Abstract
Over the past several years, analyses of data from high-throughput studies have elucidated many fundamental insights into prostate cancer biology. These insights include the identification of molecular alterations and subtypes that drive tumour progression, recurrent aberrations in signalling pathways, the existence of substantial intertumoural and intratumoural heterogeneity, Darwinian evolution in response to therapeutic pressures and the complicated multidirectional patterns of spread between primary tumours and metastatic sites. However, these concepts have not yet been fully translated into clinical tools to improve prognostication, prediction and personalization of treatment of patients with prostate cancer. The current and future clinical implications of ‘omics’ level knowledge is not only revolutionizing our understanding of prostate cancer biology, but is also shaping ongoing, and future clinical investigations and practice. In this Review, these advances are summarized, and the remaining challenges surrounding tumour heterogeneity and the ability to overcome treatment resistance are described.
Historically, the decision-making processes regarding the prognosis of patients with prostate cancer, and the optimal therapeutic approach have been driven by clinical and pathological variables such as tumour stage, Gleason score and serum PSA levels(1, 2). However, new insights into the genomic aberrations underlying prostate cancer are rapidly transforming the diagnosis and treatment of patients with this disease, from early detection through to treatment of late-stage tumours. Characterizing the molecular landscape of prostate cancer through ‘omics’ level analyses has, historically, been technically challenging owing to the small size of the available diagnostic specimens (usually core biopsy samples), the difficulties in identifying tumours grossly at the time of resection (for procurement of fresh tissue for high-quality nucleic acid isolation) and the difficulties in obtaining highly pure samples of the tumour population, owing to the presence of large amounts of stroma and normal tissue within cancer foci.
Characterizing the genomic landscape of metastatic prostate cancer that has progressed despite castration levels of serum testosterone, as induced by gonadotrophin-releasing hormone agonists (a form of androgen deprivation therapy; ADT), termed castration-resistant prostate cancer (CRPC), has been even more challenging given the lack of biopsy sampling of metastases in routine clinical practice and the difficulties in obtaining quantitatively and qualitatively adequate tissue from metastatic sites, such as bone. Nevertheless, data from multiple rigorous, large, whole genome, exome and/or transcriptome sequencing studies have revolutionized our understanding of the genomic processes underlying the development and evolution of prostate cancer(3-11). For example, DNA copy-number alterations (CNAs) and chromosomal rearrangements, compared with point mutations or small insertion and/or deletion (indel) mutations, are more frequently observed alterations in patients with prostate cancer(3). Importantly, higher burdens of CNAs and other mutations have been observed in patients with more-aggressive prostate tumours in numerous studies, and correlate with clinical outcomes(3-6, 10, 12).
Despite these advances, application of these insights into clinical decision making, particularly for patients with metastatic disease, remains in its infancy and therapeutic strategies are largely driven by the presence, and extent of routine clinical and pathological factors. Over the next decade, insights from genomics are likely to have a much more prominent role in drug development, prognostication, patient selection, monitoring treatment response and/or resistance, and clinical practice as a whole. In this Review, we outline a selected number of important genomic alterations in prostate cancer, and the clinical implications of these discoveries (Table 1).
Table 1.
Select genomic alterations and their future clinical implications
| Pathway process |
Targets | Drug development |
Potential prognostic or predictive biomarkers |
|---|---|---|---|
| AR signalling | AR NCOR1/2 FOXA1 ZBTB16 SPOP |
N-terminal domain AR inhibitors; Dual AR/GR inhibitors |
AR-V7 spice variants; AR amplification |
| Cell cycle | P53 MYC CDKN2A RB1 AURKA |
DNA-binding domain AR inhibitors; CDK4/6 inhibitors; AURKA inhibitors |
RB1 status; AR lo/independence; AURKA amplification |
| DNA repair | BRCA ATM RAD51 MSH2/6 SPOP DNAPK |
PARP inhibitors, PD- L1 inhibitors |
DNA repair defects |
| ETS Fusion | ERG ETV1 |
HDAC inhibitors, PARP inhibitors |
ETS gene fusion status |
| MAPK pathway | BRAF RAF1 HRAS |
BRAF inhibitors; MEK inhibitors |
Mutations or gene fusions |
| Wnt pathway | CTNNB1 APC ZNRF3 RNF43 RSPO2 |
Porcupine inhibitors |
Mutations or gene fusions |
| PI3K pathway | PTEN PIK3CA PI3KCB AKT1 |
pan-PI3K and dual PI3K– mTOR inhibitors; PI3KCB inhibitors |
Mutations or copy number alterations |
AKT, v−akt murine thymoma viral oncogene homologue; APC, adenomatous polyposis coli; AR, androgen receptor; BRAF, B-Raf proto-oncogene, serine/threonine kinase; BRCA, breast cancer; CTNNB1, catenin β 1; ERG, v−ets avian erythroblastosis virus E26 oncogene homologue; ETS variant 1; FOXA1, forkhead box A1; GR, glucocorticoid receptor; HDAC, histone deacetylases; HRAS, Harvey rat sarcoma viral oncogene homologue; MSX, msh homeobox; MYC, MYC proto-oncogene protein; NCOR, nuclear receptor co-repressor; PARP, poly(ADP-ribose) polymerase; PD-L1, programmed cell death 1 ligand 1; PIK3C, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit; PTEN, phosphatase and tensin homologue; RAD51, RAD51 recombinase; RAF1, Raf-1 proto-oncogene, serine/threonine kinase; RB1, retinoblastoma 1; RNF43, ring finger protein 43; RSPO2, R-spondin 2; SPOP, speckle type BTB/POZ protein; ZBTB16, zinc finger and BTB domain containing 16; ZNRF3, zinc and ring finger 3.
Molecular subtypes
Findings of gene expression profiling, exome sequencing and candidate gene-based studies have provided robust evidence supporting the existence of a range of molecular subytpes of prostate cancer, based largely on the presence or absence of gene fusions involving members of the ETS gene family of transcriptional regulators (most commonly ERG, ETV1, ETV4, ETV5 and FLI1). Fusions juxtaposing the non-coding androgen-driven promoter elements of the transmembrane protease gene TMPRSS2 to nearly full length ERG (TMPRSS2–ERG fusions) are the most common ETS gene fusions observed in prostate cancer biopsy samples, occurring in approximately 40–50% of all tumours in PSA-screened, predominantly white populations. Alterations occurring exclusively in the approximately 50-60% of prostate tumours that are ETS fusion-negative (ETS−) include overexpression of the serine peptidase inhibitor gene SPINK1, recurrent point mutations in the transcriptional repressor gene SPOP, loss and/or mutation of the DNA-binding protein CHD1, and RAS/RAF family gene fusions or mutations(10, 11, 13, 14). Data from gene expression profiling studies support the distinction of ETS fusion-positive (ETS+) and ETS− tumours; furthermore, findings of several studies15–17, including those from the The Cancer Genome Atlas (TCGA) multi-omics profiling study17, support the distinction of ETS+ tumours into those with ERG fusions (ERG+, comprising approximately 90% of all ETS+ tumours) and those with fusions involving non-ERG members of the ETS family (occurring in approximately 5–10% of all ETS+ tumours )(15-17).
Other than TMPRSS2–ERG fusions, the development and progression of most prostate cancers seems to be driven by a variety of diverse, low-frequency oncogenic events. Owing in part to this genetic diversity, research by the TCGA has resulted in a large number of primary prostate cancer molecular classes being described (seven in total). These subtypes are defined by the presence of genetic alterations that are, essentially, mutually exclusive: ERG fusions (46%), ETV1 fusions (8%), ETV4 fusions (4%), FLI1 fusions (1%), SPOP mutations (11%), FOXA1 mutations (3%) and IDH1 mutations (1%)(17). However, even within these subgroups, marked genetic diversity exists in terms of mutations, copy number alterations, gene expression and DNA methylation, and several common alterations can occur across different molecular classes (for example, chromosome 8q gain or PTEN deletion). Moreover, 26% of prostate cancers could not be classified into one of these seven subgroups17, suggesting that even more genetically distinct molecular subtypes are likely to be uncovered in the future.
Androgen signaling
The androgen receptor
Androgen signalling has been the principle focus of medical treatment of prostate cancer since the discovery by Huggins and Hodges that surgical castration of men with advanced-stage prostate cancer resulted in tumour regression(18). The androgen receptor (AR) signalling axis is the most clinically targeted pathway in patients with both untreated prostate cancer (who are castration-sensitive) and in those with CRPC. Lowering serum testosterone levels, or more specifically, dihydrotestosterone (DHT) levels, activates a feedback loop that increases transcription of the AR in prostate cancer cells(19, 20). This paradoxical increase in AR expression and signalling is hypothesized to lead to DNA strand breaks, which might be responsible for the resulting AR amplifications seen in 20–55% of CRPC samples, which is the most common mechanism of developing CRPC(21, 22). In the majority of patients with newly diagnosed CRPC, AR overexpression is driven by these DNA strand breaks, resulting in X chromosome rearrangement and subsequent focal AR copy number gain(5). High-level, focal AR amplifications are almost never (in <1% of patients) identified in patients with untreated, localized prostate cancer, but as described above, are much more common in those with CRPC, usually after prolonged exposure to ADT(3-5, 10, 11, 23-27). Mutations in the AR itself are less common than amplifications of the AR and are found in 2–18% of prostate cancer specimens in mCRPC(28, 29). The resulting overexpression and mutation of AR has important clinical implications for treatment resistance, promiscuous effects of other adrenal steroids, and even the conversion of first-generation AR antagonists to agonists(30).
Owing to the dependency of prostate cancer on AR signalling, multiple clinical attempts to further suppress AR signaling activity have been made. Most notably, CYP17A1 inhibitors (such as abiraterone) and second-generation anti-androgens (such as enzalutamide ) that reduce ligand availability or compete for the ligand-binding domain (LBD) of the AR, respectively, have been developed and shown to confer survival benefits in patients with CRPC(31-35). The added benefit derived from use of second-generation anti-androgens is not only the more complete blockade of the AR compared with first-generation anti-androgens , but also the prevention of nuclear translocation of the AR and the subsequent downstream activation of AR-target genes(36, 37). Enzalutamide, the first FDA-approved, second-generation anti-androgen has demonstrated improvement in overall survival in both the pre-docetaxel and post-docetaxel setting in patients with CRPC, and is currently being tested as a treatment of patients with early-stage prostate cancer(31, 32). A structurally similar compound, ARN-509, has also shown promise in early-phase clinical trials and is current being tested in a phase III randomized clinical trial (NCT01946204)167 in patients with non-metastatic CRPC (TABLE 2)(34, 37). A third, structurally distinct, second-generation AR antagonist, ODM-201 has also shown promise in phase I clinical trials as a treatment of both metastatic and non-metastatic CRPC(38, 39). Similar to ARN-509, ODM-201 is currently being evaluated in a phase III trial in patients with non-metastatic CRPC (NCT02200614)168.
Table 2.
Select clinical trials of novel therapies in prostate cancer
| Therapy | Mechanism of action | ClinicalTrials.gov identifier |
|---|---|---|
| AR signalling | ||
| ARN-509 | Second-generation AR antagonist |
NCT01946204, NCT02200614 |
| ARN-509 + abiraterone | Second-generation AR antagonist, CYP17 inhibitor |
NCT01792687 |
| Enzalutamide + abiraterone | Second-generation AR antagonist, CYP17 inhibitor |
NCT01650194, NCT01949337 |
| Galeterone (TOK-001) | Dual CYP17 inhibitor and AR antagonist |
NCT01709734 |
| ODM-201 | Second-generation AR antagonist |
NCT02200614 |
| VT-464 | Lyase-selective inhibitor of CYP17 |
NCT02012920 |
| EPI-001/EPI-506 | N-terminal domain AR inhibitor | NCT2606123 |
| ISIS-ARRx | AR mRNA inhibitor | NCT02144051 |
| ASO EZN-4176 | Antisense oligonucleotide | NCT01337518 |
| PI3K | ||
| GDC-0068 + abiraterone | Pan-AKT inhibitor; CYP17 inhibitor |
NCT01485861 |
| BEZ235 + abiraterone | Dual PI3K and mTOR inhibitor; CYP17 inhibitor |
NCT01634061 |
| BKM120 + abiraterone | Pan PI3K inhibitor; CYP17 inhibitor |
NCT01634061 |
| AZD8186 | PI3Kβ and PI3Kδ inhibitor | NCT01884285 |
| GSK2636771 | PI3Kβ inhibitor | NCT01458067 |
| Temsirolimus + vorinostat | mTOR inhibitor, HDAC inhibitor |
NCT01174199 |
| DNA repair | ||
| Olaparib | PARP 1 inhibitor | NCT01682772 |
| Olaparib + Abiraterone | PARP 1 inhibitor, CYP17 inhibitor |
NCT01972217 |
| Abiraterone +/− veliparib | PARP inhibitor, CYP17 inhibitor | NCT01576172 |
| ABT-888 + temozolamide | PARP inhibitor, DNA alkylating agent |
NCT01085422 |
| Neuroendocrine | ||
| Alisertib (MLN8237) | AURKA inhibitor | NCT01799278 |
| Alisertib + abiraterone | AURKA inhibitor; CYP17 inhibitor |
NCT01848067 |
| Cell cycle | ||
| Ribociclib (LEE011) + docetaxel |
CDK 4/6 inhibitor | NCT02494921 |
| AZD6738 | ATR inhibitor | NCT02223923 |
| MYC | ||
| DCR–MYC | MYC inhibitor | NCT02110563 |
| ISIS–STAT3Rx | STAT3 antisense oligonucleotide inhibitor |
NCT01563302 |
| Other | ||
| SB939 | HDAC inhibitor | NCT01075308 |
| Panobinostat + bicalutamide |
HDAC inhibitor; 1st generation antiandrogen |
NCT00878436 |
| Abiraterone +/− vemurafenib |
CYP17 inhibitor; RAF inhibitor | NCT01085422 |
AR, androgen receptor; HDAC, histone deacetylase; PARP, Poly [ADP-ribose] polymerase; MYC, MYC proto-oncogene protein; STAT3, Signal transducer and activator of transcription 3.
Despite widespread clinical use of CYP17A1 inhibitors or second-generation AR antagonists in patients with CRPC, no prospectively validated clinical biomarkers that enable accurate prediction of a response to treatment currently exist. AR amplifications and/or mutations detected in circulating cell-free DNA are associated with resistance to abiraterone and enzalutamide in patients with metastatic CRPC(40). Additionally, data from transcriptome analysis has revealed that acquired resistance to ARN-509 or enzalutamide correlates with glucocorticoid receptor (GR) upregulation(41), and high levels of GR expression have been associated with resistance to neoadjuvant androgen deprivation(42). The effectiveness of combined AR and GR inhibition is being testing in an early phase clinical trial combining enzalutamide with the GR antagonist mifepristone (NCT02012296)169. The identification of truncated AR transcript isoforms, referred to as AR splice variants, or AR-Vs is a particularly exciting discovery with high relevance to predicting a response to AR-targeted therapy(43). These isoforms are missing an LBD, but retain the abilty to code for the DNA binding and transactivation domains. Furthermore, the presence of specific AR-Vs, in comparison to the full-length AR, appears to drive the development of distinct transcriptional profiles in patients with CRPC. For example, AR-V7 has been shown to upregulate a set of cell-cycle genes independent of full-length AR signalling, whereas the full-length AR induces upregulation of gene sets related to biosynthesis, metabolism and secretion that are largely not upregulated by AR-V7(44). Furthermore, these AR-V splice variants are constitutively active and are not inhibited by standard first-generation or second-generation anti-androgens. As such, AR-V expression is associated with progression to CRPC(45, 46), resistance to anti-androgen therapy(47), and poor cancer-specific outcomes(48) compared with patients who do not express AR-Vs. For example, the presence of AR-V7 in circulating tumour cells (CTCs) is associated with resistance to abiraterone and enzalutamide, as demonstrated in a small clinical cohort study(47). These findings have demonstrated that the presence of AR-Vs in CTCs is a potentially powerful biomarker for the identification of patients that are unlikely to respond to therapies targetting the full-length AR protein. Assessments of clinical grade assays, performed prospectively in Clinical Laboratory Improvement Amendment-approved laboratories will be of critical importance to determining the predictive validity of AR-V7 detection. Likewise, whether AR-V7 expression is responsible for resistance to AR-targeted therapies, or is an indicator of other processes associated with resistance (for example, AR-V7 expression is often highest in patients with AR amplification) is unclear, and AR-V7 transcripts can be detected at low levels in both benign prostate tissue biopsy specimens, and those from patients with untreated prostate cancer(3).
Following the discovery of AR-Vs that could potentially confer treatment resistance owing to constitutive activity with a lack of a LBD, a series of new compounds that target other domains of the truncated AR protein have been developed. For instance, compounds that target the N-terminal domain (EPI-001; ESSA Pharma, Vancouver, Canada, ASO EZN-4167; Enzon pharmaceuticals, Florida, USA) and the DNA-binding domain (ISIS-ARRx, Ionis pharmaceuticals, California, USA) of the AR have demonstrated early potential as viable therapies in preclinical models(49-53) and in a phase I clinical trial49. These experimental therapies will need to demonstrate an acceptable level of specificity for the AR, with few, or no off-target effects; however, tremendous hope exists that these therapies could provide clinical benefits for patients whose tumors are no longer reliant on the LBD of the AR and are therefore resistant to anti-androgen therapy. Likewise, AR-directed therapies with combined mechanisms of action might enable targetting of both the full-length AR and AR-Vs. For example, based on data demonstrating efficacy in a small number of patients with AR-V7 expression in CTCs, a phase III randomized trial (NCT02438007)170 has been initiated to test galeterone—a CYP17A1 inhibitor that also acts as a direct AR antagonist and degrades full-length and truncated AR versus enzalutamide in men with CRPC and AR-V7 expression, as confirmed by analysis of CTCs. Importantly, this trial is the first phase III trial in patients with prostate cancer to use the presence of a molecular biomarker as an inclusion criterion.
AR co-factors
A host of co-factors modulate the expression of downstream targets of the AR(54, 55). These include, but are not limited to, the forkhead protein FOXA1, the transcription factor GATA2, and the P160 steroid receptor co-activator proteins, NCoA-1, NCoA-2 and NCoA-3. Unlike AR mutations which are almost exclusively found in CRPC biopsy samples, mutations of these AR cofactors have been detected in both primary and metastatic tumours(3, 5, 11). When combining AR alterations and AR co-factor aberrations, 71% of patients with CRPC harbour AR signalling pathway aberrations(3).
FOXA1 interacts with the AR and serves as a pioneer co-factor that is capable of specifying unique AR binding sites(56). Additionally, FOXA1 is also able to a regulate metastatic potential in an AR-independent manner(57), thus development of an inhibitor of FOXA1 might hold some promise(58). GATA2 co-localizes with FOXA1 and AR on chromatin, and a complex feedback mechanism exists between AR and GATA2 whereby GATA2 promotes expression of AR and conversely, GATA2 expression is repressed by androgen and AR(59). Furthermore, preclinical data support the efficacy of Bromodomain and Extra-Terminal motif (BET) protein inhibitors, which have been shown to downregulate AR by disrupting the AR–BRD4 interaction at the N-terminal domain of the AR, thus altering gene expression by preventing interactions of this complex with chromatin(60). These results have led to the clinical assessment of BET inhibitors as a treatment of CRPC, including a phase I trial of the BET inhibitor OTX015 (Merck, New Jersey, USA) in men with CRPC (NCT02259114)171.
The P160 SRC family genes, NCOA1, NCOA2 and NCOA3 which function as steroid receptor co-activators, are recognized as important cofactors in CRPC. Even in the absence of circulating androgens, overexpression of NCOA1 or NCOA2 can drive increased AR transactivation(61). NCOA2 is amplified in approximately 6% of patients with advanced-stage prostate cancer, and increased NCOA2 function amplifies AR pathway activity(10, 62). NCOA3 overexpression is associated with tumour proliferation in patients prostate cancer, and notably, is a key target of the ubiquitin ligase SPOP(11, 63). Targeting P160 SRC proteins might prove beneficial as a treatment of CRPC and might resensitize patients to the standard treatments of CRPC. Promisingly, 65% of patients with metastatic CRPC have a potentially targetable genetic aberration through established agents, even excluding those with alteration of the AR signalling axis(3).
TMPRSS2–ETS fusions
Stimulation of the AR can bring the TMPRSS2 and the ERG gene loci into close proximity, an effect thought to be critical for the development of the TMRPSS–ERG gene fusion(64-66). Younger men (~55% in men <50 vs. ~35% in men >75) have higher incidences of structural rearrangements and ERG gene fusions than older men, resulting in the hypothesis that androgen-activated transcription might be an early driver of prostate cancer(67).
The translational relevance of TMPRSS2–ERG fusions has been demonstrated by the introduction of a urine-based early detection test (in combination with urine levels of PCA3, which is a non-coding RNA transcript) and a diagnostic, tissue-based test (using antibodies directed against ERG)(68-71). Thus, the first clinical applications of ETS fusions are as diagnostic tools, exploiting the specificity of this gene fusion to prostate cancer, rather than as a prognostic biomarker post-treatment. The prognostic utility of ERG fusions has been extensively investigated in various contexts, particularly after radical prostatectomy, with the largest published series to date indicating no utility of the presence of these fusions or ERG overexpression for prediction of biochemical recurrence(72, 73). No clinically available methods of directly inhibiting TMPRSS2–ERG signalling currently exist; although, inducible knockdown of ERG, which is endogenously expressed in the VCaP CRPC cell line has shown that ERG drives cellular proliferation and blocks differentiation of these cells to neuroendocrine or luminal cell types, supporting the clinical utility of targeting these early, driving alterations(74). Likewise, in preclinical studies investigating targetted deletion of TMPRSS2, this protein was found to promote cancer cell invasion and metastasis(75).
Inhibition of ETS cofactors, rather than the fusion product itself, has attracted increased interest as a potential therapeutic strategy. In preclinical studies, ETS-positive tumours have been shown to be more sensitive to pharmacological inhibition of targetable cofactors, including PARP1, HDAC1, and DNAPK.(72, 76). Multiple phase I and II studies have focused on the inhibition of PARP1 in patients with CRPC, most notably a randomized study, in which patients were stratified patients based on ETS fusion status (NCT01576172)172. Likewise, two phase II studies designed to investigate the effectiveness of HDAC inhibitors in patients with CRPC have been attempted, however these have yielded disappointing results(77, 78). Several other trials assessing HDAC inhibitors in patients with CRPC are still in progress, or have been completed, with results currently pending (NCT01075308, NCT00878436 and NCT01174199). Thus, prognostic and predictive relevance of ETS fusions continues to be investigated, and the results of ongoing clinical trials are highly anticipated.
Signal transduction pathways
Molecularly targeted therapies for the treatment of prostate cancer have, historically, and given the importance and ubiquity of androgen axis signalling in the pathogenesis of this malignancy, largely focused on androgen signalling pathways. Nevertheless, the importance of other signal transduction cascades has been increasingly elucidated from the findings of genomic analyses. For example, in the setting of metastatic CRPC, nearly 50% of tumours carry PI3K abnormalities, and 18% have MAPK or WNT alterations(3). Furthermore, 100% and 90% of metastatic tumours have upregulated PI3K and MAPK signalling, respectively, according to gene expression analyses(10).
PI3K Pathway
Outside of the androgen signalling axis, the PI3K signalling cascade is the most commonly dysregulated signal transduction pathway in patients with prostate cancer. The PI3K pathway is a critical regulator of proliferation, survival, metabolism, angiogenesis, and immune function. Hyperactivation of the pathway through loss of PTEN, which encodes a lipid phosphatase that acts as a negative regulator of PI3K signalling is by far the most common PI3K aberration observed in patients with prostate cancer(3, 10). Homozygous PTEN deletions were present in 15% of primary prostate cancers in the TCGA dataset, which is one of the highest incidences of this deletion among any tumour type, and PTEN mutations were present in another 2%17. PTEN alterations are even more common in the setting of metastatic disease, with over 40% of tumours having PTEN mutation or loss(3, 5, 10, 17). PTEN loss has been associated with a poor clinical outcome in numerous studies compared with patients whose tumours express wild-type PTEN(79-83), with some(82), but not all(83), suggesting that the prognostic value of PTEN loss is dependent on ERG-fusion status. PI3K signalling and androgen signalling have been suggested to be reciprocally regulated(84), and treatment with combination therapy is, therefore, likely to be necessary to overcome intrinsic and acquired resistance to single-agent therapy, given that the findings of trials designed to investigate the efficacy of both pan-PI3K and dual PI3K–mTOR inhibitors have been disappointing to date(85, 86).
Wnt pathway
Alterations in the Wnt signalling pathway are also common in prostate cancer, both in the primary setting and in patients with metastatic CRPC, where it is present in approximately 18% of patients(3, 5, 17). Canonical Wnt signalling is an evolutionarily conserved pathway that has been implicated in cancer stem-cell maintenance, epithelial-to-mesenchymal transition, embryonic development and homeostasis in adults(87). Alterations have been observed in multiple nodes of the Wnt-signaling pathway in patients with prostate cancer, including recurrent mutations in CTNNB1, the gene encoding the central mediator of the Wnt-pathway β-catenin(3, 17), and APC, which forms part of the multiprotein deconstruction complex that regulates β-catenin levels(3, 5, 88). Also, mutations in the ubiquitin ligases RNF43, ZNRF3, and in RSPO2, an activator of Wnt signalling via LGR4-6 activation were observed in 6% of patients with mCRPC in a study published in 2015(3). The presence of mutations in these genes in patients with metastatic CRPC was mutually exclusive of APC alterations(3). By profiling paired pre-treatment and post-treatment prostate cancer biopsy samples, investigators have demonstrated that Wnt-pathway activating alterations (such as CTNNB1 mutation or amplification) are adaptive responses to therapy, similar to AR mutations/amplifications(88, 89).
Multiple Wnt-pathway targeting agents are currently in clinical development, although none are being specifically studied as treatments of prostate cancer, to date(90). These agents include the small molecule PRI-724, an inhibitor of the interaction between β-catenin and one of its co-activators in the nucleus, CREB binding protein(91), (NCT01302405, NCT01764477, NCT01606579) and LGK-974 (Novartis, Basel, Switzerland), a small molecule that inhibits PORCN, a key regulatory protein in the Wnt signalling pathway, thereby impairing Wnt ligand secretion (NCT01351103). Prostate cancers with mutations in RNF43, ZNRF3, and RSPO2 would be expected to respond to these agents(92). Several molecular therapies targeted to the Wnt-ligand-binding Frizzled receptor are also being investigated in various cancers (NCT01973309, NCT01957007, NCT02005315, NCT02069145, NCT02092363, NCT02050178, and NCT01469975).
MAPK Pathway
The MAPK/ERK pathway, which is implicated in cell survival, cell-cycle progression, tumour dissemination, and resistance to therapy in multiple cancers(93), also seems to have a role in a subset of prostate cancers, and data from preclinical research suggests an association with more aggressive disease in certain genetic contexts(94). BRAF is the most commonly altered MAPK gene in prostate cancer, with gene fusions or activating mutations present in 2–3% of tumours(3, 5, 17). Interestingly, canonical BRAFV600E mutation was not observed in prostate cancer specimens analyzed in the TCGA dataset, although this mutation has been described, rarely, in other studies(10, 17). Additionally, no activating BRAF rearrangements were found in the TCGA study17, despite these having been described previously in prostate cancer(95). Given the rarity of BRAFV600E mutations in prostate cancer, less specific BRAF or MEK inhibitors might prove to have more clinical utility in patients with prostate cancer than the currently available mutant BRAF inhibitors such as vemurafenib.
IDH1 Signaling
Approximately 1% of primary prostate cancers are defined by the presence of isocitrate dehydrogenase 1 (IDH1) ArgR132His hotspot mutations(17), this critical finding from the TCGA prostate cancer study represents a novel prostate cancer molecular subtype and confirms previous observations of these mutations in prostate cancer(88, 96-98). These IDH1 gene mutations generally result in a loss of function, owing to the conversion of isocitrate to 2-ketoglutarate. Instead, the altered enzyme adopts an abnormal function: production of D-2-hydroxyglutarate(99). These genetic changes have been found to inhibit the enzymatic function of many α-ketoglutarate-dependent dioxygenases, including histone and DNA demethylases, resulting widespread changes in the epigenome.
The presence of these IDH1 mutations appears to define a rare, unique subset of early onset prostate cancer, with relatively few copy number alterations and even higher levels of genomic hypermethylation than IDH1-mutant glioblastoma or acute myelocytic leukaemia (AML)(100, 101). Notably, in both AML and glioblastoma, patients with IDH1 mutations have a markedly better prognosis than those with wild-type IDH1(102, 103). Whether or not the presence of IDH1 mutations also portends an improved prognosis for patients with prostate cancer is worthy of future investigation, given the significant potential implications for active surveillance and treatment recommendations in men harbouring tumours with this alteration. Additionally, patients with IDH1 mutations might be candidates for treatment with one of the various IDH1-specific therapies that are currently being developed(103, 104).
Cell cycle/proliferation
Similar to many other cancers, alterations in genes encoding proteins involved in regulation of the cell cycle have an important role in prostate cancer. For example, TP53 is mutated or deleted in 8% of primary prostate cancers and up to 53% of metastatic CRPCs(3, 5, 10, 17, 88). TP53 encodes the p53 transcription factor that, in response to DNA damage, activates the G1–S cell-cycle checkpoint and the apoptotic signalling cascade(105). Furthermore, other genes encoding important inhibitors of cell-cycle progression at G1–S, including CDKN2A, CDKN2B and CDKN1B, are each lost in about 2–3% of primary prostate cancers(17). RB1, another critical negative regulator of the G1–S checkpoint that is responsible for repressing the E2F family of transcription factors(106), is also commonly lost in metastatic CRPC(3, 10, 88) (most specifically in small-cell carcinoma); whereas CCND1, the gene encoding cyclin D1, which is an activator of cell-cycle progression through G1–S transition, is recurrently amplified(3, 88). Thus, dysfunction of the G1–S checkpoint, potentially owing to a variety of different genetic aetiologies, is a frequently ocurring molecular event in patients with prostate cancer, particularly in those with metastatic CRPC.
Despite being perhaps the most commonly altered set of genes across all cancers, clinical therapeutic strategies targeting cancer cells harbouring deficiencies in cell-cycle regulation (besides non-selective chemotherapy) have been lacking . However, multiple new agents are moving into the clinic and have shown promising results in the metastatic setting. For example, the cyclin-dependent kinase (CDK) 4/6 inhibitor palbociclib in combination with anti-oestrogen therapy has demonstrated impressive activity in patients with hormone-receptor positive breast cancer(107, 108). CDK4/6 inhibitors are expected to be particularly effective in tumours harbouring inactivating CDKN2A/B mutations or CCND1 amplification, both of which are recurrent alterations present in primary and, more commonly, metastatic CRPC. Three registered clinical trials currently exist to investigate the effectiveness of these agents in patients with prostate cancer: ribociclib in combination with docetaxel in patients with metastatic CRPC (NCT02494921), enzalutamide +/− ribociclib in patients with metastatic CRPC (NCT02555189), and ADT +/− paclociclib in patients with hormone-sensitive prostate cancer, in which patients with RB1 wild-type tumours were pre-selected for recruitment, which is the first example of biomarker-driven recrutiment of patients with hormone-sensitive prostate cancer (NCT02059213).
Targeting cells with p53 deficiency has long been a goal of oncologists, but has proven difficult in clinical practice(109). Given that p53 is a fundamental regulator of the G1–S checkpoint, one strategy for targeting cells with this deficiency could be to exploit their hypothesized increased reliance on the G2–M checkpoint for DNA damage repair. Inhibitors of the G2–M regulatory proteins ATR(110, 111), Wee1(112, 113) and Chk1(114, 115) have been shown to have increased activity when used as single agents, and also promote sensitization of p53-deficient cancer cells to DNA-damaging agents. Inhibitors of each of these molecules have entered early phase clinical testing in various cancer types(116, 117) (NCT02223923) and might be promising in patients with metastatic CRPC.
In addition to G1–S checkpoint aberrations, localized prostate cancers might also harbour amplification of one of the three MYC isoforms that also promote progression from G1–S phase: MYC (also known as c-MYC), MYCL, or MYCN(8),(17). Like p53, MYC has long been considered undruggable, and direct targeting is challenging with standard pharmaceutical approaches(118). Nevertheless, in a novel approach to this challenge, DCR-MYC, a first-in-class Dicer substrate small interfering double-stranded RNA targeted to the MYC oncogene in a lipid nanoparticle suspension is currently being tested in early phase clinical trials (NCT02110563, NCT02314052). Thus far, this therapy has been well tolerated and showed some patients have had promising responses to treatment in early clinical testing(119). In addition to this direct therapeutic targeting strategy, several indirect experimental therapies have shown promising results in preclinical testing in patients with MYC-driven tumours, such as PIM inhibition(120), inhibition of the bromodomain-protein BET(121), exploiting replication stress in MYC-expressing cancer cells via ATR inhibition(122), targeting MYC-upregulating STAT proteins via an antisense nucleotide to STAT3 (NCT01563302) or direct inhibition the STAT5–SH2 binding domain(123).
DNA damage repair/heredity
DNA Repair
The DNA damage repair (DDR) response is governed by input from a diverse array of signalling cascades that are integral to the maintenance of genomic integrity(124). Many proteins involved in DDR act as tumour suppressors, thus preventing the formation and propagation of both mutations and copy number alterations. Genomic instability via loss of DDR proteins is common in prostate cancers compared with many other types of cancer, owing to both somatic and germline alterations. For example, approximately 19% of primary prostate cancers and 23% of patients with mCRPC have been reported to harbour inactivating mutations in DDR genes(3, 5, 17, 88). Notably, certain DDR deficiencies appear to be much more common in patients with meatastatic CRPC than in those with primary prostate cancer. For example, approximately 20% of meatastatic CRPC primary tumour samples have been reported to harbour germ-line and/or somatic aberrations in BRCA2, BRCA1, or ATM(3, 5, 88, 125), whereas only ~3% of tumours from those with primary prostate cancer had germline or somatic alterations in BRCA1 and BRCA2(17).
Losses or mutations of BRCA2, ATM, and BRCA1 are the most commonly reported mutations in DDR genes in patients with prostate cancer(3, 5, 17, 88). The wild-type forms of these genes are important components of homologous recombination, a high fidelity DDR process that utilizes the sister chromatid as a template during G2/M to excise and replace defective stretches of DNA in an error-free manner. Mutations in other components of the homologous recombination response have also been described, including those in PALB2, RAD51B and RAD51C(3, 17). Thus, defective homologous recombination is a recurrent motif in the development of prostate cancer, particularly in the advanced-stage or castration-resistant setting. Furthermore, data from a large pan-cancer analysis revealed a mutational signature correlating with mismatch repair deficiency in a subset of prostate cancers(126), and loss-of-function genomic lesions in mismatch repair proteins (most frequently somatic mutations and deletions in MSH2) have been described in several datasets(3, 5, 127-129). Given the high mutation load of their tumours, patients that have prostate cancer with mismatch repair deficiencies might be good candidates for investigation of immunotherapeutic approaches such as CTLA-4 or PD-1 inhibition(130, 131).
Currently, defective or lost DDR proteins cannot be directly targeted pharmacologically; although, these aberrations might create tumour specific vulnerabilities that can be exploited via the principal of synthetic lethality. This process occurs when two cellular pathways are inhibited simultaneously resulting in cell death, but inhibition of either pathway by itself is not lethal. When DNA repair function is intact, single-strand DNA breaks are repaired through the base excision repair pathway. Poly (ADP-ribose) polymerase (PARP) is a key component of the base excision repair pathway, and inhibition of this protein results in an increased frequency of single-strand DNA breaks and eventually the creation of double-strand DNA breaks during replication. Normally, these double-strand breaks are repaired by homologous recombination. However, if cells have mutations in genes that encode proteins that control homologous recombination (such as BRCA) then DNA replication is likely to fail. The combination of PARP inhibition in the presence of BRCA mutations impairs these two damage repair pathways, thus causing selective synthetic lethality(132).
The clinical relevance of synthetic lethality was originally demonstrated by the discovery that breast and ovarian cancers with BRCA deficiencies are exquisitely sensitive to PARP inhibition(133-135). These findings might have relevance for prostate cancer as well. Data from a phase II trial with a cohort that contained eight patients with germline BRCA1 or BRCA2 mutations and recurrent cancer demonstrated that half of these patients with advanced-stage prostate cancer had responses(136). Even more promisingly, following the TOPARP-A trial, investigators reported that 14 of 16 (87.5%) patients with mCRPC and homozygous deletions or deleterious mutations in DNA-damage repair genes such as BRCA1 or BRCA2, ATM, CHEK2, FANCA and PALB2 responded to olaparib, in comparison to two of 33 (6%) without these aberrations(137). This high level of specificity of genomic alterations to a targeted therapy is unprecedented in patients with prostate cancer. Serious (grade ≥3) treatment-related adverse events included anaemia (in 10 patients), fatigue (in six patients), leukopenia (in three patients), thrombocytopenia (in two patients) and neutropenia (in two patients). Multiple other trials designed to investigate the effectiveness of PARP inhibitors in patients with prostate cancer are currently underway or have been completed in the past 5 years (NCT01576172, NCT01085422, NCT01972217) and a complete response in a patient with deficiencies in DDR genes who was treated with a PARP inhibitor (with or without other therapies) has also been described(138).
In addition to predicting a response to targeted DDR inhibition, tumours with genomic deficiencies in homologous recombination or Fanconi anaemia proteins might also be more responsive to platinum-based chemotherapy(139, 140). Platinum-based chemotherapy is currently rarely used in patients with prostate cancer (except in those with small-cell or neuroendocrine carcinoma) given the lack of benefit derived from this treatment by unselected patients. However, this might be a promising approach for patients with mCRPC harbouring genetic DDR pathway alterations, thus an evaluation of the effectiveness of PARP inhibitors compared with that of chemotherapy might be warranted.
The existence of a novel interplay between DNA repair and AR signalling was described in 2013(141). Inhibition of the AR seems to supress non-homologous end joining, and to reduce the functional ability of the DNA-dependent protein kinase catalytic subunit(142). Furthermore, genotoxic stress (such as radiotherapy) not only causes DNA damage, but also upregulates AR signalling and pro-survival pathways to mediate treatment resistance(143). Further research is ongoing to better characterize how to optimally utilize this information to enable the rational combination of radiotherapy with ADT (NCT02297386). Additionally, an important interplay exists between PARP and the AR given that PARP-1 is recruited to sites of AR function and supports AR transcriptional function, suggesting that PARP should be further investigated as an treatment target in patients with prostate cancer(141, 142).
[H3] Heredity
Data from genomic studies have confirmed that, in addition to the somatic mutations present in prostate cancer, germline aberrations in DDR genes also have a critical influence on genetic predisposition to prostate cancer(3, 144). BRCA2 was one of the first mutated genes to be associated with an increased risk of prostate cancer(145-147). Furthermore, the presence of BRCA1 mutations also increases the risk of prostate cancer, albeit to a lesser extent than the presence of BRCA2 aberrations(148). Additionally, men with germline mutations in mismatch repair genes have approximately twice the risk of developing prostate cancer as unaffected individuals, with a cumulative risk of approximately 30% by the age of 80, compared with around 18% in the age-matched general population 142. Other genetic alterations have been identified that confer an increased familial risk of prostate cancer (such as HOXB13 mutations), however, these mutations usually occur at low frequencies(149).
Identifying men who have a strong genetic predisposition to prostate cancer has important clinical implications. For example, these men might be eligible for more intense screening, or lowering the serum PSA threshold that indicates a need for biopsy sampling. Unfortunately, no reliable evidence exists that pharmacological prevention, such as use of finasteride, selenium, and/or vitamins can prevent, or reduce the risk of developing high grade cancers(150),(151). Thus, no pharmacological strategy for prostate cancer prevention can currently be recommended. The role of even more aggressive risk reduction, such as prophylactic prostatectomy, is also unclear. Further study of screening and prevention of prostate cancer in men with a high genetic predisposition to prostate cancer is an important avenue for future investigation.
Tumour heterogeneity and evolution
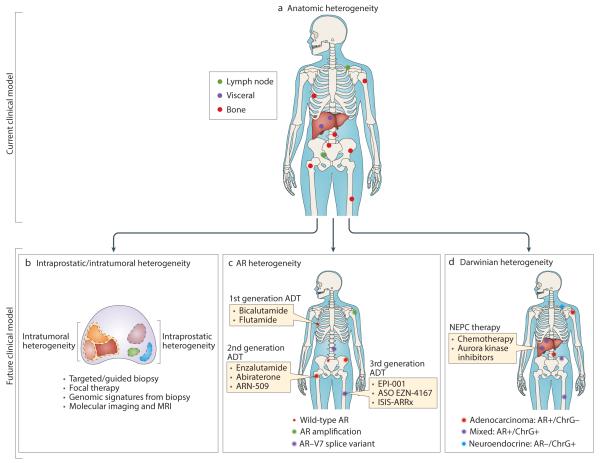
The substantial interpatient tumour heterogeneity observed among patients with prostate cancer is highlighted by the variety of different molecular aberrations present across different cancers (FIG 1)(3, 10). This interpatient heterogeneity has been a major challenge in identifying effective therapies in ‘all-comers’ in randomized trials, given that the potential benefit derived from use of a targeted treatment in selected patients harbouring a predictive genomic alteration might be masked if the alteration is only present in a small subset of those enrolled. Nevertheless, intra-patient heterogeneity might also pose hurdles to the personalization of treatment. In patients with localized prostate cancer, the majority of prostate cancers are known to be multifocal(152, 153). Furthermore, although these multifocal tumours have often been identified, using either single-gene and/or whole-exome sequencing, to be genetically distinct, suggesting an independent origin, non-driving somatic mutations might be shared between tumor foci and even among histologically ‘normal’ prostate tissue samples(7, 8, 154). Moreover, lethal metastases have been reported to arise from a minor population of subclones, including dissemination from an organ-confined, low-grade area of a bulky, high-grade primary tumour(155). Likewise, data from sequential profiling of circulating cell-free DNA has demonstrated that the basic prostate cancer molecular subtype (defined by ERG-fusion status) might change in response to selective pressure induced by anti-androgen therapy(156). Therefore, the use of targeted biopsy sampling using image guided techniques (CT, MRI, and/or ultrasound guided), both in patients with localized, and mCRPC will need to be carefully evaluated to determine whether or not this approach can capture the dominant biology of any one patient’s cancer. Additionally, the delivery of focal therapy to the prostate gland, such as focal brachytherapy or cryotherapy, might be unwise until a method to readily detect the driver foci that are responsible for metastatic spread is developed.
Figure 1. Types and implications of prostate cancer heterogeneity.
a ∣ The clinical and anatomical heterogeneity of metastatic prostate cancer fails to capture the immense biological heterogeneity of prostate cancer. b ∣ Intraprostatic/intratumoral heterogeneity, c ∣ Androgen receptor heterogeneity, and d ∣ Darwinian heterogeneity demonstrate clinical strategies and obstacles regarding treatment and diagnosis.
Given the extensive intratumoural and intertumoural heterogeneity observed in patients with prostate cancer, in line with Darwinian theories of evolution, certain subclones containing a selective growth or survival advantage in comparison to others are likely to proliferate and form the majority of the tumour population throughout the natural history of the disease, and in response to selective pressures. Eventually, in most patients with lethal prostate cancer, a subset of tumour subclones gains the ability to disseminate to distant organs. Furthermore, the introduction of therapeutic interventions, which provide new pressures on tumour selection, can radically reshape the subclonal composition of a tumour(156). Thus, the tumour genome and epigenome are in a dynamic state of selection pressure and evolutionary drift as the tumour continually divides and new mutations arise.
Some of the most interesting insights to emerge from genome sequencing projects involve tracking the evolution of prostate cancer from a locally invasive process into a disseminated malignancy. For example, data from several copy number analyses and genome sequencing studies from individual patients have suggested that, despite the multifocal and multiclonal nature of primary prostate cancers, lethal metastases might arise from a single subclone(155, 157). However, studies published in the past 2 years, with data from multiple patients, suggest that prostate cancer metastases can arise from either a single, or several subclones within the primary tumour (6, 156). Furthermore, the complexity of metastatic spread patterns of prostate cancer has been highlighted by the findings of genomic studies. In addition to the classical primary-to-metastasis model, high rates of metastasis-to-metastasis, and even metastasis-to-surgical bed, spread have been observed in heavily treated patients(6, 158). In summary, these data support the hypothesis that, rather than only unidirectional seeding from the primary to metastatic sites, metastatic CRCP is truly a systemic disease arising from a subclone or subclones with multidirectional spread of subclones between all sites of malignancy(159). This hypothesis has clinical implications for treating both the primary, as well as metastatic sites in order to limit the extent of any further metastatic progression. Given that not all metastatic spread arises from the primary tumour, ablation of oligometastases in patients in whom the primary tumour is eradicated might, in theory, decrease the reservoir of subclones containing the ability to disseminate, thereby slowing the progresion of metastatic spread.
Multiple lines of evidence suggest that tumours evolve in response to treatment(156, 158). For example, certain aberrations such as AR mutations and amplifications that are common in prostate biopsy samples from patients with mCRPC are almost never detected in similar samples from patients with hormone-naïve prostate cancer(3, 5, 6, 160). Additionally, the relative proportions of circulating DNA from the various tumour subclones markedly changes over time in response to treatment, with those containing mutations that are known to confer treatment resistance emerging during progression, whereas others regress during treatment responses, thus revealing a complex dynamic of temporal and spatial heterogeneity150 . In many cases these resistant clones are already extant in small numbers prior to treatment, rather than arising de novo, and eventually become the dominant population under selection pressure. For example, in the TCGA study, researchers detected AR splice variants, including the AR-V7 splice variant that is associated with resistance to anti-androgen therapy, and is present at low levels in biopsy samples from normal prostate tissue and from androgen-naïve prostate cancers(17). Given that diversity often exists with respect to therapeutic resistance mechanisms across different metastatic sites, a challenge can occur when a targeted therapy is adequately controlling the majority of the patient’s disease, but one or more foci are discordantly progressing. One potential method that is of clinical interest, but needs to be rigorously tested in context of clinical trials, is to maintain patients on targeted therapies whilst also controlling the emergence of metastatic tumour clones that harbour acquired resistance using ablative treatments. Stereotactic body radiotherapy (SBRT) is one form of ablative therapy that allows high doses of radiotherapy to be delivered to the tumour and avoids exposing normal tissue to radiation. This technique holds promise for treatment of patients in the metastatic setting that should be tested prospectively.
The dynamic intratumoural heterogeneity observed in patients with prostate cancer, along with its fundamental intertumoural diversity, is a fundamental challenge to the personalized management of patients with this disease. Of particular relevance, targeting early driving (‘truncal’) alterations in prostate cancer, such as ERG gene fusions or SPOP mutations, is currently not possible. Given the assumption that prostate cancer metastases might arise from a minor subclonal population, attempting to utilize molecular sequencing to improve prognostication will be extremely challenging. Hence, determining both common and rare patterns of tumour progression is a critical area of need for clinicians treating patients with prostate cancer. Such studies, which must track the emergence lethal clones from the primary tumour to ADT-refractory CRPC, through to disease that is refractory to second-line anti-androgen therapies, are complicated by the long follow-up duration that is required to obtain such samples. Likewise, studies assessing tumour tissue histology, circulating tumour cells and circulating DNA are required to determine the most informative (and clinically relevant) approaches.
The molecular diversity of metastatic prostate cancer generally indicates that achieving long-term remission from treatment with agents targeted to a single pathway is unlikely, and the clinical effectiveness of combination strategies, possibly using orthogonal targets or synergistic combinations, should be investigated(161). In addition, the dynamic nature of prostate cancer genomics supports the repeated, longitudinal evaluation of the genomic characteristics of a patient’s tumour over time, and during treatment(156). For example, all rapid autopsy studies conducted on heavily treated patients with mCRPC in the pre second-generation antiandrogen therapy era demonstrate uniform ETS fusion status within an individual patient’s metastases(5, 6, 157, 162), assessment of cell-free DNA supports dynamic ETS fusion status upon treatment with second generation anti-androgens(156). Hence, non-invasive methods (coupled with tissue-based assessments) are being actively investigated in order to capture the spectrum of a patient’s tumour heterogeneity in response to treatment in real-time through assessment of circulating tumour cells, cell-free DNA, urine and serum biomarkers, and molecular imaging.
Neuroendocrine prostate cancer
Small-cell carcinoma of the prostate can exist de novo within the prostate and patients with this type of prostate cancer typically have a shorter overall survival compared with those who have prostate adenocarcinoma(163). Additionally, prostate adenocarcinomas can also progress to lose expression of active AR signalling markers (such as serum PSA) with, or without small-cell carcinoma differentiation, thus likely reflecting a truly AR-independent phenotype and a terminally differentiated state (3, 5, 128, 164). Clinically, patients with this type of prostate cancer have metastases in atypical visceral sites, and abnormally low serum PSA levels(125, 164-166). The terminology of neuroendocrine prostate cancer (NEPC) is evolving and this type of prostate cancer is clearly a heterogeneous spectrum of de-differentiation, particularly as patients are increasingly being treated with therapies designed to target androgen signalling(125, 164, 167). The exact incidence of NEPC is unclear, and ranges from 1–32% after treatment with enzalutamide or abiraterone(3, 168).
Distinct genomic alterations are likely required to enable prostate cancer to transition from an AR-dependent adenocarcinoma to an AR-independent disease state (with or without overt neuroendocrine differentiation, as defined by marker expression). Some of the proposed and identified aberrations include loss of TP53 and RB1, and gain of AURKA, MYCN, MYCL and PEG10, a gene that encodes an anti-apoptotic signalling protein(88, 125, 164, 169-172). Co-amplification of the genes AURKA and MYCN, although not consistently observed, has been reported in up to 40% of patients with NEPC(164). Based on these findings, a phase II trial of an AURKA inhibitor MLN8237 in patients with NEPC (NCT01799278) is underway. With the continued understanding of neuroendocrine/small-cell prostate cancer, and the process of de-differentiation, the current list of target genes (such as MYCN, AURKA and PEG10) will likely expand. Owing to a lack of functional reliance on AR signalling, typical AR-based therapies are largely ineffective in patients who exclusively harbour these aggressive disease variants, and novel treatments are desperately needed.
Conclusions
In summary, improvements in multiplatform sequencing technologies have revolutionized research into prostate cancer genetics, enabling unprecedented insight into the biology of this disease. This knowledge has elucidated promising opportunities for personalized treatment interventions, and has also highlighted the formidable hurdles in managing a dynamically evolving heterogeneous disease. Developing novel avenues for applying this knowledge regarding genomics to the clinical care of patients with prostate cancer in order to increase therapeutic effectiveness while limiting adverse treatment sequelae is, undoubtedly, a major challenge to clinical oncology over the coming years. To overcome these hurdles, continued multidisciplinary integration of basic science, genomics, bioinformatics, industry research and clinical practice will be necessary in order to advance the clinical science of prostate cancer.
Acknowledgements
S.A.T. is supported by the Department of Defense (PC130652 and PC141474), the National Institutes of Health (R01 CA 183857). F.Y.F. and S.A.T. are supported by the A. Alfred Taubman Medical Research Institute, the Prostate Cancer Foundation, the Evans Foundation and the University of Michigan Prostate Cancer S.P.O.R.E. (P50 CA186786) D.E.S. is also supported by the Prostate Cancer Foundation.
Footnotes
Author contributions
D.E.S., Z.S.Z. and S.A.T. researched data for this article. All authors made a substantial contribution to discussions of content, wrote the manuscript, and reviewed and/or edited the manuscript prior to submission.
Competing interests statement
F.Y.F. has acted as a consulted for, and received honoraria from Astellas, Medivation, Celgene, Myriad, and GenomeDx Biosciences. The University of Michigan has been issued a patent on the use of ETS gene fusions in patients with prostate cancer, on which S.A.T. is listed as a co-inventor; similarly, a patent application on the use of SPINK1 in patients with prostate cancer has also been filed. The diagnostic field of use has been licensed to Gen-Probe (California, USA), which has sublicensed rights to Ventana Medical Systems (Arizona, USA). S.A.T. has consulted for, and received honoraria from AbbVie, Astellas, Jannsen and Ventana Medical Systems. S.A.T. has had a sponsored research agreement with, and has received travel support from ThermoFisher Scientific/Life Technologies. S.A.T. is a co-founder and equity holder in Strata Oncology. D.E.S. and Z.S.Z. declare no competing interests.
References
- 1.Mohler J, Armstrong A, Bahnson R. NCCN clinical practice guidelines for prostate cancer [Internet] 2015.
- 2.Spratt DE, Pei X, Yamada J, Kollmeier MA, Cox B, Zelefsky MJ. Long-term survival and toxicity in patients treated with high-dose intensity modulated radiation therapy for localized prostate cancer. International Journal of Radiation Oncology* Biology* Physics. 2013;85(3):686–92. doi: 10.1016/j.ijrobp.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, Montgomery B, Taplin M-E, Pritchard CC, Attard G. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 2015;161(5):1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M. Punctuated evolution of prostate cancer genomes. Cell. 2013;153(3):666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, Brewer DS, Kallio HM, Högnäs G, Annala M. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015 doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, Kremeyer B, Butler A, Lynch AG, Camacho N. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nature genetics. 2015;47(4):367–72. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nature genetics. 2015 doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 9.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat J-P, White TA, Stojanov P, Van Allen E, Stransky N. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature genetics. 2012;44(6):685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lalonde E, Ishkanian AS, Sykes J, Fraser M, Ross-Adams H, Erho N, Dunning MJ, Halim S, Lamb AD, Moon NC. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. The Lancet Oncology. 2014;15(13):1521–32. doi: 10.1016/S1470-2045(14)71021-6. [DOI] [PubMed] [Google Scholar]
- 13.Barbieri CE, Tomlins SA, editors. Urologic Oncology: Seminars and Original Investigations. Elsevier; 2014. The prostate cancer genome: perspectives and potential. [DOI] [PubMed] [Google Scholar]
- 14.Demichelis F, Fall K, Perner S, Andrén O, Schmidt F, Setlur S, Hoshida Y, Mosquera J, Pawitan Y, Lee C. TMPRSS2: ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26(31):4596–9. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 15.Tomlins SA, Alshalalfa M, Davicioni E, Erho N, Yousefi K, Zhao S, Haddad Z, Den RB, Dicker AP, Trock BJ. Characterization of 1577 Primary Prostate Cancers Reveals Novel Biological and Clinicopathologic Insights into Molecular Subtypes. European urology. 2015 doi: 10.1016/j.eururo.2015.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ. Integrative molecular concept modeling of prostate cancer progression. Nature genetics. 2007;39(1):41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research Network Electronic address scmo, Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–25. doi: 10.1016/j.cell.2015.10.025.. PubMed PMID: 26544944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huggins C, Hodges CV. Studies on prostatic cancer. Cancer research. 1941;1:297. [Google Scholar]
- 19.Wolf DA, Herzinger T, Hermeking H, Blaschke D, Hörz W. Transcriptional and posttranscriptional regulation of human androgen receptor expression by androgen. Molecular Endocrinology. 1993;7(7):924–36. doi: 10.1210/mend.7.7.8413317. [DOI] [PubMed] [Google Scholar]
- 20.Knuuttila M, Yatkin E, Kallio J, Savolainen S, Laajala TD, Aittokallio T, Oksala R, Häkkinen M, Keski-Rahkonen P, Auriola S. Castration induces up-regulation of intratumoral androgen biosynthesis and androgen receptor expression in an orthotopic VCaP human prostate cancer xenograft model. The American journal of pathology. 2014;184(8):2163–73. doi: 10.1016/j.ajpath.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 21.Mathas S, Misteli T. The dangers of transcription. Cell. 2009;139(6):1047–9. doi: 10.1016/j.cell.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, Rosenfeld MG. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139(6):1069–83. doi: 10.1016/j.cell.2009.11.030. PubMed PMID: 19962179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocrine reviews. 2004;25(2):276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 24.Bubendorf L, Kononen J, Koivisto P, Schraml P, Moch H, Gasser TC, Willi N, Mihatsch MJ, Sauter G, Kallioniemi O-P. Survey of gene amplifications during prostate cancer progression by high-throughput fluorescence in situ hybridization on tissue microarrays. Cancer Research. 1999;59(4):803–6. [PubMed] [Google Scholar]
- 25.Miyoshi Y, Uemura H, Fujinami K, Mikata K, Harada M, Kitamura H, Koizumi Y, Kubota Y. Fluorescence in situ hybridization evaluation of c-myc and androgen receptor gene amplification and chromosomal anomalies in prostate cancer in Japanese patients. The Prostate. 2000;43(3):225–32. doi: 10.1002/(sici)1097-0045(20000515)43:3<225::aid-pros9>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 26.Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinänen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi O-P. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nature genetics. 1995;9(4):401–6. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 27.Linja MJ, Savinainen KJ, Saramäki OR, Tammela TL, Vessella RL, Visakorpi T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer research. 2001;61(9):3550–5. [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research N The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–25. doi: 10.1016/j.cell.2015.10.025.. PubMed PMID: 26544944; PMCID: 4695400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, Beltran H, Abida W, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel EA, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis F, Robinson BD, Schiffman M, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–28. doi: 10.1016/j.cell.2015.05.001.. PubMed PMID: 26000489; PMCID: 4484602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nature medicine. 2004;10(1):33–9. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 31.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, Iversen P, Bhattacharya S, Carles J, Chowdhury S. Enzalutamide in metastatic prostate cancer before chemotherapy. New England Journal of Medicine. 2014;371(5):424–33. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cabot RC, Harris NL, Rosenberg ES, Shepard J-AO, Cort AM, Ebeling SH, McDonald EK, Scher HI, Fizazi K, Saad F. Increased survival with enzalutamide in prostate cancer after chemotherapy. New England Journal of Medicine. 2012;367(13):1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 33.De Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB, Jr, Saad F. Abiraterone and increased survival in metastatic prostate cancer. New England Journal of Medicine. 2011;364(21):1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rathkopf DE, Antonarakis ES, Shore ND, Tutrone R, Alumkal JJ, Ryan CJ, Saleh MN, Hauke RJ, Bandekar R, Maneval EC. Abstract CT239: ARN-509 in patients with metastatic castration-resistant prostate cancer (mCRPC) previously treated with abiraterone acetate (AA) Cancer Research. 2014;74(19 Supplement):CT239–CT. doi: 10.1158/1078-0432.CCR-16-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, Fizazi K, Mainwaring P, Piulats JM, Ng S. Abiraterone in metastatic prostate cancer without previous chemotherapy. New England Journal of Medicine. 2013;368(2):138–48. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324(5928):787–90. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clegg NJ, Wongvipat J, Joseph JD, Tran C, Ouk S, Dilhas A, Chen Y, Grillot K, Bischoff ED, Cai L. ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer research. 2012;72(6):1494–503. doi: 10.1158/0008-5472.CAN-11-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fizazi K, Massard C, Bono P, Jones R, Kataja V, James N, Garcia JA, Protheroe A, Tammela TL, Elliott T. Activity and safety of ODM-201 in patients with progressive metastatic castration-resistant prostate cancer (ARADES): an open-label phase 1 dose-escalation and randomised phase 2 dose expansion trial. The lancet oncology. 2014;15(9):975–85. doi: 10.1016/S1470-2045(14)70240-2. [DOI] [PubMed] [Google Scholar]
- 39.Fizazi K, Massard C, James ND, Culine S, Jones R, Oksala R, Moilanen A, Aho E, Ravanti L, Kallio P. ODM-201, a new generation androgen receptor inhibitor for castration-resistant prostate cancer: preclinical and phase I data. J Clin Oncol. 2013;31(6 suppl):65. [Google Scholar]
- 40.Azad AA, Volik SV, Wyatt AW, Haegert A, Le Bihan S, Bell RH, Anderson SA, McConeghy B, Shukin R, Bazov J. Androgen receptor gene aberrations in circulating cell-free DNA: biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clinical Cancer Research. 2015;21(10):2315–24. doi: 10.1158/1078-0432.CCR-14-2666. [DOI] [PubMed] [Google Scholar]
- 41.Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, Shah N, Cai L, Efstathiou E, Logothetis C. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155(6):1309–22. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nature Reviews Cancer. 2015 doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocrine-related cancer. 2011;18(5):R183–R96. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, Edwards J, Isaacs WB, Nelson PS, Bluemn E. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer research. 2012;72(14):3457–62. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, Han M, Partin AW, Vessella RL, Isaacs WB. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer research. 2009;69(1):16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, Page ST, Coleman IM, Nguyen HM, Sun H. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. The Journal of clinical investigation. 2010;120(8):2715. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. New England Journal of Medicine. 2014;371(11):1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hornberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A, Bergh A, Wikstrom P. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PloS one. 2011;6(4):e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto Y, Loriot Y, Beraldi E, Zhang F, Wyatt AW, Al Nakouzi N, Mo F, Zhou T, Kim Y, Monia BP. Generation 2.5 Antisense Oligonucleotides Targeting the Androgen Receptor and Its Splice Variants Suppress Enzalutamide-Resistant Prostate Cancer Cell Growth. Clinical Cancer Research. 2015;21(7):1675–87. doi: 10.1158/1078-0432.CCR-14-1108. [DOI] [PubMed] [Google Scholar]
- 50.Amin KS, Jagadeesh S, Baishya G, Rao PG, Barua NC, Bhattacharya S, Banerjee PP. A naturally derived small molecule disrupts ligand-dependent and ligand-independent androgen receptor signaling in human prostate cancer cells. Molecular cancer therapeutics. 2014;13(2):341–52. doi: 10.1158/1535-7163.MCT-13-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dalal K, Roshan-Moniri M, Sharma A, Li H, Ban F, Hessein M, Hsing M, Singh K, LeBlanc E, Dehm S. Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer. Journal of Biological Chemistry. 2014;289(38):26417–29. doi: 10.1074/jbc.M114.553818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bianchini D, Omlin A, Pezaro C, Lorente D, Ferraldeschi R, Mukherji D, Crespo M, Figueiredo I, Miranda S, Riisnaes R. First-in-human Phase I study of EZN-4176, a locked nucleic acid antisense oligonucleotide to exon 4 of the androgen receptor mRNA in patients with castration-resistant prostate cancer. British journal of cancer. 2013;109(10):2579–86. doi: 10.1038/bjc.2013.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myung J-K, Banuelos CA, Fernandez JG, Mawji NR, Wang J, Tien AH, Yang YC, Tavakoli I, Haile S, Watt K. An androgen receptor N-terminal domain antagonist for treating prostate cancer. The Journal of clinical investigation. 2013;123(7):2948. doi: 10.1172/JCI66398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chmelar R, Buchanan G, Need EF, Tilley W, Greenberg NM. Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. International Journal of cancer. 2007;120(4):719–33. doi: 10.1002/ijc.22365. [DOI] [PubMed] [Google Scholar]
- 55.Heemers HV, Regan KM, Schmidt LJ, Anderson SK, Ballman KV, Tindall DJ. Androgen modulation of coregulator expression in prostate cancer cells. Molecular Endocrinology. 2009;23(4):572–83. doi: 10.1210/me.2008-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nature Reviews Cancer. 2012;12(6):381–5. doi: 10.1038/nrc3263. [DOI] [PubMed] [Google Scholar]
- 57.Sahu B, Laakso M, Pihlajamaa P, Ovaska K, Sinielnikov I, Hautaniemi S, Jänne OA. FoxA1 specifies unique androgen and glucocorticoid receptor binding events in prostate cancer cells. Cancer research. 2013;73(5):1570–80. doi: 10.1158/0008-5472.CAN-12-2350. [DOI] [PubMed] [Google Scholar]
- 58.Gormally MV, Dexheimer TS, Marsico G, Sanders DA, Lowe C, Matak-Vinković D, Michael S, Jadhav A, Rai G, Maloney DJ. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nature communications. 2014;5 doi: 10.1038/ncomms6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He B, Lanz RB, Fiskus W, Geng C, Yi P, Hartig SM, Rajapakshe K, Shou J, Wei L, Shah SS, Foley C, Chew SA, Eedunuri VK, Bedoya DJ, Feng Q, Minami T, Mitsiades CS, Frolov A, Weigel NL, Hilsenbeck SG, Rosen DG, Palzkill T, Ittmann MM, Song Y, Coarfa C, O'Malley BW, Mitsiades N. GATA2 facilitates steroid receptor coactivator recruitment to the androgen receptor complex. Proc Natl Acad Sci U S A. 2014;111(51):18261–6. doi: 10.1073/pnas.1421415111.. PubMed PMID: 25489091; PMCID: 4280633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–82. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gregory CW, He B, Johnson RT, Ford OH, Mohler JL, French FS, Wilson EM. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer research. 2001;61(11):4315–9. [PubMed] [Google Scholar]
- 62.Agoulnik IU, Vaid A, Nakka M, Alvarado M, Bingman WE, Erdem H, Frolov A, Smith CL, Ayala GE, Ittmann MM. Androgens modulate expression of transcription intermediary factor 2, an androgen receptor coactivator whose expression level correlates with early biochemical recurrence in prostate cancer. Cancer research. 2006;66(21):10594–602. doi: 10.1158/0008-5472.CAN-06-1023. [DOI] [PubMed] [Google Scholar]
- 63.Zhou H-J, Yan J, Luo W, Ayala G, Lin S-H, Erdem H, Ittmann M, Tsai SY, Tsai M-J. SRC-3 is required for prostate cancer cell proliferation and survival. Cancer research. 2005;65(17):7976–83. doi: 10.1158/0008-5472.CAN-04-4076. [DOI] [PubMed] [Google Scholar]
- 64.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X-W, Varambally S, Cao X, Tchinda J, Kuefer R. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 65.Yu J, Yu J, Mani R-S, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer cell. 2010;17(5):443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tu JJ, Rohan S, Kao J, Kitabayashi N, Mathew S, Chen Y-T. Gene fusions between TMPRSS2 and ETS family genes in prostate cancer: frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissues. Modern Pathology. 2007;20(9):921–8. doi: 10.1038/modpathol.3800903. [DOI] [PubMed] [Google Scholar]
- 67.Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, Wuttig D, Warnatz H-J, Stehr H, Rausch T. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer cell. 2013;23(2):159–70. doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 68.Tomlins SA, Day JR, Lonigro RJ, Hovelson DH, Siddiqui J, Kunju LP, Dunn RL, Meyer S, Hodge P, Groskopf J, Wei JT, Chinnaiyan AM. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur Urol. 2015 doi: 10.1016/j.eururo.2015.04.039.. Epub 2015/05/20. doi: S0302-2838(15)00397-8 [pii] PubMed PMID: 25985884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomlins SA, Palanisamy N, Siddiqui J, Chinnaiyan AM, Kunju LP. Antibody-based detection of ERG rearrangements in prostate core biopsies, including diagnostically challenging cases: ERG staining in prostate core biopsies. Arch Pathol Lab Med. 2012;136(8):935–46. doi: 10.5858/arpa.2011-0424-OA.. Epub 2012/08/02. PubMed PMID: 22849743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald T, Srivastava A, Tewari AK, Sathyanarayana U, Nagy D, Pestano G, Kunju LP, Demichelis F, Chinnaiyan AM, Rubin MA. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12(7):590–8. doi: 10.1593/neo.10726. Epub 2010/07/24. PubMed PMID: 20651988; PMCID: 2907585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Furusato B, Tan SH, Young D, Dobi A, Sun C, Mohamed AA, Thangapazham R, Chen Y, McMaster G, Sreenath T, Petrovics G, McLeod DG, Srivastava S, Sesterhenn IA. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010;13(3):228–37. doi: 10.1038/pcan.2010.23.. Epub 2010/06/30. doi: pcan201023 [pii] PubMed PMID: 20585344; PMCID: 3010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feng FY, Brenner JC, Hussain M, Chinnaiyan AM. Molecular pathways: Targeting ETS gene fusions in cancer. Clinical Cancer Research. 2014;20(17):4442–8. doi: 10.1158/1078-0432.CCR-13-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC, Martin NE, Kunz L, Penney KL, Ligon A, Suppan C, Flavin R, Sesso H, Rider JR, Sweeney CS, Stampfer MJ, Fiorentino M, Kantoff PW, Sanda MG, Giovannucci E, Ding EL, Loda M, Mucci LA. The TMPRSS2:ERG Rearrangement, ERG Expression, and Prostate Cancer Outcomes: a Cohort Study and Meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21(9):1497–509. doi: 10.1158/1055-9965.EPI-12-0042.. Epub 2012/06/28. doi: 1055-9965.EPI-12-0042 [pii] PubMed PMID: 22736790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mounir Z, Lin F, Lin VG, Korn JM, Yu Y, Valdez R, Aina OH, Buchwalter G, Jaffe AB, Korpal M, Zhu P, Brown M, Cardiff RD, Rocnik JL, Yang Y, Pagliarini R. TMPRSS2:ERG blocks neuroendocrine and luminal cell differentiation to maintain prostate cancer proliferation. Oncogene. 2015;34(29):3815–25. doi: 10.1038/onc.2014.308.. PubMed PMID: 25263440. [DOI] [PubMed] [Google Scholar]
- 75.Lucas JM, Heinlein C, Kim T, Hernandez SA, Malik MS, True LD, Morrissey C, Corey E, Montgomery B, Mostaghel E. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer discovery. 2014;4(11):1310–25. doi: 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H, Yu J. Mechanistic rationale for inhibition of poly (ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer cell. 2011;19(5):664–78. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bradley D, Rathkopf D, Dunn R, Stadler WM, Liu G, Smith DC, Pili R, Zwiebel J, Scher H, Hussain M. Vorinostat in advanced prostate cancer patients progressing on prior chemotherapy (National Cancer Institute Trial 6862) Cancer. 2009;115(23):5541–9. doi: 10.1002/cncr.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Molife L, Attard G, Fong P, Karavasilis V, Reid A, Patterson S, Riggs C, Higano C, Stadler W, McCulloch W. Phase II, two-stage, single-arm trial of the histone deacetylase inhibitor (HDACi) romidepsin in metastatic castration-resistant prostate cancer (CRPC) Annals of oncology. 2010;21(1):109–13. doi: 10.1093/annonc/mdp270. [DOI] [PubMed] [Google Scholar]
- 79.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She Q-B, Maurer M, Koujak S, Ferrando AA, Malmström P, Memeo L. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proceedings of the National Academy of Sciences. 2007;104(18):7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferraldeschi R, Rodrigues DN, Riisnaes R, Miranda S, Figueiredo I, Rescigno P, Ravi P, Pezaro C, Omlin A, Lorente D. PTEN protein loss and clinical outcome from castration-resistant prostate cancer treated with abiraterone acetate. European urology. 2015;67(4):795–802. doi: 10.1016/j.eururo.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krohn A, Diedler T, Burkhardt L, Mayer P-S, De Silva C, Meyer-Kornblum M, Kötschau D, Tennstedt P, Huang J, Gerhäuser C. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. The American journal of pathology. 2012;181(2):401–12. doi: 10.1016/j.ajpath.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 82.Yoshimoto M, Joshua AM, Cunha IW, Coudry RA, Fonseca FP, Ludkovski O, Zielenska M, Soares FA, Squire JA. Absence of TMPRSS2: ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Modern Pathology. 2008;21(12):1451–60. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 83.Reid A, Attard G, Ambroisine L, Fisher G, Kovacs G, Brewer D, Clark J, Flohr P, Edwards S, Berney DM. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. British journal of cancer. 2010;102(4):678–84. doi: 10.1038/sj.bjc.6605554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer cell. 2011;19(5):575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwartz S, Wongvipat J, Trigwell CB, Hancox U, Carver BS, Rodrik-Outmezguine V, Will M, Yellen P, de Stanchina E, Baselga J. Feedback suppression of PI3Kα signaling in PTEN-mutated tumors is relieved by selective inhibition of PI3Kβ. Cancer cell. 2015;27(1):109–22. doi: 10.1016/j.ccell.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klempner SJ, Myers AP, Cantley LC. What a tangled web we weave: emerging resistance mechanisms to inhibition of the phosphoinositide 3-kinase pathway. Cancer discovery. 2013;3(12):1345–54. doi: 10.1158/2159-8290.CD-13-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vincan E, Barker N. The upstream components of the Wnt signalling pathway in the dynamic EMT and MET associated with colorectal cancer progression. Clinical & experimental metastasis. 2008;25(6):657–63. doi: 10.1007/s10585-008-9156-4. [DOI] [PubMed] [Google Scholar]
- 88.Hovelson DH, McDaniel AS, Cani AK, Johnson B, Rhodes K, Williams PD, Bandla S, Bien G, Choppa P, Hyland F. Development and Validation of a Scalable Next-Generation Sequencing System for Assessing Relevant Somatic Variants in Solid Tumors. Neoplasia. 2015;17(4):385–99. doi: 10.1016/j.neo.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349(6254):1351–6. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, Ivy SP. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nature Reviews Clinical Oncology. 2015 doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.El-Khoueiry AB, Ning Y, Yang D, Cole S, Kahn M, Zoghbi M, Berg J, Fujimori M, Inada T, Kouji H, editors. JOURNAL OF CLINICAL ONCOLOGY. ALEXANDRIA, VA 22314 USA: 2013. A phase I first-in-human study of PRI-724 in patients (pts) with advanced solid tumors. AMER SOC CLINICAL ONCOLOGY 2318 MILL ROAD, STE 800. [Google Scholar]
- 92.Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T, Kasibhatla S, Schuller AG, Li AG, Cheng D. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proceedings of the National Academy of Sciences. 2013;110(50):20224–9. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: A new perspective. Cancer. 2014;120(22):3446–56. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, Gleave M, Wu H. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72(7):1878–89. doi: 10.1158/0008-5472.CAN-11-3132.. PubMed PMID: 22350410; PMCID: 3319847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, Han B, Cao Q, Cao X, Suleman K. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nature medicine. 2010;16(7):793–8. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ghiam A, Cairns R, Thoms J, Dal Pra A, Ahmed O, Meng A, Mak T, Bristow R. IDH mutation status in prostate cancer. Oncogene. 2012;31(33):3826. doi: 10.1038/onc.2011.546. [DOI] [PubMed] [Google Scholar]
- 97.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, Lee JY, Yoo NJ, Lee SH. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. International journal of cancer. 2009;125(2):353–5. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 98.Mauzo SH, Lee M, Petros J, Hunter S, Chang C-M, Shu H-K, Bellail AC, Hao C, Cohen C. Immunohistochemical demonstration of isocitrate dehydrogenase 1 (IDH1) mutation in a small subset of prostatic carcinomas. Applied Immunohistochemistry & Molecular Morphology. 2014;22(4):284–7. doi: 10.1097/PAI.0b013e3182649d1c. [DOI] [PubMed] [Google Scholar]
- 99.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–44. doi: 10.1038/nature08617.. PubMed PMID: 19935646; PMCID: 2818760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18(6):553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD. Recurring mutations found by sequencing an acute myeloid leukemia genome. New England Journal of Medicine. 2009;361(11):1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–30. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schumacher T, Bunse L, Pusch S, Sahm F, Wiestler B, Quandt J, Menn O, Osswald M, Oezen I, Ott M. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014 doi: 10.1038/nature13387. [DOI] [PubMed] [Google Scholar]
- 105.Vousden KH, Lane DP. p53 in health and disease. Nature reviews Molecular cell biology. 2007;8(4):275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 106.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nature Reviews Cancer. 2008;8(9):714–24. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Turner NC, Ro J, André F, Loi S, Verma S, Iwata H, Harbeck N, Loibl S, Huang Bartlett C, Zhang K. Palbociclib in hormone-receptor–positive advanced breast cancer. New England Journal of Medicine. 2015;373(3):209–19. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 108.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, Ettl J, Patel R, Pinter T, Schmidt M. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. The Lancet Oncology. 2015;16(1):25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 109.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nature Reviews Cancer. 2009;9(12):862–73. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 110.Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nature structural & molecular biology. 2011;18(6):721–7. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reaper PM, Griffiths MR, Long JM, Charrier J-D, MacCormick S, Charlton PA, Golec JM, Pollard JR. Selective killing of ATM-or p53-deficient cancer cells through inhibition of ATR. Nature chemical biology. 2011;7(7):428–30. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 112.Bridges KA, Hirai H, Buser CA, Brooks C, Liu H, Buchholz TA, Molkentine JM, Mason KA, Meyn RE. MK-1775, a novel Wee1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Clinical cancer research. 2011;17(17):5638–48. doi: 10.1158/1078-0432.CCR-11-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aarts M, Sharpe R, Garcia-Murillas I, Gevensleben H, Hurd MS, Shumway SD, Toniatti C, Ashworth A, Turner NC. Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer discovery. 2012;2(6):524–39. doi: 10.1158/2159-8290.CD-11-0320. [DOI] [PubMed] [Google Scholar]
- 114.Origanti S, Cai S, Munir AZ, White LS, Piwnica-Worms H. Synthetic lethality of Chk1 inhibition combined with p53 and/or p21 loss during a DNA damage response in normal and tumor cells. Oncogene. 2013;32(5):577–88. doi: 10.1038/onc.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ma CX, Cai S, Li S, Ryan CE, Guo Z, Schaiff WT, Lin L, Hoog J, Goiffon RJ, Prat A. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. The Journal of clinical investigation. 2012;122(4):1541. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Do K, Wilsker D, Ji J, Zlott J, Freshwater T, Kinders RJ, Collins J, Chen AP, Doroshow JH, Kummar S. Phase I Study of Single-Agent AZD1775 (MK-1775), a Wee1 Kinase Inhibitor, in Patients With Refractory Solid Tumors. Journal of Clinical Oncology. 2015 doi: 10.1200/JCO.2014.60.4009. JCO. 2014.60. 4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Daud AI, Ashworth MT, Strosberg J, Goldman JW, Mendelson D, Springett G, Venook AP, Loechner S, Rosen LS, Shanahan F. Phase I dose-escalation trial of checkpoint kinase 1 inhibitor MK-8776 as Monotherapy and in combination with gemcitabine in patients with advanced solid tumors. Journal of Clinical Oncology. 2015 doi: 10.1200/JCO.2014.57.5027. JCO. 2014.57. 5027. [DOI] [PubMed] [Google Scholar]
- 118.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nature Reviews Cancer. 2008;8(12):976–90. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 119.Tolcher AW, Papadopoulos KP, Patnaik A, Rasco DW, Martinez D, Wood DL, Fielman B, Sharma M, Janisch LA, Brown BD, editors. Safety and activity of DCR-MYC, a first-in-class Dicer-substrate small interfering RNA (DsiRNA) targeting MYC, in a phase I study in patients with advanced solid tumors. ASCO Annual Meeting Proceedings.2015. [Google Scholar]
- 120.Kirschner AN, Wang J, van der Meer R, Anderson PD, Franco-Coronel OE, Kushner MH, Everett JH, Hameed O, Keeton EK, Ahdesmaki M. PIM Kinase Inhibitor AZD1208 for Treatment of MYC-Driven Prostate Cancer. Journal of the National Cancer Institute. 2015;107(2):dju407. doi: 10.1093/jnci/dju407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montaña MF, D'Artista L, Schleker T, Guerra C, Garcia E. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nature structural & molecular biology. 2011;18(12):1331–5. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Page BD, Khoury H, Laister RC, Fletcher S, Vellozo M, Manzoli A, Yue P, Turkson J, Minden MD, Gunning PT. Small molecule STAT5-SH2 domain inhibitors exhibit potent antileukemia activity. Journal of medicinal chemistry. 2012;55(3):1047–55. doi: 10.1021/jm200720n. [DOI] [PubMed] [Google Scholar]
- 124.Karanika S, Karantanos T, Li L, Corn P, Thompson T. DNA damage response and prostate cancer: Defects, regulation and therapeutic implications. Oncogene. 2014 doi: 10.1038/onc.2014.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, Jarosz M, Lipson D, Tagawa ST, Nanus DM. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. European urology. 2013;63(5):920–6. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pritchard CC, Morrissey C, Kumar A, Zhang X, Smith C, Coleman I, Salipante SJ, Milbank J, Yu M, Grady WM. Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nature communications. 2014;5 doi: 10.1038/ncomms5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Grasso C, Cani A, Hovelson D, Quist M, Douville N, Yadati V, Amin A, Nelson P, Betz B, Liu C. Integrative molecular profiling of routine clinical prostate cancer specimens. Annals of Oncology. 2015 doi: 10.1093/annonc/mdv134. mdv134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kumar A, White TA, MacKenzie AP, Clegg N, Lee C, Dumpit RF, Coleman I, Ng SB, Salipante SJ, Rieder MJ. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proceedings of the National Academy of Sciences. 2011;108(41):17087–92. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348(6230):124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS. Genetic basis for clinical response to CTLA-4 blockade in melanoma. New England Journal of Medicine. 2014;371(23):2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lord CJ, Tutt AN, Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annual review of medicine. 2015;66:455–70. doi: 10.1146/annurev-med-050913-022545.. PubMed PMID: 25341009. [DOI] [PubMed] [Google Scholar]
- 133.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ. Inhibition of poly (ADP-ribose) polymerase in tumors from BRCA mutation carriers. New England Journal of Medicine. 2009;361(2):123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 134.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN, Oaknin A, Loman N. Oral poly (ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. The Lancet. 2010;376(9737):245–51. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 135.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott CL, Meier W, Shapira-Frommer R, Safra T. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. The lancet oncology. 2014;15(8):852–61. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 136.Kaufman B, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, Balmaña J, Mitchell G, Fried G, Stemmer SM, Hubert A. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. Journal of Clinical Oncology. 2014 doi: 10.1200/JCO.2014.56.2728. JCO. 2014.56. 728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A, Tunariu N. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. New England Journal of Medicine. 2015;373(18):1697–708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.VanderWeele DJ, Paner GP, Fleming GF, Szmulewitz RZ. Sustained Complete Response to Cytotoxic Therapy and the PARP Inhibitor Veliparib in Metastatic Castration-Resistant Prostate Cancer–A Case Report. Frontiers in oncology. 2015;5 doi: 10.3389/fonc.2015.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, De Greve J, Lubinski J, Shanley S, Messiou C. Poly (ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. Journal of Clinical Oncology. 2010;28(15):2512–9. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 140.Ceccaldi R, O'Connor KW, Mouw KW, Li AY, Matulonis UA, D'Andrea AD, Konstantinopoulos PA. A Unique Subset of Epithelial Ovarian Cancers with Platinum Sensitivity and PARP Inhibitor Resistance. Cancer research. 2015;75(4):628–34. doi: 10.1158/0008-5472.CAN-14-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Polkinghorn WR, Parker JS, Lee MX, Kass EM, Spratt DE, Iaquinta PJ, Arora VK, Yen W-F, Cai L, Zheng D. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer discovery. 2013;3(11):1245–53. doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, Ma T, Den RB, Dicker AP, Feng FY. A hormone–DNA repair circuit governs the response to genotoxic insult. Cancer discovery. 2013;3(11):1254–71. doi: 10.1158/2159-8290.CD-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Spratt DE, Evans MJ, Davis BJ, Doran MG, Lee MX, Shah N, Wongvipat J, Carnazza KE, Klee GG, Polkinghorn W. Androgen receptor upregulation mediates radioresistance after ionizing radiation. Cancer research. 2015 doi: 10.1158/0008-5472.CAN-15-0892. canres. 0892.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Eeles R, Goh C, Castro E, Bancroft E, Guy M, Al Olama AA, Easton D, Kote-Jarai Z. The genetic epidemiology of prostate cancer and its clinical implications. Nature Reviews Urology. 2014;11(1):18–31. doi: 10.1038/nrurol.2013.266. [DOI] [PubMed] [Google Scholar]
- 145.Ostrander EA, Udler MS. The role of the BRCA2 gene in susceptibility to prostate cancer revisited. Cancer Epidemiology Biomarkers & Prevention. 2008;17(8):1843–8. doi: 10.1158/1055-9965.EPI-08-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tischkowitz M, Eeles R. Mutations in BRCA1 and BRCA2 and predisposition to prostate cancer. The Lancet. 2003;362(9377):80. doi: 10.1016/S0140-6736(03)13823-8. [DOI] [PubMed] [Google Scholar]
- 147.Liede A, Karlan BY, Narod SA. Cancer risks for male carriers of germline mutations in BRCA1 or BRCA2: a review of the literature. Journal of Clinical Oncology. 2004;22(4):735–42. doi: 10.1200/JCO.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 148.Thompson D, Easton DF, Consortium BCL Cancer incidence in BRC A1 mutation carriers. Journal of the National Cancer Institute. 2002;94(18):1358–65. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 149.Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, Wiley KE, Isaacs SD, Johng D, Wang Y. Germline mutations in HOXB13 and prostate-cancer risk. New England Journal of Medicine. 2012;366(2):141–9. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Thompson IM, Jr, Goodman PJ, Tangen CM, Parnes HL, Minasian LM, Godley PA, Lucia MS, Ford LG. Long-term survival of participants in the prostate cancer prevention trial. New England Journal of Medicine. 2013;369(7):603–10. doi: 10.1056/NEJMoa1215932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Klein EA, Thompson IM, Tangen CM, Crowley JJ, Lucia MS, Goodman PJ, Minasian LM, Ford LG, Parnes HL, Gaziano JM. Vitamin E and the risk of prostate cancer: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) Jama. 2011;306(14):1549–56. doi: 10.1001/jama.2011.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Djavan B, Susani M, Bursa B, Basharkhah A, Simak R, Marberger M. Predictability and significance of multifocal prostate cancer in the radical prostatectomy specimen. Techniques in urology. 1999;5(3):139–42. [PubMed] [Google Scholar]
- 153.Wise AM, Stamey TA, McNeal JE, Clayton JL. Morphologic and clinical significance of multifocal prostate cancers in radical prostatectomy specimens. Urology. 2002;60(2):264–9. doi: 10.1016/s0090-4295(02)01728-4. [DOI] [PubMed] [Google Scholar]
- 154.Lindberg J, Klevebring D, Liu W, Neiman M, Xu J, Wiklund P, Wiklund F, Mills IG, Egevad L, Grönberg H. Exome sequencing of prostate cancer supports the hypothesis of independent tumour origins. European urology. 2013;63(2):347–53. doi: 10.1016/j.eururo.2012.03.050. [DOI] [PubMed] [Google Scholar]
- 155.Haffner MC, Mosbruger T, Esopi DM, Fedor H, Heaphy CM, Walker DA, Adejola N, Gürel M, Hicks J, Meeker AK. Tracking the clonal origin of lethal prostate cancer. The Journal of clinical investigation. 2013;123(11):4918. doi: 10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Carreira S, Romanel A, Goodall J, Grist E, Ferraldeschi R, Miranda S, Prandi D, Lorente D, Frenel J-S, Pezaro C. Tumor clone dynamics in lethal prostate cancer. Science translational medicine. 2014;6(254):254ra125–254ra125. doi: 10.1126/scitranslmed.3009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, Chen L, Ewing CM, Eisenberger MA, Carducci MA. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nature medicine. 2009;15(5):559–65. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hong MK, Macintyre G, Wedge DC, Van Loo P, Patel K, Lunke S, Alexandrov LB, Sloggett C, Cmero M, Marass F. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nature communications. 2015;6 doi: 10.1038/ncomms7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Comen E, Norton L, Massague J. Clinical implications of cancer self-seeding. Nature reviews Clinical oncology. 2011;8(6):369–77. doi: 10.1038/nrclinonc.2011.64. [DOI] [PubMed] [Google Scholar]
- 160.Taplin M-E, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, Keer HN, Balk SP. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. New England Journal of Medicine. 1995;332(21):1393–8. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 161.Bozic I, Reiter JG, Allen B, Antal T, Chatterjee K, Shah P, Moon YS, Yaqubie A, Kelly N, Le DT. Evolutionary dynamics of cancer in response to targeted combination therapy. Elife. 2013;2:e00747. doi: 10.7554/eLife.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Holcomb IN, Young JM, Coleman IM, Salari K, Grove DI, Hsu L, True LD, Roudier MP, Morrissey CM, Higano CS. Comparative analyses of chromosome alterations in soft-tissue metastases within and across patients with castration-resistant prostate cancer. Cancer research. 2009;69(19):7793–802. doi: 10.1158/0008-5472.CAN-08-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Beltran H, Mosquera JM, Rubin MA. Prostate Cancer: A Comprehensive Perspective. Springer; 2013. Neuroendocrine prostate cancer; pp. 277–82. [Google Scholar]
- 164.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, Wang Y, Sheikh KL, Terry S, Tagawa ST. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer discovery. 2011;1(6):487–95. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Beltran H, Tagawa ST, Park K, MacDonald T, Milowsky MI, Mosquera JM, Rubin MA, Nanus DM. Challenges in recognizing treatment-related neuroendocrine prostate cancer. Journal of Clinical Oncology. 2012;30(36):e386–e9. doi: 10.1200/JCO.2011.41.5166. [DOI] [PubMed] [Google Scholar]
- 166.Pezaro CJ, Omlin A, Lorente D, Rodrigues DN, Ferraldeschi R, Bianchini D, Mukherji D, Riisnaes R, Altavilla A, Crespo M. Visceral disease in castration-resistant prostate cancer. European urology. 2014;65(2):270–3. doi: 10.1016/j.eururo.2013.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Epstein JI, Amin MB, Beltran H, Lotan TL, Mosquera J-M, Reuter VE, Robinson BD, Troncoso P, Rubin MA. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. The American journal of surgical pathology. 2014;38(6):756. doi: 10.1097/PAS.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Small E, Youngren J, Alumkal J, Evans C, Ryan C, Lara P, Beer T, Witte O, Baertsch R, Stuart J. 760PDNEUROENDOCRINE PROSTATE CANCER (NEPC) IN PATIENTS (PTS) WITH METASTATIC CASTRATION RESISTANT PROSTATE CANCER (MCRPC) RESISTANT TO ABIRATERONE (ABI) OR ENZALUTAMIDE (ENZ): PRELIMINARY RESULTS FROM THE SU2C/PCF/AACR WEST COAST PROSTATE CANCER DREAM TEAM (WCDT) Annals of Oncology. 2014;25(suppl 4):iv258–iv. [Google Scholar]
- 169.Tan H-L, Sood A, Rahimi HA, Wang W, Gupta N, Hicks J, Mosier S, Gocke CD, Epstein JI, Netto GJ. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clinical Cancer Research. 2014;20(4):890–903. doi: 10.1158/1078-0432.CCR-13-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen H, Sun Y, Wu C, Magyar CE, Li X, Cheng L, Yao JL, Shen S, Osunkoya AO, Liang C. Pathogenesis of prostatic small cell carcinoma involves the inactivation of the P53 pathway. Endocrine-related cancer. 2012;19(3):321–31. doi: 10.1530/ERC-11-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Akamatsu S, Wyatt A, Lin D, Lysakowski S, Zhang F, Kim S, Fazli L, Beltran H, Rubin M, Zoubeidi A. MP31-09 IDENTIFICATION OF A RETRO-TRANSPOSON DERIVED GENE ASSOCIATED WITH PROGRESSION TO NEUROENDOCRINE PROSTATE CANCER. The Journal of Urology. 2014;191(4):e325. [Google Scholar]
- 172.Kadakia KC, Tomlins SA, Sanghvi SK, Cani AK, Omata K, Hovelson DH, Liu C-J, Cooney KA. Comprehensive serial molecular profiling of an “N of 1” exceptional non-responder with metastatic prostate cancer progressing to small cell carcinoma on treatment. Journal of hematology & oncology. 2015;8(1):1–7. doi: 10.1186/s13045-015-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]