Abstract
Guanine-nucleotide dissociation inhibitors (GDI) are negative regulators of Rho family GTPases that sequester the GTPases away from the membrane. Here we ask how GDI-Cdc42 interaction regulates localized Cdc42 activation for cell motility. The sensitivity of cells to overexpression of Rho family pathway components led us to a new biosensor design (GDI.Cdc42 FLARE), in which Cdc42 was modified with a FRET ‘binding antenna’ that selectively reported Cdc42 binding to endogenous GDI. Similar antennae could also report GDI-Rac1 and GDI-RhoA interaction. Through computational multiplexing and simultaneous imaging, we determined the spatiotemporal dynamics of GDI-Cdc42 interaction and Cdc42 activation during cell protrusion and retraction. This revealed a remarkably tight coordination of GTPase release and activation on a time scale of 10 seconds, suggesting that GDI-Cdc42 interactions are a critical component in the spatiotemporal regulation of Cdc42 activity, and not merely a mechanism for global sequestration of an inactivated pool of signaling molecules.
Introduction
Rho GTPases are critical in numerous cell functions but especially well characterized in the regulation of cytoskeletal dynamics1-3. GTPases are activated via binding of a GTP nucleotide and deactivated via hydrolysis of GTP to GDP. Molecular mechanisms and effector pathways have been determined through structural analysis, biochemical approaches and imaging. However, only through implementation of biosensors that report GTPase activity in situ has it become clear that GTPase cycles are modulated on length and time scales of single microns and seconds 4-9.
Three families of regulatory proteins modulate Rho GTPase activity: Guanine-nucleotide exchange factors (GEFs) facilitate removal of GDP and binding of GTP, and GTPase activating proteins (GAPs) accelerate the hydrolysis of bound GTP to GDP. Guanine-nucleotide dissociation inhibitors (GDI) sterically block effector interactions, and extract GTPases from the membrane to prevent activation by GEFs. More than 80 GEFs and GAPs act on Rho GTPases10. This diversity confers specificity, enabling a wide variety of input signals to generate different cell behaviors via a relatively smaller number of Rho GTPases (22 Rho family members in mammals, of which Cdc42, Rac1, and RhoA have been most prominently implicated in cytoskeletal regulation). Initial evidence using GTPase biosensors and other approaches suggests that GEFs and GAPs generate this wide variety of cell behaviors by controlling the location and kinetics of GTPase activation.
In contrast to the GEFs and GAPs, there are only three genes encoding RhoGDIs, and each of their protein products interacts with several GTPases. Among them, RhoGDI1 (or RhoGDIα, henceforth referred to simply as GDI) is the most abundant and ubiquitously expressed isoform, and it interacts with all three canonical GTPases. Accordingly, it has been assumed that GDI-GTPase interactions were less specific and less spatially regulated, functioning primarily to provide a uniform cytosolic pool of inactive GTPases that can be accessed by localized regulatory proteins. However, some studies suggested that there is a certain degree of spatiotemporal regulation of GDI-GTPase interactions, mediated by lipids11-13 and kinases that control GDI-GTPase affinity by phosphorylating RhoGDI14-17 or the GTPase18-21. Therefore, it is plausible that modulation of GDI-GTPase interactions also contributes to the fine-tuned spatiotemporal organization of Rho GTPase signaling.
To address the question of how much GDI-GTPase interactions contribute to the regulation of GTPase activity, we focused on developing biosensors that could report the localization and concentration of GDI-GTPase complexes in living cells. We developed a new biosensor design based on a ‘binding antenna’, a single chain containing two fluorescent proteins (FPs) attached to the GTPase of interest. The fluorescence resonance energy transfer (FRET) efficiency of the binding antenna was altered when GDI bound to the GTPase. An important advantage of this design was its ability to reduce the need to introduce exogenous protein. The biosensor reports interaction between modified Cdc42 and endogenous GDI. With the GDI-GTPase biosensor, we explored changes in the distribution of GDI-Cdc42 complex during motility. Far from being a uniform reservoir of inactive GTPase, the GDI-GTPase complex concentrated in protrusions at precise times and positions relative to edge movement and to nearby Cdc42 activation. To test how GDI-Cdc42 complex formation affects Cdc42 activation we modified our existing dye-based biosensor of Cdc42 activity (meroCBD) 6 and imaged GDI-Cdc42 interaction and Cdc42 activity concurrently in the same cell. This paper presents the first spatiotemporally resolved map of GDI-mediated Cdc42 regulation.
Results
A fluorescent biosensor of GDI-Cdc42 binding
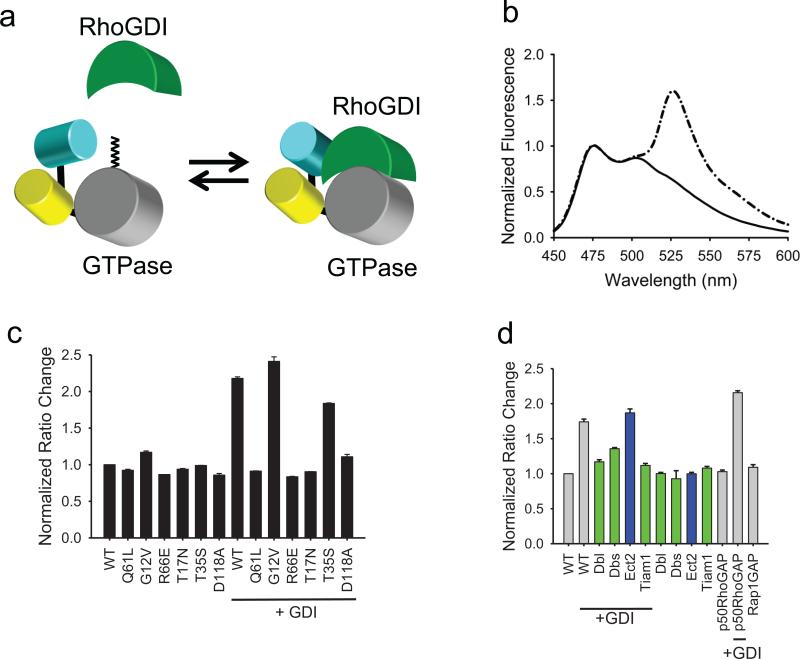
The design of the new GDI-Cdc42 interaction biosensor stemmed from biosensor optimization studies in which we discovered that FRET between two FPs attached to the N terminus of Cdc42 responded strongly to interaction with wild type, “unlabeled” GDI (Fig. 1a). The “binding antenna” attached to Cdc42's N terminus consisted of monomeric Cerulean (mCer)22, attached via an optimized short linker to circularly permutated Venus (cpVen)23. The mCer contained an A206K mutation to reduce interactions between the FPs 24. By optimizing the antenna structure using different linkers and circular permutants (see Supplementary Results, Supplementary Fig. 1), we were able to generate a structure that effectively responded to GDI, but not to GEFs, GAPs or Cdc42 effectors. GTPases require post-translational modification with a lipid moiety at the C-terminus for GDI binding (the lipid binds to the immunoglobulin-like β sandwich of the GDI) 25. Thus, to maintain normal GDI functions it was essential to keep the GTPase C-terminus intact. We named the new biosensor GDI.Cdc42 FLARE. Although we focused here on Cdc42-GDI interactions, we also produced biosensors for Rac1- and RhoA-GDI interactions (see Supplementary Fig. 1&2); biological studies with these will be pursued in the future.
Fig.1.
Design and validation of GDI.Cdc42 FLARE, a genetically encoded biosensor that reports localization of Cdc42-GDI complexes. a, The FRET of a “binding antenna” on Cdc42 is altered when Cdc42 binds endogenous GDI. b, A representative, normalized emission spectra of GDI.Cdc42 FLARE in HEK 293T cells (excitation 433 nm; normalized at the 474 nm emission peak). Solid line, wild-type biosensor; dash-dotted line, wild-type biosensor plus excess GDI. c, Normalized FRET/CFP emission ratios of GDI.Cdc42 FLARE mutants, expressed with and without excess GDI. d, Normalized FRET/CFP emission ratios of wildtype GDI.Cdc42 FLARE with co-expressed GEFs or GAPs, in the presence or absence of excess GDI. Green bars indicate Cdc42-interacting GEFs. Blue bars indicate GEFs that do not interact with Cdc42. For (b)–(d), Results are the mean of three independent measurements. Error bars represent ±SEM.
We assayed the specificity and fluorescence response of the biosensor by obtaining fluorescence spectra of biosensor mutants co-expressed with different Cdc42 upstream regulators in suspensions of HEK293T cells. Using increasing ratios of regulator to biosensor cDNA revealed plateaus in fluorescence change, indicating where binding and the resulting effects on fluorescence saturated, similar to previously results 5. We observed a 218% increase in FRET/CFP emission ratio when we increased GDI from no expression to a 4:1 GDI:biosensor cDNA ratio. Figures 1b and 1c show the fluorescence of the biosensor and its mutants with or without excess-GDI co-expression. Only mutants capable of binding to GDI showed effects of GDI co-expression. The R66E mutant, deficient in GDI-binding, showed no change in FRET with or without the excess-GDI. The D118 mutant, unable to release GEFs, showed no fluorescence change with or without excess-GDI, indicating that the biosensor did not respond to GEF binding. This mutation also prevented response to GDI, consistent with previous data indicating competition between GEFs and GDI for the same binding regions on GTPases 2,10. Effector binding did not affect FRET, as the T35S mutant, which blocks effector binding26,27, did not alter response to GDI. Furthermore, there was no response to either co-expressed GEFs or to downstream effectors (see Supplementary Fig. 3a&b).
The co-expression of exogenous, activated GEFs abrogated FRET response to excess GDI (Fig. 1d), but only for GEFs that affect Cdc42 (the RhoA GEF Ect2 does not bind Cdc42). Reduction of the FRET response caused by 4-fold excess-GDI required a 4-fold excess of GEF over biosensor cDNA, consistent with GEF-Cdc42 affinities and previously published results 5. Excess GEFs alone had no effect on biosensor fluorescence, nor did GAPs (Fig. 1d). For studies in Figure 1d we used constitutively active DH-PH fragments of GEFs. Similar results were obtained with full-length GEFs (see Supplementary Fig. 3a&b), using T17N and D118A Cdc42 mutants that are known to tightly sequester GEFs 28.
The biosensor's remarkable selectivity for GDI is consistent with published crystal structures. The cp49Ven FP extends from Cdc42 at a position that is not part of the known binding sites for GEFs, GAPs, GDI or effectors (see Supplementary Fig. 4). There was almost no FRET when the biosensor was free of GDI, as indicated by biosensor FRET with and without GDI co-expression (Fig. 1b), and by the dependence of FRET on intracellular concentration (see Supplementary Fig. 5a; biosensor FRET plateaued at a low value when increasing expression level reduced binding to endogenous GDI). Low FRET was not due to proteolysis, as shown by gel electrophoresis (see Supplementary Fig. 5b). The low FRET in the GDI-free state indicated that the two FPs adopted a relatively fixed conformation, where their distance and/or orientation produced little FRET. The two FPs could be in a low FRET conformation due to steric and torsional factors, or because mCer bound to a portion of the Cdc42 surface accessible only when the sensor was GDI-free (see Supplementary Fig. 4). mCer may interact with the region where GDI binds to the C terminal tail of Cdc42, and where GEFs, GAPs and effectors do not bind. FRET increased upon GDI binding, but this FRET diminished when we reduced interactions between the FPs by adding an A206K mutation24 into cpVenus (see Supplementary Fig. 5c). This suggested that interactions between the FPs enhanced FRET in the GDI-bound state. Interaction of the FPs with GDI may also induce a high FRET conformation.
Dynamics of the GDI-Cdc42 complex in cell protrusions
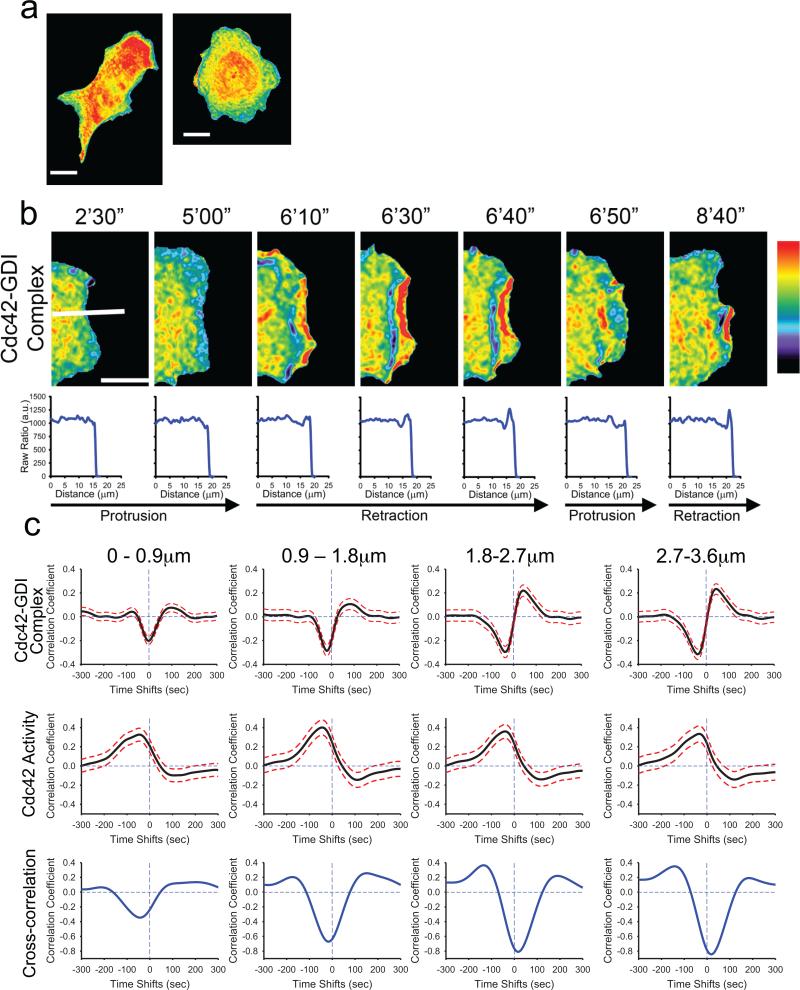
We stably expressed the GDI.Cdc42 FLARE biosensor in mouse embryonic fibroblasts (MEF/3T3) under control of a tetracycline-inducible promoter (tet-OFF) as described previously 5. The cells were sensitive to overexpression of the biosensor, as they are to overexpression of Cdc42 6. We therefore established stable, inducible cells with expression levels below those producing effects on cell morphology or motile behavior (see Supplementary Fig. 6). The biosensor showed elevated binding of GDI to Cdc42 throughout most of the cell body, but cell edges showed reduced FRET/donor ratio, indicating that the majority of the GDI-Cdc42 complex occupied the bulk cytosolic space in the cell body (Fig. 2a). Live-cell imaging of the GDI-Cdc42 complex revealed a strikingly dynamic behavior at the cell edge during protrusion/retraction cycles (Fig. 2b; see Supplementary Videos 1-4). Linescans perpendicular to the cell edge showed reduced FRET ratios within 2 μm of the edge during protrusion, whereas the ratio elevated during retraction, with the highest values associated with ruffles undergoing retrograde movement (Fig.2b). This indicated a reduced GDI-Cdc42 complex specifically at the edge during protrusion, and concentrated in rearward moving ruffles during retraction. The ruffles showed reduction in GDI-Cdc42 complex on the side distal from the edge.
Fig.2.
Cdc42-GDI complex localization in living cells. a, MEFs expressing the GDI.Cdc42 FLARE biosensor. Note the relatively uniform distribution of Cdc42-GDI complex except at the cell edges. b, Localization of GDI-Cdc42 at a cell edge undergoing protrusion/retraction cycles. Below each image is the fluorescence ratio measured along the white line shown in the first image GDI-Cdc42 complex is reduced at the cell edge during protrusion and increased during retraction. c, TOP: Correlation of Cdc42-GDI localization and edge velocity, measured using the GDI.Cdc42 FLARE biosensor. Correlation curves are computed from n = 704 individual windows in 8 cells (see Supplementary Fig. 6). MIDDLE: Correlation of Cdc42 activity and edge velocity measured in the wild-type MEFs using the MeroCBD biosensor. Correlation curves are computed from n = 420 individual windows in 7 cells. BOTTOM: Cross-correlation of the correlation curves shown in the top and middle rows. Panels (a) and (b) pseudocolor range: 1.0 = black; 1.4 = red. White bar indicates 10 μm.
To investigate the dynamics of GDI-Cdc42 binding more quantitatively we utilized cross-correlation between the FRET/donor ratio and cell edge motion, as used previously to analyze coupling of GTPase activities with cell edge velocity4. We first verified that the average period of protrusion-retraction cycles in the wild type and GDI.Cdc42 FLARE (wildtype) biosensor cells were statistically indistinguishable (see Supplementary Fig. 6). This confirmed that expressing the biosensor did not perturb the regulation of basal protrusion-retraction cycles. Consistent with the linescan analysis in Figure 2b, the coupling between GDI-Cdc42 and edge motion in sampling windows sub-adjacent to the leading edge (0–0.9 μm) showed a significant negative cross-correlation (–0.203 ±0.034) at –2s ±13s time lag (Fig. 2c). This suggested that overall GDI-Cdc42 binding reduced concurrently with edge protrusion and increased during retraction. The strength of this negative correlation increased at the next sampling window position (0.9–1.8 μm from the edge), while the negative time lag also increased to significant values (–20s ±9s). This indicated that the modulation of GDI-Cdc42 interaction slightly lagged the modulation of edge velocity. In sampling windows even further away from the edge, a significant positive cross-correlation peak appeared at +42 ±23s. The strength of this positive peak increased with the distance to the cell edge, indicating increased GDI-Cdc42 binding prior to protrusion and/or GDI-Cdc42 release prior to retraction. Together these analyses revealed distinct, fine-tuned regulation of GDI-Cdc42 binding for different regions at the cell edge.
Next, we wished to understand GDI-Cdc42 dynamics relative to Cdc42 activation. We decided to approach this first by computational multiplexing (see Supplementary Fig. 7), in which we measured the kinetics of biosensor activities in separate cells and determined their relationship to cell edge motion, which was used as a common fiduciary4. As previously published6, we monitored Cdc42 activity in wildtype MEFs using meroCBD. Consistent with our previous analyses4, we found significant correlation between Cdc42 activation and edge velocity at a time lag of –46s ±31s (Fig. 2c) indicating a delayed Cdc42 activation relative to cell protrusion. Also consistent with our previous findings, the correlation between edge velocity and Cdc42 activity was maximal in the window row 0.9 – 1.8 μm from the cell edge, suggesting that Cdc42 likely activated at the location of mature focal adhesions, which were, on average, 2–3 μm back from the cell edge (Fig. 2c). Beyond the second row of windows, the correlation strength decreased. Qualitatively, these Cdc42 activation characteristics matched well with our observation of a negative correlation peak between edge motion and GDI-Cdc42 interaction at –20s to –40s. This suggested that the negative correlation between edge motion and GDI-Cdc42 binding predominantly associated with a release of Cdc42 from GDI shortly after protrusion onset. To confirm this relationship more systematically we computed the cross-correlation between two correlation functions: velocity to GDI-Cdc42 interaction (using GDI.Cdc42 FLARE) and velocity to Cdc42 activity (using meroCBD) (Fig. 2c). As expected, these two functions correlated strongly and negatively, with a time lag of ~45s at the leading edge (0–0.9 μm); i.e. the reduction in GDI-Cdc42 interaction preceded the activation of the Cdc42 signal by ~45s. In locations further away from the edge, the strength of negative cross-correlation increased while the time lag converged to zero or even slightly positive values. This suggested a reduction in time between Cdc42 release and activation with greater distance from the cell edge. However, we could not quantify the exact reduction in time lag as the positive lobe in the velocity to GDI-Cdc42 interaction correlation function at positive lag times biased the position of the cross-correlation minimum. Overall, these results showed that the new GDI.Cdc42 FLARE biosensor reported signals that were tightly coordinated with the activation of Cdc42 and the relationship between GDI-Cdc42 complex localization and Cdc42 activation.
Re-engineering a Cdc42 biosensor for multiplexed imaging
To analyze the relations between localization of the GDI-Cdc42 complex and Cdc42 activation more directly, we strived for concurrent imaging of both events in the same cell. Our previously described meroCBD biosensor 6 was based on a fragment of the Wiskott Aldrich Syndrome Protein (WASP) that binds selectively to active Cdc42. We covalently derivatized this fragment with mero87, a dye whose fluorescence responds to changes in solvent environment 29. Dye fluorescence increased when the WASP fragment bound active Cdc42, revealing Cdc42 activation dynamics. Enhanced green fluorescent protein (eGFP) was attached to this biosensor to enable “ratio imaging”, a technique for eliminating effects of cell morphology, biosensor concentration, or uneven illumination on biosensor readouts6. eGFP wavelengths overlap those of the GDI.Cdc42 FLARE biosensor, so for the current study we replaced eGFP with maltose binding protein (a nonfluorescent protein that enhances water solubility as does eGFP) and site-specifically labeled the biosensor with Atto-700 dye for ratio imaging (taking advantage of selective deprotonation of the WASP fragment N-terminus at pH 5.2, see Supplementary Fig. 8a). We attached the mero87 dye 6,29 selectively to a cysteine inserted at position 271 in the WASP fragment 6, using a cysteine-selective iodoacetamido derivative (neither the maltose binding protein nor the WASP domain had other exposed cysteines). The two dyes attached with a 1:1 ratio and labeling efficiencies of >95% .
The mero87 dye on the new meroCBD biosensor showed a 325% increase in fluorescence upon binding to Cdc42 (see Supplementary Fig. 8b). The Atto-700 emission at 705 nm also increased by 162%, enabling ratiometric measurements of Cdc42 activation (change in ratio of mero87/Atto700 emission = 200%). The 705 nm emission of the Atto dye not only eliminated bleed-through into the CFP-YFP FRET channels, but was also compatible with the laser-autofocus systems used in several commercial imaging systems. The Atto-700 dye showed photostability superior to other dyes used previously (see Supplementary Fig. 8c).
Dynamics of Cdc42 activation and GDI-Cdc42 localization
To study the relative dynamics of Cdc42 activation and the localization of Cdc42-GDI complex in the same cell, we microinjected the modified meroCBD biosensor into Swiss 3T3 mouse embryonic fibroblasts (MEF) stably expressing the GDI.Cdc42 FLARE biosensor. Although each biosensor could be used at effective concentrations without altering motility 6, we observed effects on cell morphology and reduced adhesion when we used similar concentrations of the two biosensors 26,27. At concentrations where we observed no perturbation, low signal/noise precluded correlation analysis. Therefore, we included a T35S mutation in the GDI.Cdc42 FLARE biosensor to reduce effector binding and enable use of higher biosensor concentrations without changing cell behavior. This mutation could potentially generate dominant negative effects, so we stably expressed the T35S GDI.Cdc42 biosensor in mouse embryonic fibroblasts (MEF) under control of the tet-OFF system30 and FACS-sorted the MEFs on the basis of GDI.Cdc42 FLARE brightness prior to injection. We determined brightness levels that did not perturb normal motile behavior or Cdc42 activation dynamics (see Supplementary Fig. 9).
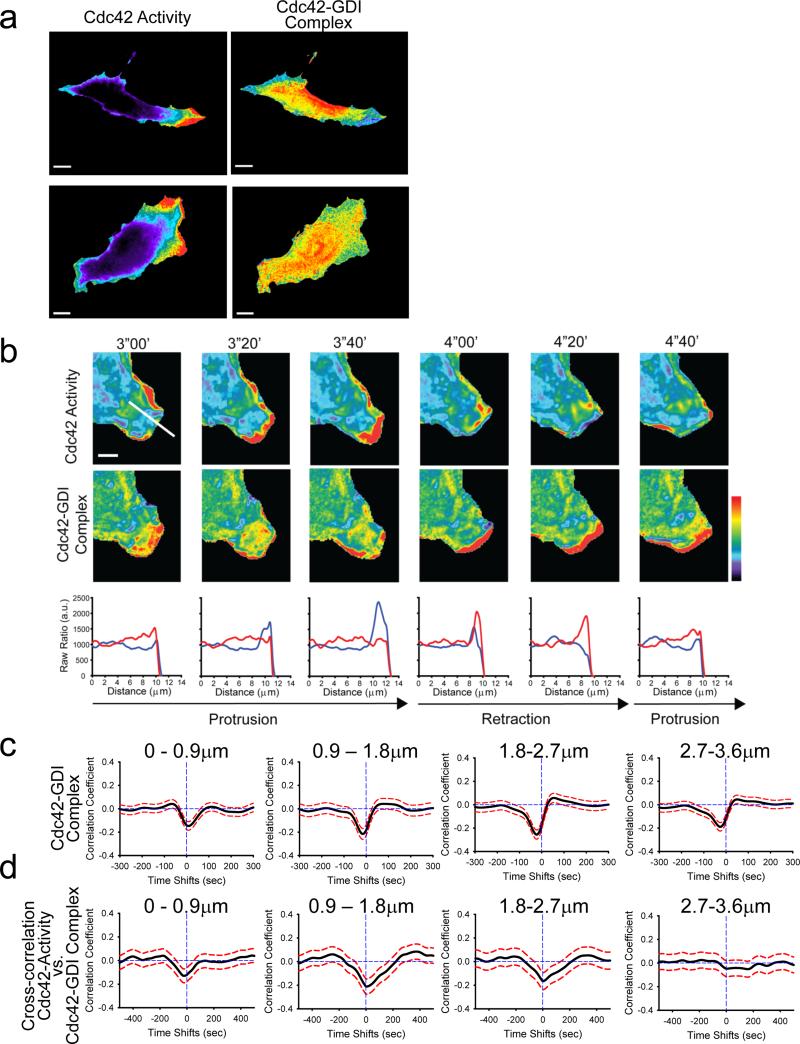
The co-imaging of the T35S GDI.Cdc42 FLARE and meroCBD biosensors in living fibroblasts revealed a relatively homogenous distribution of low Cdc42 activity corresponding with elevated GDI-Cdc42 binding throughout most of the cell. This was consistent with the data generated by the wildtype sensor and with the notion of GDI maintaining a pool of inactive Cdc42 (Fig. 3a). Again, we observed transient and localized changes in the GDI-Cdc42 complex at the cell edge (Fig. 3a&b; see Supplementary Videos 5-7), especially in regions with active retraction and protrusion. As with the wildtype GDI.Cdc42 FLARE sensor, cross-correlation of the T35S GDI.Cdc42 FLARE signal with cell edge velocity consistently displayed a negative peak in all window rows between 0–4 μm (Fig. 3c), which likely associated with the release of Cdc42 from GDI during protrusion events. Data from wildtype and T35S mutant biosensors differed quantitatively, but not qualitatively, in the time lags of the peak, suggesting these two biosensors had somewhat different kinetics due to the influence of effector binding. Moreover, the mutant sensor did not produce a significant positive correlation peak with the velocity at positive time lags, neither at the cell edge nor further inside the cell (Fig. 3c&2c). Most probably, this secondary correlation peak disappeared because the absence of effector binding changed the dynamics of GDI-Cdc42 complex formation prior to protrusion events, or because the higher noise level masked more subtle secondary effects detected by the WT sensor.
Fig.3.
Relationship between Cdc42 activation and GDI-Cdc42 localization monitored in the same cell. a, MEFs expressing the T35S mutant version of the GDI.Cdc42 FLARE biosensor and injected with the modified meroCBD biosensor. Left column: Cdc42 activity, Right Column: GDI-Cdc42 localization. (Pseudocolor scales: for Cdc42, Black=1.0, Red=7.46 (top cell), 2.82 (bottom cell); for GDI-Cdc42: Black=1.0, Red=1.51 (top cell), 1.97 (bottom cell). Bar = 20 μm). b, Cdc42 activation (TOP) and Cdc42-GDI localization (MIDDLE) in the same cell over time. (Pseudocolor scales: Cdc42 1.0–2.3, Cdc42-GDI 1.0–1.53. Bar = 5 μm). BOTTOM: Profile of Cdc42 activation (blue) and GDI-Cdc42 (red) along the line shown in the first panel of the top row. c, Correlation of Cdc42-GDI and edge velocity, measured using the T35S mutant of the GDI.Cdc42 FLARE biosensor. Correlation curves are computed from n = 886 individual windows in 7 cells. d, Correlation of Cdc42 activity and Cdc42-GDI monitored in the same cell, using the T35S mutant version of GDI.Cdc42 FLARE and the modified meroCBD. Correlation curves are computed from n = 420 individual windows in 7 cells.
Figure 3b displays Cdc42 activation and GDI-Cdc42 localization, observed by concurrent imaging of both FRET sensors during a representative episode of protrusion–retraction–protrusion. As predicted by the computational multiplexing analysis using wildtype GDI.Cdc42 FLARE this data showed a strong reduction in Cdc42.GDI signal during protrusion. Moreover, reduction in Cdc42-GDI complex accompanied Cdc42 activation as reported by the reengineered meroCBD. During retraction, we observed the opposite signals: The Cdc42 activation dropped, whereas localized GDI-Cdc42 complex sharply increased. This suggested that GDI promptly and efficiently sequestered deactivated Cdc42 during retraction. The profiles in Figure 3b also showed that these inverse relations between Cdc42 activity and GDI-Cdc42 binding concentrated to the very leading edge of a cell.
To quantify the coupling between Cdc42 activity and localization of the GDI-Cdc42 complex, we again applied correlation analysis. With simultaneous imaging of both activities, we could for the first time map the spatial organization of the relationship between Cdc42 activation and GDI-Cdc42 complex formation. In sampling windows immediately adjacent to the cell edge, the correlation function displayed a significant negative peak with a time lag of approx. –20s, indicating that GDI released Cdc42 before Cdc42 activation (Fig. 3d). Qualitatively, this time lag between release and activation of the GTPase matched the prediction of a GTPase activation delay by computational multiplexing (Fig. 2c). Also consistent between these two analyses was that the negative peaks of the correlation function moved to zero in sampling windows 0.9–1.8 μm and 1.8–2.7 μm from the cell edge. As before, this data is consistent with concentration of active Cdc42-GEFs a few μm behind the cell edge, a proposition we made before based on the cross-correlation between Cdc42 activities and velocity alone 4. Contrary to the predictions from the computational multiplexing suggesting a significant inverse relationship between Cdc42 activity and GDI-Cdc42 localization in sampling windows 2.7–3.6 μm from the cell edge, correlation analysis from simultaneous imaging of the two biosensors indicated that GDI-Cdc42 binding lacked significant influence on the regulation of Cdc42 beyond a ~3 μm wide band from the cell edge. Again, this could be due to the elimination of effector binding in the mutant biosensor, but overall these analyses showed that in qualitative terms the mutated T35S GDI.Cdc42 FLARE and the wildtype GDI.Cdc42 FLARE reported the same spatiotemporal coordination of GDI binding and GTPase activation, and that the antenna can be used together with dye-based biosensors for multiplexed imaging.
Src regulation of GDI-Cdc42 interaction
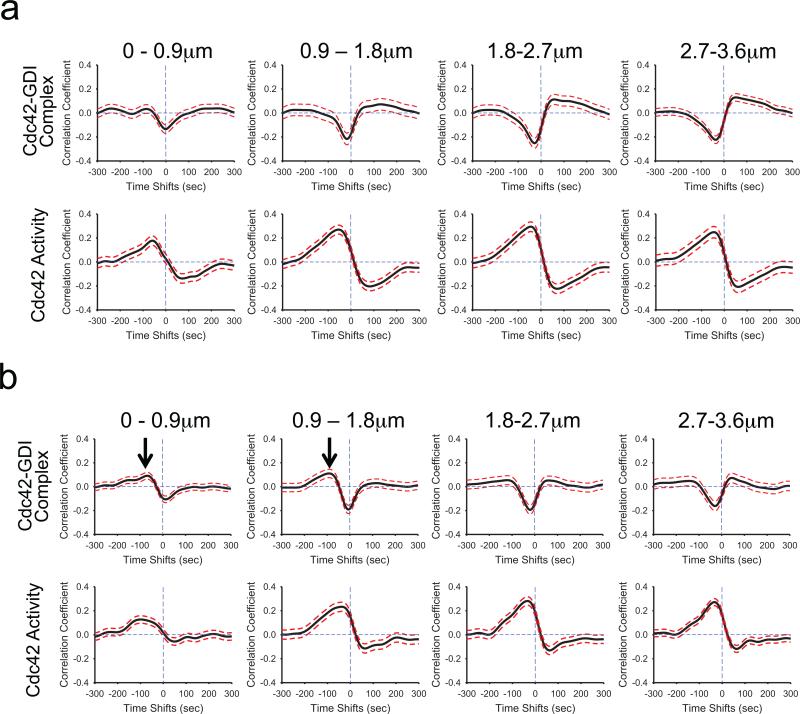
Equipped with tools to directly observe and quantify the relationships between GDI-Cdc42 complex and Cdc42 activation we sought to validate and expand biochemical models of the regulation of GDI-GTPase interactions in the context of living cells. Previous biochemical studies show that phosphorylation of GDI by Src at Y156 largely abolishes GDI's affinity for Cdc42, Rac1 and RhoA 15. Moreover, this phosphorylation significantly impacts the ability of GDI to extract RhoA from isolated plasma membrane 15. Thus, Src may regulate the localization and timing of GDI-Cdc42 interactions during protrusion. To test this in living cells, we established an optimized knockdown/rescue procedure to replace endogenous GDI with either Y156F (phosphorylation-deficient) or Y156E (phosphomimetic) GDI, and examined the effects of these changes on the leading edge correlations (Fig. 4). Previously, Y156F GDI mutant shows cellular distribution and phenotype similar to the wildtype GDI, while Y156E GDI accumulates at the leading edge and within edge ruffles 15. While such accumulation was observed in fixed-cells, in vivo evidence for binding of this mutant GDI to Cdc42 at the edge ruffles in living cells remains elusive, especially in light of the severely abrogated affinity of the Y156E GDI mutant for GTPases 15. We used shRNA against endogenous GDI (see Supplementary Fig. 10) followed by stable, viral transduction with GDI mutants resistant to the shRNA.2,15,31 For these experiments we used the T35S GDI.Cdc42.FLARE biosensor to probe GDI-Cdc42 localization and imaged Cdc42 activity in separate cells to avoid overloading cells harboring GDI mutations with two biosensors (see Supplementary Videos 8–11). As expected, cross-correlations between edge motion and Cdc42 activity in cells with GDI Y156F were identical to those using wildtype GDI (Figs. 4a&2c). In contrast, GDI Y156E produced a small but measureable positive cross-correlation between GDI-binding and edge motion at negative time lags for the first two window rows (Fig. 4b). From this observation, we concluded that at these locations the mutant GDI bound to the plasma membrane-localized Cdc42 and was thus confined within its activation compartment. Previous observations by immunofluorescence 15 shows an accumulation of theY156E GDI mutant in this region, supporting the notion that the reduced affinity of this GDI mutant for GTPase prevents transfer of the prenylated C-terminus of the GTPase out of the plasma membrane into the β-sandwich of the GDI structure. Hence, this mutation may prevent extraction of the GTPase from the plasma membrane. Our results here showed that a significant fraction of GDI actually bound and remained bound to Cdc42 at the edge, likely because of its reduced affinity. At distances further away from the cell edge the correlation functions were qualitatively the same as those of the non-mutated biosensor (Fig. 4b). This indicated that activation of Cdc42 and localization of the Cdc42-Y156E GDI complex were concurrent/concomitant only within the edge region (0–1.8 μm). Furthermore, the formation of Cdc42-GDI complex and Cdc42 activation at those locations occurred with the same dynamics (Fig. 4b). Both lagged changes in protrusion velocity (Cdc42 (0–0.9 μm): –87s ±47.5s; GDI-binding (0–0.9 μm): –67s ±28s; t-test between Cdc42 activation and GDI-binding at 0–0.9 μm: p=0.358425; Cdc42 (0.9–1.8 μm): −45s ±42.5s; GDI-binding (0.9–1.8 μm): –92s ±40.5s; t-test between Cdc42 activation and GDI-binding at 0.9–1.8 μm: p=0.21733). As in previous observations, the GDI Y156E mutant produced slightly faster leading edge turnover rates (see Supplementary Fig. 5) 15. Together, these data demonstrated that in living cells, phosphorylation of GDI at Y156 by Src did not regulate the ability of GDI to bind to GTPase but rather the ability of GDI to extract the GTPases from the plasma membrane.
Fig.4.
Src-mediated phosphorylation of GDI at Y156 regulates the coordination of Cdc42-GDI localization and Cdc42 activity in regions close to the edge. a, A phosphorylation-deficient mutant of GDI (Y156F) had no effect on the timing of Cdc42 activation or modulation of GDI-Cdc42 localization. TOP: Correlation of Cdc42-GDI and edge velocity, using the T35S mutant of the GDI.Cdc42 FLARE biosensor expressed in cells containing Y156F GDI. Correlation curves are computed from n = 902 individual windows in 11 cells. BOTTOM: Correlation of Cdc42 activity and edge velocity using the MeroCBD biosensor in cells containing Y156F GDI. Correlation curves are computed from n = 373 individual windows from 5 cells. b, A phosphomimetic mutant of GDI (Y156E) produced positive correlation with edge velocity at negative time lags (i.e. after protrusion onset) similar to those of the correlation maxima of Cdc42 activity with edge velocity in regions close to the edge (0–1.8 μm; Black arrows). TOP: Cross-correlation of Cdc42-GDI and edge velocity using the T35S mutant of the GDI.Cdc42 FLARE biosensor expressed in cells containing the Y156E mutant GDI. Correlation curves are computed from n = 979 individual windows from 11 cells. Black arrows in regions 0–1.8 μm from the edge indicate the appearance of a positive cross correlation peak in GDI-Cdc42 complex localization. BOTTOM: Cross-correlation of Cdc42 activity and edge velocity, using the MeroCBD biosensor in cells containing GDI Y156E. Correlation curves are computed from n = 491 individual windows in 4 cells.
Discussion
We described an approach to sense protein interaction in living cells by modifying only one of two interacting proteins. A ‘binding antenna’ was placed on Rho GTPases where its FRET was affected by GDI binding, but not by other ligands. Use of a binding antenna can substantially reduce cell perturbation because only one protein in an interacting pair needs modification. By attaching binding antennae to affinity reagents that interact selectively with activated target proteins (e.g. the WASP domain in the meroCBD biosensor used here) it should be possible to make genetically encoded biosensors that sense activation of endogenous proteins.
Through multiplexed imaging of GDI-Cdc42 interaction and Cdc42 activation, we determined the relative kinetics of these two events in the context of cell protrusions. We showed that localized buildups of Cdc42-GDI complex and Cdc42 activation were inversely related within protrusions. Mapping this relation revealed a remarkably tight coordination on a time scale of 10 seconds. This suggests that GDI-Cdc42 interactions are a critical component in the spatiotemporal regulation of Cdc42 activity, and not merely a mechanism for global sequestration of an inactive pool of GTPase. Extrapolating from the time scale of cell protrusion, regulation of GDI-Cdc42 interaction may in fact be rate-limiting in GTPase activation during this process: After release of signaling molecules into the plasma membrane, sufficient GEFs are already primed or rapidly activated to induce GDP to GTP exchange on a time scale of seconds. It would follow that the characteristic timing of Cdc42 activation during protrusion events is at least in part mediated by localized release of GTPase from GDI. Indeed, the cross-correlation between appearance of the Cdc42-GDI complex and edge velocity shows a remarkably narrow spike over the time lag interval of ~25s, suggesting that Cdc42 is released shortly after the protrusion edge velocity has reached maximal values. The resulting positive correlation of Cdc42 with velocity stretches over a 6-fold longer time lag interval. This observation is again consistent with a mechanism in which GEFs rapidly activate Cdc42 at the protruding cell edge, and activated molecules then diffuse over a broader region in which they remain active by repeated interactions with GEFs, or by diminished GAP activity. This example illustrates that understanding GTPase signaling will require correlating the activity of GDI, GEFs and GAPs. With the present work, we have taken a first step in this direction.
Our spatiotemporal analysis of the GDI-Cdc42 complex also revealed precisely localized decreases in Cdc42-GDI complex concentrations. This was especially surprising for an interaction that has thus far been associated with global sequestration of GTPases. While several cues could modulate the interaction, our findings now add to this data the requirement that the cues themselves operate with precise localization and timing. As a proof of principle, we demonstrated here how the Cdc42-GDI interaction responded to Src-kinase activity, impacting the ability of GDI to extract GTPase from the plasma membrane. We picked Src among the GDI-phosphorylating kinases because its activity is precisely coupled to the formation and force-mediated activation of adhesions 32,33, and because of its documented roles in activating GEFs 34. As other kinases could be specific for different GTPases 15-17,35, further investigation in this area using biosensors would be valuable.
In summary, our work proposes a hitherto unappreciated complexity in the regulation of Rho GTPase signaling activity. Besides the well-established roles of GEFs and GAPs as spatiotemporal sculptors of the GTPase signaling landscape, this study adds GDI interactions as an additional factor. The described biosensor provides a valuable means to dissect these regulatory mechanisms in the context of diverse cell functions, and the antenna readout mechanism provides a tool to examine protein-protein interactions with modification of only one interacting partner.
Online Methods
Biosensor expression constructs
The G/C FLARE biosensor was cloned by connecting a hexaHis tag, monomeric Cerulean fluorescent protein (FP) 22 and a circularly permutated (cp49) Venus FP 23. Monomeric Cerulean and cp49 Venus FPs were connected by an GSGS linker, which was flanked by HindIII and NotI restriction sites on the 5’ and 3’ ends, respectively. The construct was cloned into a pTriEX-HisMyc4 backbone (Novagen) at NcoI/EcoRI sites. RhoGTPases were cloned by polymerase chain reaction (PCR) using primer sets 5’-CCAAGGAATTCATGGCTGCCATCCGGAAG-3’ and 5’-GGTGATGGTGGTGCTCGAGTTATCACAAGAC-3’ for RhoA, 5’-CAAGGAATTCATGCAGGCCATCAAGTGTGTGG-3’ and 5’-GGTGATGGTGGTGCTCGAGTTATTACAACAGC-3’ for Rac1, and 5’-CCCCCGGAATTCATGCAGACAATTAAGTGTG-3’ and 5’-GGTGATGGTGGTGCTCGAGTTATCATAGCAG-3’ for Cdc42. Primers encoded an EcoRI site at the 5’ end of the fragment and an XhoI site after the stop codon on the 3’ end. They were inserted in frame at the C-terminus of the Antenna module in the pTriEx-HisMyc4 backbone. The Quikchange mutagenesis kit (Stratagene) was used to introduce point mutations into the fragment encoding the GTPase, producing different mutant versions of the biosensor. To produce the tetracycline-inducible retroviral construct, the cDNA encoding the GDI biosensor was cut out as a single cassette using NcoI/XhoI sites, blunt-ended using an end-filling reaction using DNA Polymerase I Large (Klenow) fragment (New England Biologicals) and blunt-end ligated into the multiple cloning site at HpaI in the pBabe-sin-Puro-tet-CMV backbone 36. The GP2-293 cell line (Clontech) was used to produce the pantropic retrovirus by cotransfecting the pVSVg vector (Clontech). Infected mouse embryonic fibroblasts which stably expressed the tetracycline transactivating element (tet-OFF MEF/3T3; Clontech) were cultured in DMEM supplemented with 10% fetal bovine serum. They were selected for stable incorporation of the biosensor by puromycin treatment (10μg/ml), and FACS sorted to collect cell fractions expressing different levels of the biosensor. Doxycycline (10μg/ml) was maintained in the culture medium to repress the expression of biosensor during normal cell culture. 48hrs prior to imaging experiments, doxycycline was removed by detaching cells by brief trypsinization and replating into fresh culture medium at 10–4 cells per 10cm TC dish.
Western blot, reagents and antibodies
Whole cell lysates were prepared by lysing cells in ice-cold NP-40 lysis buffer (NaCl, 150 mM, NP-40, 1.0%, Tris-Cl, 50 mM, pH 8.0) with protease inhibitor cocktail (Sigma). Lysates were resolved by SDS-PAGE and transferred to PVDF membrane. After blocking in 5% BSA in TBST, blots were incubated overnight at 4°C in primary antibody at 1:1000 dilution, followed by secondary antibody at 1:10000 dilution. Blots were analyzed using the Odyssey Infrared Imaging System (LI-COR). For quantification, the integrated values of background-corrected band densities were measured with Metamorph software (Molecular Devices). The following primary antibodies were used for the Western analysis and immunofluorescence (primary antibodies at 1:1000 dilution): mouse monoclonal anti-RhoA (Santacruz biotechnology), clone 26C4; mouse monoclonal anti-Rac1 (Millipore), clone 23A8; rabbit polyclonal anti-Cdc42 (Millipore); rabbit polyclonal anti-GDI-α (Santacruz biotechnology) A-20; mouse monoclonal anti-β-actin (Santa Cruz biotechnology), clone AC-15; mouse monoclonal anti-HA (Covance), clone 16B12; mouse monoclonal anti-myc (Cell Signalling), clone 9B11; rabbit polyclonal anti Src pY418 (Invitrogen); mouse monoclonal anti-Src (Millipore), clone GD11; and mouse monoclonal anti-GFP (Roche) clone 7.1 and 13.1. Secondary antibodies (used at 1:10000 dilution): goat anti- mouse IRDye800CW and goat anti-rabbit IRDye800CW (Li-Cor); goat anti-mouse Alexa-594 (Molecular Probes); and donkey anti-rabbit IRDye680LT (Li-Cor). Mammalian expression cDNA constructs for myc-WASP and HA-Tiam1 were gifts from Dr. Dianne Cox. Myc-Intersectin1L was a gift from Dr. Henry Bourne. Myc-mDia2 was from Dr. Shuh Narumiya. PAK1 was from Dr. Yi Wu.
In vitro fluorometry assay
Fluorometry assays were performed following published protocols 5. Briefly, in experiments in which the biosensor was co-transfected with negative regulators (GDI or GAP), the ratio of biosensor to regulator cDNA was 1/4. In experiments using GEFs with excess GDI, the DNA ratio for biosensor/RhoGDI/GEF was 1/4/1–5. In experiments with GEFs and GAP without excess GDI, the ratio of biosensor to regulator cDNA was 1/4. In experiments with effectors, the biosensor/effector DNA ratio was 1/4. The total amount of transfected DNA was kept to 4–5μg per well in a 6-well plate, using a total fluid volume of 2 ml per well. HEK 293T (ATCC) cells were plated in 6 well plates overnight at 1.25×10–6 cells per well. DNA was transfected using the Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's protocols. 48hrs post-transfection, cells were trypsinized, detached, and suspended in cold PBS. The cell suspension was immediately measured in a Fluorolog SPEX MF2 spectrofluorometer (Horiba Jobin Yvon). Emission was measured from 450~600 nm, with 433 nm excitation. Emission counts at 525 nm were then divided by the emission counts at 470 nm to normalize for biosensor concentration.
shRNA-knockdown/rescue
The shRNA for GDI was as designed previously 15. cDNA encoding the shRNA (and a scrambled control) 5’-TGCGCGCTATGTAGGATTCGTTCAAGAGACGAATCCTACATAGCGCGCTTTTTTC-3’) was cloned into pLL5.5CMV 37 at HpaI/XhoI sites. Lentivirus incorporating the shRNA expression construct was produced using the GP2-293 packaging cell line together with pVSVg envelope system following the manufacturer's protocols (Clontech). In order to produce an efficient knockdown and rescue using the shRNA resistant versions of the wildtype and mutant GDI 15, a retroviral expression system was used to stably incorporate the Y156E and Y156F mutant versions of the GDI using the pQXIH system (Clontech) and selected with 100μg/ml hygromycin B. In the endogenous wildtype GDI background, stable expressions of these mutant versions of GDI did not appear to affect cell health. For the production of wildtype GDI KD/rescue, we utilized a pRetro-X-Hygro backbone (Clontech) to suppress the expression of the wildtype GDI using the tet-OFF system. MEF stably incorporating the GDI biosensor were plated onto tissue culture dishes plated with poly-L-lysine and allowed to induce the expression of the biosensor for 48hrs through the removal of doxycycline. Lentivirus containing the shRNA for GDI was applied and, for most effective KD efficiency, the cells were allowed to recover for 96hrs prior to imaging experiments.
Microscopy and live-cell imaging
To quantify two biosensor activities in the same cell, we combined the GC-FLARE biosensor, based on CFP-YFP FRET, with a modification of the MeroCBD Cdc42 biosensor published earlier 4,6. The wavelengths of MeroCBD were altered to produce fluorescence orthogonal to that of G/C FLARE by using a version of the far-red dye, Atto-700-NHS (Sigma) with improved photostability. We also removed the EGFP from MeroCBD because of spectral overlap between EGFP and the CFP-YFP FRET pair in G/C FLARE. Removal of the GFP greatly reduced the water solubility of the dye-labeled CBD domain, so maltose binding protein (MBP) was attached to the C-terminus of CBD using a GSGSGS linker. The N-terminal amine of CBD-MBP was labelled site-specifically with Atto-700-NHS succinimidyl ester at pH 5.6. Modification of the original MeroCBD biosensor was prompted by two factors: 1) photobleacing of the original Alexa750 (Invitrogen) dye was very fast and 2) our multi-channel imaging system 38 used a laser-based autofocus system that precluded the use of wavelengths above 750nm.
MEF/3T3 cells induced to express the GDI biosensor by removal of tetracycline were plated directly on glass coverslips for five to six hours prior to microinjection and imaging. Coverslips were cleaned by ultrasonication followed by soaking in 70% ethanol prior to use in these studies. Modified MeroCBD was microinjected and cells were allowed to recover for 30 minutes before imaging. Cells were imaged in Ham's F-12K medium without phenol red (Biosource) sparged with Argon to remove dissolved oxygen, and supplemented with 3% fetal bovine serum, 10mM HEPES, 10 μg/ml Oxyfluor reagent (Oxyrase Inc.) and 10mM dl-lactate. Cells were imaged at 37°C in a closed chamber. Images were obtained using a 40× 1.3 N/A PLANAPO UIS2 DIC objective (Olympus) and an IX81 ZDC inverted microscope (Olympus) with a beam splitter for simultaneous acquisitions of both FRET and CFP channels using two CoolsnapESII CCD cameras (Roper Photometrics). For the acquisitions of mero87 and Atto-700 fluorescence, a CoolsnapHQ2 (Roper Photometrics) camera was attached to the bottom port of the microscope via a filterwheel that changed the emission filters between mero87 and Atto700 emission wavelengths during acquisition at each time point. Metamorph software version 7.1.7 was used to control the microscope and acquire image sets at 10s time intervals The imaging system has been described in detail 38.
Image processing and analysis
Metamorph ver 7.1.7 was used to perform image processing and data analysis. All images were flat-field corrected and background subtracted 39. Both CFP/FRET and mero87/Atto700 images were intensity thresholded to generate a binary mask with a value of zero outside the cell and a value of one inside the cell. Shading-corrected and background-subtracted images were multiplied by the corresponding binary masks to set areas outside the cell uniformly to zero, to minimize noise and other artefacts. These images were then aligned computationally, using previously described procedures 40,41 to ascertain optimal registration with subpixel accuracy. Masked FRET or mero87 images were divided by the masked CFP or Atto700 images respectively, to yield a ratio reflecting GDI-GTPase interaction or Cdc42 activation. A linear pseudocolor lookup table was applied to the ratiometric images. In every data set, raw fluorescent images were carefully inspected to verify that all portions used to create the ratio image had sufficient signal/noise ratio. This was especially important in thin parts of the cell where fluorescence was low. In time-lapse experiments, various fluorophores bleached at different rates. The ratio was corrected for bleaching using a previously published method 42.
Cell edge tracking
Cell edges were derived from the binary mask mentioned above. The changing position of edges over time was tracked computationally using previously described software written in Matlab (Mathworks) 43. Rates of protrusion and retraction (velocity maps) were calculated using the finite differences of positions in consecutive frame triplets at T–1, T, and T+1.
Definition of sampling windows
Due to the subcellular heterogeneity of protrusion and retraction states, relationships between GDI–Cdc42 binding, Cdc42 activation, and cell edge motion had to be identified locally. As in previous work 4 we determined small probing windows of 1–2 microns width and 0.9 micron depths that moved with the edge (see Supplementary Fig. 6). The width of the windows was set in agreement with the FWHM (full width at half maximum) of ~4 microns of the average spatial autocorrelation of edge velocity fluctuations along the cell edge; i.e. along the cell edge signaling and motion activity are sampled at Nyquist or better. The depth of the windows was set based on our previous studies revealing changes in Cdc42 signaling away from the cell edge over distances as short as 1 micron 4,6. Windows were propagated over time such that they maintained a fixed geometric relationship with a particular location at the cell edge. For windows at the cell edge protrusion/retraction vectors were averaged to report a mean velocity over the edge segment covered by a particular window. For all windows the values of the normalized FRET values were averaged over all pixels contained by a particular window. This allowed the construction of space-time activity maps that were cell-shape invariant and amenable to statistical analysis of the temporal relations between the two signaling activities and edge motion.
Correlation analysis
Temporal cross-correlations between two variables sampled in a particular window were determined by Pearson's correlation coefficient. For windows located away from the cell edge correlations between signaling activity and edge motion were calculated by extracting the time series of the former with the time series of the edge motion measured in the closest window located at the cell periphery. The correlation between two variables at a particular distance from the edge was obtained by averaging the correlation function over all sampling windows at the set distance.
To investigate the cell-to-cell heterogeneity of the cross-correlation a common cross-correlation was estimated by computing a smoothing spline for the ensemble of average correlation data from different cells. The variance of the smoothing spline approximation and hence of the location of the maximum correlation was calculated by a non-parametric bootstrap method 44,45: From the residuals of the approximated spline 2000 bootstrap samples were taken to calculate local variation of the spline about the mean, variation in maximum correlation coefficient, and variation in location of the maximum correlation (time lag). The 95% confidence interval for the estimated common cross-correlation was obtained in each location as the interval containing 95% of the bootstrapped spline samples.
Representing the biosensor using crystal structure data
The hypothetical structure of the GDI.Cdc42 FLARE biosensor in Supplementary Figure 4 was constructed using the crystal structures of Cdc42 in complex with GAP(pdb id: 2NGR), GEF(pdb id: 1KZ7), and GDI(pdb id: 1DOA). PyMOL software v1.7.4.4 (Schrödinger LLC) was used to visualize these structures and to edit the relative locations of FPs. We assumed cp49Venus and Cerulean would form a dimer similar to GFP wild type dimer, which was used as a template (pdb id: 2B3Q) to show the dimer of cp49Venus and Cerulean in the high-FRET state.
Statistical analysis
All p-values were determined using a Student t-test. No statistical methods were used to pre-determine the sample size, no vertebrate animals were involved. No randomizations were used. The investigators were not blinded to allocation during experiments and outcome assessment. Statistical tests used are stated on every figure legend with p-values as appropriate. Data distribution should meet the normal distribution requirements. No estimate of variation. No pre-established criteria were used to determine data inclusion/exclusion.
Supplementary Material
Acknowledgements
We thank the National Institute of General Medical Sciences (P01-GM103723 and T-R01 GM090317 to GD and KH; GM099837 to CD) and the National Cancer Institute (CA181838 to LH) for funding. OD is a Howard Hughes Medical Institute International Student Research Fellow.
Footnotes
Author contributions
L.H., ‘conceived the biosensor’, ‘optimized and built the biosensor’, ‘designed and interpreted correlation experiments’, ‘performed biological experiments’, ‘performed the computational analysis’, and ‘wrote the manuscript with input from all authors’; D.S., ‘designed and interpreted correlation experiments’ and ‘performed biological experiments’; M.S-G., ‘performed the computational analysis’; O.D., ‘performed structural analysis and interpreted studies to examine the antennae mechanism’; C.D., ‘subcloned the shRNA expression constructs and gave critical feedback’; G.D., ‘designed and interpreted correlation experiments’ and ‘wrote the manuscript with input from all authors’; K.H., ‘conceived the biosensor’, ‘performed structural analysis and interpreted studies to examine the antennae mechanism’, and ‘wrote the manuscript with input from all authors’.
Code availability
Matlab codes are available upon request.
Authors have no competing financial interests.
References for main text
- 1.Jaffe AB, Hall A. Rho GT Pases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Mata R, Boulter E, Burridge K. The 'invisible hand': regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12:493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawson CD, Burridge K. The on-off relationship of Rho and Rac during integrin-mediated adhesion and cell migration. Small GTPases. 2014;5:e27958. doi: 10.4161/sgtp.27958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–72. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 6.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–9. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 7.Tsukada Y, et al. Quantification of local morphodynamics and local GTPase activity by edge evolution tracking. PLoS Comput Biol. 2008;4:e1000223. doi: 10.1371/journal.pcbi.1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurokawa K, Matsuda M. Localized RhoA activation as a requirement for the induction of membrane ruffling. Mol Biol Cell. 2005;16:4294–303. doi: 10.1091/mbc.E04-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh RE, et al. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–91. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 11.Chuang TH, Bohl BP, Bokoch GM. Biologically active lipids are regulators of Rac.GDI complexation. J Biol Chem. 1993;268:26206–11. [PubMed] [Google Scholar]
- 12.Del Pozo MA, et al. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat Cell Biol. 2002;4:232–9. doi: 10.1038/ncb759. [DOI] [PubMed] [Google Scholar]
- 13.Ugolev Y, Berdichevsky Y, Weinbaum C, Pick E. Dissociation of Rac1(GDP).RhoGDI complexes by the cooperative action of anionic liposomes containing phosphatidylinositol 3,4,5-trisphosphate, Rac guanine nucleotide exchange factor, and GTP. J Biol Chem. 2008;283:22257–71. doi: 10.1074/jbc.M800734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michaelson D, et al. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J Cell Biol. 2001;152:111–26. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DerMardirossian C, Rocklin G, Seo JY, Bokoch GM. Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling. Mol Biol Cell. 2006;17:4760–8. doi: 10.1091/mbc.E06-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dovas A, et al. Serine 34 phosphorylation of rho guanine dissociation inhibitor (RhoGDIalpha) links signaling from conventional protein kinase C to RhoGTPase in cell adhesion. J Biol Chem. 2010;285:23296–308. doi: 10.1074/jbc.M109.098129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DerMardirossian C, Schnelzer A, Bokoch GM. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Molecular Cell. 2004;15:117–27. doi: 10.1016/j.molcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Lang P, et al. Protein kinase A phosphorylation of RhoA mediates the morphological and functional effects of cyclic AMP in cytotoxic lymphocytes. EMBO J. 1996;15:510–9. [PMC free article] [PubMed] [Google Scholar]
- 19.Tkachenko E, et al. Protein kinase A governs a RhoA-RhoGDI protrusion-retraction pacemaker in migrating cells. Nat Cell Biol. 2011;13:660–7. doi: 10.1038/ncb2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellerbroek SM, Wennerberg K, Burridge K. Serine phosphorylation negatively regulates RhoA in vivo. J Biol Chem. 2003;278:19023–31. doi: 10.1074/jbc.M213066200. [DOI] [PubMed] [Google Scholar]
- 21.Forget MA, Desrosiers RR, Gingras D, Beliveau R. Phosphorylation states of Cdc42 and RhoA regulate their interactions with Rho GDP dissociation inhibitor and their extraction from biological membranes. Biochem J. 2002;361:243–54. doi: 10.1042/0264-6021:3610243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizzo MA, Piston DW. High-contrast imaging of fluorescent protein FRET by fluorescence polarization microscopy. Biophys J. 2005;88:L14–6. doi: 10.1529/biophysj.104.055442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A. 2004;101:10554–9. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F, Moss LG, Phillips GN., Jr. The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–51. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 25.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–63. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 26.White MA, et al. Multiple Ras Functions Can Contribute to Mammalian Cell Transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 27.Ellis S, Mellor H. The novel Rho-family GTPase rif regulates coordinated actin-based membrane rearrangements. Curr Biol. 2000;10:1387–90. doi: 10.1016/s0960-9822(00)00777-6. [DOI] [PubMed] [Google Scholar]
- 28.Takai Y, et al. Rho small G protein and cytoskeletal control. Princess Takamatsu Symp. 1994;24:338–50. [PubMed] [Google Scholar]
- 29.Toutchkine A, Nguyen DV, Hahn KM. Merocyanine dyes with improved photostability. Org Lett. 2007;9:2775–7. doi: 10.1021/ol070780h. [DOI] [PubMed] [Google Scholar]
- 30.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc.Natl.Acad.Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boulter E, et al. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat Cell Biol. 2010;12:477–83. doi: 10.1038/ncb2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–5. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 33.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aghazadeh B, Lowry WE, Huang XY, Rosen MK. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell. 2000;102:625–33. doi: 10.1016/s0092-8674(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 35.Qiao J, et al. Phosphorylation of GTP dissociation inhibitor by PKA negatively regulates RhoA. Am J Physiol Cell Physiol. 2008;295:C1161–8. doi: 10.1152/ajpcell.00139.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–96. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–29. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spiering D, Hodgson L. Multiplex Imaging of Rho Family GTPase Activities in Living Cells. Methods in Molecular Biology. 2012;827:215–34. doi: 10.1007/978-1-61779-442-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodgson L, Shen F, Hahn K. Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr Protoc Cell Biol. 2010 doi: 10.1002/0471143030.cb1411s46. Chapter 14, Unit 14 11 1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiering D, Bravo-Cordero JJ, Moshfegh Y, Miskolci V, Hodgson L. Quantitative Ratiometric Imaging of FRET-Biosensors in Living Cells. Methods Cell Biol. 2013;114:593–609. doi: 10.1016/B978-0-12-407761-4.00025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen F, et al. Functional proteometrics for cell migration. Cytometry A. 2006;69:563–72. doi: 10.1002/cyto.a.20283. [DOI] [PubMed] [Google Scholar]
- 42.Hodgson L, Nalbant P, Shen F, Hahn K. Imaging and photobleach correction of Mero-CBD, sensor of endogenous Cdc42 activation. Methods Enzymol. 2006;406:140–56. doi: 10.1016/S0076-6879(06)06012-5. [DOI] [PubMed] [Google Scholar]
- 43.Machacek M, Danuser G. Morphodynamic Profiling of Protrusion Phenotypes. Biophys. J. 2006;90:1439–1452. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Efron B, Tibshirani R. An Introduction to the bootstrap. Vol. 436. Chapman & Hall; New York: 1993. [Google Scholar]
- 45.Efron B. Bootstrap Methods: Another Look at the Jackknife. Annals of Statistics. 1979;7:1–26. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.