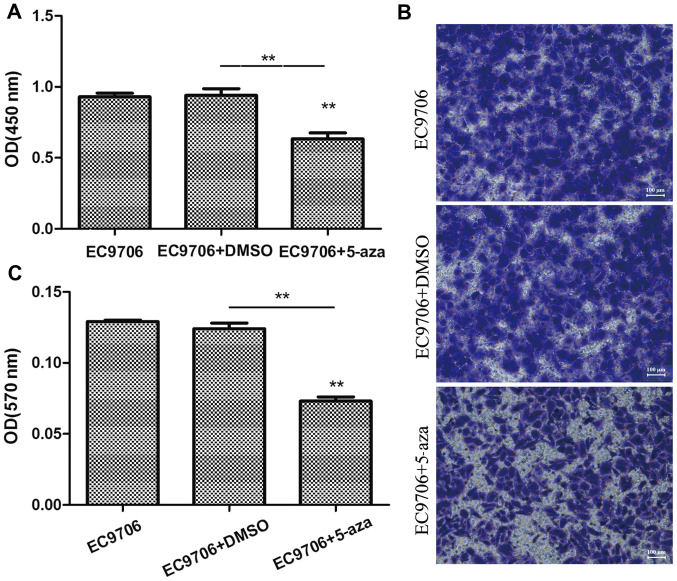
To examine the effects of 5-azacytidine on the growth of esophageal cancer cells, the EC9706 cells were treated with 50 µM 5-azacytidine for 72 h, and CCK-8 cell viability assay and Transwell assay were then performed to determine the effects of 5-azacytidine on cell proliferation and invasion. The results of CCK-8 assay revealed that the viability of the EC9706 cells was markedly suppressed following treatment with 5-azacytidine (Fig. 1A). In addition, the results of Transwell invasion assay indicated that the invasive ability of the EC9706 cells was significantly decreased following treatment with 5-azacytidine (Fig. 1B and C).
Figure 1.
Suppression of the proliferation and invasion of EC9706 cells by 5-azacytidine. 5-Azacytidine inhibited the proliferation and metastasis of EC9706 cells. (A) Analysis of EC9706 cell viability using the cell counting kit-8 (CCK-8). Compared with the DMSO vehicle control group, EC9706 cell viability was significantly inhibited 72 h following treatment with 5-azacytidine (50 µM) (**P<0.01). (B) Transwell assay of the invasion of EC9706 cells. (C) Statistical analysis of Transwell invasion assay. The invasive ability of the EC9706 cells treated with 5-azacytidine was significantly inhibited (**P<0.01). 5-aza, 5-azacytidine.
Treatment with 5-azacytidine upregulates CDH1 and SOX17 expression in EC9706 cells
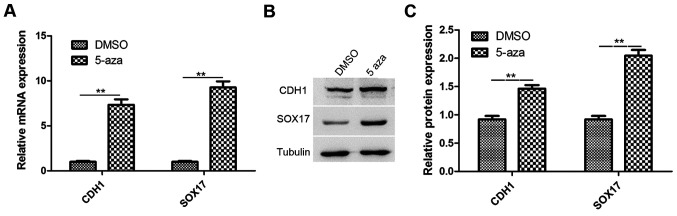
Studies have reported that CDH1 and SOX17 are involved in the inhibition of cancer metastasis and growth, respectively. CDH1 and SOX17 are downregulated in esophageal cancer (22,32). Therefore, in this study, RT-qPCR and western blot analysis were performed to analyze the expression of CDH1 and SOX17 in the EC9706 cells treated with 5-azacytidine (50 µM) for 72 h. The results revealed that the CDH1 and SOX17 expression levels were significantly increased in the 5-azacytidine treatment group, in comparison to the DMSO vehicle control (Fig. 2A). The results of western blot analysis also revealed that the CDH1 and SOX17 protein expression levels were significantly upregulated by 5-azacytidine treatment (Fig. 2B).
Figure 2.
Upregulation of cadherin 1 (CDH1) and SRY-box containing gene 17 (SOX17) expression in EC9706 cells by 5-azacytidine treatment. The expression of CDH1 and SOX17 was upregulated following treatment with 5-azacytidine in EC9706 cells. (A) RT-qPCR analysis of CDH1 and SOX17 expression. Compared with the vehicle control (DMSO-treated cells), the expression levels of CDH1 and SOX17 were increased approximately 7.33- and 9.30-fold, respectively (**P<0.01) in the 5-azacytidine-treated EC9706 cells. (B and C) Western blot analysis of the protein expression of CDH1 and SOX17. The expression levels of CDH1 (**P<0.01) and SOX17 (**P<0.01) was significantly upregulated by 5-azacytidine. 5-aza, 5-azacytidine.
5-Azacytidine inhibits EC9706 cell metastasis via the upregulation of CDH1
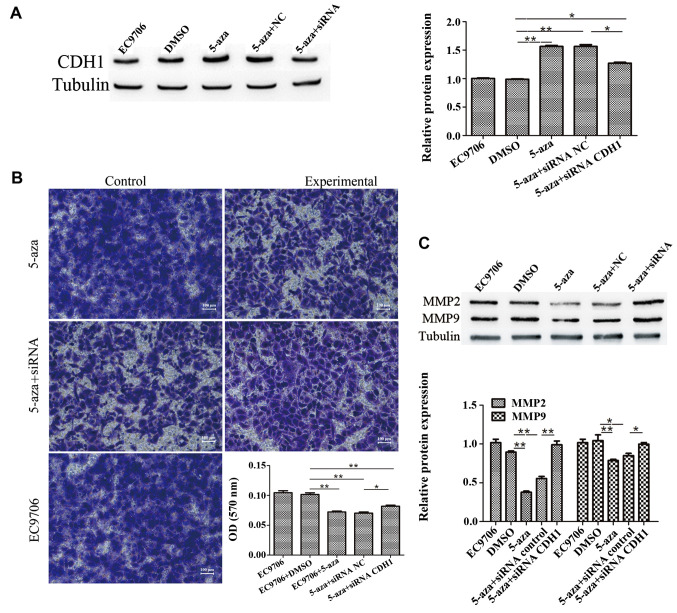
To further confirm the involvement of CDH1 in the inhibition of EC9706 cell invasion induced by 5-azacytidine, the cells were transfected with CDH1 siRNA to knockdown CDH1 expression. The results of western blot analysis revealed that transfection with CDH1 siRNA suppressed the upregulation of CDH1 that was induced by 5-azacytidine (Fig. 3A). The results of Transwell invasion assay demonstrated that the inhibitory effects of 5-azacytidine on EC9706 cell invasion were impaired by transfection with CDH1 siRNA (Fig. 3B). Western blot analysis also revealed that 5-azacytidine significantly downregulated the expression of matrix metalloproteinase (MMP)2 and MMP9. The suppressive effects of 5-azacytidine on the expression of MMP2 and MMP9 were attenuated by transfection with CDH1 siRNA (Fig. 3C). These results indicate the involvement of CDH1 in the suppression of EC9706 cell metastasis by 5-azacytidine.
Figure 3.
5-Azacytidine inhibits EC9706 cell metastasis via the upregulation of cadherin 1 (CDH1). CDH1 siRNA attenuated the inhibitory effects of 5-azacytidine on the metastasis of EC9706 cells. (A) Western blot analysis was used to detect the expression of CDH1 in EC9706 cells. Compared with the negative control (DMSO-treated cells), the expression of CDH1 was increased following treatment with 5-azacytidine (**P<0.01), whereas the expression of CDH1 was significantly reduced following CDH1-targeted siRNA transfection (*P<0.05). (B) Transwell assay was used to detect the invasion of EC9706 cells. EC9706 cell invasion was significantly inhibited by 5-azacytidine (**P<0.01), but not by DMSO, whereas the invasive ability of the cells was increased by CDH1 siRNA transfection (**P<0.01). The panels correspond to the following groups: top left panel, DMSO-treated vehicle control; top right panel, 5-azacytidine-treated cells; middle left panel, cells treated with 5-azacytidine and transfected with the control siRNA; middle right panel, cells treated with 5-azacytidine and transfected with CDH1 siRNA; bottom panel, untreated cells. (C) Western blot analysis was used to detect matrix metalloproteinase (MMP)2 and MMP9 expression. The results revealed that the expression of MMP2 and MMP9 was significantly reduced following treatment with 5-azacytidine in the EC9706 cells (**P<0.01 and *P<0.05), while the expression of MMP2 and MMP9 was increased by CDH1 siRNA transfection (**P<0.01 and *P<0.05). 5-aza, 5-azacytidine.
5-Azacytidine inhibits EC9706 cell proliferation through the upregulation of SOX17
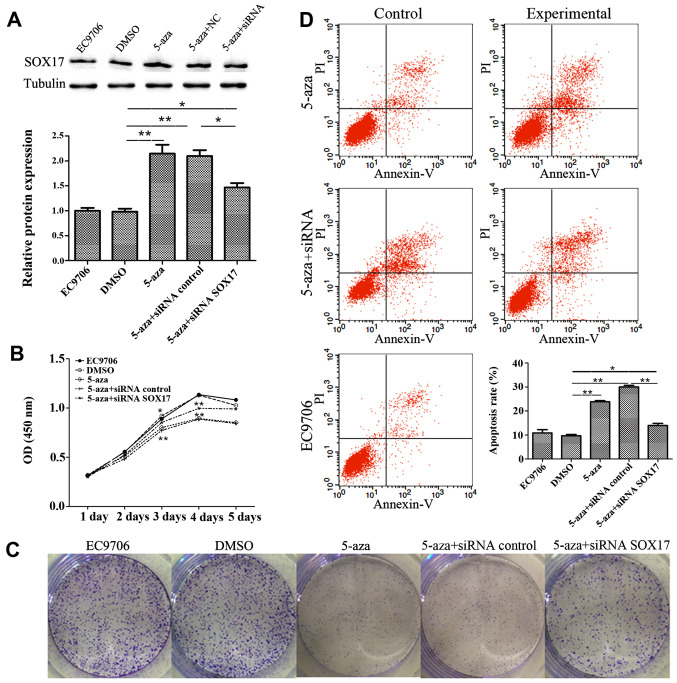
To verify the involvement of SOX17 in the inhibition of EC9706 cell proliferation by 5-azacytidine, SOX17 siRNA was used to knockdown SOX17 expression. The results of western blot analysis revealed that transfection with SOX17 siRNA suppressed SOX17 expression which had been increased by 5-azacytidine (Fig. 4A). CCK8 growth curves proved that SOX17 siRNA suppressed the 5-azacytidine-induced growth inhibition (Fig. 4B). This finding was also supported by the results of colony formation assay (Fig. 4C). Flow cytometry displayed the effects of 5-azacytidine and SOX17 on cell apoptosis. The apoptotic rate in the 5-azacytidine-treated EC9706 cells increased by 13.32% compared to the negative control (DMSO-treated cells). Transfection with SOX17 siRNA abolished the 5-azacytidine-induced apoptosis of EC9706 cells (Fig. 4D). These results indicate the involvement of SOX17 in the inhibition of EC9706 cell proliferation by 5-azacytidine.
Figure 4.
5-Azacytidine inhibits EC9706 cell proliferation via the upregulation of SRY-box containing gene 17 (SOX17). The downregulation of SOX17 by siRNA attenuatd the inhibitory effects of 5-azacytidine on cell proliferation. (A) Western blot analysis was used to detect the expression of SOX17 in the cells treated with 5-azacytidine and transfected with SOX17 siRNA. Compared with the DMSO vehicle control group, the protein expression of SOX17 was increased by 5-azacytidine treatment (**P<0.01). siRNA targeting SOX17 significantly attenuated the 5-azacytidine-induced upregulation of SOX17 (**P<0.01). (B) Cell counting kit-8 (CCK-8) assay of EC9706 cell viability. EC9706 cell viability was inhibited by 5-azacytidine treatment on days 3 and 4 (*P<0.05 and **P<0.01). SOX17 siRNA significantly impaired the effects of 5-azacytidine on days 3 and 4 (**P<0.01). (C) Cell colony-formation assay of EC9706 cells. 5-Azacytidine inhibited EC9706 cell colony formation, while SOX17 siRNA hampered the effects of 5-azacytidine. (D) Flow cytometry was carried out to examine the apoptosis of EC9706 cells. Compared with the negative control (DMSO-treated cells), 5-azacytidine promoted the apoptosis of EC9706 cells (**P<0.01). The 5-azacytidine-induced apoptosis was significantly inhibited by SOX17 siRNA (*P<0.05). 5-aza, 5-azacytidine.
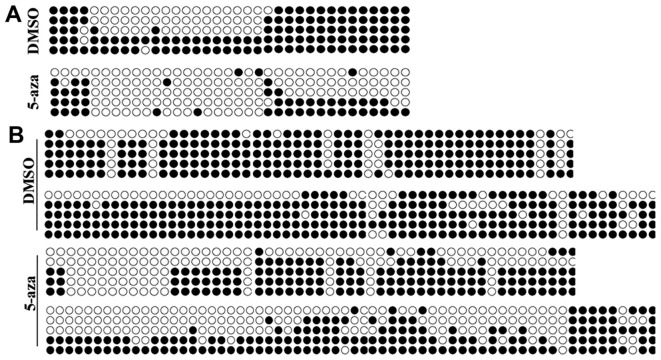
Methylation of CDH1 and SOX17 promoters is decreased by 5-azacytidine in EC9706 cells
Studies have confirmed that 5-azacytidine is able to regulate gene expression by decreasing the methylation of their promoters (14). We hypothesized that 5-azacytidine upregulates the expression of CDH1 and SOX17 via their promoter methylation. BSP methylation analysis was performed to evaluate the methylation of the CDH1 and SOX17 promoters. The results revealed that the promoters of CDH1 and SOX17 were hypermethylated in the EC9706 cells. Yet, the methylation levels in the EC9706 cell were significantly reduced following treatment with 5-azacytidine (Fig. 5).
Figure 5.
Methylation of cadherin 1 (CDH1) and SRY-box containing gene 17 (SOX17) promoters is decreased by 5-azacytidine in EC9706. Methylation level analysis of CDH1 and SOX17 promoters. (A) Methylation analysis of CDH1 promoter in 5-azacytidine-treated EC9706 cells. Methylation level of the CDH1 promoter was decreased significantly by 5-azacytidine. (B) Methylation analysis of SOX17 promoter in 5-azacytidine-treated EC9706 cells. Methylation level of SOX17 promoter was decreased significantly by 5-azacytidine. 5-aza, 5-azacytidine.
Discussion
Recently, the incidence of esophageal cancer has gradually increased. Esophageal cancer has become the second cause of cancer-related mortality in China (36). To date, the most common treatment for esophageal cancer is surgery, chemotherapy and radiotherapy, in combination. However, the curative effects of 5-fluorouracil (5-FU) and cisplatin (DDP), two basic clinical chemotherapeutic drugs for esophageal cancer, is unsatisfactory in patients with highly metastatic esophageal cancer (37,38). It has been reported that epigenetic behaviors, such as DNA methylation, play important roles in the development and metastasis of esophageal cancer. Previous studies have found that tumorigenesis and the metastasis of esophageal cancer result from the downregulation of the expression of multiple tumor suppressor genes and the abnormal hypermethylation of their promoters (33-35). Studies have shown that 5-azacytidine, a clinical chemotherapeutic drug used in the treatment of various types of cancer, inhibits the methylation of the promoter of multiple genes and affects the expression of these genes (39,40). In this study, we used EC9706 cells to assess the anticancer effects of 5-azacytidine in esophageal cancer.
This study confirmed that the proliferation and invasion of EC9706 cells were inhibited by 5-azacytidine, suggesting that 5-azacytidine is effective for the treatment of esophageal cancer. To investigate the mechanisms of action of 5-azacytidine, we screened numbers of genes that could be related to the proliferation and metastasis of cancer cells by western blot analysis. We found that the expression levels of SOX17 and CDH1 were significantly upregulated in the EC9706 cells by 5-azacytidine. Combined with the findings of previous studies, we hypothesized that SOX17 and CDH1 may participate in the 5-azacytidine-mediated inhibition of cell proliferation and metastasis. To further confirm our hypothesis, siRNAs were used to knockdown the expression of SOX17 and CDH1 in the EC9706 cells. We found that the siRNA-mediated downregulation of CDH1 was greatly impaired by 5-azacytidine, while the 5-azacytidine-induced inhibition of EC9706 cell growth and the induction of apoptosis were significantly attenuated by the siRNA-mediated downregulation of SOX17. These results support our hypothesis that SOX17 and CDH1 are involved in the 5-azacytidine-induced inhibition of the proliferation and metastasis of EC9706 cells, respectively.
To examine the mechanisms responsible for the regulation of CDH1 and SOX17 expression by 5-azacytidine, the methylation of the SOX17 and CDH1 promoters was analyzed. Our results revealed that the methylation of the SOX17 and CDH1 promoters was significantly decreased by 5-azacytidine treatment in the EC9706 cells. This may be an important regulatory pattern for 5-azacytidine to regulate SOX17 and CDH1 expression.
In conclusion, the findings of our study confirm that 5-azacytidine inhibits esophageal cancer cell proliferation and metastasis by exerting inhibitory effects on the methylation of SOX17 and CDH1 promoters. Our findings may prove to be beneficial to clinicians in selecting appropriate chemotherapeutic regimens and enhancing the therapeutic effects.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81101813) and the Qingdao Outstanding Health Professional Development Fund (V-YQ2014Y14).
Abbreviations
- SOX17
SRY-box containing gene 17
- CDH1
cadherin-1
References
- 1.Wu C, Kraft P, Zhai K, Chang J, Wang Z, Li Y, Hu Z, He Z, Jia W, Abnet CC, et al. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat Genet. 2012;44:1090–1097. doi: 10.1038/ng.2411. [DOI] [PubMed] [Google Scholar]
- 2.Brown LM, Devesa SS, Chow WH. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst. 2008;100:1184–1187. doi: 10.1093/jnci/djn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamangar F, Malekzadeh R, Dawsey SM, Saidi F. Esophageal cancer in Northeastern Iran: A review. Arch Iran Med. 2007;10:70–82. [PubMed] [Google Scholar]
- 4.Rubenstein JH, Shaheen NJ. Epidemiology, diagnosis, and management of esophageal adenocarcinoma. Gastroenterology. 2015;149:302–317. doi: 10.1053/j.gastro.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai J, Yang F, Zhang W, Wang Y, Xu J, Song W, Huang G, Gu J, Guan X. TAp73 and ΔNp73 have opposing roles in 5-aza-2'-deoxycytidine-induced apoptosis in breast cancer cells. Mol Cells. 2014;37:605–612. doi: 10.14348/molcells.2014.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Baroudi M, La Sala D, Cinti C, Capobianco E. Pathway landscapes and epigenetic regulation in breast cancer and melanoma cell lines. Theor Biol Med Model. 2014;11 (Suppl 1)(S8) doi: 10.1186/1742-4682-11-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Momparler RL. Epigenetic therapy of cancer with 5-aza-2'-deoxycytidine (decitabine) Semin Oncol. 2005;32:443–451. doi: 10.1053/j.seminoncol.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Lemaire M, Momparler LF, Bernstein ML, Marquez VE, Momparler RL. Enhancement of antineoplastic action of 5-aza-2'-deoxycytidine by zebularine on L1210 leukemia. Anticancer Drugs. 2005;16:301–308. doi: 10.1097/00001813-200503000-00009. [DOI] [PubMed] [Google Scholar]
- 9.Momparler RL, Bovenzi V. DNA methylation and cancer. J Cell Physiol. 2000;183:145–154. doi: 10.1002/(SICI)1097-4652(200005)183:2<145::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 10.Ahn JS, Kim YK, Min YH, Cheong JW, Jang JH, Jung CW, Kim IH, Yoon HJ, Lee HG, Sohn SK, et al. Azacitidine pre-treatment followed by reduced-intensity stem cell transplantation in patients with higher-risk myelodysplastic syndrome. Acta Haematol. 2015;134:40–48. doi: 10.1159/000368711. [DOI] [PubMed] [Google Scholar]
- 11.Maurillo L, Venditti A, Spagnoli A, Gaidano G, Ferrero D, Oliva E, Lunghi M, D'Arco AM, Levis A, Pastore D, et al. Azacitidine for the treatment of patients with acute myeloid leukemia: Report of 82 patients enrolled in an Italian Compassionate Program. Cancer. 2012;118:1014–1022. doi: 10.1002/cncr.26354. [DOI] [PubMed] [Google Scholar]
- 12.Sudan N, Rossetti JM, Shadduck RK, Latsko J, Lech JA, Kaplan RB, Kennedy M, Gryn JF, Faroun Y, Lister J. Treatment of acute myelogenous leukemia with outpatient azacitidine. Cancer. 2006;107:1839–1843. doi: 10.1002/cncr.22204. [DOI] [PubMed] [Google Scholar]
- 13.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 14.Cho YH, Yazici H, Wu HC, Terry MB, Gonzalez K, Qu M, Dalay N, Santella RM. Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Res. 2010;30:2489–2496. [PMC free article] [PubMed] [Google Scholar]
- 15.Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59:67–70. [PubMed] [Google Scholar]
- 16.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 17.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky SA. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 18.Mikhailenko DS, Kushlinskii NE. The somatic mutations and aberrant methylation as potential genetic markers ofurinary bladder cancer. Klin Lab Diagn. 2016;61:78–83. (In Russian) [PubMed] [Google Scholar]
- 19.Vaissière T1, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, Ferro G, Paliwal A, Hainaut P, Brennan P, et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009;69:243–252. doi: 10.1158/0008-5472.CAN-08-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaz AM, Grady WM. Epigenetic biomarkers in esophageal cancer. Cancer Lett. 2014;342:193–199. doi: 10.1016/j.canlet.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bussemakers MJ, van Bokhoven A, Mees SG, Kemler R, Schalken JA. Molecular cloning and characterization of the human E-cadherin cDNA. Mol Biol Rep. 1993;17:123–128. doi: 10.1007/BF00996219. [DOI] [PubMed] [Google Scholar]
- 22.Yu Q, Guo Q, Chen L, Liu S. Clinicopathological significance and potential drug targeting of CDH1 in lung cancer: A meta-analysis and literature review. Drug Des Devel Ther. 2015;9:2171–2178. doi: 10.2147/DDDT.S78537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karayiannakis AJ, Syrigos KN, Chatzigianni E, Papanikolaou S, Alexiou D, Kalahanis N, Rosenberg T, Bastounis E. Aberrant E-cadherin expression associated with loss of differentiation and advanced stage in human pancreatic cancer. Anticancer Res. 1998;18:4177–4180. [PubMed] [Google Scholar]
- 24.Zheng Z, Pan J, Chu B, Wong YC, Cheung AL, Tsao SW. Downregulation and abnormal expression of E-cadherin and beta-catenin in nasopharyngeal carcinoma: Close association with advanced disease stage and lymph node metastasis. Hum Pathol. 1999;30:458–466. doi: 10.1016/s0046-8177(99)90123-5. [DOI] [PubMed] [Google Scholar]
- 25.Oka H, Shiozaki H, Kobayashi K, Inoue M, Tahara H, Kobayashi T, Takatsuka Y, Matsuyoshi N, Hirano S, Takeichi M, et al. Expression of E-cadherin cell adhesion molecules in human breast cancer tissues and its relationship to metastasis. Cancer Res. 1993;53:1696–1701. [PubMed] [Google Scholar]
- 26.Cui H, Wang L, Gong P, Zhao C, Zhang S, Zhang K, Zhou R, Zhao Z, Fan H. Deregulation between miR-29b/c and DNMT3A is associated with epigenetic silencing of the CDH1 gene, affecting cell migration and invasion in gastric cancer. PLoS One. 2015;10(e0123926) doi: 10.1371/journal.pone.0123926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Sun J, Wang B, Ren JC, Su W, Zhang T. MicroRNA-10b triggers the epithelial-mesenchymal transition (EMT) of laryngeal carcinoma Hep-2 cells by directly targeting the E-cadherin. Appl Biochem Biotechnol. 2015;176:33–44. doi: 10.1007/s12010-015-1505-6. [DOI] [PubMed] [Google Scholar]
- 28.Viotti M, Nowotschin S, Hadjantonakis AK. SOX17 links gut endoderm morphogenesis and germ layer segregation. Nat Cell Biol. 2014;16:1146–1156. doi: 10.1038/ncb3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Glöckner SC, Guo M, Machida EO, Wang DH, Easwaran H, Van Neste L, Herman JG, Schuebel KE, Watkins DN, et al. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 2008;68:2764–2772. doi: 10.1158/0008-5472.CAN-07-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia Y, Yang Y, Liu S, Herman JG, Lu F, Guo M. SOX17 antagonizes WNT/β-catenin signaling pathway in hepatocellular carcinoma. Epigenetics. 2010;5:743–749. doi: 10.4161/epi.5.8.13104. [DOI] [PubMed] [Google Scholar]
- 31.Yin D, Jia Y, Yu Y, Brock MV, Herman JG, Han C, Su X, Liu Y, Guo M. SOX17 methylation inhibits its antagonism of Wnt signaling pathway in lung cancer. Discov Med. 2012;14:33–40. [PMC free article] [PubMed] [Google Scholar]
- 32.Fu D, Ren C, Tan H, Wei J, Zhu Y, He C, Shao W, Zhang J. Sox17 promoter methylation in plasma DNA is associated with poor survival and can be used as a prognostic factor in breast cancer. Medicine (Baltimore) 2015;94(e637) doi: 10.1097/MD.0000000000000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo IY, Chang JM, Jiang SS, Chen CH, Chang IS, Sheu BS, Lu PJ, Chang WL, Lai WW, Wang YC. Prognostic CpG methylation biomarkers identified by methylation array in esophageal squamous cell carcinoma patients. Int J Med Sci. 2014;11:779–787. doi: 10.7150/ijms.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, Chen Z, Zhao X, Wang J, Ding D, Wang Z, Tan F, Tan X, Zhou F, Sun J, et al. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2012;421:640–645. doi: 10.1016/j.bbrc.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 35.Lee EJ, Lee BB, Han J, Cho EY, Shim YM, Park J, Kim DH. CpG island hypermethylation of E-cadherin (CDH1) and integrin alpha4 is associated with recurrence of early stage esophageal squamous cell carcinoma. Int J Cancer. 2008;123:2073–2079. doi: 10.1002/ijc.23598. [DOI] [PubMed] [Google Scholar]
- 36.Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y, Ueda J, Wei W, Inoue M, Tanaka H. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23:233–242. doi: 10.2188/jea.je20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leonard GD, Reilly EM. Post-operative chemotherapy improves disease-free survival, but not overall survival in people with oesophageal squamous cell carcinoma. Cancer Treat Rev. 2004;30:473–477. doi: 10.1016/j.ctrv.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 38.Chen HX, Wang Z. Retrospective study of adjuvant chemotherapy effects on survival rate after three-field lymph node dissection for stage IIA esophageal cancer. Asian Pac J Cancer Prev. 2015;16:5169–5173. doi: 10.7314/apjcp.2015.16.13.5169. [DOI] [PubMed] [Google Scholar]
- 39.Yun H, Damm F, Yap D, Schwarzer A, Chaturvedi A, Jyotsana N, Lübbert M, Bullinger L, Döhner K, Geffers R, et al. Impact of MLL5 expression on decitabine efficacy and DNA methylation in acute myeloid leukemia. Haematologica. 2014;99:1456–1464. doi: 10.3324/haematol.2013.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Öz S, Raddatz G, Rius M, Blagitko-Dorfs N, Lübbert M, Maercker C, Lyko F. Quantitative determination of decitabine incorporation into DNA and its effect on mutation rates in human cancer cells. Nucleic Acids Res. 2014;42(e152) doi: 10.1093/nar/gku775. [DOI] [PMC free article] [PubMed] [Google Scholar]