Summary
The rice pathogens Xanthomonas oryzae pathovar (pv.) oryzae and pv. oryzicola produce numerous transcription activator-like (TAL) effectors that increase bacterial virulence by activating expression of host susceptibility genes. Rice resistance mechanisms against TAL effectors include polymorphisms that prevent effector binding to susceptibility gene promoters, or that allow effector activation of resistance genes. This study identifies, in the heirloom variety Carolina Gold Select, a third mechanism of rice resistance involving TAL effectors. This resistance manifests through strong suppression of disease development in response to diverse TAL effectors from both X. oryzae pathovars. The resistance can be triggered by an effector with only 3.5 central repeats, is independent of the composition of the repeat variable diresidues that determine TAL effector binding specificity, and is independent of the transcriptional activation domain. We determined that the resistance is conferred by a single dominant locus, designated Xo1, that maps to a 1.09 Mbp fragment on chromosome 4. The Xo1 interval also confers complete resistance to the strains in the African clade of X. oryzae pv. oryzicola, representing the first dominant resistance locus against bacterial leaf streak in rice. The strong phenotypic similarity between the TAL effector triggered resistance conferred by Xo1 and that conferred by the tomato resistance gene Bs4 suggests that monocots and dicots share an ancient or convergently evolved mechanism to recognize analogous TAL effector epitopes.
Keywords: TAL effector, Carolina Gold Select, Xanthomonas oryzae, Bs4, Oryza sativa, bacterial leaf blight, bacterial leaf streak
Introduction
The bacterial species Xanthomonas oryzae comprises pathovar (pv.) oryzae, which causes bacterial blight (BB) of rice (Oryza sativa L.), and pv. oryzicola, which causes bacterial leaf streak of rice (BLS). Both diseases are globally distributed and cause significant yield losses. Numerous resistance (R) genes have been identified that confer complete resistance to BB; however, only one recessive, race-specific resistance locus to BLS, tentatively named bls1, has been mapped (He et al. 2012). BLS is an important crop threat in Asia and Africa (Verdier et al. 2012b), and sources of resistance are highly sought.
The genomes of African and Asian strains of X. oryzae contain between eight and 26 genes encoding transcription activator-like (TAL) effectors (Bogdanove et al. 2011, White et al. 2009). TAL effectors bind host DNA to activate gene transcription, and DNA binding specificity is mediated by a central repeat region (CRR) made up of a variable number of repeats, each 34 amino acids in length. Some repeats from the X. gardneri effector AvrHah1 have 35 amino acids. The sequence of each repeat is nearly identical to the others except in the repeat variable residue (RVD), the pair of hypervariable residues at positions 12 and 13. Each RVD specifies a corresponding nucleotide in the host promoter, such that the RVD composition determines which nucleotide sequences are targeted in the host (Boch et al. 2009, Moscou and Bogdanove 2009). A C-terminal activation domain (AD) activates host gene transcription.
Several TAL effectors play a role in X. oryzae pv. oryzae virulence (Boch and Bonas 2010). Among these, TAL effectors that activate transcription of a group of sugar transporter genes called SWEET genes are particularly effective as virulence factors, hypothetically by increasing the extracellular concentration of carbohydrates to favor bacterial growth (Chen et al. 2012). A TAL effector can also act as an avirulence factor in rice varieties where a corresponding binding site lies upstream of a dominant resistance (R) gene. The rice resistance genes Xa10, Xa23, and Xa27 are expressed after activation by corresponding specific TAL effectors, resulting in hypersensitive response (HR) and cell death (Gu et al. 2005, Hopkins et al. 1992, Wang et al. 2015). A second mechanism of rice resistance to TAL effectors is genetically recessive and results from escape of effector-induced susceptibility, occurring if both copies of a host promoter have polymorphisms that prevent TAL effector binding. For example, a mutation in the TAL effector binding site upstream of the SWEET gene OsSWEET11 prevents the increase in susceptibility mediated by the corresponding TAL effector, PthXo1 (Yang et al. 2006).
In addition to the two mechanisms of TAL effector-targeted resistance characterized in rice, a third mechanism, conferred by the nucleotide-binding leucine-rich repeat (NLR) resistance protein Bs4 in tomato, is independent of direct gene activation by the effector. Unlike Xa10 and Xa27, Bs4 recognizes even a severely truncated version of its corresponding effector, AvrBs4, containing the N-terminal signal sequence and only three repeats of the CRR, without the AD and nuclear localization signals (Schornack et al. 2004). Bs4 also exhibits broad specificity, triggering defense in response to the effectors Hax3 and Hax4 (Kay et al. 2005), in addition to the effector AvrBs3 expressed to high levels in planta (Schornack et al. 2005). Thus, it was hypothesized that Bs4 recognizes a general feature of the TAL effector CRR in the cytoplasm. The mechanism of TAL effector targeted resistance exemplfied by Bs4 is so far rare: to date, no other R protein has been found that recognizes inactive TAL effectors.
Although TAL effectors are central to the virulence of X. oryzae pathovars, strains of X. oryzae isolated from rice in the United States have none. The US X. oryzae strains form a clade phylogenetically distinct from X. oryzae pv. oryzae and X. oryzae pv. oryzicola, and cause weak BB-like symptoms (Jones et al. 1989, Triplett et al. 2011). Heterologous expression of selected TAL effectors in US X. oryzae strain X11-5A caused a substantial increase in virulence on 14 of 21 diverse rice varieties, supporting the hypothesis that the lack of TAL effectors underlies the weak pathogenicity of the US strains (Verdier et al. 2012a). Given the strong fitness advantage conferred by TAL effectors and the widespread distribution of X. oryzae in the rice-producing world, it is intriguing that TAL effector-producing X. oryzae strains have never been identified in the US. It is possible that populations of US X. oryzae have been geographically isolated from populations of TAL effector-harboring bacteria, preventing horizontal transfer. Alternately, TAL effectors might have conferred a virulence disadvantage on rice genotypes originally introduced to the US, restricting their acquisition or causing their loss.
In this paper, we describe the identification and mapping of a resistance gene in Carolina Gold Select, a genetically purified line of the heirloom rice variety Carolina Gold, that might explain the absence of TAL effectors in US strains of X. oryzae. Carolina Gold is thought to be one of the earliest varieties of rice cultivated in colonial America, grown from the early 18th century (Schulze 2012). Valued for disease resistance and grain cooking quality traits, it was the dominant US variety for well over a century, playing a significant role in the early American economy and cuisine (Schulze 2012). Carolina Gold is the progenitor of several modern rice cultivars, including Dawn, Lemont and Gulfmont, as well as the aromatic hybrid Charleston Gold (Durham and Avant 2011). We found that diverse X. oryzae TAL effectors, including variants lacking an activation domain or lacking all but 3.5 repeats, triggered resistance on Carolina Gold Select rice when expressed and delivered heterologously by X. oryzae X11-5A. The Bs4-like resistance is conferred by a single dominant locus that maps to a 1.09 Mb interval on chromosome 4 and co-segregates with complete resistance to isolates of X. oryzae pv. oryzicola from Africa, indicating its potential for deployment there to control bacterial leaf streak.
Results
Diverse TAL effectors from X. oryzae pv. oryzae and X. oryzae pv. oryzicola, but not from X. gardneri or X. axonopodis, trigger resistance on Carolina Gold Select rice
Our previous work demonstrated that diverse TAL effectors are expressed and translocated from the US strain X. oryzae X11-5A, and that several TAL effectors augment virulence in this strain in many rice varieties (Verdier et al. 2012a). To determine whether TAL effectors confer a selective disadvantage to strains inoculated to US heirloom rice, Carolina Gold Select was inoculated with X. oryzae X11-5A and X11-5A transformants expressing constructs based on nine different X. oryzae TAL effectors, cloned into the high-copy vectors pKEB31 or pSKX1. Three of these constructs, avrXa7CRR, avrXa10CRR, and pthXo1CRR, consist of the CRR-containing SphI fragment of the effector gene of interest cloned in the context of the tal1C effector gene of X. oryzae pv. oryzicola strain BLS256. The other six constructs express either the full native TAL effector gene or the BamHI fragment, encoding all but 17 C-terminal amino acids of the native TAL effector, cloned into tal1C. These constructs will be heretofore referred to by the names of the TAL effector genes (Table 1). The TAL effectors tested in X11-5A have CRRs with diverse origins and target specificities and include those of pthXo1, avrXa7, and avrXa10 from Asian X. oryzae pv. oryzae, talC and tal5 from African X. oryzae pv. oryzae, and tal1c, tal2a, tal8, and tal2g from X. oryzae pv. oryzicola (Table 1).
Table 1.
Bacterial strains and plasmids used in this study.
| Strains | Relevant characteristicsa | Source |
|---|---|---|
| X. oryzae | ||
| BLS256 | pv. oryzicola from Asia (Philippines) | CFBP7109b |
| PXO99A | pv. oryzae from Asia (Philippines) | (Mew et al. 1992) |
| PXO86 | pv. oryzae from Asia (Philippines) | (Mew et al. 1982) |
| CFBP1947 | pv. oryzae from Africa (Cameroon) | CFBP |
| CFBP1951 | pv. oryzae from Africa (Mali) | CFBP |
| MAI10 | pv. oryzicola from Africa (Mali) | CFBP7331 |
| MAI3 | pv. oryzicola from Africa (Mali) | CFBP7326 |
| BAI5 | pv. oryzicola from Africa (Burkina Faso) | LMG 25979c |
| X11-5A | no pv. designation; wild type; Apr | (Triplett et al. 2011) |
| X11-5A(avrXa7CRR) | X11-5A derivative containing pKEB31-avrXa7CRR | (Verdier et al. 2012a) |
| X11-5A(avrXa10CRR) | X11-5A derivative containing pKEB31-avrXa10CRR | This study |
| X11-5A(pthXo1CRR) | X11-5A derivative containing pKEB31-pthXo1CRR | (Verdier et al. 2012a) |
| X11-5A(talΔCRR) | X11-5A derivative containing pKEB31-talΔCRR | This study |
| X11-5A(pthXo1CRRΔAD) | X11-5A derivative containing pKEB31-pthXo1CRRΔAD | This study |
| X11-5A(tal1c) | X11-5A derivative containing pKEB31-tal1c | (Verdier et al. 2012a) |
| X11-5A(tal2a) | X11-5A derivative containing pKEB31-tal2a | (Verdier et al. 2012a) |
| X11-5A(tal2g) | X11-5A derivative containing pKEB31-tal2g | (Verdier et al. 2012a) |
| X11-5A(tal8) | X11-5A derivative containing pKEB31-tal8 | (Verdier et al. 2012a) |
| X11-5A(talC) | X11-5A derivative containing pSKX1-2-talC | (Verdier et al. 2012a) |
| X11-5A(tal5) | X11-5A derivative containing pSKX1-tal5 | This study |
| X11-5A(avrHah1) | X11-5A derivative containing pKEB31-native avrHah1 | This study |
| X11-5A(avrHah1S) | X11-5A derivative containing pKEB31-avrHah1S | This study |
| X11-5A(avrHah1A) | X11-5A derivative containing pKEB31-avrHah1A | This study |
| X11-5A (pK107) | X11-5A derivative containing pK107 (avrXa7) | This study |
| X11-5A(pK110) | X11-5A derivative containing pK110 (avrXa10) | This study |
| X11-5A (pHM1-AD) | X11-5A derivative containing pHM1-pthXo1ΔAD | This study |
| X11-5A(pHM1) | X11-5A derivative containing pHM1 | This study |
| X. campestris | ||
| Xcv 81-23 | Race 2 strain of pv. vesicatoria | Stall, R.E. |
| Xcv 82-8 | Race 1 strain of pv. vesicatoria | Stall, R.E. |
|
| ||
| Plasmids | ||
| pKEB31 | pDD62 derivative containing Gateway destination vector cassette (Invitrogen) between XbaI and BamHI sites; Tcr | (Cermak et al. 2011) |
| pKEB31-talΔCRR | pKEB31 containing tal1c gene of X. oryzae pv. oryzicola BLS256 without repeat-containing SphI fragment; Tcr | (Verdier et al. 2012a) |
| pKEB31-tal1c (pCS472) | pKEB31 containing tal1c gene of X. oryzae pv. oryzicola BLS256; Tcr | (Verdier et al. 2012a) |
| pKEB31-avrXa10CRR (pCS481) | pKEB-tal1c with repeat-containing SphI fragment replaced by that of avrXa10 gene of X. oryzae pv. oryzae PXO86; Tcr | This study |
| pKEB31-avrXa7CRR (pCS718) | pKEB-tal1c with repeat-containing SphI fragment replaced by that of avrXa7 gene of X. oryzae pv. oryzae PXO86; Tcr | (Verdier et al. 2012a) |
| pKEB31-pthXo1CRR (pMP45) | pKEB-tal1c with repeat-containing SphI fragment replaced by that of pthXo1 gene of X. oryzae pv. oryzae PXO99A ; Tcr | (Verdier et al. 2012a) |
| pKEB31-pthXo1CRRΔAD (pAH338) | pMP45 derivative with a stop codon inserted upstream of the activation domain; Tcr | This study |
| pKEB31-tal2a (pCS695) | pKEB-tal1c with repeat-containing BamHI fragment replaced by that of tal2a gene of X. oryzae pv. oryzicola BLS256; Tcr | (Verdier et al. 2012a) |
| pKEB31-tal2g (pCS587) | pKEB-tal1c with repeat-containing BamHI fragment replaced by that of tal2g gene of X. oryzae pv. oryzicola BLS256; Tcr | (Verdier et al. 2012a) |
| pKEB31-tal8 (pCS696) | pKEB-tal1c with repeat-containing BamHI fragment replaced by that of tal8 gene of X. oryzae pv. oryzicola BLS256; Tcr | (Verdier et al. 2012a) |
| pSKX1-2-talC | pSKX1-2 derivative containing the talC gene of X. oryzae pv. oryzae BAI3; Gmr | (Verdier et al. 2012a) |
| pSKX1-tal5 | pSKX1 derivative containing the tal5 gene of X. oryzae pv. oryzae MAI1; Gmr | (Streubel et al. 2013) |
| pKEB31-avrHah1 | pKEB-tal1c with repeat-containing BamHI fragment replaced by that of avrHah1 from X. gardneri, Tcr | (Cermak et al. 2011) |
| pKEB31-avrHah1A | pKEB31 containing a pthXo1-based gene with avrHah1 RVDs, Tcr | (Cermak et al. 2011) |
| pKEB31-avrHah1S | pKEB-tal1c with repeat-containing SphI fragment replaced by that of avrHah1, Tcr | This study |
| pCS495 | Gateway starter plasmid containing tal1C N- and C-termini for cloning designer TAL effectors, Kmr (see Table S3) | This study |
| pCS503 | Single repeat module plasmid, RVD=HD, Apr (see Table S3) | This study |
| pCS510 | Double repeat module plasmid, RVD=HD-NI, Apr (see Table S3) | This study |
| pCS541 | Triple repeat module plasmid, RVD=HD-NI-NG, Apr (see Table S3) | This study |
| pCS553 | Triple repeat module plasmid, RVD=HD-NG-NG, Apr (see Table S3) | This study |
| pSPC03 | pCS495 containing the PspXI-XhoI fragment of pCS503 (1.5 repeats) | This study |
| pSPC04 | pCS495 containing the PspXI-XhoI fragment of pCS510 (2.5 repeats) | This study |
| pSPC07 | pCS495 containing the PspXI-XhoI fragment of pCS541(3.5 repeats) | This study |
| pSPC09 | pSPC07 with an extra NG repeat added (4.5 repeats) | This study |
| pSPC01 | 0.5 repeat TAL effector gene from pCS495 in pKEB31 | This study |
| pSPC05 | 1.5 repeat TAL effector gene from pSPC03 in pKEB31 | This study |
| pSPC06 | 2.5 repeat TAL effector gene from pSPC04 in pKEB31 | This study |
| pSPC08 | 3.5 repeat TAL effector gene from pSPC07 in pKEB31 | This study |
| pSPC10 | 4.5 repeat TAL effector gene from pSPC09 in pKEB31 | This study |
| pHM1 | Broad host-range cosmid derivative of pRI40, Spr, Smr | (Hopkins et al. 1992) |
| pK107 | avrXa7 gene in pHM1, Spr, Smr | (Hopkins et al. 1992) |
| pK110 | avrXa10 gene in pHM1, Spr, Smr | (Bai et al. 2000) |
| pHM1- pthXo1ΔAD | pHM1 containing the HindIII fragment from pAH338 | This study |
Tcr, tetracycline resistance; Gmr, gentamycin resistance; Apr, ampicillin resistance; Kmr, kanamycin resistance; Spr, spectinomycin resistance; Smr, streptomycin resistance.
CFBP: in the collection of plant-associated bacteria at the Institut National de la Recherche Agronomique (INRA), Angers, France
LMG: in the the Belgium Co-ordinated Collections of Microorganisms, Ghent, Belgium
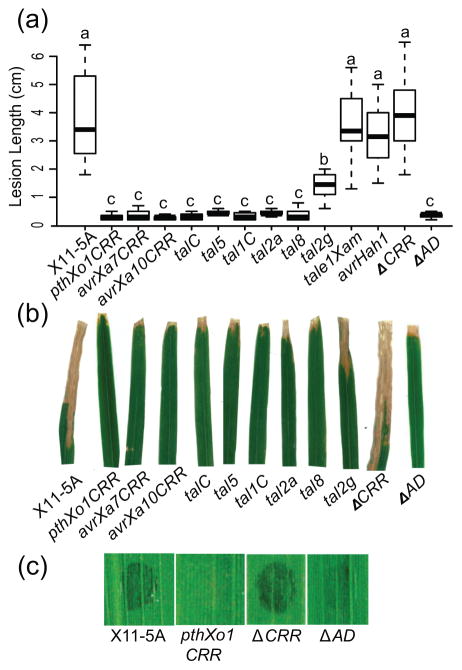
Following clip inoculation, wild-type X. oryzae X11-5A produces short lesions on Carolina Gold Select in conditions of high humidity (>85%, Figure 1A); a test at 70% humidity did not yield lesions. Eight of the nine X. oryzae effector constructs caused a drastic decrease in the length of lesions caused by X11-5A at high humidity; tal2g caused an intermediate decrease (Figure 1A and 1B). However, effector genes avrHah1 from X. gardneri and tale1Xam from X. axonopodis pv. manihotis did not cause a significant change. tal1c missing the central repeat region likewise did not cause a change in lesion length, but a truncation of pthXo1CRR missing the region encoding the C-terminal activation domain still did (Figure 1A and 1B). Thus, the AD is not required for resistance. No dark coloration indicative of a HR was observed in clip inoculation experiments (Figure 1B). In a leaf infiltration assay, pthXo1CRR and pthXo1CRR(ΔAD) inhibited the watersoaking normally caused by X11-5A by 48 h (Figure 1C), although constructs lacking the CRR did not. Two rice varieties related to Carolina Gold, Dawn and Lemont, were also inoculated, but no TAL effectors triggered resistance in these lines (Figure S1). Together, these results indicate that Carolina Gold Select has a mechanism of resistance triggered by X. oryzae TAL effectors independently of their transcription activation activity.
Figure 1.
Response of rice variety Carolina Gold Select to US X. oryzae strain X11-5A and X11-5A transformed with diverse TAL effectors. (a) Lesion lengths on Carolina Gold Select rice 14 d after inoculation. Boxes represent first through third quartiles of at least 12 independent leaves, and bars show the range of observed values. Letters denote significance groups (α = 0.05). Data were combined from two independent experiments that included the same group of treatments. (b) Representative inoculated leaves, taken 21 d after inoculation, show light-colored inoculation sites in resistant lines. Leaf scans were taken separately and combined into one image. (c) Watersoaking responses at the inoculation site 48 h after infiltration with selected strains.
Carolina Gold Select resistance triggered by pthXo1CRR(ΔAD) inhibits bacterial growth
Decreased X. oryzae lesion length is often, but not always, associated with decreased bacterial population growth in the rice leaves (Verdier et al. 2012a). To determine whether the TAL effector-triggered resistance restricts bacterial multiplication, we measured the populations of wild-type X11-5A and X11-5A expressing TAL effector variants on Carolina Gold Select rice, in the topmost 5 cm of clip-inoculated leaves. X11-5A(pthXo1CRR) and X11-5A(pthXo1CRRΔAD) multiplied in the leaf, but reached population sizes significantly smaller than X11-5A or X11-5A(tal1CΔCRR) (Figure 2). The population size of X11-5A(tal1CΔCRR) was slightly smaller than that of X11-5A after 7 days, but not after 14 days. Whether that is due to a plant response to the CRR-less effector, which does not affect lesion development, or to a slight negative effect of expression of the effector on the doubling rate of the bacterium, is not clear. Nevertheless, the data indicate that the TAL effector-triggered resistance in Carolina Gold Select restricts population growth, and that this is AD-independent.
Figure 2.
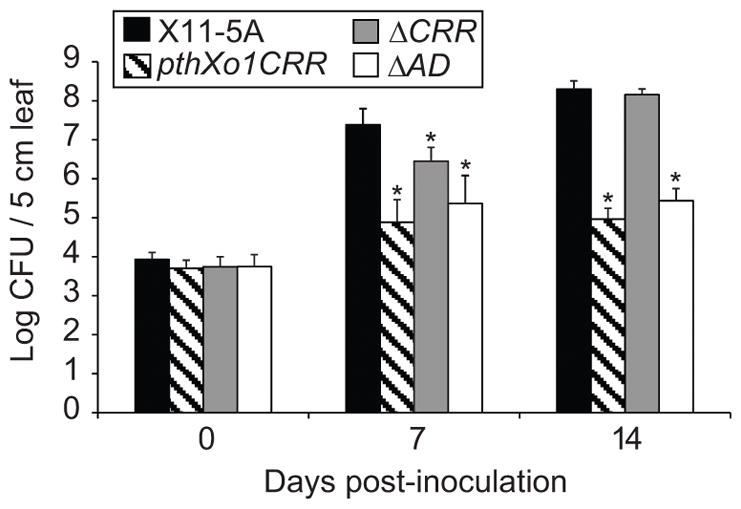
Bacterial population growth in leaves of Carolina Gold Select rice inoculated with US X. oryzae strain X11-5A, X11-5A(pthXo1CRR), X11-5A(tal1CΔCRR), and X11-5A(pthXo1CRRΔAD). Population sizes were measured in the leaf segment up to 5 cm from the inoculation site. Error bars represent standard deviation of four independent leaves, and asterisks denote treatments significantly different from WT X11-5A (p<0.01).
Resistance does not depend on the composition of RVDs
As shown in Figure 1, in contrast to the X. oryzae effectors, avrHah1 from X. gardneri does not trigger resistance in Carolina Gold Select. Because AvrHah1 is more distantly related to Xo TAL effectors than they are to one another, we hypothesized that sequence differences between X. oryzae TAL effectors and AvrHah1 might be critical for determining whether resistance is triggered. The domain swap construct avrHah1S, consisting of the CRR of avrHah1 cloned into the N- and C-terminal context of tal1C, did not trigger resistance when expressed from X11-5A (Figure 3A). X11-5A(avrHah1S) triggered resistance mediated by the AvrHah1 target gene Bs3 in pepper, demonstrating that AvrHah1S is expressed and secreted from X11-5A and is a functional TAL effector in planta (Figure S2). The lack of avrHah1S activity in triggering resistance in Carolina Gold Select suggests that the composition of the TAL effector CRR is critical for this phenotype. To determine whether the RVD or non-RVD portions of the CRR are key, we introduced an engineered avrHah1 “analog” (avrHah1A)(Cermak et al. 2011) into X11-5A. The avrHah1A construct expresses an effector of which the repeat sequences match the consensus for PthXo1, except for the RVD composition, which is that of AvrHah1. X11-5A (avrHah1A) triggered full resistance in Carolina Gold Select (Figure 3A), and was also able to trigger Bs3 in pepper (Figure S2). This confirms that the RVD composition is not relevant to the ability of X. oryzae TAL effectors to trigger resistance in Caroina Gold Select.
Figure 3.
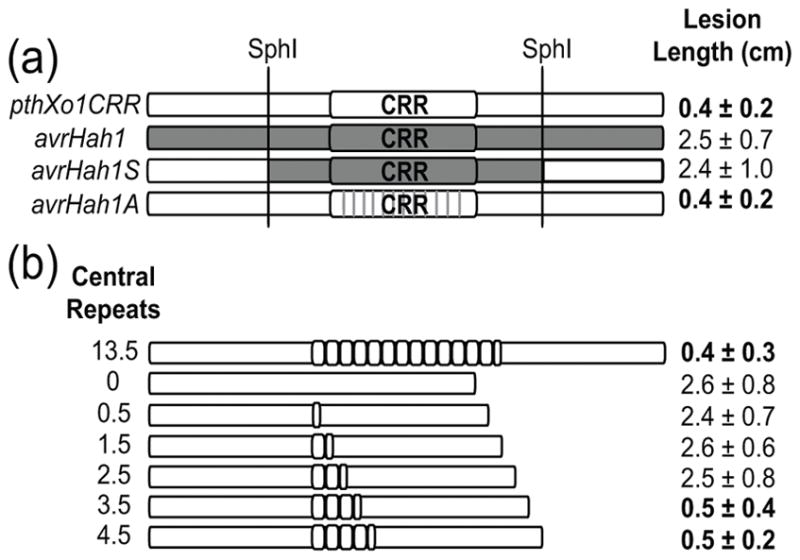
Lesion lengths elicited on Carolina Gold Select by US X. oryzae strain X11-5A carrying domain swap or internal deletion variants of TAL effector genes. (a) Lesion lengths elicited by X11-5A transformed with the X. oryzae effector construct pthXo1CRR, the X. gardneri effector gene avrHah1, the SphI fragment of avrHah1 cloned into pthXo1CRR (avrHah1S), or the RVD codons of avrHah1 in the pthXo1CRR context (avrHah1A). (b) Lesion lengths of X11-5A transformed with pthXo1 (encoding 13.5 central repeats) and internal deletions engineered to encode 0.5-4.5 central repeats. Values are means and standard deviations of values from nine inoculated leaves. Resistant responses are shown in bold.
A TAL effector with only 3.5 repeats is sufficient to trigger resistance in Carolina Gold Select
The above experiments demonstrate that TAL effector activity is not required for triggering resistance in Carolina Gold Select, but suggest that at least part of the CRR is required. To ascertain whether a long CRR is required to trigger resistance, we used a modular gene construction approach (Supporting Methods S1) to design pthXo1CRR-based TAL effectors with small numbers of central repeats. We constructed effector genes with full N- and C-terminal coding regions, but containing only the first 0.5, 1.5, 2.5, 3.5, and 4.5 repeats of the pthXo1 CRR. In X11-5A, the TAL effector constructs encoding 0, 0.5, 1.5 or 2.5 repeats did not cause a reduction in lesion length on Carolina Gold Select, but the effector constructs encoding 3.5 or 4.5 repeats triggered the resistance response (Figure 3B). This demonstrates that a TAL effector with 3.5 repeats is sufficient to trigger resistance on Carolina Gold Select.
Carolina Gold Select is resistant to strains of X. oryzae from Africa, but not from Asia
Because diverse X. oryzae strains express TAL effectors, we hypothesized that Carolina Gold Select might have broad-spectrum resistance to strains of both X. oryzae pathovars. To test this, Carolina Gold Select plants were inoculated with African and Asian strains of X. oryzae pv. oryzicola and X. oryzae pv. oryzae. African X. oryzae pv. oryzicola strains BAI3, MAI5, and MAI10 triggered a strong HR, while the Asian strain BLS256 produced lesions (Figure 4A). The Asian X. oryzae pv. oryzae strains PXO86 and PXO99A caused long lesions indicative of full susceptibility; however, the African strains CFBP1947 and CFBP1951 produced lesions shorter than 5 cm, a limit conventionally used to classify varieties as resistant to a strain (Gonzalez et al. 2007). The two African strains produced long lesions indicative of full susceptibility on another US tropical japonica variety, Lemont (Figure 4B). To determine the breadth of resistance to African X. oryzae strains, Carolina Gold Select was inoculated with 12 strains each of X. oryzae pv. oryzicola and X. oryzae pv. oryzae, respectively, collected from rice fields throughout Mali (Table S1). All X. oryzae pv. oryzicola strains tested triggered the HR. Carolina Gold Select was also resistant to all African X. oryzae pv. oryzae strains, although like CFBP1947 and CFBP1951 these formed short lesions and did not trigger HR (Table S1). Thus, the resistance of Carolina Gold Select triggered by TAL effectors heterologously expressed from X11-5A is not effective against many Asian X. oryzae strains, but may account for the resistance of Carolina Gold Select to a broad range of African X. oryzae isolates.
Figure 4.
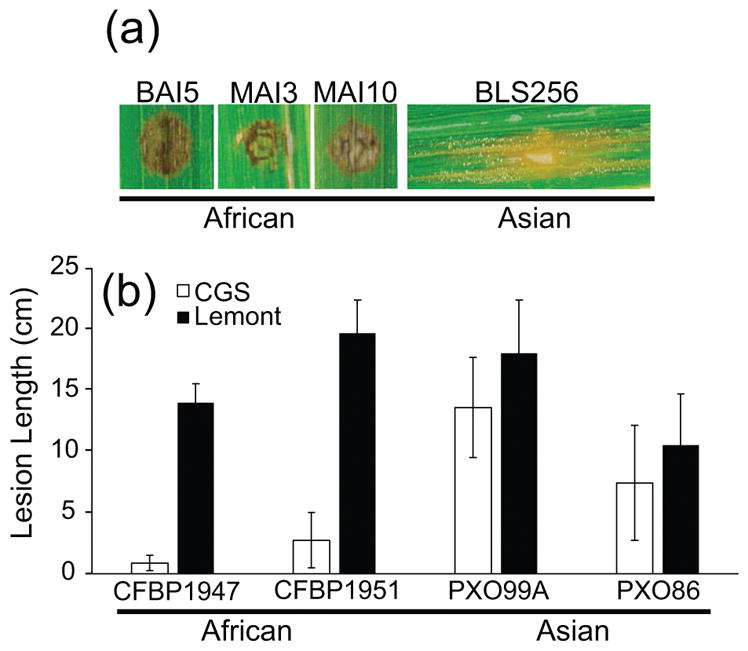
Carolina Gold Select is resistant to African strains of X. oryzae. (a) Carolina Gold Select leaves infiltrated with African X. oryzae pv. oryzicola strains BAI5, MAI3, and MAI10 caused a strong hypersensitive response, while Asian X. oryzae pv. oryzicola strain BLS256 produced disease symptoms. Images were collected at 7 dpi. (b) Carolina Gold Select (CGS) is resistant (i.e., <5 cm lesions) to African X. oryzae pv. oryzae strains CFBP1947 and CFBP1951, but susceptible to Asian strains PXO99A and PXO86. Inoculation on a related rice variety, Lemont, was performed for comparison. Values and error bars represent the means and standard deviations of at least nine biological replicates. Experiments were repeated two additional times with similar results. Refer to Table S1 for lesion lengths of additional African strains.
Because Carolina Gold Select was not resistant to Asian strains, in which the TAL effectors are encoded in the chromosome, we next tested the possibility that the resistance we observed in response to TAL effectors expressed from pKEB31 or pSKX1 in strain X11-5A is due to relative overexpression of the TAL effectors from these plasmids. To do this, we tested effector genes avrXa7 and avrXa10 cloned in the low-copy plasmid pHM1. Expressed from pHM1 in X11-5A, both triggered full resistance on Carolina Gold Select (Figure S3A). Conversely, introduction of pthXo1CRR cloned in the high-copy vector pKEB31 into Asian X. oryzae pv. oryzae strains PXO86 and PXO99A did not result in strong resistance to these strains (Figure S3B). Introduction of pthXo1CRR(ΔAD) on this vector into PXO99A did cause a small reduction in lesion length (Figure S3B), but since this was not observed for PXO86, we conclude that this is likely due to pthXo1CRR(ΔAD) interfering with activation of OsSWEET11 by the native PthXo1 protein of PXO99A by competing at the binding site. In sum, the results demonstrate that the resistance is independent of TAL effector gene copy number and suggest that Asian X. oryzae strains in some way suppress or overcome the TAL effector-triggered resistance of Carolina Gold Select.
Carolina Gold Select resistance to TAL effectors is controlled by a dominant locus that co-segregates with resistance to African isolates of X. oryzae pv. oryzicola
Toward identifying the genetic determinant(s) of the resistance triggered by X. oryzae TAL effectors and of the resistance to African X. oryzae pv. oryzicola, we crossed Carolina Gold Select with Kitaake, a Japonica variety in which susceptibility to X11-5A is enhanced by PthXo1 (Verdier et al. 2012a). A total of 157 F2 individuals from 4 independent hybrid lines were tested for susceptibility to X11-5A, X11-5A(pthXo1CRR), and the African X. oryzae pv. oryzicola strain MAI10 on separate leaves. Due to the limited number of testable leaves per plant, resistance to African X. oryzae pv. oryzae was not assayed in this study. In both the Kitaake and Carolina Gold Select parent lines, X11-5A caused short lesions; X11-5A(pthXo1CRR) caused long lesions in Kitaake and very short lesions in Carolina Gold Select (Figure 5A). The phenotypes of F2 hybrids were similar either to the Kitaake or Carolina Gold Select parent, with the TAL effector-expressing strain eliciting lesions either longer or much shorter than the X11-5A control. About 80% of the F2 individuals were resistant to X11-5A(pthXo1CRR) (Figure 5A). Hybrid plant responses to X. oryzae pv. oryzicola strain MAI10 also segregated into complete resistance (the Carolina Gold Select parent phenotype) and susceptibility (the Kitaake parent phenotype), and these showed 100% co-segregation in 157 lines with the resistant and susceptible responses to X11-5A (pthXo1CRR), respectively. Unlike Carolina Gold Select, F2 hybrids did not develop a clear HR to MAI10. Instead, qualitative resistance in the F2 lines manifested in a complete lack of lesion formation. To determine whether a weak HR was occurring, 14 additional F2 hybrids and four additional Carolina Gold Select individuals were inoculated with MAI10 in a growth chamber under high-intensity light conditions that favor HR development. After 72 hours, dark color development was visible to a variable degree at the inoculation sites of resistant lines, but this phenotype was weak compared to the response of Carolina Gold Select (Supplemental Figure S4). This suggests that additional loci in the Carolina Gold Select genome contribute to development of a strong HR. Together, these results demonstrate that the TAL effector-triggered resistance phenotype in Carolina Gold Select is governed by a single dominant locus, and that this locus is also the source of resistance to the African strain MAI10. Because the TAL-effector triggered resistance activity is not a typical bacterial leaf blight resistance gene, and is triggered by and effective against genes from both X. oryzae pathovars, we name this locus Xo1.
Figure 5.
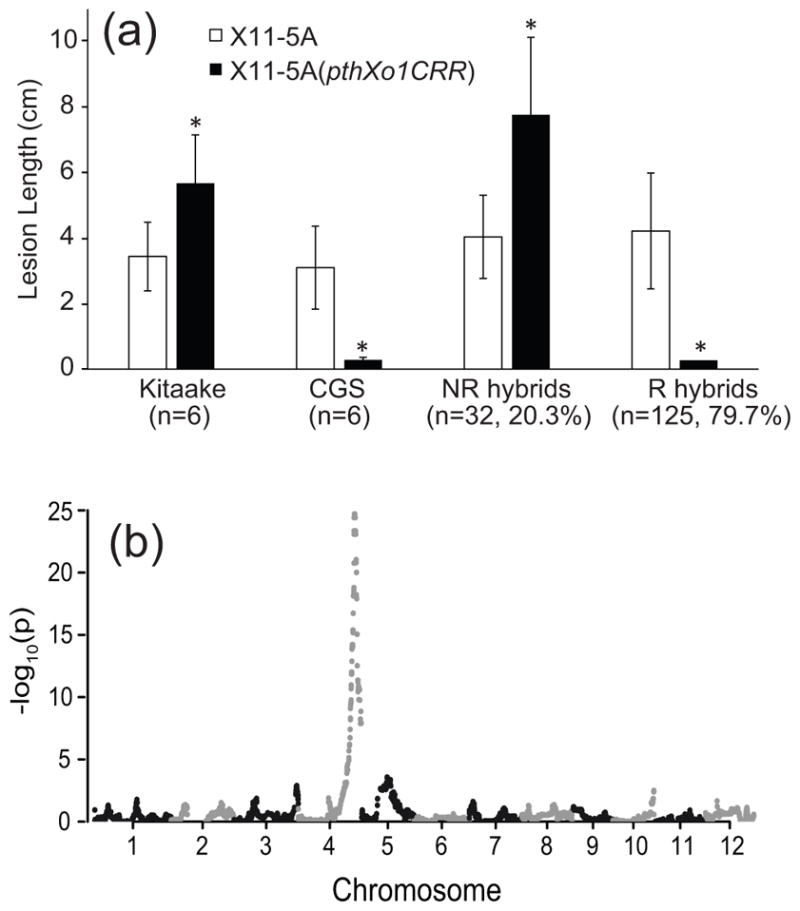
Resistance triggered by TAL effectors and African X. oryzae pv. oryzicola is conferred by a single locus on Chromosome 4. (a) Lesion lengths of X11-5A with and without the TAL effector gene pthXo1CRR inoculated onto hybrid parent lines Kitaake and Carolina Gold Select, 32 F2 hybrid lines lacking resistance to African X. oryzae strain MAI10 (NR hybrids), and 125 lines resistant to MAI10 (R hybrids). All lines resistant to MAI10 were also resistant to X11-5A (pthXo1CRR). Values and error bars represent the means and standard deviations of 2 inoculated leaves per plant, or 2n, measured at 14 d after inoculation. Asterisks represent values significantly different from X11-5A for each group (p<0.001). Resistance to MAI10 was judged by the absence of lesions in all of four inoculation sites on a single leaf at 7 dai. (b) TASSEL plot of SNPs significantly associated with resistance in 110 F2 hybrid individuals, indicating a single causative locus on Chromosome 4.
Xo1 maps to a 1.09 Mbp segment of the long arm of Chromosome 4
We used a genotyping-by-sequencing approach to map Xo1. DNA from 110 phenotyped F2 hybrids and three individuals of each parental line was sequenced to low coverage from RsaI digestion sites. From an initial set of 207,303 non-reference calls, 18,696 high-quality SNPs were recovered after filtering and data imputation. SNPs in a single region of chromosome 4 correlated strongly with the resistance to X11-5A(pthXo1) and MAI10, placing the locus in the interval from 31,358,156 bp to 32,448,509 bp (Figure 5B). This interval contains approximately 275 annotated genes in Nipponbare (Kawahara et al. 2013), including 10 full or partial NLR genes; one of these is bacterial leaf blight resistance gene Xa1 (Loc_Os04g53120). Five other dominant resistance genes for bacterial leaf blight, Xa12, Xa14, Xa2, Xa30(t), and Xa31(t), have been mapped to intervals near the Xo1 locus of chromosome 4, demonstrating that this is an important genomic region for blight resistance (reviewed in (Xia et al. 2012)). Further research is underway to determine whether Xo1 is conferred by an allele of Xa1 or by another NLR gene.
Discussion
Rice production has increased substantially in West Africa in recent years, but with limited yields and increases in consumption, many countries still rely heavily on foreign imports (http://www.fao.org/3/a-i4337e.pdf). Intensified production and the lack of resistant germplasm has facilitated a rapid spread of BLS to multiple new rice-growing areas, affecting up to 80% of the plants in fields surveyed (Afolabi et al. 2015, Afolabi et al. 2014, Wonni et al. 2011). With yield losses from BLS reaching up to 30% (Mew 1993), identification of effective genetic resistance will be a critical step toward increasing rice production in the region. Here, we report that Carolina Gold Select rice has a resistance locus, Xo1, conferring resistance triggered by X. oryzae TAL effectors with different DNA binding specificities and from both pathovars, independent of the ability of the TAL effector to directly activate gene expression. The Xo1 locus confers complete resistance to African strains of X. oryzae pv. oryzicola. TAL effector-related resistance in rice was previously only known to hinge on the ability of a particular TAL effector to activate a gene, deriving either from an activation trap that turns on an R gene or a binding site polymorphism that prevents activation of a susceptibility gene. The broad efficacy of Xo1 against African strains of X. oryzae pv. oryzicola makes it a promising resource for breeding efforts against BLS. Interestingly, strong resistance specific to African strains of X. oryzae pv. oryzicola was recently identified in two African varieties of the cultivar indica (Wonni et al. 2015); further gene characterization will determine whether these varieties share Xo1.
Our findings suggest that Xo1 could encode a receptor that triggers resistance upon direct recognition of the TAL effector, similar to the way Bs4 has been hypothesized to recognize AvrBs4 in tomato (Schornack et al. 2006). Both Xo1 and Bs4 are characterized by activity against closely related TAL effectors with diverse RVD composition, activity against effectors containing only a few central repeats, and independence from the transcriptional activation domain. Together, these similarities suggest that Bs4 and Xo1 could both be mechanisms for plants to recognize epitopes of the highly conserved TAL effector structure.
Bs4 encodes a member of the TIR-NLR family of resistance proteins, which has only been observed in dicots (Tarr and Alexander 2009). Bs4 signals through the EDS1 pathway (Schornack et al. 2004), distinct from the mechanism used by monocot CC-NLR-type resistance proteins (Heidrich et al. 2012). The phenotypic similarities between Bs4 and Xo1 raise the intriguing possibility that rice and tomato independently evolved NLRs that recognize activity-less TAL effectors. Convergent evolution of R proteins has been observed within dicots: Arabidopsis and bean have non-orthologous R proteins that each recognize the Pseudomonas effector AvrB through its activity on RIN4 (Ashfield et al. 2004). In the case of Xo1 and Bs4, it is striking that plant phyla diverging 150 million years ago recognize the same class of pathogen effector, independent of any activity of those effectors on a conserved plant target. Identifying the gene(s) responsible for Xo1 activity will help determine whether Xo1 and Bs4 are mechanistically similar to one another.
Our discovery of Xo1 in rice and its functional similarity to Bs4 in tomato raises the question of whether resistance triggered by RVD-independent recognition of TAL effectors is prevalent in plants, or other organisms. As TAL effectors are rapidly being adapted for diverse biotechnological and therapeutic applications, this question may be increasingly important. Of 24 diverse rice varieties we have tested for TAL effector responses ((Verdier et al. 2012a) and this work), Carolina Gold Select is the only variety demonstrating resistance triggered by multiple TAL effectors and gene activation-deficient TAL effectors. Bs4 activity was detected in 15 of 17 tomato accessions tested, however (Ballvora et al. 2001). Our results with the Asian X. oryzae strains also suggest that bacteria can suppress Xo1 activity, possibly through other TAL or non-TAL effectors, just as Asian X. oryzae pv. oryzicola strains BS303 and BLS256 suppress Xa10 and Xa7-mediated resistance in a Type III-dependent manner (Makino et al. 2006). Widespread suppressive ability would obscure identification of other TAL effector-detection resistance loci through traditional race typing studies. Additionally, effectors could avoid (e.g., AvrHah1) or partially escape (e.g., Tal2g) detection through polymorphisms in the recognized epitope. Understanding the basis of Xo1 suppression by Asian strains of X. oryzae will be important to determine how widespread Xo1-like resistance mechanisms may be, and how broadly they affect TAL effector evolution. Understanding these recognition mechanisms in both tomato and rice could lead to strategies for engineering receptors that recognize a broad range of pathogen TAL effectors.
For Xo1, additional research is first needed to determine whether the response to TAL effectors and the resistance to African X. oryzae pv. oryzicola strains are controlled by one or by different genes at the locus. While Carolina Gold Select exhibits a strong hypersensitive response upon infiltration with African X. oryzae pv. oryzicola, little to no HR phenotype was observed following infiltration with X11-5A transformed with single TAL effectors. However, the broad efficacy of the resistance of Carolina Gold Select against African X. oryzae pv. oryzicola isolates from different sites might be consistent with its resulting from the non-specific TAL effector recognition conferred by Xo1. Determining the genetic basis of the resistance to African X. oryzae pv. oryzicola strains and testing a wider collection strains will help predict whether Xo1 can provide durable resistance that will be useful for rice breeders fighting BLS in Africa.
Experimental Procedures
DNA cloning and bacterial culture techniques
Bacterial strains and plasmids are listed in Table 1. Strains of E. coli were cultured on LB media containing the appropriate antibiotics, and X. oryzae was cultured on PSA (Karganilla et al. 1973). pCS503, pCS510, pCS541, and pCS553 were constructed as part of a comprehensive series of vectors with inserts of 1, 2, or 3 TAL effector repeats in every possible RVD combination (described in Methods S1, Table S2, and Table S3). TAL effector CRR truncation clones pSPC03, pSPC04, and pSPC07 were generated by subcloning the PspXI-XhoI fragments from pCS503, pCS510, and pCS541 into the XhoI site of pCS495. pSPC09 was generated during attempts to clone the PspXI-XhoI fragment from pCS553 into pSPC07 to total 6.5 repeats, but this instead resulted in a construct with 4.5 in-frame repeats with RVDs of NN-HD-NG-NG-NG, possibly due to an unexpected restriction site in pCS553. To generate Xanthomonas expression clones pSPC01, 05, 06, 08, and 10, inserts from pCS495-derived vectors were recombined into the Gateway destination vector pKEB31 (Cermak et al. 2011) using LR Clonase Enzyme Mix II (Life Technologies, lifetechnologies.com) according to the manufacturer’s instructions. pKEB31-avrHah1S was constructed by cloning the SphI fragment from pKEB31-avrHah1 into the SphI site of pCS466 (Verdier et al. 2012a). pAH338 was constructed by PCR site-directed mutagenesis of pCS466 to change the codon encoding residue W1334, immediately upstream of the activation domain, to a stop codon. This was followed by recombination into pKEB31 and insertion of the SphI fragment from pthXo1. TAL effector gene inserts were sequenced at the Colorado State University Proteomics and Metabolomics facility. TAL effector constructs were transformed into X. oryzae X11-5A by electroporation, and confirmed by colony PCR.
Plant inoculations
Experiments were performed on Carolina Gold Select (GRIN accession CSOR301024), a Carolina Gold line purified using genetic markers (Duitama et al. 2015), obtained from Dr. Anna McClung. Rice accessions Lemont (CSOR301093) and Kitaake (GSOR 300108) from the Leach lab seed collection, and Dawn (CIor9534), obtained through the USDA-ARS National Plant Germplasm System, were also used. Lines of pepper (Capsicum annuum) ECW-10R and ECW-30R were obtained through the USDA-ARS U.S. National Plant Germplasm System as part of the Pepper Bacterial Spot Differential Host Set (accessions CPPSIH_1_02 and 1_04).
For inoculations with X. oryzae pv. oryzae or the US X. oryzae strain X11-5A, fully-expanded leaves of 6 week-old plants were inoculated with bacterial suspensions of 108 cfu/mL using a standard scissor-clip inoculation technique (Kauffman et al. 1973). A leaf infiltration technique was used for inoculations with X. oryzae pv. oryzicola (Reimers and Leach 1991). Lesion lengths were measured at 14 d post-inoculation for X. oryzae pv. oryzae and US X. oryzae and at 7 d for X. oryzae pv. oryzicola. Figure 1A represents combined data from two experiments performed in different months; two prior experiments yielded similar results but did not include all of the X. oryzae TAL effector genes. Inoculations depicted in Figures 2, 3, and 5 show results from one representative experiment out of three experiments yielding similar results.
Inoculation of pepper differential lines ECW-10R and ECW-30R was performed by infiltration of leaves on six-week-old plants with fresh cultures of 108 cfu/mL. Leaves were imaged five days after inoculation. Decoloring was performed in a 19:1 ethanol/glycerol solution.
For population count experiments, a 5 cm segment of each leaf terminus was macerated in 2 ml sterile water, followed by serial dilution plating at 7 and 14 days as previously described (Verdier et al. 2012a). All inoculations for lesion length analysis were performed in a greenhouse kept at >85% humidity. Inoculations for bacterial population growth analyses were performed in a growth chamber (28° days, 24° nights, 16 h days, 85% humidity).
Carolina Gold Select (maternal line) was crossed with Kitaake (paternal line) using a standard procedure (Herrera and Coffman 1975). F1 plants were selected as likely crosses based on short (<65 d) flowering time. 38 to 42 F2 individuals were grown from each of 4 independent lines, for a total of 157 plants. For phenotyping of individual F2 segregants, five fully expanded leaves of each 6-week-old plant were inoculated: two were clip-inoculated with X. oryzae X11-5A, two were clip-inoculated with X11-5A (pthXo1CRR), and one leaf was inoculated by infiltration in four spots with X. oryzae pv. oryzicola MAI10. X11-5A lesions were measured at 14 d, and MAI10 presence or absence of lesions was observed at 7 d post-inoculation.
Data analyses and image generation
For the inoculations in Figure 1A, differences among means were determined using one-way ANOVA and Tukey’s HSD test. For all other figures, pairwise differences with the X11-5A wild type treatment were determined using Welch’s T-Test. Statistical analyses were conducted in R version 3.2.1 (http://www.r-project.org/), using the package agricolae for Tukey’s HSD (http://tarwi.lamolina.edu.pe/~fmendiburu/). For all lesion and infiltration images, leaf scans or photographs were taken separately of separate leaves and combined into one figure. Any adjustments in size, brightness, and contrast were then made equally on the combined images.
Genotyping-by-Sequencing
Leaf tissue was collected from both parents and 110 Carolina Gold Select X Kitaake F2 plants that originated from a single F1 plant, and all plants were phenotyped as described above. Genomic DNA was extracted via standard protocols (Dellaporta 1994). Genomic DNA was used to generate GBS libraries as previously described (Heffelfinger et al. 2014) using RsaI as the restriction enzyme. Libraries were pooled, then sequenced on 40% of one lane of an Illumina HiSeq 2500 at the Yale Center for Genome Analysis, yielding a total of 60,437,829 reads across all samples.
Data processing
Raw reads were aligned to the MSU 7.0 rice reference genome (Kawahara et al. 2013) using NovoAlign (www.novocraft.com) under default parameters. Variants were then called from aligned reads using GATK (McKenna et al. 2010). Sites that failed to meet the following criteria were removed: a quality depth of at least 2, at least two non-reference and reference allele calls, a Fisher strand-bias score of at least 60, a fraction of heterozygous calls between 0.1 and 0.9, a haplotype score less than or equal to 10, mapping quality of at least 40, mapping quality rank-sum score of at least −12.5, read position rank sum of at least −8, Phred score of at least 40, and overall heterozygosity (the ratio of reference to non-reference alleles) between 0.1 and 0.9. Following these hard filters, non-independent sites were collapsed into single markers. A site was declared non-independent if it was identified from the same sequencing read as another polymorphic site. Finally, retained variants were required to have a mean r2 correlation of at least 0.05 with the five variants immediately upstream and downstream. These filters were performed using custom scripts.
Following initial filtering, data imputation was performed using an impute-filter-impute process. First, missing markers within the parents were imputed. Next, variants were filtered such that only variants that were homozygous within and polymorphic between the parents were retained. Finally, offspring markers were imputed. Imputation was performed using LB-Impute (Heffelfinger et al., submitted) under default settings.
Trait mapping
The Xo1 locus was mapped from the filtered and imputed data via a generalized linear model using TASSEL 5.2.11 (Bradbury et al. 2007) under default settings. Data were visualized via the qqman package in R (Turner 2014).
Gene name registration
Xo1 was registered and approved as a rice gene name according to the guidelines of the Committee on Gene Symbolization, Nomenclature, and Linkage (CGSNL) of the Rice Genetics Cooperative (McCouch 2008).
Supplementary Material
Response of US tropical japonica varieties Lemont and Dawn to US X. oryzae strain X11-5A and TAL effectors.
Response of near-isogenic Bs1 and Bs3 pepper lines to strain X11-5A and derivatives expressing domain-swap and artificial avrHah1 constructs.
Response of Carolina Gold Select to TAL effectors expressed from high and low-copy vectors.
Responses of Carolina Gold Select X Kitaake F2 hybrid individuals to infiltration with African X. oryzae pv. oryzicola strain MAI10.
Experimental procedures describing construction of TAL effector cloning modules.
Response of Carolina Gold Select to inoculation with with X. oryzae pv. oryzae and X. oryzae pv. oryzicola strains from Mali.
Oligos used for module construction.
Plasmids for Modular Assembly of Designer TAL Effectors.
Acknowledgments
This project is supported by the Agriculture and Food Research Initiative competitive grant no. 2014-67013-21564 to Triplett and Leach from the USDA National Institute of Food and Agriculture. Cohen was supported by a teaching assistantship from Colorado State University. Verdier was supported by a Marie Curie Fellowship (EU Grant PIOF-GA-2009-235457). GBS sequencing and Heffelfinger were supported by an NSF and Bill and Melinda Gates Foundation BREAD grant NSF0965420 to Dellaporta. Schmidt was supported by an NIH grant (R01GM9886101) to Bogdanove. Mariko Alexander, Aaron Hummel, and Emily Delorean provided technical assistance with crosses, cloning, and inoculation studies. DNA samples and GBS libraries were prepared by Yingchun Tong. Computational analyses were performed on the Yale University Biomedical High Performance Computing Cluster, which is supported by National Institutes of Health grants RR19895 and RR029676-01. The “Rice: Research to Production” short course at the International Rice Research Institute, funded by the NSF Developing Country Collaborations in Plant Genome Research (DCC-PGR), helped make this work possible by providing valuable training to Triplett. We thank Dr. Anna McClung for providing Carolina Gold Select seed, and the USDA-ARS National Plant Germplasm System for providing seed of Dawn.
Contributor Information
Stephen P. Cohen, Email: stephen.cohen@colostate.edu.
Christopher Heffelfinger, Email: christopher.heffelfinger@yale.edu.
Clarice L. Schmidt, Email: clarice@iastate.edu.
Cheick Tekete, Email: teketecherif@yahoo.fr.
Valerie Verdier, Email: valerie.verdier@ird.fr.
Adam J. Bogdanove, Email: ajb7@cornell.edu.
Jan E. Leach, Email: jan.leach@colostate.edu.
References
- Afolabi O, Milan B, Amoussa R, Koebnik R, Poulin L, Szurek B, Habarugira G, Bigirimana J, Silue D. First report of Xanthomonas oryzae pv. oryzicola causing bacterial leaf streak of rice in Burundi. Appl Environ Microbiol. 2015;81:688–698. doi: 10.1094/PDIS-05-14-0504-PDN. [DOI] [PubMed] [Google Scholar]
- Afolabi O, Milan B, Poulin L, Ongom J, Szurek B, Koebnik R, Silue D. First report of Xanthomonas oryzae pv. oryzicola causing bacterial leaf streak of rice in Uganda. Plant Dis. 2014;98:1579–1579. doi: 10.1094/PDIS-07-14-0745-PDN. [DOI] [PubMed] [Google Scholar]
- Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. Convergent evolution of disease resistance gene specificity in two flowering plant families. The Plant cell. 2004;16:309–318. doi: 10.1105/tpc.016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Choi SH, Ponciano G, Leung H, Leach JE. Xanthomonas oryzae pv. oryzae avirulence genes contribute differently and specifically to pathogen aggressiveness. Mol Plant Microbe Interact. 2000;13:1322–1329. doi: 10.1094/MPMI.2000.13.12.1322. [DOI] [PubMed] [Google Scholar]
- Ballvora A, Pierre M, Van Den AG, Schornack S, Rossier O, Ganal M, Lahaye T, Bonas U. Genetic mapping and functional analysis of the tomato Bs4 locus governing recognition of the Xanthomonas campestris pv. vesicatoria AvrBs4 protein. Mol Plant Microbe Interact. 2001;14:629–638. doi: 10.1094/MPMI.2001.14.5.629. [DOI] [PubMed] [Google Scholar]
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Koebnik R, Lu H, Furutani A, Angiuoli SV, Patil PB, Van Sluys MA, Ryan RP, Meyer DF, Han SW, Aparna G, Rajaram M, Delcher AL, Phillippy AM, Puiu D, Schatz MC, Shumway M, Sommer DD, Trapnell C, Benahmed F, Dimitrov G, Madupu R, Radune D, Sullivan S, Jha G, Ishihara H, Lee SW, Pandey A, Sharma V, Sriariyanun M, Szurek B, Vera-Cruz CM, Dorman KS, Ronald PC, Verdier V, Dow JM, Sonti RV, Tsuge S, Brendel VP, Rabinowicz PD, Leach JE, White FF, Salzberg SL. Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J Bacteriol. 2011;193:5450–5464. doi: 10.1128/JB.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung M, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2012;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S. The Maize Handbook. Springer; 1994. Plant DNA miniprep and microprep: versions 2.1–2.3; pp. 522–525. [Google Scholar]
- Durham S, Avant S. ARS and CGIAR: Working To Provide International Food Security. Agricultural Research. 2011;59:4. [Google Scholar]
- Gonzalez C, Szurek B, Manceau C, Mathieu T, Sere Y, Verdier V. Molecular and pathotypic characterization of new Xanthomonas oryzae strains from West Africa. Mol Plant Microbe Interact. 2007;20:534–546. doi: 10.1094/MPMI-20-5-0534. [DOI] [PubMed] [Google Scholar]
- Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, Yin Z. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- Heffelfinger C, Fragoso CA, Moreno MA, Overton JD, Mottinger JP, Zhao H, Tohme J, Dellaporta SL. Flexible and scalable genotyping-by-sequencing strategies for population studies. BMC Genomics. 2014;15:979. doi: 10.1186/1471-2164-15-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich K, Blanvillain-Baufumé S, Parker JE. Molecular and spatial constraints on NB-LRR receptor signaling. Curr Opin Plant Biol. 2012;15:385–391. doi: 10.1016/j.pbi.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Herrera R, Coffman R. Emasculation of rice by vacuum extraction. 5. Scientific Meeting of the Crop Science Society of the Philippines; Naga City (Philippines). 16 May 1974; CSSP; 1975. [Google Scholar]
- Hopkins CM, White FF, Choi SH, Guo A, Leach JE. Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact. 1992;5:451–459. doi: 10.1094/mpmi-5-451. [DOI] [PubMed] [Google Scholar]
- Jones RK, Barnes LW, Gonzalez CF, Leach JE, Alvarez AM, Benedict AA. Identification of low virulence strains of Xanthomonas campestris pv. oryzae from rice in the United States. Phytopathology. 1989;79:984–990. [Google Scholar]
- Karganilla A, Paris-Natural M, Ou SH. A comparative study of culture media for Xanthomonas oryzae. Philipp Agric. 1973;57:141–152. [Google Scholar]
- Kauffman H, Reddy A, Hsiek S, Merca S. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S, Boch J, Bonas U. Characterization of AvrBs3-like effectors from a Brassicaceae pathogen reveals virulence and avirulence activities and a protein with a novel repeat architecture. Mol Plant Microbe Interact. 2005;18:838–848. doi: 10.1094/MPMI-18-0838. [DOI] [PubMed] [Google Scholar]
- Makino S, Sugio A, White F, Bogdanove AJ. Inhibition of resistance gene-mediated defense in rice by Xanthomonas oryzae pv. oryzicola. Mol Plant Microbe Interact. 2006;19:240–249. doi: 10.1094/MPMI-19-0240. [DOI] [PubMed] [Google Scholar]
- McCouch SR. Gene Nomenclature System for Rice. Rice. 2008;1:72–84. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mew T, Cruz V, Reyes R. Interaction of Xanthomonas campestris pv. oryzae and a Resistant Rice Cultivar. Phytopathology. 1982;72:786–789. [Google Scholar]
- Mew TW. Xanthomonas oryzae pathovars on rice: Cause of bacterial blight and bacterial leaf streak. In: Swings JG, Civerolo EL, editors. Xanthomonas. London: Chapman & Hall; 1993. pp. 30–40. [Google Scholar]
- Mew TW, Vera C, CM, Medalla ES. Changes in race frequency of Xanthomonas oryzae pv. oryzae in response to rice cultivars planted in the Philippines. Plant Dis. 1992;76:1029. [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Reimers PJ, Leach JE. Race-specific resistance to Xanthomonas oryzae pv. oryzae conferred by bacterial blight resistance gene Xa-10 in rice Oryza sativa involves accumulation of a lignin-like substance in host tissues. Physiol Mol Plant Pathol. 1991;38:39–55. [Google Scholar]
- Schornack S, Ballvora A, Gürlebeck D, Peart J, Ganal M, Baker B, Bonas U, Lahaye T. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 2004;37:46–60. doi: 10.1046/j.1365-313x.2003.01937.x. [DOI] [PubMed] [Google Scholar]
- Schornack S, Meyer A, Römer P, Jordan T, Lahaye T. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. Journal of Plant Physiology. 2006;163:256–272. doi: 10.1016/j.jplph.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Schornack S, Peter K, Bonas U, Lahaye T. Expression Levels of avrBs3-like genes affect recognition specificity in tomato Bs4- but not in pepper Bs3-mediated perception. Mol Plant Microbe Interact. 2005;18:1215–1225. doi: 10.1094/MPMI-18-1215. [DOI] [PubMed] [Google Scholar]
- Schulze R. Carolina Gold Rice: The Ebb and Flow History of a Lowcountry Cash Crop. Charleston, SC: The History Press; 2012. [Google Scholar]
- Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 2013;200:808–819. doi: 10.1111/nph.12411. [DOI] [PubMed] [Google Scholar]
- Tarr DEK, Alexander HM. TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders. BMC research notes. 2009;2:197. doi: 10.1186/1756-0500-2-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett LR, Hamilton JP, Buell CR, Tisserat NA, Verdier V, Zink F, Leach JE. Genomic analysis of Xanthomonas oryzae isolates from rice grown in the United States reveals substantial divergence from known X. oryzae pathovars. Appl Environ Microbiol. 2011;77:3930–3937. doi: 10.1128/AEM.00028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SD. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. 2014 bioRxiv. [Google Scholar]
- Verdier V, Triplett LR, Hummel AW, Corral R, Cernadas RA, Schmidt CL, Bogdanove AJ, Leach JE. Transcription activator-like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector-deficient strain of Xanthomonas oryzae. New Phytol. 2012a;196:1197–1207. doi: 10.1111/j.1469-8137.2012.04367.x. [DOI] [PubMed] [Google Scholar]
- Verdier V, Vera Cruz C, Leach JE. Controlling rice bacterial blight in Africa: needs and prospects. J Biotechnol. 2012b;159:320–328. doi: 10.1016/j.jbiotec.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Wang C, Zhang X, Fan Y, Gao Y, Zhu Q, Zheng C, Qin T, Li Y, Che J, Zhang M. XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol Plant. 2015;8:290–302. doi: 10.1016/j.molp.2014.10.010. [DOI] [PubMed] [Google Scholar]
- White FF, Potnis N, Jones JB, Koebnik R. The type III effectors of Xanthomonas. Mol Plant Pathol. 2009;10:749–766. doi: 10.1111/j.1364-3703.2009.00590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonni I, Djedatin G, Ouédraogo L, Verdier V. Evaluation of Rice Germplasm against Bacterial Leaf Streak Disease Reveals Sources of Resistance in African Varieties. Journal of Plant Pathology & Microbiology 2015 [Google Scholar]
- Wonni I, Ouedraogo L, Verdier V. First Report of Bacterial Leaf Streak Caused by Xanthomonas oryzae pv. oryzicola on Rice in Burkina Faso. Plant Dis. 2011;95:72–73. doi: 10.1094/PDIS-08-10-0566. [DOI] [PubMed] [Google Scholar]
- Xia C, Chen H, Zhu X. Identification, mapping, isolation of the genes resisting to bacterial blight and application in rice. Molecular Plant Breeding. 2012:3. [Google Scholar]
- Yang B, Sugio A, White FF. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci USA. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Response of US tropical japonica varieties Lemont and Dawn to US X. oryzae strain X11-5A and TAL effectors.
Response of near-isogenic Bs1 and Bs3 pepper lines to strain X11-5A and derivatives expressing domain-swap and artificial avrHah1 constructs.
Response of Carolina Gold Select to TAL effectors expressed from high and low-copy vectors.
Responses of Carolina Gold Select X Kitaake F2 hybrid individuals to infiltration with African X. oryzae pv. oryzicola strain MAI10.
Experimental procedures describing construction of TAL effector cloning modules.
Response of Carolina Gold Select to inoculation with with X. oryzae pv. oryzae and X. oryzae pv. oryzicola strains from Mali.
Oligos used for module construction.
Plasmids for Modular Assembly of Designer TAL Effectors.