Abstract
The prototypical second messenger cAMP regulates a wide variety of physiological processes. It can simultaneously mediate diverse functions by acting locally within independently-regulated microdomains. In mammalian cells, two types of adenylyl cyclase generate cAMP; G protein regulated transmembrane adenylyl cyclases and bicarbonate- calcium- and ATP-regulated soluble adenylyl cyclase (sAC). Because each type of cyclase regulates distinct microdomains, understanding cAMP signaling demands methods to distinguish between them. We developed a mass spectrometry based adenylyl cyclase assay which we used to identify a novel sAC-specific inhibitor, LRE1. LRE1 binds to the bicarbonate activator binding site and inhibits sAC via a unique allosteric mechanism. LRE1 prevents sAC-dependent processes in cellular and physiological systems and facilitates exploration of the therapeutic potential of sAC inhibition.
INTRODUCTION
Cyclic AMP is the prototypical second messenger. It has been implicated in many physiological processes, including cell proliferation, apoptosis, and differentiation. In recent years it has become clear how this single second messenger can mediate so many diverse, sometimes seemingly contradictory, processes. It is now appreciated that cAMP acts locally within independently-regulated, spatially-restricted microdomains (reviewed in1).
Cyclic AMP is produced from ATP by adenylyl cyclases (AC). Traditionally, cAMP had been thought to be produced exclusively via G protein-regulated, transmembrane adenylyl cyclases (tmACs) in response to hormones signaling via G protein coupled receptors (GPCRs). It is now known that mammalian cells contain a second type of AC, soluble adenylyl cyclase (sAC; ADCY10)2. sAC is a widely distributed source of cAMP3, which unlike tmACs, is localized throughout the cell and targeted to intracellular compartments and cellular organelles, including inside the nucleus and the mitochondrial matrix4. Consistent with its localization, sAC defines multiple, independently-regulated, intracellular cAMP signaling microdomains. Because signaling occurs in spatially restricted microdomains, understanding cAMP biology requires methods to distinguish between sAC-dependent and tmAC-dependent cascades.
sAC is also distinct from tmACs in its regulation. Unlike tmACs, sAC is not regulated by heterotrimeric G proteins5; instead, it is directly regulated by bicarbonate (HCO3−)6,7, calcium (Ca2+)8,9, and due to its millimolar Km for its substrate, ATP9, sAC is a physiological ATP sensor10. Inside cells, HCO3− levels are dynamically regulated by ubiquitously expressed carbonic anhydrases (CA), which equilibrate carbon dioxide (CO2) and intracellular pH (pHi) with HCO3−. Due to this nearly instantaneous equilibrium, mammalian sAC, and its HCO3−-regulated orthologs throughout the kingdoms of life, serve as Nature’s physiological CO2/HCO3−/pHi sensors (Reviewed in3,11). In response to changes in the CO2/HCO3−/pHi equilibrium, sAC regulates sperm activation and motility12,13; ciliary beat frequency in airway14,15; luminal pH in the epididymis16; the mitochondrial electron transport chain17; apoptosis18–20; activity dependent feeding of neurons in the brain21; glucose-stimulated insulin release from β cells of the pancreas10; and intraocular pressure in the eye22.
The physiological roles of sAC have been identified genetically, using two different sAC knockout (KO) mouse strains12,13,23 and sAC-specific siRNA24–26, and pharmacologically. Two sAC inhibitors useable in cellular systems have thus far been identified: catechol derivatives of estrogen (CEs) and KH72,27. CEs are natural products which were the first known inhibitors of sAC28; they inhibit sAC by binding to a hydrophobic cleft distinct from the active site and chelating essential metal cofactors29. While CEs have been useful for demonstrating sAC’s role in cAMP dependent processes, CEs can also inhibit tmACs29, which limits their utility. The second pharmacological sAC inhibitor, KH7, was identified in a small molecule screen against purified sAC protein13. In contrast to CEs, KH7 is specific for sAC relative to tmACs27, and it has grown into the most widely used reagent for identifying cAMP signaling functions mediated by sAC. KH7’s mechanism of action remains unknown, and it exhibits non-specific cellular effects30,31.
To identify additional sAC specific inhibitors to use as chemical probes and if sufficiently safe, for potential therapeutic applications, we developed a novel mass-spectrometry-based high throughput screening method. Using this screen, we identified an improved sAC-specific inhibitor LRE1. LRE1 inhibits sAC via a unique mechanism; it occupies the binding site of the physiological activator HCO3− and allosterically inhibits sAC. LRE1 effectively inhibits sAC-mediated functions in sperm and mitochondria, and it is non-toxic to cells. Identification of LRE1 as a second sAC-specific inhibitor facilitates studies of the physiological roles played by sAC-defined intracellular and intraorganellar cAMP microdomains and fosters a greater understanding of CO2/HCO3−/pHi chemosensing. Furthermore, because it has more favorable properties, LRE1 may enable development of first-of-their-kind sAC inhibition therapeutic strategies.
RESULTS
Development of a novel adenylyl cyclase screening assay
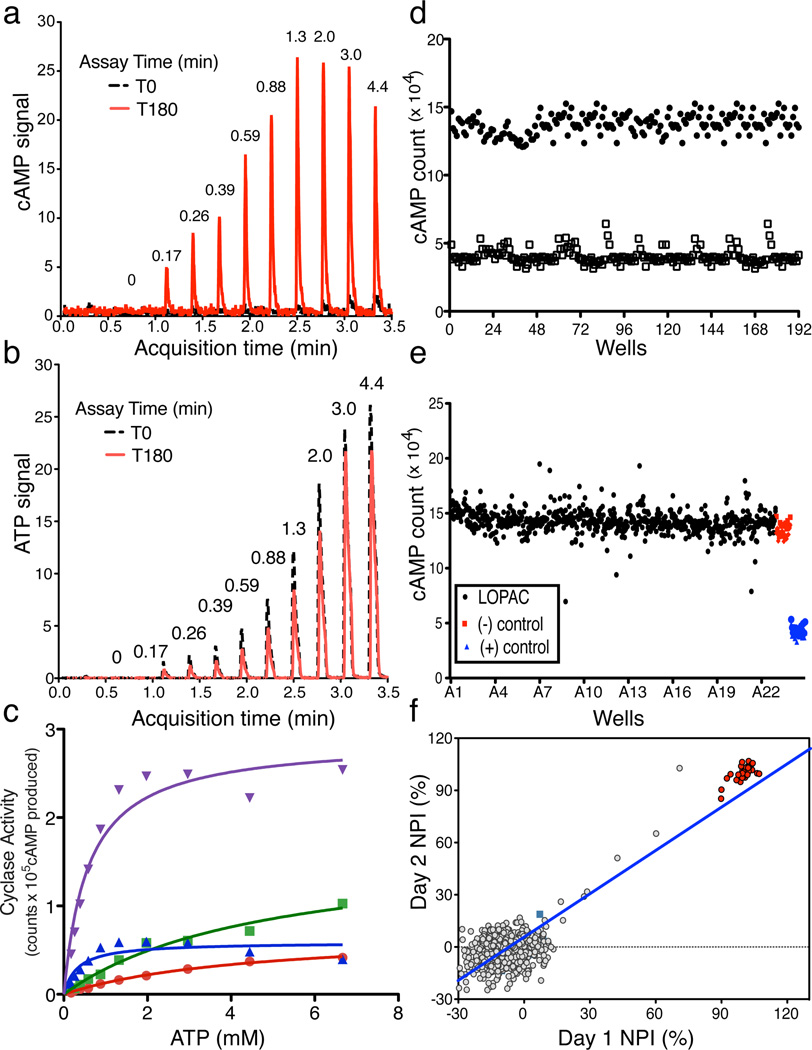
We developed a mass spectrometry (MS) based assay that accurately quantifies levels of both product cAMP and substrate ATP in a single sample over several orders of magnitude. Thus, by correlating cAMP production (Fig.1A) with reduction of input ATP (Fig. 1B), this MS-based assay provides assurance that the measured product derives from substrate similar to traditional, radioactivity-based assays where [α32P] ATP is converted into [32P] cAMP32. The RapidFire 365 High-throughput MS System (Agilent Technologies; RF-MSS) can process samples every 15 seconds allowing analysis of a 384 well plate in under two hours; thus, RF-MSS provides a platform for high throughput MS screening which can simultaneously measure both cAMP produced and ATP consumed in an individual sAC assay. Using the RF-MSS we observed no appreciable sample carryover between assay samples, and we were able to detect both a sAC-dependent increase in cAMP signal (Fig. 1A) and decrease in ATP signal (Fig. 1B) over time. Activity of sAC measured using this RF-MSS based adenylyl cyclase assay was consistent with sAC’s known kinetics9. When measured in the presence of increasing concentrations of substrate ATP in the presence of Mg2+ as the sole divalent cation, HCO3− stimulated sAC activity by increasing the Vmax with little effect on the apparent Km for substrate ATP-Mg2+ (Fig. 1C). Addition of Ca2+ stimulated sAC activity by decreasing its apparent Km for ATP, and addition of both Ca2+ and HCO3− synergistically activated sAC. These data confirm that using RF-MSS as a tool for measuring in vitro adenylyl cyclase activity is comparable to the classic, radioactivity based, “two-column” method32 for measuring adenylyl cyclase activity.
Fig. 1. Validation of RapidFire Mass Spectrometry System (RF-MSS) cyclase assay and high throughput screening conditions.
An example of a RF-MS chromatogram showing (a) the product cAMP as extracted ion intensity (EIC) × 103, and (b) substrate ATP as EIC × 104. Peaks derive from increasing input ATP (in mM) recorded at time “0” (black) and after 180 minutes incubation with sAC (red). (c) Human sAC activity measured using RF-MSS as a function of substrate of ATP-Mg2+ for 120 min in the presence of excess MgCl2 (20 mM). Shown are curves for Mg2+ alone (red dots), Mg2+/40 mM HCO3− (green squares), Mg2+/10 mM Ca2+ (blue triangles), and Mg2+/40 mM HCO3−/10 mM Ca2+ (purple triangles). Determinations are representative of at least two independent experiments and curves are nonlinear fits generated by Prism. (d–f) RF-MSS screen assaying sAC in the presence of 1 mM ATP/5 mM MgCl2/5 mM CaCl2/40 mM NaHCO3. (d) Comparison of active (black dots) and denatured (open squares) sAC enzyme: Z’ score = 0.7. (e) LOPAC library (1280 compounds) pilot screen (black); DMSO control (red); denatured sAC protein (blue). (f) Results of screening LOPAC library twice. LOPAC compounds (gray); DMSO control (green square); denatured sAC protein (red). (R2 = 0.81).
For screening, we chose in vitro assay conditions which reflect physiologically stimulated enzyme (i.e., in the presence of Mg2+, Ca2+, and HCO3−) to maximize our chances for identifying therapeutically useful sAC inhibitors. Under these conditions, the cAMP generated in the presence of active human sAC protein compared to the RF-MSS signal for cAMP in the presence of inactive sAC yields a Z’ score of ~0.7 (Fig. 1D). Thus, to screen for inhibitors, we used denatured sAC protein as “positive control.” Under these conditions, a pilot screen of the LOPAC reference library (Sigma) containing small organic compounds with well-documented pharmacological activities also yielded a Z’ factor of ~0.7 (Fig. 1E). Repeated screening of individual plates of the LOPAC library confirmed reproducibility with an R2 value of 0.81 (Fig. 1F).
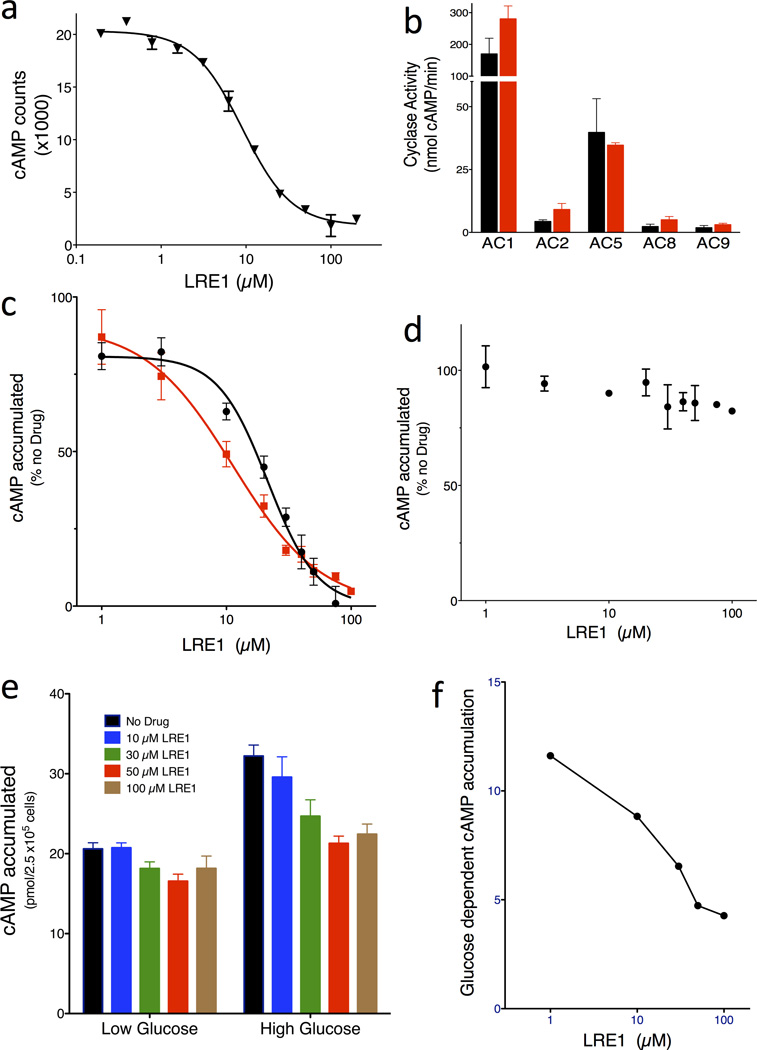
A total of 33,135 compounds, selected (as described in Methods) from a larger set of 7.87 million commercially available screening compounds to maximize coverage of chemical space and “drug-likeness” properties, were screened at 10 µM for their ability to inhibit in vitro human sAC activity (details summarized in Supplementary Results, Supplementary Table 1). From the 33,135 compounds screened, 46 compounds which inhibited sAC activity by ≥ 35% (0.14% hit rate) were tested for reproducibility and concentration-dependency. To avoid assay dependent artifacts, reproducibility and concentration-dependency were simultaneously assessed in the RF-MSS cyclase assay as well as in an independent colorimetric competitive enzyme immunoassay (i.e., ELISA-based Correlate-EIA Direct cAMP Assay). Four (4) of the 46 compounds proved to be concentration-dependent sAC inhibitors in both types of assay (Supplementary Table 2). Two compounds, RU-0204277 (1) and RU-0207148 (2), reproducibly had IC50s in the low µM range, which is comparable to KH713,27. We decided to focus on RU-0204277 due to its lower molecular weight, better Q.E.D. score, and unique structural features relative to KH7. The systematic name of RU-0204277 is 6-chloro-N4-cyclopropyl-N4-[(thiophen-3-yl)methyl]pyrimidine-2,4-diamine; hereafter, we refer to it as LRE1. The purity and identity of the “cherry picked” compounds were confirmed by LC-MS, and both commercially obtained LRE1 and independently synthesized LRE1 inhibited sAC in vitro activity with similar potency as the “cherry picked” compound. The concentration-response of independently synthesized LRE1 on sAC protein is shown in Fig. 2A.
Fig. 2. LRE1 is a potent and selective inhibitor of sAC in vitro and in cells.
(a) Concentration-response curve of LRE1 on sAC protein in the presence of 1 mM ATP/5 mM MgCl2/5 mM CaCl2/40 mM NaHCO3. Values are averages of triplicate determinations with SEM. (b) AC activities of 293 cells overexpressing each of the indicated tmACs in the absence (black bars) or presence of 50 µM LRE1 (red bars). Shown are representative assays repeated at least three times; values represent averages of quintuple determinations of tmAC specific activity with SEM. (c) Concentration-response of LRE1 (red squares) and KH7 (black circles) on cellular accumulation of cAMP in sAC overexpressing 4-4 cells. (d) Concentration-response of LRE1 on cellular accumulation of cAMP in forskolin-stimulated sAC knockout fibroblasts. (c,d) Representative assays repeated at least three times; values are means ± SEM of triplicate determinations normalized to activity in the absence of any compound. (e) Concentration-response of LRE1 on glucose-induced cAMP production in INS-1E cells in 2.5 mM (low) or 16 mM (high) glucose with 0 (black), 10 (blue), 30 (cyan), 50 (red), or 100 (brown) µM LRE1. Shown are means ± SEM of triplicate determinations of at least three independent experiments. (f) The glucose-induced cAMP, calculated from panel (e) by subtracting the cAMP accumulated in low glucose from the cAMP accumulated in high glucose, in the presence of the indicated concentration of LRE1.
LRE1 is specific for sAC
LRE1 is not a “frequent hitter;” it was negative in 17 other in vitro and cell-based screens against various targets including other nucleotidyl cyclases, proteases, ion channels, and receptors. In mammals, the enzymes most related to sAC are the G protein-regulated transmembrane adenylyl cyclases (tmACs)5. Nine genes encode tmAC isozymes in mammals, but among these, tmACs I, III, and VIII define a highly homologous and biochemically related subclass; tmACs II, IV, and VII define a second subclass; and tmACs V and VI define a third33. Therefore, to assess LRE1’s selectivity for sAC relative to other mammalian adenylyl cyclases, we tested whether LRE1 affected the in vitro G protein stimulated activities of heterologously expressed tmACs I, II, V, VIII, and IX. At 50 µM, which completely inhibited sAC, LRE1 did not inhibit any tmAC (Fig 2B).
LRE1 is efficacious in cellular systems
We next tested whether LRE1 could inhibit sAC in intact cells. Cellular levels of cAMP reflect a balance between its synthesis by adenylyl cyclases and its catabolism by phosphodiesterases (PDEs). When cells are grown in the presence of a cocktail of PDE inhibitors, they accumulate cAMP which reflects the activity of endogenous adenylyl cyclases. To assess adenylyl cyclase inhibitor efficacy in cellular systems, we used 4-4 cells, which stably overexpress sAC in a HEK293 cell background. The cAMP accumulation in 4-4 cells in the presence of PDE inhibitors is almost exclusively due to sAC10,27. LRE1 inhibited cAMP accumulation in 4-4 cells with an IC50 of 11 µM (Fig. 2C), which is comparable to KH7 and which is in good agreement with LRE1’s ≤ 10 µM IC50 on sAC protein (Fig. 2A).
To confirm LRE1’s specificity for sAC relative to tmACs in a cellular context, we used a second cell line, immortalized mouse embryo fibroblasts derived from sAC knockout mice (sAC KO MEFs). sAC KO MEFs are devoid of sAC, and they express a mixture of tmAC isoforms, specifically tmAC Types I, III, IV, VI, VII, VIII, and IX27. LRE1 was inert towards forskolin-stimulated cAMP accumulation in sAC KO MEFs (Fig. 2D). Together, these assays identify LRE1 as a sAC-specific inhibitor suitable for use in cell-based assays.
We demonstrated LRE1’s ability to inhibit a physiological sAC-dependent cAMP response. Pancreatic β cells sense serum glucose and respond to elevated glucose levels by releasing insulin. We previously demonstrated that sAC is essential for glucose sensing24 and for glucose dependent insulin secretion10. INS-1E cells are an insulinoma cell line which recapitulate β cell glucose sensing34, and we previously showed that while tmACs are largely responsible for the basal cAMP production in INS-1E cells growing in low glucose, sAC is responsible for the increased cAMP production in cells grown in high glucose24. Consistent with it being a sAC specific inhibitor, LRE1 effectively blocks the glucose induced cAMP response in INS-1E cells, while having little effect on the basal, tmAC-dependent cAMP synthesized in low glucose (Fig. 2E,F).
LRE1 inhibits sAC allosterically
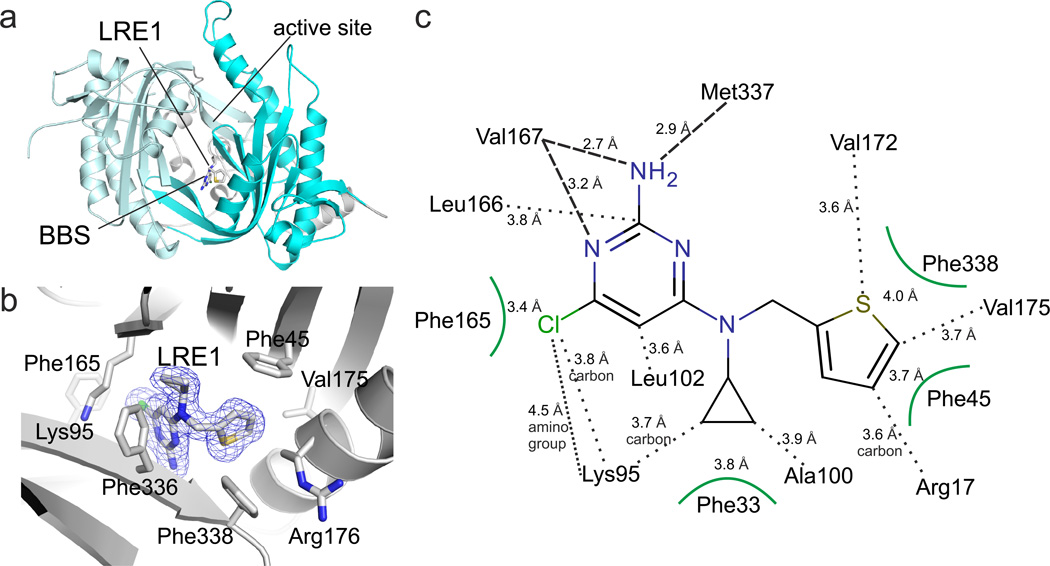
To obtain molecular insights into the LRE1 binding site and inhibition mechanism, we solved the crystal structure of a sAC/LRE1 complex. LRE1 was soaked into sAC-cat apo crystals, and excellent additional electron density for the LRE1 ligand was visible after molecular replacement phasing of the 1.79 Å resolution diffraction data (Supplementary Table 3). The final sAC/LRE1 complex showed good refinement statistics (R/Rfree = 16.1/20.3%) and a ligand geometry well-defined by electron density (Fig. 3A,B; Supplementary Fig. 1A). LRE1 occupies the HCO3− binding site (BBS) between Lys95 and Arg176, both of which are essential for HCO3− regulation7. The inhibitor extends into a channel connecting the BBS to the active site (see below). The overall B-value of 20.7 Å2 for LRE1 corresponds to the B-values of its protein environment and indicates a tightly bound ligand. The substituted pyrimidine ring of LRE1 occupies a rather hydrophobic pocket formed by Phe165, Leu166, Leu102, Val167, Met337 and Lys95 (Fig. 3B,C). Hydrogen bonds are formed by the backbone oxygens of Val167 (2.7 Å) and Met337 (2.9 Å) to the amino group of the LRE1 pyrimidine, and by the Val167 α-amino group to a ring nitrogen. The chlorine substituent points into a highly hydrophobic pocket formed by Leu102, Val167, Phe165, Leu166, and the side chain carbons of Lys95, and it might for a weak dipole-charge interaction with the Lys95 amino group. The LRE1 cyclopropene ring is accommodated in a hydrophobic pocket lined by Phe336 and Phe45/Ala97. Phe45 also interacts, through a T-shaped π-stacking interaction, with the LRE1 thiophene ring, which is further surrounded by Val175/172, Phe338, and Arg176. This Arg is a “trigger arm,” which links BBS and substrate binding site7. In the LRE1 complex, the Arg176 side chain is in a unique orientation; it points away from active site and BBS and is fixed in this orientation through polar interactions with Asp339 and Asn180.
Fig. 3. Crystal structure of sAC/LRE1 complex.
(a) Overall structure of the sAC/LRE1 complex, with C1 in grey and C2 in cyan. LRE1 is shown in stick representation, and active site and BBS are indicated. (b) Close-up of the BBS with ligand and interacting residues as sticks colored according to atom type. LRE1 is overlaid with 2Fo-Fc electron density (blue) contoured at 1σ. (c) Interaction scheme for sAC and LRE1. Interactions of side chains are indicated by black dots and by arcs for aromatic residues, and backbone interactions by dashed lines.
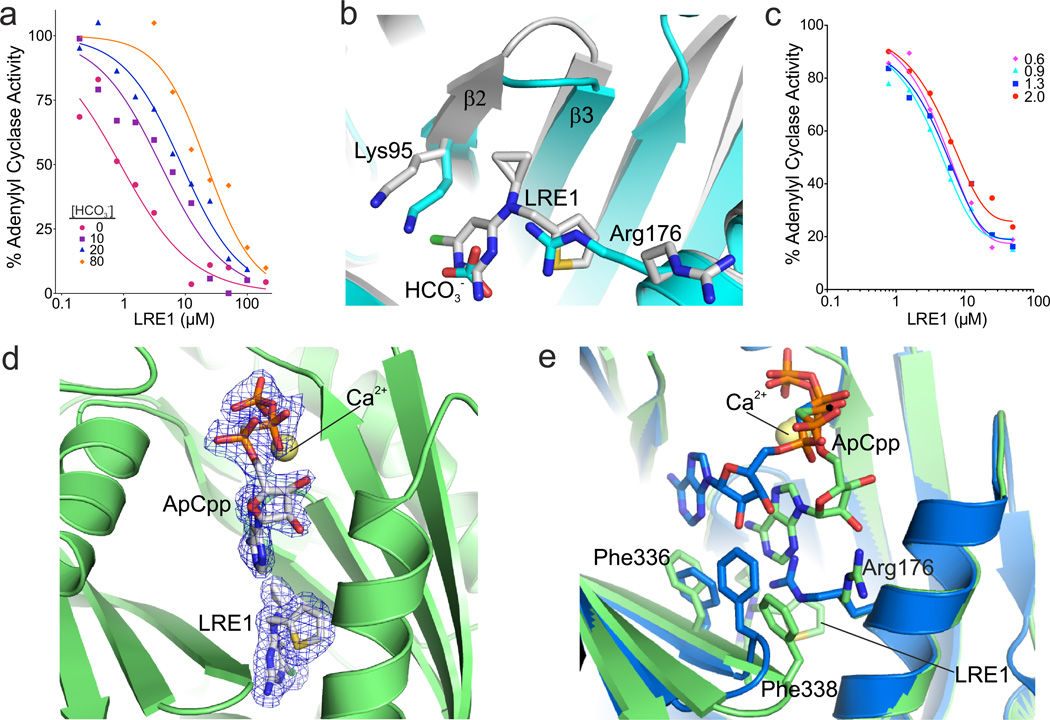
Consistent with LRE1 occupying the allosteric regulatory region mediating HCO3−-dependent sAC activation, kinetic experiments revealed that LRE1 inhibition is competitive with HCO3− (Fig. 4A). Overlaying the sAC/LRE1 complex with a sAC complex in the presence of HCO3− (PDB code 4cll)7 shows that part of the LRE1 pyrimidine ring assumes the positions of the HCO3− atoms (Fig. 4B) and appears to simulate their polarity pattern. However, the bulky LRE1 is not compatible with the Arg176 conformation assumed to be relevant for sAC activation7. For its interaction with the LRE1 thiophene, Phe338 from the β2–β3 loop is pulled ~3 Å toward the thiophene pocket, away from its normal position next to the active site. The LRE1 thiophene itself and the rearranged Phe338 block the region accommodating Arg176 during HCO3− binding. Due to the size of LRE1, Lys95, which forms the BBS border opposite from Arg176, is also shifted ~2.1 Å away from its normal position. These distortions explain why occupying the BBS with LRE1 does not stimulate sAC activity.
Fig. 4. Mechanistic characterization of sAC inhibition by LRE1.
(a) LRE1 is competitive with HCO3−. sAC activity, measured by RF-MSS in the presence of the indicated concentration of LRE1 and 1 mM ATP/5 mM MgCl2/5 mM CaCl2 with 0 (red); 10 (purple squares); 20 (blue triangles); or 80 (orange diamonds) mM NaHCO3. Data represent means of triplicate determinations of an experiment repeated twice. (b) Overlay of sAC/LRE1 (grey) with a sAC/bicarbonate complex (cyan, RMSD 0.3 Å5 for 352 Cα atoms). The ligands and the two key residues for bicarbonate binding are shown as sticks colored according to atom type. (c) LRE1 does not compete with ATP. sAC activity, measured by RF-MSS in the presence of the indicated concentration of LRE1 and either 0.6 (pink diamonds), 0.9 (cyan triangles), 1.3 (blue squares), or 2.0 (red dots) mM ATP and 5 mM MgCl2/5 mM CaCl2/40 mM NaHCO3. Data represent individual points of a serial dilution of an experiment repeated three times. (d) Crystal structure of a sAC/ApCpp/LRE1 complex. The substrate analog ApCpp in the active site and the inhibitor in the BBS are shown as sticks colored according to atom type and overlaid with 2Fo-Fc electron density contoured at 1σ (blue). Ca2+ is shown as yellow sphere. (e) Overlay of sAC/ApCpp/LRE1 (green) with a sAC/ApCpp complex (blue, RMSD 0.31 Å2 for Cα atoms). Ligands and key interacting residues are shown as sticks, and Ca2+ as a yellow sphere.
To investigate how LRE1 inhibits basal sAC activity (i.e., in the absence of HCO3−), we compared the inhibitor complex to a sAC-cat complex with the substrate analog α,β-methylene-ATP (ApCpp; PDB code 4clk)7. The closest distance between inhibitor and substrate analog would be a 6.4 Å gap between the LRE1 cyclopropene and the ApCpp ribose (Supplementary Fig. 1B). Thus, the inhibitor does not appear to overlap with the substrate binding site. Consistently, kinetic experiments revealed LRE1 inhibition to be non-competitive with the substrate ATP (Fig. 4C). We thus solved a sAC structure after soaking with both substrate analog ApCpp and the inhibitor LRE1. In this structure, solved at 1.86 Å resolution (Supplementary Table 3), both ligands are well defined by electron density (Fig. 4D). The inhibitor occupies an identical position as it occupies in the binary sAC/LRE1 complex, while part of ApCpp binds differently in the trimeric complex relative to how it binds in the sAC/ApCpp complex (Fig. 4E). The adenosine moiety of the substrate analog is shifted toward the area vacated by the Arg176 reorientation, and it directly interacts with the inhibitor. Phe336 and Phe338, which are shifted due to direct interactions with the inhibitor, appear to be major factors contributing to the adenosine relocation upon LRE1 binding. Phe336 is shifted into the original adenosine binding site which prevents regular ATP binding, and Phe338 is moved away from the active site and tears the directly interacting substrate base away from its original position. Thus, while LRE1 is not competitive with substrate binding (Fig. 4C), its presence dramatically affects ATP’s position in the active site forcing it into an orientation inconsistent with catalysis.
Comparison of sAC/LRE1 with a tmAC/forskolin structure (Supplementary Figure 1C; PDB code 1cjt) illustrates why LRE1 is specific for sAC. The bulky tmAC activator forskolin fits into the much deeper and wider regulator binding site in tmACs, but it does not fit in the tighter sAC activation site7. The position of LRE1 in sAC overlaps only with the small portion of the forskolin site in tmACs which directly corresponds to the BBS. LRE1 extends into the access channel toward the BBS, and the relatively small LRE1 is well suited for this tight channel. In contrast, LRE1 would be unable to fill most of the large activator site in tmACs, thus lacking sufficient interactions for tight binding to tmACs.
LRE1 inhibits sperm and mitochondrial functions of sAC
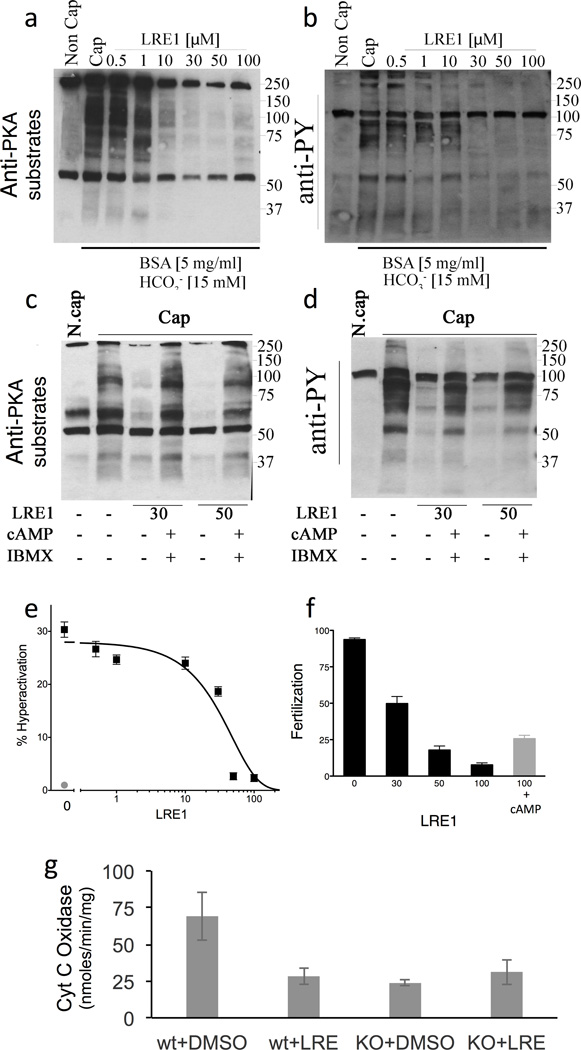
To assess its efficacy as a physiologically relevant inhibitor, we tested whether LRE1 could inhibit known sAC-dependent functions. The first identified role of sAC was in sperm35. Mammalian sperm acquire fertilization capacity as they transit through the reproductive tract in a process known as capacitation. At the molecular level, an early event in capacitation is HCO3−-induced activation of cAMP synthesis by sAC followed by the consequent activation of the main cAMP effector, Protein Kinase A (PKA). Activation of sAC and PKA precedes stimulation of a prototypical pattern of tyrosine (tyr) phosphorylation, induction of a “hyperactivated” form of motility, and finally competence to fertilize35. sAC activity has been genetically13,36 and pharmacologically13 demonstrated to be essential for these physiological changes sperm undergo during capacitation.
In sperm, LRE1 concentration-dependently blocks PKA dependent phosphorylation of its substrates (Fig. 5A; Supplementary Figure 2A), as well as the downstream increase in tyr phosphorylation (Fig. 5B; Supplementary Figure 2B), hyperactivation (Fig. 5C), and fertilization (Fig. 5D). Consistent with its effective concentration at inhibiting sAC in isolated cells (Fig. 2B,D), LRE1 inhibits these processes with an IC50 of ~10 µM (Supplementary Figure 2). KH7 also blocked each of these sAC-dependent functions13, but because the inhibitory activity of KH7 is quenched by the presence of BSA, which is an important component of capacitation-media37,38, KH7’s effect on capacitation had to be studied using other cholesterol-binding compounds (e.g., beta cyclodextrin)13. In contrast to KH7, LRE1 inhibition was unaffected by the presence of BSA in the incubation media. Finally, confirming that LRE1 mediates its effects exclusively via inhibition of sAC, these effects of LRE1 can be rescued by addition of exogenous membrane permeable cAMP (Fig. 5A,B,D).
Fig. 5. LRE1 inhibits sAC dependent processes in sperm and mitochondria.
(a–d) Sperm experiments. (a) Western blot using anti-PKA substrates antibodies of mouse cauda sperm activated by incubation in capacitation (Cap) media in the presence of the shown amounts of LRE1 in presence (+) or absence (−) of dibutyryl cAMP (1 mM) and IBMX (100 µM). N.Cap = non capacitated negative control. (b) Western blot using anti-phospho tyrosine antibodies of the same blot as in (a). For a,b, shown are representative Western blots of experiments repeated three times using independent sperm preparations from different mice. The complete gels used for these images are shown in Supplementary Figures 4 and 5, respectively. (c) The percentage of hyperactivated motility in sperm capacitated in the presence of the indicated concentration of LRE1 (black). Non capacitated sperm (red). Values are averages (± S.E.M.) of three independent sperm preparations from three different mice collected and analyzed on separate days. (d) Percentage of fertilized eggs from sperm capacitated in the presence of the indicated concentration of LRE1 (black bars) or sperm capacitated in the presence of 100 µM LRE1 + 1 mM dibutyryl cAMP (red bar). Values are averages (± S.E.M.) of four independent sperm preparations from four different mice collected and analyzed on separate days. (e) Cytochrome c oxidase (COX) activities in cells from WT and sAC KO MEFs treated with DMSO or 50 µM LRE1 for 30 min. Values are averages (± S.D.); N=5. COX activity was statistically different in WT cells ± LRE1 (P=0.037) by t-test.
A second well established function of sAC is regulation of mitochondrial respiration. sAC resides in the mitochondrial matrix where it senses Krebs cycle generated CO2 and hormonally stimulated calcium signals to regulate the activity of the electron transport chain39. Among other potential sites of regulation40, sAC generated cAMP stimulates the activity of cytochrome c oxidase (COX)41. LRE1 inhibits COX activity in WT MEFs, and consistent with LRE1’s effects being solely mediated via inhibition of sAC, COX activity in sAC KO MEFs was unaffected by LRE1 (Fig. 5E).
LRE1 is non-toxic and does not uncouple mitochondria
Reliable chemical probes and effective therapeutic compounds should have limited toxicity to cells with as few off-target effects as possible. Thus far, pharmacologic experiments exploring sAC function with the widely-used sAC specific inhibitor KH7 have focused on short-term assays or single doses in vivo22. In our own experience, we have observed non-sAC mediated toxicity in cells treated with KH7 for prolonged periods of time. We used the Cell-Titer Glo high throughput viability assay (Promega Inc.) to directly compare cellular toxicity of LRE1 and KH7. While neither compound exhibited significant toxicity after a single day in culture (Supplementary Figure 3A), after 2 days, KH7 was toxic to MEFs derived from WT C57Bl/6 mice at low µM concentrations (Supplementary Figure 3B). This toxicity was independent of sAC because KH7 exhibited a similar concentration-dependent toxicity in MEFs derived from sAC KO mice (Supplementary Figure 3C). In contrast, LRE1 exhibited no significant toxicity after two days in culture (Supplementary Figure 3B,C) at concentrations (i.e., up to 50 µM) which effectively inhibited sAC in cellular systems (Fig. 2C, and Fig. 5). These data identify LRE1 as the first pharmacological tool suitable for probing sAC functions in long-term assays.
KH7 has been reported to lead to mitochondrial uncoupling in a sAC-independent manner30. Under conditions which reproduced KH7’s uncoupling effect, LRE1 exhibited no uncoupling of isolated mouse brain mitochondria (Supplementary Figure 3D) and did not appreciably perturb bilayer properties (O.S. Andersen, personal communication). These studies suggest that LRE1 is a chemically inert nontoxic compound that does not interfere with essential cellular pathways.
DISCUSSION
Historically, adenylyl cyclase activity has been studied using two different types of in vitro assays. The classical method for measuring adenylyl cyclase activity involves supplying [α32P] ATP and measuring the amount of [32P] cAMP produced after separating formed product from input substrate using cyclic nucleotide separation chromatography methods32. This assay has exquisite specificity (the only way to generate radiolabelled product cAMP is from the input substrate ATP) and a broad dynamic range. It has been the ‘standard’ for measuring in vitro cyclase activity for the past ~40 years. Unfortunately, this method is not suitable for high throughput screening. For screening, numerous methods, which are dependent upon antibodies or other binding proteins which discriminate cAMP from other nucleotides, including input ATP, are available to quantitate cAMP formed after a cellular or in vitro reaction42. These methods are well-suited for high throughput screening, and while they can be extremely sensitive, they suffer from narrow dynamic ranges limiting their utility, especially for kinetic studies. Additionally, because sAC has a ~ 20 fold higher Km for its substrate ATP relative to other mammalian adenylyl cyclases (its Km for ATP is 1 mM under physiological conditions)9,10, in vitro sAC assays are performed with mM concentrations of ATP. When input ATP is this high, the assays we have tested suffer from high backgrounds due to cross reactivity with input ATP which masked the concentrations of cAMP produced.
The RF-MSS cyclase assay we have developed, based upon MS determination of both cAMP produced and ATP consumed, combines the specificity and broad dynamic range of the radioactive substrate assay with the high throughput adaptability of more modern methods. By measuring both product formation and substrate consumption, the RF-MSS assay substantiates that product is derived from input substrate. While the sensitivity and dynamic range of the assay will depend upon the particular Mass Spectrometer used, the 15 second per sample cycle time of the RapidFire delivery system provides high throughput capability. In addition, the RF-MSS is non-radioactive which provides for a safer assay with less environmental impact, and it is not dependent upon consumable and often non-renewable reagents such as antisera. Thus, the RF-MSS assay has the potential to replace previously existing adenylyl cyclase assays.
Using the RF-MSS assay, we identified a new sAC-specific inhibitor, LRE1. Among the different adenylyl cyclase inhibitors known, those structurally studied and mechanistically understood target the substrate binding region (e.g., P site inhibitors) or its immediate environment (e.g., catechol estrogens, DIDS)2,7. Crystallographic and kinetic data reveal that LRE1 specifically inhibits sAC via a novel allosteric mechanism induced through binding to its unique bicarbonate binding site (BBS). The corresponding site in tmACs is where the tmAC-specific activator forskolin binds43. Forskolin is considerably larger than bicarbonate (Supplementary Fig. 1C), and it does not fit in sAC’s BBS7. This difference in size between the forskolin binding site of tmACs and the BBS of sAC allows highly specific ligand interactions explaining the selective inhibition of sAC by LRE1. LRE1 not only occupies the BBS, it also extends into the channel connecting the BBS to the active site which provides additional favorable interactions for specific and tight ligand binding. This channel is also the binding site of the structure-based designed inhibitor, ASI-844 (Supplementary Fig. 1D), and it is postulated to be the site of KH7 inhibition of sAC2. However, both KH7 and ASI-8 are long compounds which would extend into the active site and inhibit, at least partially, via competition with the substrate. In fact, bithionol, a cytotoxic organochlorine, was recently found to inhibit sAC by binding to the BBS and this connecting channel45, similar to ASI-8. And bithionol indeed inhibits sAC by at least partially competing with substrate ATP. In contrast, LRE1 only occupies the connecting channel with its short cyclopropane moiety and does not reach the active site. Consistently, LRE1 is non-competitive with ATP (Fig. 4C); thus, LRE1 is the only known, exclusively allosteric inhibitor of sAC. Despite this shorter cyclopropane extension (as compared to ASI-8 and KH7), LRE1 still exhibits high affinity. Presumably, its affinity is augmented by additional interactions from the branching thiophene moiety, which exploits an additional sAC pocket (Supplementary Fig. 1D). These data identify LRE1 as the first potent and selective pharmacological modulator which acts allosterically via BBS binding. Thus, our study reveals that drug-like ligands can bind tightly to the BBS and allosterically inhibit sAC, and it raises the possibility that pharmacological ligands could mimic the stimulating effect of the small physiological sAC activator bicarbonate.
Understanding cAMP signaling in mammalian cells requires methods to distinguish between the two different sources of cAMP, tmACs and sAC. We previously identified pharmacological tools and conditions for selectively inhibiting tmAC-dependent cAMP generation using the P site inhibitor 2’5’ dideoxyadenosine and for specifically inhibiting sAC using KH727. Unfortunately, because KH7 exhibits an intrinsic fluorescence, it has limited utility for studying intracellular cAMP microdomains using fluorescence-based live cell cAMP sensors. In contrast, LRE1 does not appear to have intrinsic fluorescence. Thus, our identification of LRE1 as an improved sAC-specific inhibitor provides an essential chemical probe for the study of cAMP microdomains.
Finally, the many identified adenylyl cyclase inhibitors, in particular those affecting sAC, lack high potency and/or selectivity or possess non specific cellular toxicity2. In contrast, LRE1 combines high potency and selectivity with stability, solubility and, most importantly, lack of cytotoxicity. In particular, LRE1 exhibits less non-specific toxicity than KH7, the widely used sAC specific inhibitor2,27, and bithionol, a recently characterized sAC selective inhibitor45. Therefore, LRE1 is an ideal candidate to explore the therapeutic possibilities of sAC inhibition.
In summary, we have exploited the RapidFire mass spectrometry system (RF-MSS) to develop a safe and robust adenylyl cyclase assay suitable for a wide variety of applications. Using this RF-MSS cyclase assay, we identified an improved sAC specific inhibitor, LRE1, which occupies the regulatory “activator” site to allosterically inhibit the enzyme, and which exhibits pharmacologically favorable characteristics.
METHODS
Chemicals and Cell Lines
All chemicals were purchased from Sigma Aldrich unless otherwise noted. KH7 was synthesized by The Milstein Synthetic Chemistry Core Facility at Weill Cornell Medical College. LRE1 was purchased from Nanosyn and verified by The Milstein Synthetic Chemistry Core Facility by LC/MS (Supplementary Figure 8) and NMR (Supplementary Figure 9). Small molecules were purchased as powder and suspended in DMSO. Anti-phosphotyrosine (anti-PY) monoclonal antibody (clone 4G10) was obtained from Millipore, and rabbit monoclonal anti-phosphoPKA substrates (clone 100G7E) was purchased from Cell Signaling.
4-4 cells, sAC KO MEFs, and WT MEFs were generated and functionally authenticated in our laboratory as previously described27 and grown in DMEM + 10% FBS. INS-IE cells34 were obtained directly from P. Antinozzi and C. Wollheim and functionally authenticated in our laboratory10,24. They were cultured in RPMI media containing 10% heat-inactivated FBS, 10 mM HEPES, and 1 mM sodium pyruvate, 50 µM β-mercaptoethanol, and passaged every 2–3 days. All cells were maintained at 37°C in 5% CO2 and are periodically checked for mycoplasma contamination.
RapidFire Mass Spectroscopy
Analyses of cyclase assays were performed on an Agilent Rapid Fire 365 high throughput Mass Spectroscopy System (RF-MSS), equipped with an Agilent 6520 TOF accurate mass spectrometer as a detection system (Agilent Technologies). The Agilent 6520 TOF MS has a theoretical limit of high femtogram sensitivity and up to 5 orders of magnitude dynamic range. This instrument also includes a Zymark Twister robotic arm that handles microtiter plates and a purification SPE system. Sample (35 µl) was aspirated from each well of a 384-well reaction microtiter plate and injected onto a graphitized carbon SPE column extraction cartridge. Columns were washed with aqueous alkaline buffer (5 mM ammonium acetate, pH 10) and eluted using an alkaline/organic solvent (25% acetonitrile, 25% acetone in 5 mM ammonium acetate, pH 10) onto the electrospray-MS, where the mass spectra were collected. The RapidFire sipper was washed between sample injections using alkaline/organic and aqueous alkaline solvents. RapidFire mass spectrometry screening data were processed and analyzed using Agilent MassHunter Software.
For in vitro cyclase assays, human sACt protein9 was assayed in the presence of 50 mM Tris pH 7.5, 2 mM DTT, 5 mM MgCl2, and the indicated concentration of ATP in the presence or absence of 5 mM CaCl2 and/or 40 mM NaHCO3. cAMP (MW 328.0412) produced and ATP (MW 505.9816) consumed were determined by RF-MSS in comparison with standards of known concentrations, and for all quantitative cAMP measurements, we confirmed there was a corresponding decrease in input ATP. Mammalian tmAC isozymes tmAC I (ADCY1; bovine), tmAC II (ADCY2; rat), tmAC V (ADCY5; rat), tmAC VIII (ADCY8; rat), and tmAC IX (ADCY9; mouse) were transfected and expressed in HEK293 cells using the CMV promoter and whole cell extracts were assayed in the presence of 100 µM GTPγS via the classical two column radioassay measuring conversion of [α32P] ATP into [32P] cAMP as described previously46. To determine tmAC-specific activities, the activities of empty vector transfected lysates were subtracted. The activity in vector transfected HEK293 lysates in the presence of 100 µM GTPγS alone was 5.5 ± 0.8 nmol cAMP/min; its activity in the presence of 100 µM GTPγS + 50 µM LRE1 was 7.7 ± 1.7 nmol cAMP/min.
Small Molecule Library
33,135 compounds were selected from a larger set of 7.87 million commercially available screening compounds. The strategy for purchasing these compounds was to maximize coverage of chemical space and “drug-likeness” properties based on quantitative scores, while simultaneously minimizing cost. Pipeline Pilot software (Dassault Systemes Biovia Corp.) was used to execute a published algorithm for scoring compounds based on drug-likeness, called the quantitative exponent of drug-likeness (Q.E.D.) score47. From the 7.87 million commercially available compounds, 3.05 million compounds had a weighted Q.E.D. score greater than 0.7. The “choose diverse” component of the Pipeline Pilot software utilizes ECFP6 fingerprint descriptors to cluster the compounds into structurally related groups and chooses a single representative of each cluster, based on Tanimoto distances, called “cluster centers”. The “choose diverse” component identified 100,000 relatively diverse compounds from among these 3.05 million structures. From these 100,000 “cluster centers,” 33,135 could be purchased from Enamine Ltd. in 1 mg quantities at a price which was within our budgetary limits. These compounds were formatted in 10 copies of 5 mM DMSO stocks in 384-well deep well plates for screening.
High-throughput Screening
The primary screen (summarized in Supplementary Table 1) was performed in polystyrene 384-well microplates (Greiner Bio One International AG). 33,135 compounds were screened at 10 µM. The reagents were dispensed using a liquid handling instrument Multidrop Combi. Assay buffer (50 mM Tris pH 7.5, 5 mM CaCl2, 5 mM MgCl2, 40 mM NaHCO3, 2 mM DTT) was added first on the 384-well plate (Greiner-Bio One) followed by the compounds. The small molecule compounds were dispensed with a nanohead containing a 384-well array of Norgen Kloehn nanosyringes, and each plate was bar-coded. The human sACt protein9 was added next followed by the remainder of the assay buffer. The stop reaction buffer (1% formic acid) was added to the last column of each plate (denatured protein; T0). Reactions were started by addition of ATP at a final concentration of 1 mM. Reactions were incubated for 3 hours at 37°C, and stopped with the addition of 1% formic acid.
Cellular cAMP accumulation assays
sAC KO MEFs or 4-4 cells (2.0 × 106 cells/ml) in suspension were transferred to 1.5 ml tubes and incubated at 37°C, 5% CO2 for one hour. A time zero value for each condition was determined by adding 100 µl of cells directly into 100 l stop solution (0.2 M HCl). To measure cAMP accumulation, cells in suspension were incubated for the indicated period of time in the presence of 500 M IBMX (± LRE1 or ± KH7) at 37°C after which 100 l of cells were transferred to a fresh tube containing stop solution. Intracellular cAMP content was determined using Correlate-EIA Direct Assay (Assay Designs, Inc). INS-1E insulinoma cells were incubated in 2.5 mM glucose Krebs-Ringer buffer (pH 7.5) supplemented with 2 mM sodium bicarbonate, 10 mM HEPES, and 0.1% BSA for 2 h before start of the experiment. At time zero for each experiment, media was switched to Krebs-Ringer buffer containing 2.5 mM glucose or 16 mM glucose in the presence of 500 µM IBMX and inhibitor at the shown concentrations. After 10 min cells were lysed in 200 µl 0.1 M HCl. Intracellular cAMP contents were determined using Correlate-EIA Direct Assay (Assay Designs, Inc). Accumulated cAMP quantitated after 10 minutes are presented as pmol cAMP accumulated per 2.5 × 105 cells.
Crystal structure determination of sAC complexes
Protein for crystallization comprised the catalytic domains of human sAC (residues 1–469; sAC-cat), corresponding to the native sACt isoform, and a C-terminal his-tag. sAC-cat was expressed in Hi5 insect cells using the BIIC-method and purified by nickel affinity chromatography, anion exchange and size exclusion chromatography as described previously48. Apo sAC-cat was crystallized using the hanging drop method at 4 °C as described48. Crystals were soaked with LRE1 by transferring them into cryo solution (100 mM sodium acetate pH 4.8, 200 mM tri-sodium citrate, 18% (w/v) PEG 4000, 20% (v/v) glycerol) supplemented with 20 mM LRE1 alone or together with 150 mM ApCpp, 75 mM MgCl2/CaCl2 and incubation for 24 h at 4 °C. Crystals were flash-frozen in liquid nitrogen, and a complete dataset (space group P63) was collected from one crystal at 100 K at the Berlin Electron Storage Ring society for Synchrotron Radiation beamline 14.1 (BESSY BL14.1) operated by Helmholtz-Zentrum Berlin49. All diffraction data were processed with XDSAPP50, with the resolution limit set automatically using CC* 51. Molecular replacement phasing with the apo sAC structure (pdb code 4cll) as a search model, manual model building and refinement were done as described for previous sAC/ligand complexes7. Structure figures were generated with PyMol (www.pymol.org).
Cheminformatics and Data Handling
Data from all screening studies are archived and analyzed using the CDD Vault from Collaborative Drug Discovery (www.collaborativedrug.com). MarvinSketch (ChemAxon, version 14.9.8.0) was used for drawing and naming chemical structures. Instant Jchem for Excel (ChemAxon, Budapest Hungary, version 15.8.24.0) was also used for creating structure spreadsheets and tables. Compound similarity search was performed using Pipeline Pilot version 9.2 (Dassault Systemes Biovia Corp.). Graphpad PRISM (www.graphpad.com, version 6.05) was used for curve fitting.
Sperm Assays
Mouse sperm were isolated as described52. Sperm were incubated in Toyoda–Yokoyama–Hosi (standard TYH) medium buffered with 25 mM bicarbonate in 5 % CO2 for sperm capacitation and fertilization assays. Western blot and motility assays were performed in Hepes-buffered TYH (H-TYH). For capacitation, 15 mM NaHCO3 and 5 mg/ml BSA was added, and sperm incubated at 37 °C for at least 1 hour. For LRE1 treatment, sperm were pretreated for 10 min with LRE1 before addition of capacitation medium and/or dibutyryl cAMP. Anti-PKA substrates and anti-pY western blots, in vitro fertilization (IVF), and sperm motility analysis using a CEROS computer-assisted semen analysis (CASA) system (Hamilton Thorne Research) were performed as described52. At least 20 microscopy fields corresponding to a minimum of 200 sperm were analyzed in each motility experiment. For fertilization, wells containing 25–40 eggs were inseminated with sperm that had been incubated for 1 h and 20 min in capacitation supporting Whitten’s medium with the shown amounts of LRE1. Sperm in the presence of 100 µM LRE1 were also incubated in the presence of 1 mM dibutyryl cAMP. After 4 h of insemination, eggs were washed and put in fresh Whitten’s media. The eggs were evaluated 24 h post-insemination. To assess fertilization, the following criteria were considered: (1) the formation of the male and female pronuclei, (2) the emission of the second polar body, and (3) two-cell stage events.
Mitochondrial COX activity
Wild type and sAC KO MEFs were plated in P10 dishes. At 70% of confluence cells were incubated at 37°C with DMSO or LRE1 50µM for 30 min in serum free medium. Subsequently the cells were scraped in PBS and centrifuged for 5 min at 1000 rpm. The pellet was snap frozen in liquid nitrogen and stored at −80° C. Before the experiment the pellets were resuspended in PBS, COX activity was measured as described previously53 and normalized to protein concentration.
Cytotoxicity Assays
Cells were plated in 384- or 96-well plates. The next day, compounds were added (ten concentrations), and the plates were incubated for 24 or 48 hrs at 37°C/ 5% CO2, and viability was quantified by measuring cellular ATP using luminescence (CellTiter-Glo, Promega).
Measurement of the membrane potential in isolated mouse brain mitochondria
Mouse brain mitochondria were isolated by the Percoll gradient method as described54 and re-suspended in 90 µl of isolation medium containing 225 mM mannitol, 75 mM sucrose, 20 mM HEPES-KOH (pH 7.4), 1 mM EGTA, and 0.2 mg/ml fatty acid-free bovine serum albumin (BSA). The membrane potential changes were assessed from the changes in fluorescence of a permeating cation Safranin O55–57 at 495 nm excitation and 586 nm emission wavelength with Hitachi F-7000 spectrofluorimeter (Hitachi). The incubation medium was composed of 125 mM KCl, 20 mM HEPES (pH 7.4), 4 mM KH2PO4, 0.5 mM EGTA, 0.2 mg/ml BSA, 2 µM Safranin O (membrane potential probe), 5 mM Pyruvate, 2.5 mM malate, and 0.1 mg/ml mitochondria protein.
Supplementary Material
Acknowledgments
We thank Dr. David Warren and The Milstein Synthetic Chemistry Core Facility at Weill Cornell Medical College and Drs. Steven S. Gross and Qiuying Chen. CS and SK thank the BESSY staff and the Helmholtz Protein Sample Production Facility (PSPF) for technical assistance. L.R.E. and F.G. thank the HTSRC staff, the Helmsley Trust for funding of the Rapid-Fire Automated SPE TOF Mass-Spectrometry Instrument, and Agilent staff, especially to Peter Rye and Lauren Frick, for technical assistance. This work was supported, in whole or in part, by National Institutes of Health grants GM107442 and EY025810 and by the Weill Cornell Medicine CTSC, NIH/NCATS Grant UL1TR00457 (to L.R.L. and J.B.); NIH grants HD38082 and HD44044 (to P. E. V.), R01GM088999 (to G.M.), and K08 CA160657 (to J.H.Z.); Melanoma Research Alliance Team Science and Clinique Clinical Scholars Awards (to J.H.Z.); and Deutsche Forschungsgemeinschaft grant STE1701/11 (to C.S.).
Footnotes
Accession Codes
Crystal structures and diffraction data have been deposited with the worldwide protein data bank (wwPDB) under accession codes 5IV4 (sAC/LRE1) and 5IV3 (sAC/LRE1/ApCpp).
Author contributions
L.R.E., S.K., C.S., F.G., L.R.L., and J.B. designed the research project and analyzed data; L.R.E. and C.A. performed the HTS; S.K. and C.S. solved the crystal structure; J.v.H. expressed protein; L.R.E. performed the kinetic and cellular inhibition studies; H.B. performed the assays of individual tmACs; F.A.N., A.A., and P.E.V. performed the sperm studies; F.V., A.S., and G.M. performed the mitochondrial studies; and L.R.E. and J.H.Z. performed the cytotoxicity studies. All authors contributed specific parts of the manuscript, with L.R.E., L.R.L. and J.B. assuming responsibility for the manuscript in its entirety.
Conflict of interest
Drs. Buck, Levin and Zippin own equity interest in CEP Biotech which has licensed commercialization of a panel of monoclonal antibodies directed against sAC. All other authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Lefkimmiatis K, Zaccolo M. cAMP signaling in subcellular compartments. Pharmacology & Therapeutics. 2014;143:295–304. doi: 10.1016/j.pharmthera.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steegborn C. Structure, mechanism, and regulation of soluble adenylyl cyclases - similarities and differences to transmembrane adenylyl cyclases. Biochim Biophys Acta. 2014;1842:2535–2547. doi: 10.1016/j.bbadis.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Levin LR, Buck J. Physiological roles of acid-base sensors. Annu Rev Physiol. 2015;77:347–362. doi: 10.1146/annurev-physiol-021014-071821. [DOI] [PubMed] [Google Scholar]
- 4.Desman G, Waintraub C, Zippin JH. Investigation of cAMP microdomains as a path to novel cancer diagnostics. Biochim Biophys Acta. 2014;1842:2636–2645. doi: 10.1016/j.bbadis.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck J, Sinclair ML, Schapal L, Cann MJ, Levin LR. Cytosolic adenylyl cyclase defines a unique signaling molecule in mammals. Proc Natl Acad Sci U S A. 1999;96:79–84. doi: 10.1073/pnas.96.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, et al. Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science. 2000;289:625–628. doi: 10.1126/science.289.5479.625. [DOI] [PubMed] [Google Scholar]
- 7.Kleinboelting S, et al. Crystal structures of human soluble adenylyl cyclase reveal mechanisms of catalysis and of its activation through bicarbonate. Proc Natl Acad Sci U S A. 2014;111:3727–3732. doi: 10.1073/pnas.1322778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaiswal BS, Conti M. Calcium regulation of the soluble adenylyl cyclase expressed in mammalian spermatozoa. Proc Natl Acad Sci U S A. 2003;100:10676–10681. doi: 10.1073/pnas.1831008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Litvin TN, Kamenetsky M, Zarifyan A, Buck J, Levin LR. Kinetic properties of "soluble" adenylyl cyclase. Synergism between calcium and bicarbonate. The Journal of Biological Chemistry. 2003;278:15922–15926. doi: 10.1074/jbc.M212475200. [DOI] [PubMed] [Google Scholar]
- 10.Zippin JH, et al. CO2/HCO3(−)- and calcium-regulated soluble adenylyl cyclase as a physiological ATP sensor. J Biol Chem. 2013;288:33283–33291. doi: 10.1074/jbc.M113.510073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tresguerres M, Buck J, Levin LR. Physiological carbon dioxide, bicarbonate, and pH sensing. Pflugers Archiv : European journal of physiology. 2010;460:953–964. doi: 10.1007/s00424-010-0865-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esposito G, et al. Mice deficient for soluble adenylyl cyclase are infertile because of a severe sperm-motility defect. Proc Natl Acad Sci U S A. 2004;101:2993–2998. doi: 10.1073/pnas.0400050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hess KC, et al. The "soluble" adenylyl cyclase in sperm mediates multiple signaling events required for fertilization. Dev Cell. 2005;9:249–259. doi: 10.1016/j.devcel.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, et al. A soluble adenylyl cyclase form targets to axonemes and rescues beat regulation in soluble adenylyl cyclase knockout mice. Am J Respir Cell Mol Biol. 2014;51:750–760. doi: 10.1165/rcmb.2013-0542OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmid A, et al. Soluble adenylyl cyclase is localized to cilia and contributes to ciliary beat frequency regulation via production of cAMP. J Gen Physiol. 2007;130:99–109. doi: 10.1085/jgp.200709784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastor-Soler N, et al. Bicarbonate-regulated adenylyl cyclase (sAC) is a sensor that regulates pH-dependent V-ATPase recycling. The Journal of Biological Chemistry. 2003;278:49523–49529. doi: 10.1074/jbc.M309543200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acin-Perez R, et al. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell Metab. 2009;9:265–276. doi: 10.1016/j.cmet.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Appukuttan A, et al. Type 10 adenylyl cyclase mediates mitochondrial Bax translocation and apoptosis of adult rat cardiomyocytes under simulated ischaemia/reperfusion. Cardiovascular research. 2012;93:340–349. doi: 10.1093/cvr/cvr306. [DOI] [PubMed] [Google Scholar]
- 19.Flacke JP, et al. Type 10 soluble adenylyl cyclase is overexpressed in prostate carcinoma and controls proliferation of prostate cancer cells. The Journal of Biological Chemistry. 2013;288:3126–3135. doi: 10.1074/jbc.M112.403279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Kostin S, Flacke JP, Reusch HP, Ladilov Y. Soluble adenylyl cyclase controls mitochondria-dependent apoptosis in coronary endothelial cells. The Journal of Biological Chemistry. 2009;284:14760–14768. doi: 10.1074/jbc.M900925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi HB, et al. Metabolic Communication between Astrocytes and Neurons via Bicarbonate-Responsive Soluble Adenylyl Cyclase. Neuron. 2012;75:1094–1104. doi: 10.1016/j.neuron.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee YS, et al. Regulation of Anterior Chamber Drainage by Bicarbonate-sensitive Soluble Adenylyl Cyclase in the Ciliary Body. The Journal of Biological Chemistry. 2011;286:41353–41358. doi: 10.1074/jbc.M111.284679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Martinez J, Milner TA, Buck J, Levin LR. Neuronal expression of soluble adenylyl cyclase in the mammalian brain. Brain Res. 2013;1518:1–8. doi: 10.1016/j.brainres.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos LS, Zippin JH, Kamenetsky M, Buck J, Levin LR. Glucose and GLP-1 stimulate cAMP production via distinct adenylyl cyclases in INS-1E insulinoma cells. J Gen Physiol. 2008;132:329–338. doi: 10.1085/jgp.200810044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stessin AM, et al. Soluble adenylyl cyclase mediates nerve growth factor-induced activation of Rap1. The Journal of Biological Chemistry. 2006;281:17253–17258. doi: 10.1074/jbc.M603500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu KY, et al. Soluble adenylyl cyclase is required for netrin-1 signaling in nerve growth cones. Nature Neuroscience. 2006;9:1257–1264. doi: 10.1038/nn1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bitterman JL, Ramos-Espiritu L, Diaz A, Levin LR, Buck J. Pharmacological distinction between soluble and transmembrane adenylyl cyclases. J Pharmacol Exp Ther. 2013;347:589–598. doi: 10.1124/jpet.113.208496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun T. Inhibition of the soluble form of testis adenylate cyclase by catechol estrogens and other catechols. Proc Soc Exp Biol Med. 1990;194:58–63. doi: 10.3181/00379727-194-43055. [DOI] [PubMed] [Google Scholar]
- 29.Steegborn C, et al. A novel mechanism for adenylyl cyclase inhibition from the crystal structure of its complex with catechol estrogen. The Journal of Biological Chemistry. 2005;280:31754–31759. doi: 10.1074/jbc.M507144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Benedetto G, Scalzotto E, Mongillo M, Pozzan T. Mitochondrial Ca(2+) Uptake Induces Cyclic AMP Generation in the Matrix and Modulates Organelle ATP Levels. Cell Metab. 2013;17:965–975. doi: 10.1016/j.cmet.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Tian G, Sandler S, Gylfe E, Tengholm A. Glucose- and Hormone-Induced cAMP Oscillations in {alpha}- and {beta}-Cells Within Intact Pancreatic Islets. Diabetes. 2011;60:1535–1543. doi: 10.2337/db10-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salomon Y. Adenylate cyclase assay. Advances in Cyclic Nucleotide Research. 1979;10:35–55. [PubMed] [Google Scholar]
- 33.Cooper DMF, Mons N, Karpens JW. Adenylyl cyclases and the interaction between calcium and cAMP signalling. Nature. 1995;374:421–424. doi: 10.1038/374421a0. [DOI] [PubMed] [Google Scholar]
- 34.Merglen A, et al. Glucose Sensitivity and Metabolism-Secretion Coupling Studied during Two-Year Continuous Culture in INS-1E Insulinoma Cells. Endocrinology. 2004;145:667–678. doi: 10.1210/en.2003-1099. [DOI] [PubMed] [Google Scholar]
- 35.Buffone MG, Wertheimer EV, Visconti PE, Krapf D. Central role of soluble adenylyl cyclase and cAMP in sperm physiology. Biochim Biophys Acta. 2014;1842:2610–2620. doi: 10.1016/j.bbadis.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie F, et al. Soluble adenylyl cyclase (sAC) is indispensable for sperm function and fertilization. Dev Biol. 2006;296:353–362. doi: 10.1016/j.ydbio.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 37.Visconti PE, et al. Cholesterol efflux-mediated signal transduction in mammalian sperm. beta-cyclodextrins initiate transmembrane signaling leading to an increase in protein tyrosine phosphorylation and capacitation. J Biol Chem. 1999;274:3235–3242. doi: 10.1074/jbc.274.5.3235. [DOI] [PubMed] [Google Scholar]
- 38.Visconti PE, et al. Cholesterol efflux-mediated signal transduction in mammalian sperm: cholesterol release signals an increase in protein tyrosine phosphorylation during mouse sperm capacitation. Dev Biol. 1999;214:429–443. doi: 10.1006/dbio.1999.9428. [DOI] [PubMed] [Google Scholar]
- 39.Valsecchi F, Konrad C, Manfredi G. Role of soluble adenylyl cyclase in mitochondria. Biochim Biophys Acta. 2014;1842:2555–2560. doi: 10.1016/j.bbadis.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Rasmo D, et al. Intramitochondrial adenylyl cyclase controls the turnover of nuclear-encoded subunits and activity of mammalian complex I of the respiratory chain. Biochim Biophys Acta. 2015;1853:183–191. doi: 10.1016/j.bbamcr.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Acin-Perez R, Gatti DL, Bai Y, Manfredi G. Protein phosphorylation and prevention of cytochrome oxidase inhibition by ATP: coupled mechanisms of energy metabolism regulation. Cell Metab. 2011;13:712–719. doi: 10.1016/j.cmet.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conley JM, et al. Development of a high-throughput screening paradigm for the discovery of small-molecule modulators of adenylyl cyclase: identification of an adenylyl cyclase 2 inhibitor. J Pharmacol Exp Ther. 2013;347:276–287. doi: 10.1124/jpet.113.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsalpha.GTPgammaS. Science. 1997;278:1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- 44.Saalau-Bethell SM, et al. Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket. ChemMedChem. 2014;9:823–832. doi: 10.1002/cmdc.201300480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleinboelting S, et al. Bithionol potently inhibits human soluble adenylyl cyclase through binding to the allosteric activator site. J Biol Chem. 2016 doi: 10.1074/jbc.M115.708255. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 46.Levin LR, et al. The Drosophila learning and memory gene rutabaga encodes a Ca2+/Calmodulin-responsive adenylyl cyclase. Cell. 1992;68:479–489. doi: 10.1016/0092-8674(92)90185-f. [DOI] [PubMed] [Google Scholar]
- 47.Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL. Quantifying the chemical beauty of drugs. Nat Chem. 2012;4:90–98. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleinboelting S, et al. Expression, purification, crystallization and preliminary X-ray diffraction analysis of a mammalian type 10 adenylyl cyclase. Acta Crystallogr F Struct Biol Commun. 2014;70:467–469. doi: 10.1107/S2053230X14004014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mueller U, et al. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J Synchrotron Radiat. 2012;19:442–449. doi: 10.1107/S0909049512006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krug M, Weiss MS, Heinemann U, Mueller U. XDSAPP : a graphical user interface for the convenient processing of diffraction data using XDS. J. Appl. Crystallogr. 2012;45:568–572. [Google Scholar]
- 51.Diederichs K, Karplus PA. Better models by discarding data? Acta Crystallogr. D Biol. Crystallogr. 2013;69 doi: 10.1107/S0907444913001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Navarrete FA, et al. Biphasic role of calcium in mouse sperm capacitation signaling pathways. J Cell Physiol. 2015;230:1758–1769. doi: 10.1002/jcp.24873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods in Cell Biology. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- 54.Sims NR. Rapid isolation of metabolically active mitochondria from rat brain and subregions using Percoll density gradient centrifugation. J Neurochem. 1990;55:698–707. doi: 10.1111/j.1471-4159.1990.tb04189.x. [DOI] [PubMed] [Google Scholar]
- 55.Akerman KE, Wikstrom MK. Safranine as a probe of the mitochondrial membrane potential. FEBS Lett. 1976;68:191–197. doi: 10.1016/0014-5793(76)80434-6. [DOI] [PubMed] [Google Scholar]
- 56.Figueira TR, Melo DR, Vercesi AE, Castilho RF. Safranine as a fluorescent probe for the evaluation of mitochondrial membrane potential in isolated organelles and permeabilized cells. Methods Mol Biol. 2012;810:103–117. doi: 10.1007/978-1-61779-382-0_7. [DOI] [PubMed] [Google Scholar]
- 57.Zanotti A, Azzone GF. Safranine as membrane potential probe in rat liver mitochondria. Arch Biochem Biophys. 1980;201:255–265. doi: 10.1016/0003-9861(80)90510-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.