Abstract
Dystroglycan is a highly glycosylated extracellular matrix receptor with essential functions in skeletal muscle and the nervous system. Reduced matrix binding by α-dystroglycan (α-DG) due to perturbed glycosylation is a pathological feature of several forms of muscular dystrophy. Like-acetylglucosaminyltransferase (LARGE) synthesizes the matrix-binding heteropolysaccharide [-glucuronic acid-β1,3-xylose-α1,3-]n. Using a dual exoglycosidase digestion, we confirm that this polysaccharide is present on native α-DG from skeletal muscle. The atomic details of matrix binding were revealed by a high-resolution crystal structure of laminin G-like (LG) domains 4–5 of laminin α2 bound to a LARGE-synthesized oligosaccharide. A single glucuronic acid-β1,3-xylose disaccharide repeat straddles a Ca2+ ion in the LG4 domain, with oxygen atoms from both sugars replacing Ca2+-bound water molecules. The chelating binding mode accounts for the high affinity of this protein-carbohydrate interaction. These results reveal a novel mechanism of carbohydrate recognition and provide a structural framework for elucidating the mechanisms underlying muscular dystrophy.
Dystroglycan is a widely expressed transmembrane glycoprotein that requires complex post-translational processing to function as an extracellular matrix (ECM) receptor1–3. The protein is involved in a variety of physiological and developmental processes, including maintenance of skeletal muscle function, as well as formation and function of the central nervous system3–6. Synthesized as a single polypeptide, dystroglycan is cleaved autoproteolytically to yield a cell-surface α-subunit and a transmembrane β-subunit7. The extensively glycosylated α-dystroglycan (α-DG) acts as a receptor for laminin G (LG) domain-containing ECM proteins that are major constituents of the basement membrane of skeletal muscle and other tissues: laminin, agrin, and perlecan8. Abnormalities in the post-translational processing of α-DG that disrupt the interaction with the basement membrane result in various congenital and limb-girdle muscular dystrophies, referred to collectively as secondary dystroglycanopathies9. The ECM-binding modification of α-DG also mediates interactions with the LG domains of neurexins10, pikachurin11 and Slit12, thereby contributing to synapse formation and axon guidance. Finally, α-DG serves as a cellular receptor for Old World arenaviruses, including the highly pathogenic Lassa fever virus, and the requirements for virus entry mirror those for LG domain binding13,14.
At least 17 gene products, many of which are glycosyltransferases, are involved in biosynthesis of the functional α-DG modification15. The modification is initiated by a unique O-linked trisaccharide, GalNAc-β1,3-GlcNAc-β1,4-Man-Ser/Thr, which is phosphorylated at position 6 of the mannose residue16,17. The GalNAc is linked, via a tandem ribitol-5-phosphate moiety18, to a heteropolysaccharide that is comprised of alternating glucuronic acid (GlcA) and xylose (Xyl) residues and synthesized by the bifunctional like-acetylglucosaminyltransferase (LARGE)19. LARGE is essential for skeletal muscle function in mice20,21 and humans22. The LARGE-synthesized [-GlcA-β1,3-Xyl-α1,3-]n heteropolysaccharide has been termed matriglycan for its ability to bind LG domain-containing proteins in vitro15,20. However, direct evidence for the presence of matriglycan on native α-DG, as expressed in vivo in skeletal muscle, is currently lacking.
Within basement membranes, the heterotrimeric laminins form cell-associated networks that have both mechanical and signaling functions8. Laminin interactions with cell surface receptors, including α-DG, are mediated by five tandem LG domains at the C-terminus of the laminin α chain8. Each LG domain consists of ~200 residues folded into a β-sandwich23. LG domains that bind α-DG contain a Ca2+ ion bound to one rim of the β-sandwich, consistent with the strict Ca2+-dependence of the interaction24,25. How the Ca2+ site of the LG domain mediates the specific recognition of α-DG is a major unresolved question.
In this study, we developed a novel dual exoglycosidase digestion protocol for matriglycan. Using this protocol, we show that native α-DG from skeletal muscle contains the unmodified heteropolysaccharide [-GlcA-β1,3-Xyl-α1,3-]n, and that its presence confers laminin binding. Crystal structures of laminin α2 LG4-5 with a LARGE-synthesized oligosaccharide reveal that the Ca2+ ion in LG4 is chelated by a single GlcA-β1,3-Xyl disaccharide. This binding mode, which is unprecedented among animal lectins, is predicted to be conserved in other LG domains. Our study thus provides the first atomic-resolution insights into the ECM receptor function of dystroglycan.
RESULTS
Exoglycosidase digestion of native α-DG
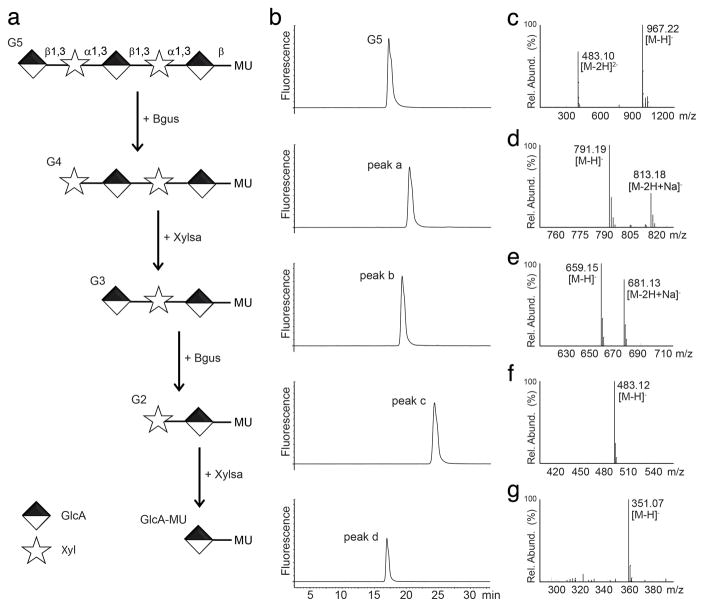
Direct evidence for the presence of GlcA-Xyl repeats is not available for native or recombinant α-DG, and the presence of GlcA or Xyl has only been demonstrated for α-DG produced in LARGE-expressing human embryonic kidney 293 cells19. To confirm that this polysaccharide structure is also present in native tissues, we identified two exoglycosidases that collectively can hydrolyze glycans on native α-DG: β-glucuronidase (Bgus) from T. maritima26, and α-xylosidase (Xylsa) from S. solfataricus27. We first tested these enzymes with a chemically defined substrate, the LARGE-synthesized pentasaccharide G5 (Fig. 1). Alternating treatment of G5 with the two enzymes removed one sugar at a time from the non-reducing end, demonstrating that Bgus and Xylsa cleave β-linked GlcA and α-linked Xyl, respectively, without measurable endoglycosidase activity.
Figure 1. Enzymatic digestion of oligosaccharide G5.
(a) Schematic of the step-wise digestion of G5 with β-glucuronidase (Bgus) and α-xylosidase (Xylsa). MU, 4-methyl-umbelliferone. (b) HPLC profile of G5 and its digestion products. Peaks a, b, c, and d indicate the products of each step of digestion. (c) MS spectrum of G5 (molecular mass: 968). (d) MS spectrum of peak a, indicating that the product of the first step is G4 (molecular mass: 792). (e) MS spectrum of peak b, indicating that the product of the second step is G3 (molecular mass: 660). (f) MS spectrum of peak c, indicating that the product of the third step is G2 (molecular mass: 484). (g) MS spectrum of peak d, indicating that the product of the fourth step is GlcA-MU (molecular mass: 352). The data shown are from a single experiment. This experiment was replicated three times using separately prepared samples.
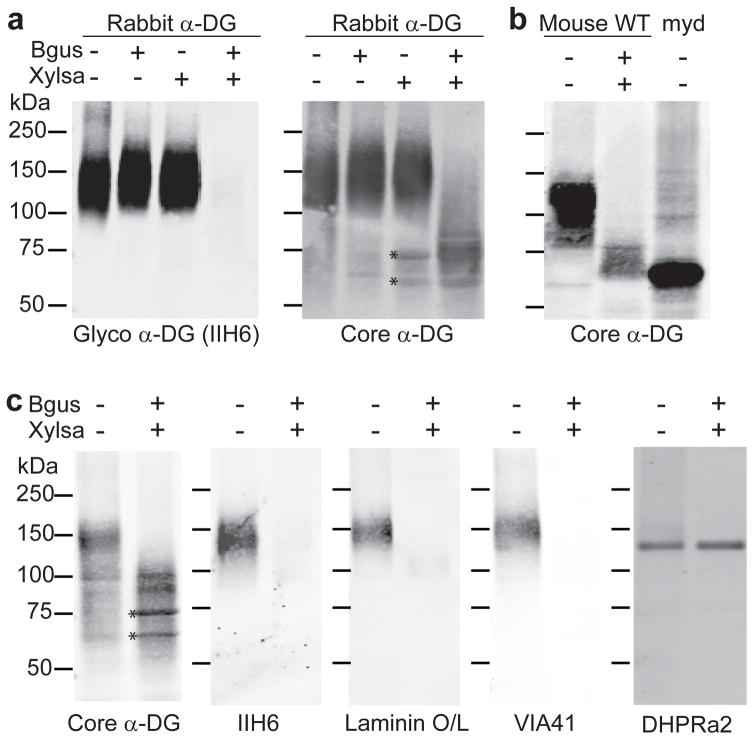
The simultaneous treatment of rabbit skeletal muscle α-DG with Bgus and Xylsa dramatically reduced the apparent molecular mass of α-DG, as assessed by SDS-PAGE, and abolished reactivity with the matriglycan-specific antibody IIH6, whereas treatment of α-DG with either exoglycosidase alone had no such effect (Fig. 2a, Supplementary Results, Supplementary Fig. 1). Similar results were obtained with mouse skeletal muscle α-DG: wild-type mouse α-DG treated with both Bgus and Xylsa migrated similarly to α-DG from Largemyd mice, which have an inactivating mutation in the Large gene21 (Fig 2b), and failed to react with matriglycan-specific antibodies IIH6 and VIA41, as well as with laminin (Fig. 2c). These findings demonstrate that native α-DG in skeletal muscle of rabbit and mouse is modified with multiple [-GlcA-β1,3-Xyl-α1,3-] repeats, and that this modification is required for the ability of α-DG to bind ligand.
Figure 2. Enzymatic digestion of skeletal muscle α-DG.
(a) Rabbit skeletal muscle α-DG was treated with either β-glucuronidase (Bgus) or α-xylosidase (Xylsa), or both enzymes simultaneously. The digestion product was analyzed by immunobloting with anti-α-DG glycan antibody IIH6 (DSHB, University of Iowa) and anti-α-DG core antibody (AF6868, R&D Systems). (b) Wild-type mouse skeletal muscle α-DG was treated with both Bgus and Xylsa, or no enzymes. The digestion product, and Largemyd mouse skeletal muscle α-DG, were analyzed by immunoblotting with anti-α-DG core antibody (AF6868, R&D Systems). (c) Wild-type mouse skeletal muscle α-DG was treated with both Bgus and Xylsa (or no enzymes) and analyzed by: immunoblotting with anti-α-DG core antibody; immunoblotting with anti-α-DG glycan antibodies IIH6 and VIA41 (DSHB, University of Iowa); and overlay assay with laminin-111 (Life Technologies)40. Immunoblotting with a rabbit polyclonal antibody against the α2 subunit of the DHPR Ca2+ channel (Campbell lab) was used for normalizing sample loading and as a marker for a control glycoprotein20. Bands labeled with asterisks that were detected by the polyclonal antibody against core α-DG are likely contaminants of the Xylsa preparation, as the same bands were observed in samples containing only the enzyme. This experiment was replicated three times using separately prepared samples.
Structure of a GlcA-Xyl oligosaccharide bound to laminin
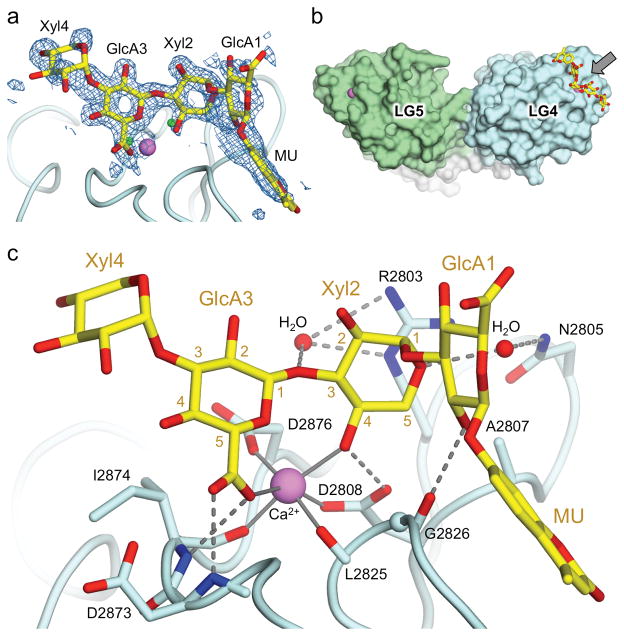
A major binding partner for α-DG in skeletal muscle is the LG4-5 region of the laminin α2 chain28,29, specifically the Ca2+ site in the LG4 domain25. To reveal the structural determinants of α-DG binding, we soaked a mixture of LARGE-synthesized hexa- and heptasaccharide (G6/7; the only oligosaccharide available at the time) into crystals of laminin α2 LG4-5 with an unobstructed LG4 Ca2+ site. Form C crystals soaked in 250 μM G6/7 diffracted to a resolution of 1.4 Å (Supplementary Table 1). The difference electron density around the LG4 Ca2+ site unambiguously defines a GlcA-Xyl disaccharide (Fig. 3a, Supplementary Fig. 2). There is weaker density for the two sugars flanking the Ca2+-bound disaccharide, as well as a flat electron density feature situated on a crystallographic dyad that we interpreted as the 4-methylumbelliferone (MU) group of G6/7. The Ca2+-bound disaccharide accordingly is assigned as GlcA3-Xyl2. We cannot rule out that the Ca2+-bound disaccharide is GlcA5-Xyl4, or a superposition of multiple binding modes, but this ambiguity does not affect any of our conclusions. All of the glycosidic bonds assume stereochemically favorable torsion angles (Supplementary Table 2). Form I crystals soaked in 850 μM G6/7 diffracted to 2.0 Å resolution (Supplementary Table 1). The four sugars defined by the electron density bind in essentially the same way as in form C crystals, but the MU group is positioned differently as a result of its stacking against a symmetry mate (Supplementary Fig. 3). Thus, the conformation of the MU group appears to be determined largely by the crystal lattice, and not by specific interactions with laminin α2 LG4-5.
Figure 3. Crystal structure of laminin α2 LG4-5 bound to oligosaccharide G6/7.
(a) Electron density for the G6/7 ligand in crystal form C. Shown is an unbiased (Fobs,lig – Fobs,apo), αcalc,apo difference electron density map (lig, G6/7 complex; apo, ligand-free structure) contoured at 2.0 σ superimposed onto the final model of G6/7 The sugars beyond Xyl4 are not defined by the electron density and are presumed to be disordered. The small green spheres indicate the positions of two water molecules in the native, ligand-free structure. The MU group is situated on a crystallographic dyad, and the G6/7 ligand is bound with 50% occupancy to each of to the two symmetry-related binding sites. (b) Surface representation of laminin α2 LG4-5 (light blue, LG4; green, LG5, pink, Ca2+) with G6/7 shown in atomic detail (yellow and red sticks). The arrow indicates the direction of the view used in panel c. (c) Atomic interactions between G6/7 (yellow carbon atoms) and laminin α2 LG4 (light blue carbon atoms) in crystal form C. The ring carbon atoms of Xyl2 and GlcA3 are numbered. Coordination bonds to the Ca2+ ion are indicated by solid lines. Two bridging water molecules are shown as small red spheres. Hydrogen bonds are indicated by dashed lines.
The G6/7 oligosaccharide is bound to LG4 in a shallow depression centered on the Ca2+ site, burying 200 Å2 of solvent-accessible protein surface (Fig. 3b). The interaction involves almost exclusively a single disaccharide repeat, GlcA3-Xyl2, which coordinates the Ca2+ ion identically in both crystal forms (Fig. 3c). The GlcA3 carboxylate group is locked into place by two hydrogen bonds with the protein backbone, and one of its oxygen atoms coordinates the Ca2+ ion (2.5 Å distance). The ring oxygen atom of GlcA3 is positioned 3.0 Å from the Ca2+ ion; this distance is longer than a typical Ca2+-O coordination bond, but likely short enough for a favorable contribution to the binding energy. Ca2+ coordination by the GlcA ring oxygen atom has been described30. The Xyl2 4-OH group coordinates the Ca2+ ion (2.4 Å) and forms a hydrogen bond with the Ca2+ ligand Asp2808. Excluding the long bond to the GlcA ring oxygen atom, the resulting Ca2+ coordination geometry is octahedral, with four coordinating atoms provided by the protein and two by the carbohydrate (Fig. 3c). Another notable feature is the stacking of Xyl2 against Arg2803, a residue that is essential for α-DG binding25. In form C crystals, Xyl2 C5 is in close contact with the side chains of Ala2807 and Asp2808 (3.7 and 3.3 Å, respectively), and the GlcA1 2-OH group forms a hydrogen bond to the protein backbone (Fig. 3c). In form I crystals, the conformation of the GlcA1-MU portion of G6/7 differs from that in form C crystals, resulting in a slight rotation of Xyl2 and concomitant disruption of the close contacts of Xyl2 C5 (Supplementary Fig. 3). Even in the more relaxed conformation observed in form I crystals, however, a hexose in place of Xyl2 would clash with Arg2803 and/or Ala2807. The steric exclusion of 5-substituted sugars likely explains why laminins do not bind hyaluronic acid, an abundant extracellular matrix polysaccharide consisting of GlcA-β1,3-GlcNAc repeats (Supplementary Fig. 4).
Laminin affinity for GlcA-Xyl oligosaccharides
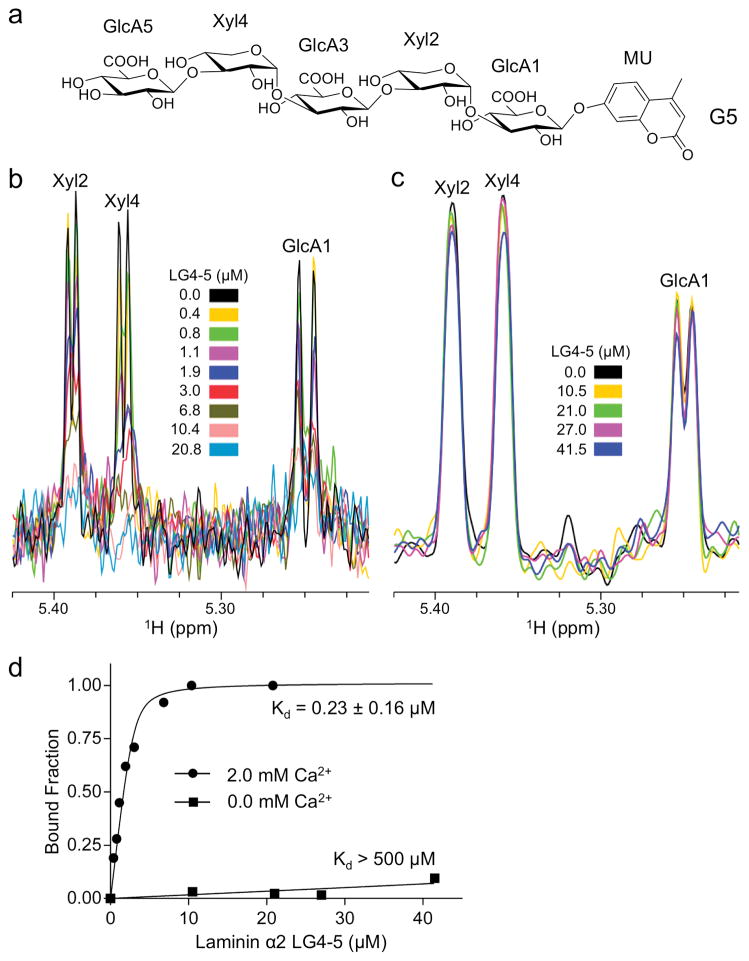
To determine the affinity of laminin α2 LG4-5 for LARGE-synhesized oligosaccharides, we used NMR spectroscopy. Because the G6/7 mixture used in the crystallographic experiments was not suitable for NMR experiments, we studied the pentasaccharide G5 (Fig. 4a). The peaks of three anomeric protons of G5 were resolved in the presence of LG4-5, and their intensities were found to decrease with increasing LG4-5 protein concentration (Fig. 4b). In agreement with an earlier study of α-DG binding to LG4-5 protein25, the interaction with G5 was observed only in the presence of Ca2+ (Fig. 4c). A dissociation constant of 0.23 μM was obtained by fitting the intensity changes of the Xyl4 peak, which was most sensitive to titration with LG4-5 (Fig. 4d). Interestingly, the peaks of the aromatic MU protons remained intense and sharp even in the presence of saturating LG4-5 concentrations, indicating that the MU group is mobile and does not interact with the protein (Supplementary Fig. 5). We therefore believe that the MU group makes only a negligible contribution to the measured affinity for G5.
Figure 4. NMR analysis of binding of oligosaccharide G5 to laminin α2 LG4-5.
(a) The chemical structure of G5. (b) 1D 1H NMR spectra of the anomeric region of 3.0 μM G5 in the presence of 2.0 mM CaCl2 acquired in Tris-d11 buffer and various concentrations of laminin α2 LG4-5 as indicated. (c) 1D 1H NMR spectra of the anomeric region of 7.5 μM G5 acquired in Tris-d11 buffer containing 10 mM EDTA and various concentrations of laminin α2 LG4-5 as indicated. The anomeric peaks in b and c are labeled. (d) Determination of dissociation constants based on the changes in intensity of the anomeric peak of Xyl4, which exhibits the greatest decrease upon addition of laminin α2 LG4-5 in the presence of Ca2+. The standard deviation from data fitting is reported.
Similar experiments with the trisaccharide GlcA-Xyl-GlcA-MU (G3) and the disaccharide GlcA-Xyl-MU (X2) gave dissociation constants of 3.7 and 47 μM, respectively (Supplementary Figs. 6 and 7). The comparatively weak binding of X2 is most likely the result of steric hindrance by the MU group. The 16-fold stronger binding of G5 compared with G3 can be explained partially by the statistical effect of multiple binding sites in G531,32. In addition, the different titration behavior of the Xyl4 and Xyl2 peaks of G5 (Supplementary Fig. 5) suggests that GlcA5-Xyl4 interacts more strongly than GlcA3-Xyl2 with the Ca2+ site. Taken together, the NMR results demonstrate a high-affinity interaction and confirm the conclusion from the crystal structures that a single GlcA-Xyl repeat of matriglycan is sufficient for Ca2+-dependent binding to laminin α2 LG4-5.
α-DG binding by other LG domains
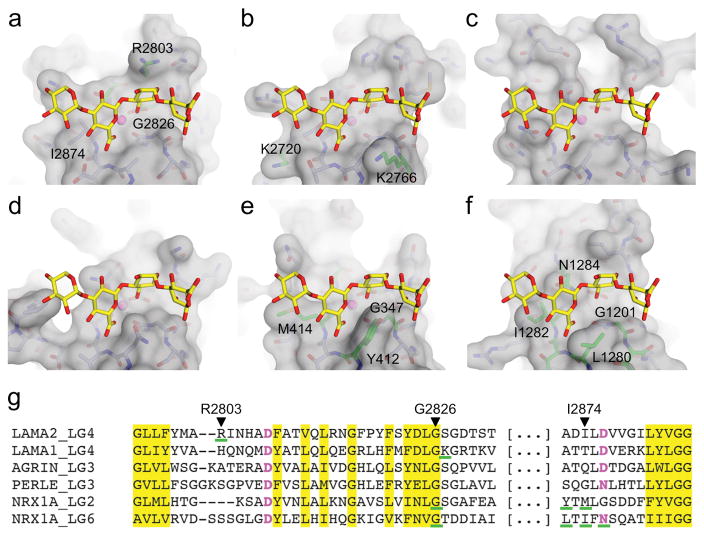
Ca2+-dependent α-DG binding is a function of many LG domain-containing proteins10–12,33. Structural comparison predicts that all these LG domains bind GlcA-Xyl repeats similarly to laminin α2 LG4, despite their divergent sequences (Fig. 5). The key determinants appear to be the Ca2+ ion and the conserved backbone structure around the ion, which strictly requires a glycine at the tip of one of the loops (Gly2826 in laminin α2). Mutation of this conserved glycine in neurexin LG2 or LG6 abolishes α-DG binding, as do mutations in the adjacent loop corresponding to laminin α2 residues 2872–2876 (ref. 41). Lysine and arginine residues in laminin α1 and α2 LG4-5 have been implicated in α-DG binding25,34. These basic residues are not conserved in other LG domains and likely interact non-specifically with the polyanionic carbohydrates on α-DG. Although some LG domains with an intact Ca2+ site (e.g. LG5 of laminins α1 and α2) have been reported not to bind α-DG29, it is possible that the solid-phase assays used in these experiments were unable to detect interactions of lower affinity with matriglycan.
Figure 5. Structural comparison of α-DG-binding LG domains.
(a–f) Molecular surfaces of LG domains with G6/7 positioned according to a local superposition of the protein backbones. Residues whose mutation abolishes α-DG binding25,34,41 are shown in green. (a) Laminin α2 LG4 (this study). (b) Laminin α1 LG4 (PDB entry 2JD4)34. (c) Agrin LG3 (1PZ8)42. (d) Perlecan LG3 (3SH5)43. (e) α-Neurexin LG2 (2H0B)44. (f) α-Neurexin LG6 (3QCW)45. (g) Partial sequence alignment of the LG regions interacting with G6/7. Residues coordinating the Ca2+ ion are in magenta.
DISCUSSION
This study defines the structural basis of LG domain binding to matriglycan, the LARGE-synthesized polysaccharides on dystroglycan. Our exoglycosidase digestion experiments demonstrate that [-GlcA-β1,3-Xyl-α1,3-] repeats are present on native α-DG from skeletal muscle, and that they are directly responsible for LG domain binding. Our structural results show that LG domains are Ca2+-dependent lectins that primarily recognize a single GlcA-β1,3-Xyl repeat. The linkage-specific recognition of a disaccharide by a Ca2+ binding site represents a novel principle of carbohydrate recognition. In typical C-type lectins, such as mannose-binding protein, a Ca2+ ion binds the vicinal hydroxyl groups of one monosaccharide35. This binding mode is inherently of low (millimolar) affinity, and C-type lectins amplify their affinity for ligand either by extending the interaction surface beyond the Ca2+ site or by forming oligomeric assemblies35,36. The GlcA-Xyl repeats of the LARGE glycan achieve a remarkably high affinity for laminin α2 LG4-5 by acting as perfectly matched chelators of the LG4 Ca2+ site. Three features may contribute to reducing the entropic cost of binding: (1) no structural changes in the protein are required to accommodate the ligand; (2) the GlcA-Xyl disaccharide that chelates the Ca2+ ion is bound in a low-energy conformation that likely represents the solution state; and (3) two functional groups of the disaccharide replace Ca2+-bound water molecules in the ligand-free structure (Fig. 3a). The electrostatic interactions formed by the GlcA carboxylate group are not sufficient for tight binding: the C form crystals were obtained in the presence of 120 mM GlcA, yet the electron density at the LG4 Ca2+ site does not indicate any bound GlcA. Thus, only GlcA that is β1,3-linked to Xyl is bound with high affinity.
The presence of multiple GlcA-Xyl repeats in matriglycan is predicted to increase the apparent affinity for LG domains by favoring rapid rebinding after dissociation32. Notably, LG domains are often arranged in tandem. In agrin and perlecan, for instance, no single LG domain is sufficient for high-affinity α-DG binding, yet 2–3 LG domains together bind with apparent dissociation constants in the low nanomolar range29,37. The laminin α2 chain also has at least two α-DG binding sites28,29. The number of GlcA-Xyl repeats within a single matriglycan polysaccharide is unknown but likely to be large, given the ~60 kDa shift in electrophoretic mobility upon enzymatic digestion (Fig. 2) and the small number of sites modified by LARGE38,39. 100 GlcA-Xyl repeats (~30 kDa) would span ~150 nm and could potentially bind more than one molecule of laminin, agrin, or perlecan. The assembly of multiprotein complexes on a single matriglycan chain would explain the graded effect of LARGE-dependent glycosylation on the structure of the basement membrane and the effectiveness of muscle function20.
In conclusion, we have elucidated the atomic mechanism whereby laminin recognizes the unique post-translational modification of α-DG. The laminin-dystroglycan interaction was discovered over 20 years ago and has since been shown to be critically important for skeletal muscle function. The structural insights presented here will facilitate a deeper understanding of the mechanisms that underlie the dystroglycanopathies, as well as other physiological and pathological functions of dystroglycan.
ONLINE METHODS
Purification of α-DG from skeletal muscle
Rabbit α-DG or mouse α-DG was enriched from skeletal muscle (Pel-Freez Biologicals) using DEAE-cellulose and WGA-agarose columns as described46. α-DG was eluted from the WGA-agarose column using 50 mM Tris (pH 7.5), 300 mM NaCl, 0.1% Triton X-100, 300mM GlcNAc, and heated to 99°C for 10 min in the presence of 10 mM 2-mercaptoethanol. The sample was buffer-exchanged using a PD-10 column equilibrated with 150 mM sodium acetate (pH 5.5) and stored at −80°C. Mouse α-DG was enriched from wild-type and Largemyd skeletal muscle using a WGA-agarose column as described47. Samples were heated to 99°C for 10 min in the presence of 10 mM 2-mercaptoethanol and buffer-exchanged into 150 mM sodium acetate (pH 5.0) using Amicon Ultra 0.5 ml centrifugal filters with a 10 kDa molecular weight cut-off.
Synthesis of extended Xyl-GlcA-β-MU products (G3, G5, G6/7)
LARGEdTM was purified using TALON metal affinity resin (Clontech) as described19. To generate Xyl-GlcA-MU (G2), LARGEdTM attached to TALON beads was added to 5 mM UDP-xylose (CarboSource Services) and 5 mM glucuronic acid-βMU (Sigma) in 100 mM sodium acetate (pH 5.5) containing 10 mM MgCl2 and 10 mM MnCl2 in a total volume of 0.5 ml. The solution was incubated for 4–5 days at 37°C with rotation. The reaction product was purified using reverse-phase HPLC on a Beckman Gold system (SUPELCOSIL LC-18 column, 25 cm × 4.6 mm, 5 micron) with HPLC buffer A (50 mM ammonium formate, pH 4.0) and HPLC buffer B (80% acetonitrile in buffer A). Using a 10% buffer B isocratic gradient and a flow rate of 1 ml/min, the G2 product gave a fluorescence signal (325 nm excitation, 380 nm emission) at ~18 min. The collected peak fractions were lyophilized, dissolved in ultra-pure water, and quantitated using fluorescence and GlcA-MU as a standard. GlcA-Xyl-GlcA-MU (G3) was then produced from the G2 sample (1–2 mM in 0.5 ml of water) in a reaction with fresh LARGEdTM beads and 5 mM UDP-GlcA (Sigma), in 100 mM MES (pH 6.0) containing 10 mM MgCl2 and 10 mM MnCl2. This solution was incubated overnight at 37°C with rotation, and the reaction product purified using reverse-phase HPLC as described above. The procedure was repeated twice to generate G5, using fresh LARGEdTM beads at each step. For the synthesis of G6, the same beads were used for multiple steps and ES-MS and NMR analysis of the purified reaction product later revealed a ~50/50 mixture of G6 and G7. This sample, termed G6/7, was used in the crystallographic experiments.
Synthesis of GlcA-Xyl-α-MU (X2)
LARGEdTM attached to TALON beads was added to 20 mM UDP-glucuronic acid and 10 mM xylose-α-MU (Carbosynth Limited, HPLC-purified before use) in 100 mM MOPS (pH 6.0) containing 10 mM MgCl2 and 10 mM MnCl2 in a total volume of 0.5 ml. The mixture was incubated at 37°C for 20 days with shaking at 1200 rpm. The reaction product was purified using reverse-phase HPLC as described above. The collected peak samples were lyophilized, dissolved in water, and analyzed by ESI-MS.
Digestion of G5 with exoglycosidases
T. maritima β-glucuronidase26 (Bgus) bearing a His-tag was overexpressed in E. coli, and purified using TALON metal affinity resin. S. solfataricus α-xylosidase (Xylsa) was purified as described27 with some modifications. Briefly, the cell pellet was resuspended in 20 mM sodium phosphate buffer (pH 7.3), 150 mM NaCl, 1% (v/v) Triton X-100, and homogenized by French press. After centrifugation (30 min at 40,000 × g), the crude extract was incubated with Benzonase (Novagen) for 1 h at room temperature and then heat-fractionated for 30 min at 55, 65, and 75°C. The supernatant was dialyzed against sodium phosphate buffer (25 mM, pH 7.5) and applied to a HiLoad 16/10 Q-sepharose column (GE Healthcare). The enzyme was eluted at 750 mM NaCl by using a linear gradient up to 1 M NaCl with a flow rate of 3 ml/min. G5 (100 μM) was incubated with Bgus (0.2 U) and 150 mM sodium acetate (pH 5.5), in a total volume of 200 μl, for 3 hours at 65°C. The reaction mixture was centrifuged at 13000 g for 10 min, and the supernatant was analyzed using reverse-phase HPLC (solvent A was 50 mM ammonium formate pH 4.0, and solvent B was 80% acetonitrile in solvent A), and monitored by fluorescence detection (325 nm for excitation, and 380 nm for emission). The collected peak samples were lyophilized and dissolved in 50 μl of H2O. An aliquot of purified product was analyzed by ESI-MS, and the rest was used for the second step of digestion. The product of the first step of G5 digestion was incubated with Xylsa (0.2 U) and 150 mM sodium acetate (pH 5.5), in a total volume of 100 μl, for 3 hours at 65°C. The reaction mixture was analyzed using reverse-phase HPLC and ESI-MS as described above. The third and fourth digestion with Bgus and Xylsa, respectively, were carried out analogously in a total volume of 100 μl.
Digestion of α-DG with exoglycosidases
Enriched rabbit α-DG (100 μl of the 150 mM sodium acetate (pH 5.5) solution) was mixed with Bgus (0.45 U) and/or Xylsa (0.09 U), or no enzymes, and incubated overnight at 65°C. Samples were then run on SDS-PAGE, transferred to PVDF-FL (Millipore), and probed with anti-α-DG core antibody (AF6868) and anti-α-DG glycan antibody (IIH6). Enriched rabbit α-DG (100 μl of the 150 mM sodium acetate (pH 5.5) solution) was mixed with Bgus (0.45 U) and/or Xylsa (0.09 U), or no enzymes, and incubated overnight at 65°C. Samples were then run on SDS-PAGE, transferred to PVDF-FL (Millipore), and probed with anti-α-DG core antibody (AF6868), rabbit polyclonal antibody against the α2 subunit of the DHPR Ca2+ channel41 (Campbell lab) and anti-α-DG glycan antibodies (IIH6, VIA41). The laminin overlay assay was performed as described40.
Solid-phase binding assay
GlcA-MU biotinylated at C3 of the MU group (custom synthesis by Sussex Research Laboratories, Ottawa, Canada) was incubated with LARGEdTM, 10 mM UDP- GlcA, 10 mM UDP-Xyl, 10 mM MgCl2 and 10 mM MnCl2 in 100 mM MES buffer (pH 6.5) at 37°C for 18 h. The enzymatic reaction was terminated by boiling in the presence of 50 mM EDTA. After centrifugation (20,000g for 10 min), the supernatant was fractionated using gel-filtration chromatography (Superdex peptide 10/300GL, GE Healthcare). The fractions corresponding to LARGE product of 10–200 kDa molecular mass were pooled. A streptavidin ELISA plate (Thermo Fisher) was coated with 10 μg of biotinylated hyaluronic acid, MW 200 kDa (Creative PEGWorks) and biotinylated (GlcA-Xyl)n-GlcA-MU overnight at 4°C. All subsequent incubations were performed at room temperature. The coated plate was washed with laminin binding buffer LBB (746 mM triethanolamine, 140 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, pH 7.6) and blocked with 3% bovine serum albumin (BSA) in LBB for 2 h. A series of laminin-111 (mouse laminin from EHS sarcoma, Life Technologies) dilutions was prepared from 0 to 10 nM in 1% BSA, 2 mM CaCl2 and added to the plate for one hour. Wells were washed with 1% BSA-LBB and incubated for 1 h with anti-laminin antibody (Sigma L9393) diluted 1:10,000 in 3% BSA-LBB. The plate was washed three times with 1% BSA-LBB before adding anti-rabbit IgG-HRP (Millipore) in 3% BSA-LBB for 30 minutes. After washing as before, the plate was developed with o-phenylenediamine dihydrochloride and H2O2, reactions were stopped with 2 N H2SO4, and absorbance at 490 nm was measured using a microplate reader. The data are reported as mean values ± standard deviation (n = 3).
Protein production for crystallography
DNA encoding residues 2730–3118 of the mouse laminin α2 chain (UniProt Q60675) was amplified and ligated into pCEP-Pu48. Human embryonic kidney HEK293 c18 cells (American Type Culture Collection) were used for protein production as described49. The combined conditioned media from 1 HYPERFlask (Corning) were concentrated and buffer-exchanged into 50 mM HEPES (pH 7.5), 2 mM CaCl2 using a VivaFlow 200 cross-filtration device (Sartorius Stedim Biotech) with a 30 kDa molecular weight cut-off. The laminin α2 LG4-5 protein was purified using heparin affinity chromatography on an Äkta Purifier (GE Healthcare) and eluted at ~240 mM NaCl. The protein was then concentrated to 5 mg/ml, flash-frozen in liquid nitrogen, and stored at −80° for future use. Upon thawing, the protein was further purified using size exclusion chromatography on a Superdex 200 Increase column (GE Healthcare) running in 20 mM HEPES (pH 7.5), 130 mM NaCl, 2 mM CaCl2. This material was used for all biochemical experiments and for the production of crystal form C. To obtain crystal form I, an amino-terminally truncated construct spanning residues 2742–3118 (α2 LG4-5 ΔN) was made and purified as described for the longer construct.
Protein crystallization and ligand soaking
Crystals were grown by hanging drop vapor diffusion. For crystal form C, 2 μl of a 9 mg/ml solution of laminin α2 LG4-5 protein in 10 mM HEPES (pH 7.5), 65 mM NaCl, 2 mM CaCl2 were mixed with an equal volume of precipitant solution composed of HEPES/MOPS (pH 7.5), 120 mM glucuronic acid and 30% of the polyethylene glycol 8000/glycerol mixture of the Morpheus screen (Molecular Dimensions). For crystal form I, 1 μl of a 7 mg/ml solution of α2 LG4-5 ΔN protein in 10 mM HEPES (pH 7.5), 65 mM NaCl, 2 mM CaCl2 was mixed with an equal volume of 100 mM dibasic ammonium citrate (pH 5.3), 18% polyethylene glycol 3350. Crystals of both forms appeared within two days. A 1 mM solution of G6/7 in precipitant solution was added to the crystallization drops in small quantities over the course of days (typically 0.2 μl/day) until the desired concentration was reached (250 and 850 μM for crystal forms C and I, respectively). Form C crystals did not require further cyroprotection before flash-freezing in liquid nitrogen. Form I crystals were briefly soaked in mother liquor supplemented with 20% ethylene glycol before freezing.
X-ray data collection and processing
Diffraction data were collected at beamlines I02 (λ = 0.979 Å; form C, apo; form C, G6/7; form I, G6/7) and I04-1 (λ = 0.920 Å; form I, apo) of the Diamond Light Source (Oxfordshire, UK) and processed using the xia2 pipeline50. The resolution limits were determined using the CC1/2 ≥ 0.5 criterion51. Phases were obtained by molecular replacement using Phaser52 with the laminin α2 LG4-5 structure53 as a search model. Manual rebuilding and refinement were done using Coot54 and PHENIX55. Target values for the glycosidic bond stereochemistry were derived from high-resolution structures of protein-glycosaminoglycan complexes in the Protein Data Bank and from polysaccharide structures in the Cambridge Structural Database (C-O bonds 1.41 ± 0.05Å, C-O-C angles 114.8 ± 2.5°). In form C crystals the MU group of the G6/7 ligand is located on a crystallographic dyad, resulting in statistical disorder between the two symmetry-related orientations (Supplementary Fig. 3). The refinement protocol assumed a 50/50 mixture of ligand-bound and free sites. Two Ca2+-bound water molecules from the ligand-free structure were included with occupancies of 0.5. Refinement of the G6/7 ligand with an occupancy of 0.5 resulted in in an average B-factor of 42.3 Å2 for Xyl4-GlcA3-Xyl2-GlcA1, compared with 37.1 Å2 for the Ca2+ ion. In form I crystals, the G6/7 ligand bound to LG4 of molecule B was refined with full occupancy. Data collection and refinement statistics are summarized in Supplementary Table 1. The final crystallographic models have excellent Ramachandran statistics (>95% favored; 0% outliers in form C crystals; 0.13% outliers in form I crystals)56.
NMR spectroscopy
1D 1H NMR spectra of the oligosaccharide samples in the absence and presence of laminin α2 LG4-5 protein were acquired on a Bruker Avance II 800 MHz spectrometer equipped with a cryoprobe at 25°C using a 50 ms T1ρ filter consisting of a train of spin-lock pulses to eliminate the broad resonances from the protein57. The laminin α2 LG4-5 titrations in the presence of Ca2+ were performed in 25 mM Tris-d11 (pH 7.5), 50 mM NaCl, and 2 mM CaCl2 in 100% D2O. The titrations in the absence of of Ca2+ were performed in 25 mM Tris-d11 (pH 7.5), 50 mM NaCl, and 10 mM EDTA in 100% D2O. The 13C and 1H resonances of oligosaccharides G5 and G3 were reported previously19. The 13C and 1H resonances of disaccharide X2 were assigned in this study using 1H homonuclear two-dimensional DQF-COSY, TOCSY, and ROESY experiments, as well as 1H/13C heteronuclear two-dimensional HMQC and H2BC experiments58. The 1H chemical shifts are referenced to 2,2-dimethyl-2-silapentane-5-sulfonate. The NMR spectra were processed using NMRPipe59 and analyzed using NMRView60. For the resolved anomeric oligosaccharide peaks, the bound fraction was calculated by measuring the difference in the peak intensity in the absence (free form) and presence (bound form) of laminin α2 LG4-5, and then dividing by the peak intensity of the free form. To obtain dissociation constants, the data were fitted to the standard quadratic equation using GraphPad Prism (GraphPad Software). The standard deviation from data fitting is reported.
Supplementary Material
Acknowledgments
We thank Stephen G. Withers for a gift of T. maritima β-glucuronidase, and Christine M. Blaumueller for critical reading of the manuscript. We acknowledge Diamond Light Source for time on beamlines I02 and I04-1 under proposal MX9424. This work was funded by a Wellcome Trust Senior Investigator Award to E.H. (101748/Z/13/Z) and a Paul D. Wellstone Muscular Dystrophy Cooperative Research Center grant to K.P.C. (1U54NS053672). K.P.C. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Accession codes. Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5IK4 (form C, apo), 5IK5 (form C, G6/7 complex), 5IK7 (form I, apo), 5IK8 (form I, G6/7 complex).
Author contributions
D.C.B. co-designed the project, carried out the crystallographic experiments, analyzed the data, and co-wrote the manuscript. T.Y.-M. produced xylosidase and glucuronidase, and performed enzyme digestions and binding assays. D.V. generated reagents G3, G5 and G6/7. M.A. performed enzyme digestions and Western blotting. T.Z. generated reagents X2, produced xylosidase and glucuronidase, and performed the G5 digestion experiment. A.S. and M.M. provided well characterized xylosidase. L.Y. carried out the NMR experiments and analyzed the data. E.H. and K.P.C. co-designed the project, co-wrote the manuscript, and supervised the research. All authors discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibraghimov-Beskrovnaya O, et al. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 3.Michele DE, et al. Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature. 2002;418:417–422. doi: 10.1038/nature00837. [DOI] [PubMed] [Google Scholar]
- 4.Cohn RD, et al. Disruption of DAG1 in differentiated skeletal muscle reveals a role for dystroglycan in muscle regeneration. Cell. 2002;110:639–648. doi: 10.1016/s0092-8674(02)00907-8. [DOI] [PubMed] [Google Scholar]
- 5.Moore SA, et al. Deletion of brain dystroglycan recapitulates aspects of congenital muscular dystrophy. Nature. 2002;418:422–425. doi: 10.1038/nature00838. [DOI] [PubMed] [Google Scholar]
- 6.Satz JS, et al. Distinct functions of glial and neuronal dystroglycan in the developing and adult mouse brain. J Neurosci. 2010;30:14560–14572. doi: 10.1523/JNEUROSCI.3247-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barresi R, Campbell KP. Dystroglycan: from biosynthesis to pathogenesis of human disease. J Cell Sci. 2006;119:199–207. doi: 10.1242/jcs.02814. [DOI] [PubMed] [Google Scholar]
- 8.Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol. 2011;3:a004911. doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godfrey C, Foley AR, Clement E, Muntoni F. Dystroglycanopathies: coming into focus. Curr Opin Genet Dev. 2011;21:278–285. doi: 10.1016/j.gde.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Sugita S, et al. A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 2001;154:435–445. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato S, et al. Pikachurin, a dystroglycan ligand, is essential for photoreceptor ribbon synapse formation. Nat Neurosci. 2008;11:923–931. doi: 10.1038/nn.2160. [DOI] [PubMed] [Google Scholar]
- 12.Wright KM, et al. Dystroglycan organizes axon guidance cue localization and axonal pathfinding. Neuron. 2012;76:931–944. doi: 10.1016/j.neuron.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao W, et al. Identification of α-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 14.Jae LT, et al. Deciphering the glycosylome of dystroglycanopathies using haploid screens for lassa virus entry. Science. 2013;340:479–483. doi: 10.1126/science.1233675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida-Moriguchi T, Campbell KP. Matriglycan: a novel polysaccharide that links dystroglycan to the basement membrane. Glycobiology. 2015;25:702–713. doi: 10.1093/glycob/cwv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida-Moriguchi T, et al. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science. 2013;341:896–899. doi: 10.1126/science.1239951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida-Moriguchi T, et al. O-mannosyl phosphorylation of α-dystroglycan is required for laminin binding. Science. 2010;327:88–92. doi: 10.1126/science.1180512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanagawa M, et al. Identification of a post-translational modification with ribitol-phosphate and its defect in muscular dystrophy. Cell Rep. 2016;14:2209–2223. doi: 10.1016/j.celrep.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Inamori K, et al. Dystroglycan function requires xylosyl- and glucuronyltransferase activities of LARGE. Science. 2012;335:93–96. doi: 10.1126/science.1214115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goddeeris MM, et al. LARGE glycans on dystroglycan function as a tunable matrix scaffold to prevent dystrophy. Nature. 2013;503:136–140. doi: 10.1038/nature12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grewal PK, Holzfeind PJ, Bittner RE, Hewitt JE. Mutant glycosyltransferase and altered glycosylation of alpha-dystroglycan in the myodystrophy mouse. Nat Genet. 2001;28:151–154. doi: 10.1038/88865. [DOI] [PubMed] [Google Scholar]
- 22.Longman C, et al. Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of α-dystroglycan. Hum Mol Genet. 2003;12:2853–2861. doi: 10.1093/hmg/ddg307. [DOI] [PubMed] [Google Scholar]
- 23.Rudenko G, Hohenester E, Muller YA. LG/LNS domains: multiple functions – one business end? Trends Biochem Sci. 2001;26:363–368. doi: 10.1016/s0968-0004(01)01832-1. [DOI] [PubMed] [Google Scholar]
- 24.Hohenester E, Tisi D, Talts JF, Timpl R. The crystal structure of a laminin G-like module reveals the molecular basis of α-dystroglycan binding to laminins, perlecan, and agrin. Mol Cell. 1999;4:783–792. doi: 10.1016/s1097-2765(00)80388-3. [DOI] [PubMed] [Google Scholar]
- 25.Wizemann H, et al. Distinct requirements for heparin and α-dystroglycan binding revealed by structure-based mutagenesis of the laminin α2 LG4-LG5 domain pair. J Mol Biol. 2003;332:635–642. doi: 10.1016/s0022-2836(03)00848-9. [DOI] [PubMed] [Google Scholar]
- 26.Salleh HM, et al. Cloning and characterization of Thermotoga maritima β-glucuronidase. Carbohydr Res. 2006;341:49–59. doi: 10.1016/j.carres.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Moracci M, et al. Identification and molecular characterization of the first α-xylosidase from an archaeon. J Biol Chem. 2000;275:22082–22089. doi: 10.1074/jbc.M910392199. [DOI] [PubMed] [Google Scholar]
- 28.Smirnov SP, et al. Contributions of the LG modules and furin processing to laminin-2 functions. J Biol Chem. 2002;277:18928–18937. doi: 10.1074/jbc.M201880200. [DOI] [PubMed] [Google Scholar]
- 29.Talts JF, Andac Z, Göhring W, Brancaccio A, Timpl R. Binding of the G domains of laminin α1 and α2 chains and perlecan to heparin, sulfatides, α-dystroglycan and several extracellular matrix proteins. EMBO J. 1999;18:863–870. doi: 10.1093/emboj/18.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeLucas L, Bugg CE. Calcium binding to D-glucuronate residues: crystal structure of a hydrated calcium bromide salt of D-glucuronic acid. Carbohydr Res. 1975;41:18–29. doi: 10.1016/s0008-6215(00)87003-2. [DOI] [PubMed] [Google Scholar]
- 31.Feinberg H, Castelli R, Drickamer K, Seeberger PH, Weis WI. Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins. J Biol Chem. 2007;282:4202–4209. doi: 10.1074/jbc.M609689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagae M, Yamaguchi Y. Three-dimensional structural aspects of protein-polysaccharide interactions. Int J Mol Sci. 2014;15:3768–3783. doi: 10.3390/ijms15033768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timpl R, et al. Structure and function of laminin LG modules. Matrix Biol. 2000;19:309–317. doi: 10.1016/s0945-053x(00)00072-x. [DOI] [PubMed] [Google Scholar]
- 34.Harrison D, et al. Crystal structure and cell surface anchorage sites of laminin α1LG4-5. J Biol Chem. 2007;282:11573–11581. doi: 10.1074/jbc.M610657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weis WI, Drickamer K. Structural basis of lectin-carbohydrate recognition. Annu Rev Biochem. 1996;65:441–473. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- 36.Somers WS, Tang J, Shaw GD, Camphausen RT. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLeX and PSGL-1. Cell. 2000;103:467–479. doi: 10.1016/s0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- 37.Gesemann M, et al. Alternative splicing of agrin alters its binding to heparin, dystroglycan, and the putative agrin receptor. Neuron. 1996;16:755–767. doi: 10.1016/s0896-6273(00)80096-3. [DOI] [PubMed] [Google Scholar]
- 38.Hara Y, et al. Like-acetylglucosaminyltransferase (LARGE)-dependent modification of dystroglycan at Thr-317/319 is required for laminin binding and arenavirus infection. Proc Acad Natl Sci USA. 2011;108:17426–17431. doi: 10.1073/pnas.1114836108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vester-Christensen MB, et al. Mining the O-mannose glycoproteome reveals cadherins as major O-mannosylated glycoproteins. Proc Acad Natl Sci USA. 2013;110:21018–21023. doi: 10.1073/pnas.1313446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanagawa M, et al. Molecular recognition by LARGE is essential for expression of functional dystroglycan. Cell. 2004;117:953–964. doi: 10.1016/j.cell.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 41.Reissner C, et al. Dystroglycan binding to α-neurexin competes with neurexophilin-1 and neuroligin in the brain. J Biol Chem. 2014;289:27585–27603. doi: 10.1074/jbc.M114.595413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stetefeld J, et al. Modulation of agrin function by alternative splicing and Ca2+ binding. Structure. 2004;12:503–515. doi: 10.1016/j.str.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Le BV, et al. Crystal structure of the LG3 domain of endorepellin, an angiogenesis inhibitor. J Mol Biol. 2011;414:231–242. doi: 10.1016/j.jmb.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 44.Sheckler LR, Henry L, Sugita S, Südhof TC, Rudenko G. Crystal structure of the second LNS/LG domain from neurexin 1α: Ca2+ binding and the effects of alternative splicing. J Biol Chem. 2006;281:22896–22905. doi: 10.1074/jbc.M603464200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen F, Venugopal V, Murray B, Rudenko G. The structure of neurexin 1α reveals features promoting a role as synaptic organizer. Structure. 2011;19:779–789. doi: 10.1016/j.str.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Combs AC, Ervasti JM. Enhanced laminin binding by α-dystroglycan after enzymatic deglycosylation. Biochem J. 2005;390:303–309. doi: 10.1042/BJ20050375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beedle AM, Nienaber PM, Campbell KP. Fukutin-related protein associates with the sarcolemmal dystrophin-glycoprotein complex. J Biol Chem. 2007;282:16713–16717. doi: 10.1074/jbc.C700061200. [DOI] [PubMed] [Google Scholar]
- 48.Kohfeldt E, Maurer P, Vannahme C, Timpl R. Properties of the extracellular calcium binding module of the proteoglycan testican. FEBS Lett. 1997;414:557–561. doi: 10.1016/s0014-5793(97)01070-3. [DOI] [PubMed] [Google Scholar]
- 49.Paracuellos P, Briggs DC, Carafoli F, Loncar T, Hohenester E. Insights into collagen uptake by C-type mannose receptors from the crystal structure of Endo180 domains 1–4. Structure. 2015;23:2133–2142. doi: 10.1016/j.str.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winter G, Lobley CM, Prince SM. Decision making in xia2. Acta Crystallogr D Biol Crystallogr. 2013;69:1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tisi D, Talts JF, Timpl R, Hohenester E. Structure of the C-terminal laminin G-like domain pair of the laminin α2 chain harbouring binding sites for α-dystroglycan and heparin. EMBO J. 2000;19:1432–1440. doi: 10.1093/emboj/19.7.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 58.Nyberg NT, Duus JO, Sorensen OW. Editing of H2BC NMR spectra. Magn Reson Chem. 2005;43:971–974. doi: 10.1002/mrc.1698. [DOI] [PubMed] [Google Scholar]
- 59.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 60.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.