Abstract
Collective migration of epithelial cells is an integral part of embryonic development, wound healing, tissue renewal and carcinoma invasion. While previous studies have focused on cell-extracellular matrix adhesion as a site of migration-driving, traction force-transmission, cadherin mediated cell-cell adhesion is also capable of force-transmission. Using a soft elastomer coated with purified N-cadherin as a substrate and a Hepatocyte Growth Factor-treated, transformed MDCK epithelial cell line as a model system, we quantified traction transmitted by N-cadherin-mediated contacts. On a substrate coated with purified extracellular domain of N-cadherin, cell surface N-cadherin proteins arranged into puncta. N-cadherin mutants (either the cytoplasmic deletion or actin-binding domain chimera), however, failed to assemble into puncta, suggesting the assembly of focal adhesion like puncta requires the cytoplasmic domain of N-cadherin. Furthermore, the cytoplasmic domain deleted N-cadherin expressing cells exerted lower traction stress than the full-length or the actin binding domain chimeric N-cadherin. Our data demonstrate that N-cadherin junctions exert significant traction stress that requires the cytoplasmic domain of N-cadherin, but the loss of the cytoplasmic domain does not completely eliminate traction force transmission.
Keywords: N-cadherin, MDCK, traction force, actin cytoskeleton, collective cell migration, Hepatocyte Growth Factor
Introduction
A unique feature of epithelial cell migration is their ability to move together as a cohesive unit. Collective cell migration is an essential feature in both physiological and pathological processes [1, 2]. Previous studies have focused on migration-driving, force-transmission through cell-extracellular matrix adhesion, yet, physical contacts between neighboring cells can also serve as a site of traction force-transmission. Recent studies uncovered an important role of molecular linkers between the cadherin complex and the actin cytoskeleton. Similar to the integrin complex at focal adhesions, the cadherin complex is regulated by force-sensitive proteins thought to control traction force necessary for collective cell migration [3].
Collective cell migration depends on adhesive properties at the sites of cell-extracellular matrix and cell-cell adhesion. For example, Madin-Darby canine kidney (MDCK) epithelial cells with abundant junctional proteins migrate as collective units on a 2D surface [4–16]. The addition of Hepatocyte Growth Factor (HGF), however, induces MDCK cells to rapidly adapt a mesenchymal phenotype and migrate as single cells on a 2D surface [6, 17–20]. These data suggest that traction force onto cell-extracellular matrix is sufficient to move away from neighboring cells and disrupt cell-cell contacts. In contrast, HGF-treated MDCK cells collectively migrate while maintaining cell-cell contacts in a 3D matrix [17, 21–23]. This is in part due to the soft 3D matrix preventing efficient traction force transmission to the matrix, thus limiting cells from moving away from each other. Instead, HGF-treated MDCK cells maintain cell-cell contacts while invading a 3D matrix.
Interestingly, HGF-treated MDCK cells migrate more efficiently as a cell cluster, which requires N-cadherin adhesion proteins [17, 24]. Unlike brief exposure of HGF to MDCK cells, prolonged exposure of HGF to MDCK cells induces complete Epithelial-to-Mesenchymal Transition (EMT), which decreases the level of epithelial markers (e.g., E-cadherin) and increases the level of mesenchymal markers (e.g., N-cadherin) [24]. In the absence of the full-length N-cadherin, the transformed MDCK cells do not migrate and remain single cells in a 3D matrix, while in the absence of the extracellular domain of N-cadherin, the cells migrate but as single cells [24]. Therefore, the adhesive activity of the N-cadherin extracellular domain is required for 3D collective migration of the transformed MDCK cells. One possible explanation for the efficient collective cell migration in the presence of N-cadherin is N-cadherin’s ability to transmit traction forces along neighboring cells which propels individual cells along the cell cluster. Using the transformed MDCK cells and traction force sensing substrates, we sought to analyze traction forces transmitted via N-cadherin junctions.
Materials and Methods
Cell culture and reagents
MDCK epithelial cells were treated with HGF to induce EMT [24]. As a result, these transformed cells down-regulated E-cadherin and up-regulated N-cadherin as the primary cell-cell adhesion protein. The cells expressing mutant cadherins were generated by transfecting both shRNA against canine N-cadherin and tandem dimer Tomato tagged, shRNA resistant mouse N-cadherin. The detail characterization of these N-cadherin mutant expressing cells were described previously [24]. The actin filaments were stained with Alexa 488 phalloidin (Invitrogen) after 3 % formaldehyde fixation in PBS. Purification of the N-cadherin extracellular domain tagged with Fc domain was described previously [25].
Preparation of traction force-detecting substrate
The substrate for traction force analysis is based on CY52-276A/B elastomer (Dow Chemicals) [26, 27]. Briefly, the A and B parts were mixed at 1:1 ratio and degassed in a vacuum to remove trapped bubbles. The coverslip was thinly coated (~ 20 μm) with the mixed elastomer using a coverslip spinner. Using a P35 dish with the bottom cutout, the elastomer-coated coverslip was then attached to the P35 dish with the elastomer-coated side facing inside of the dish, and cured for ~ 1 hour at 50 °C. The surface of elastomer was derivatized with 3-aminopropyltriethoxysilane (Pierce) using vapor deposition for overnight, then UV sterilized for 1 hour. The elastomer-coated substrates were used within a few weeks. Prior to plating cells, either purified N-cadherin extracellular domain or fibronectin and tracer particles (0.1 micron carboxylated Alexa 488 beads, Invitrogen) were crosslinked to the surface using hetero-bi-functional cross linker: 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride, EDC (Pierce).
Rheological analysis of substrate
The rheological properties of CY52-276 elastomer were characterized using a stain-controlled Discovery HR2 Hybrid Rheometer (TA Instruments). Briefly, CY52-276 elastomer was cured on the rheometer plates while the temperature was set to 50 °C and minimal strain (1% at 10 rad/s) was applied to monitor the gelation process (Figure 2B). Then, the sample was tested for frequency dependence at 1% strain at 37 °C (Figure 2B).
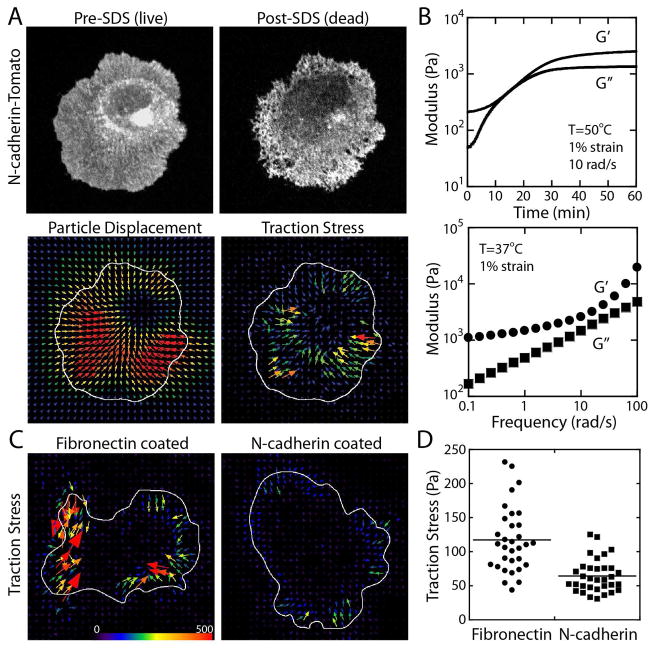
Figure 2. Traction force analysis using deformable substrates.
(A) Morphology of adhered cell on elastomer substrate before and after the SDS treatment, particle displacements and resulting traction stress vectors. (B) Viscoelastic properties of elastomer. The elastic (G′) and viscous (G″) moduli during the time sweep (top) and frequency sweep (bottom) experiments. (C) Traction stress of MDCK cell (HGF-treated, with exogenous full-length N-cadherin) adhered to fibronectin or N-cadherin coated surface. The arrows indicate the direction and magnitude of traction stress. (D) Average traction stress of MDCK cells (HGF-treated, wildtype cells without exogenous N-cadherin) adhered to fibronectin (117.21 ± 48.02 Pa, average ± standard deviation) or N-cadherin (64.32 ± 24.29 Pa) coated surface.
Microscopy
For traction force analysis, cells were briefly trypsinized using a standard trypsin solution containing EDTA, followed by longer incubation with trypsin solution containing 1.8 mM calcium. This minimizes the digestion of cadherin proteins on cell surface. Cells were then plated on the protein-coated elastomer substrate. The cells were imaged within a few hours of plating to minimize the non-specific adhesion of cells to the substrate. All images were collected using a Zeiss AxioObserver equipped with a Yokogawa CSU-10 spinning disk confocal system, 40x or 10x objectives, 488 and 561 nm solid-state lasers, and a Photometrics CoolSNAP HQ camera. The microscope system was controlled by Slidebook software (Intelligent Imaging Innovations). For live-cell imaging, the temperature was set to 37 °C by a custom microscope heating chamber. For traction force experiments, first, images of live cells and underlying particles were taken, followed by the addition of a small volume of 2 % SDS to release cell traction, then images traction-free cells and particles were taken.
Traction force analysis
Traction force was calculated based on the tracer particle displacement and substrate material properties using ImageJ plugins by Dr. Qingzong Tseng (https://sites.google.com/site/qingzongtseng/) [28]. Briefly, the live and fixed images were aligned based on the static particles, then particle displacements were calculated using Particle Image Velocimetry plugin, and finally, traction stress was calculated using Traction Force FTTC plugin. Cell boundary was defined using either bright field or fluorescence (DsRed) image, and used to calculate traction stress within the cell boundary.
Results
Cell spreading on a purified N-cadherin coated surface
Traction force generated by the actin-myosin network is transmitted via adhesion receptors. Transformed MDCK epithelial cells express both N-cadherin and fibronectin receptors [24]. To specify the adhesion molecules responsible for traction force transmission, we coated an elastomer surface with fibronectin or the extracellular domain of N-cadherin, and plated the transformed epithelial cells in the presence or absence of calcium. Homophilic, trans-interactions of cadherins strongly depend on calcium concentration in the media. On the fibronectin coated surface, the transformed cells bound to the fibronectin coating and spread out on the surface regardless of calcium concentration (Figure 1A), while no cells were bound to the outside of fibronectin coated regions (marked by the presence of surface bound particles, Figure 1A).
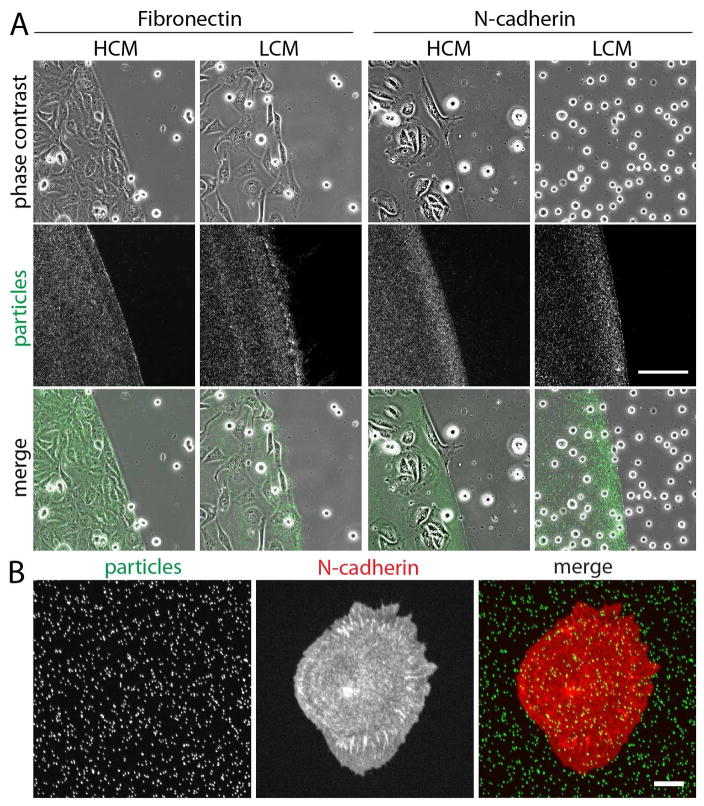
Figure 1. Cell adhesion on fibronectin and N-cadherin coated elastomer surface.
(A) Cell adhesion on fibronectin or purified N-cadherin coated elastomer in high calcium media (HCM) or low calcium media (LCM). The area of fibronectin or N-cadherin coating was marked by the presence of fluorescent particles. The MDCK cells (HGF-treated) adhere to the protein-coated surface, except in the absence of calcium, where the cells did not adhere on the N-cadherin coated surface. Scale bar 100 μm. (B) Localization of DsRed tomato tagged N-cadherins on purified N-cadherin coated surface. N-cadherins accumulate as puncta similar to focal adhesions. Scale bar 10 μm.
On the purified N-cadherin coated surface, the transformed cells adhered to the extent indistinguishable from those cells on the fibronectin coated surface in the presence of calcium, but no cell adhesion or spreading was observed in the absence of calcium (Figure 1A). These results suggest that the surface bound, purified N-cadherins promote cell adhesion of N-cadherin expressing transformed cells. Cell adhesion to the purified N-cadherin coated surface recruited cell surface N-cadherins to focal adhesion like puncta (Figure 1B). Similar to focal adhesions, these cadherin puncta co-localized with the tips of actin bundles (see Figure 4A), suggesting that N-cadherin junctions are likely force-bearing.
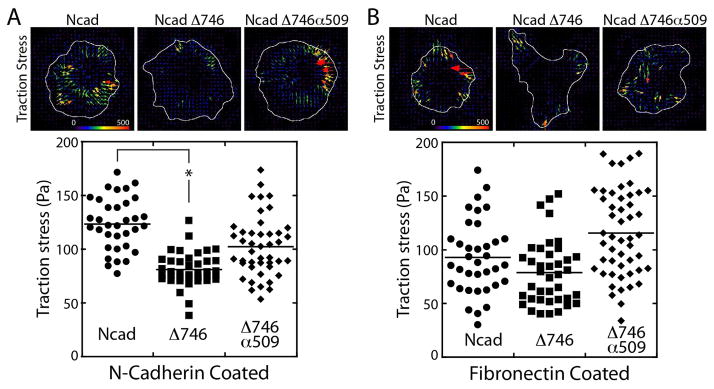
Figure 4. Traction stress generated by mutant N-cadherin expressing cells.
(A) Traction force exerted on purified N-cadherin-coated surface by the full-length (123.48 ± 24.28 Pa, average ± standard deviation), the cytoplasmic deleted (81.07 ± 15.84 Pa), or α-catenin chimeric (102.50 ± 27.97 Pa) N-cadherin expressing cells. The vector map of traction stress (top) and the average traction stresses from individual cells (bottom). Results were analyzed using a one-way ANOVA; significance was determined using Dunnett’s post hoc test. Results were considered significant with P<0.0001. (B) Traction force exerted on fibronectin-coated surface by the full-length (92.97 ± 34.33 Pa, average ± standard deviation), the cytoplasmic deleted (78.81 ± 30.15 Pa), or α-catenin chimeric (115.55 ± 40.36 Pa) N-cadherin expressing cells The vector map of traction stress (top) and the average traction stresses from individual cells (bottom).
Traction force analysis using a soft elastomer
To analyze traction force generated by N-cadherin expressing transformed epithelial cells, these cells were plated on a CY52-276 elastomer film. With elastic modulus of ~1000 Pa at low frequencies (Figure 2B), the surface deformation by adhered cells was easily detectable. The surface deformation was detected by the difference in the positions of fiducially particle markers between live cells and SDS detergent treated dead cells (Figure 2A). Using the particle displacements (Figure 2A) and the elastomer elastic modulus (Figure 2B), traction stress generated by the cells was calculated (Figure 2C and D – see also Materials and Methods). On fibronectin coated elastomer substrate, the transformed cells exerted greater average traction stress than on N-cadherin coated substrate (Figure 2C and D).
N-cadherin mutants expressing transformed MDCK cells
The cytoplasmic domain of N-cadherin is thought to be indispensable for the cytoskeletal linkage necessary for traction force transmission. To test the roles of the cytoplasmic domain of N-cadherin in generation of traction force, transformed epithelial cells expressing mutant N-cadherins: the full-length (Ncad), the cytoplasmic deleted (NcadΔ746), and the α-catenin actin binding domain fused (NcadΔ746α509) N-cadherins (Figure 3A) [24] were analyzed. Transfected cells also expressed a canine specific N-cadherin shRNA to reduce the endogenous level of N-cadherin (Figure 3B) [24]. All N-cadherin constructs were tagged with a tandem dimer tomato at the C-terminus (red circle, Figure 3A). In Western blots using the N-cadherin cytoplasmic domain specific antibody (Ncad) and DsRed antibody (DsR), the level of full-length N-cadherin were significantly higher than that of the endogenous level of N-cadherin (Figure 3B, Ncad blot). Furthermore, the levels of the full-length, the cytoplasmic deleted and the chimeric N-cadherins were comparable, suggesting that all exogenous N-cadherins were expressed at higher levels than endogenous N-cadherin. All N-cadherin mutant expressing cells were able to adhere to the N-cadherin coated elastomer, but not on non-coated elastomer surface (Figure 3C), suggesting that the extracellular domain of N-cadherin was sufficient for adhesion to the purified N-cadherin coated surface.
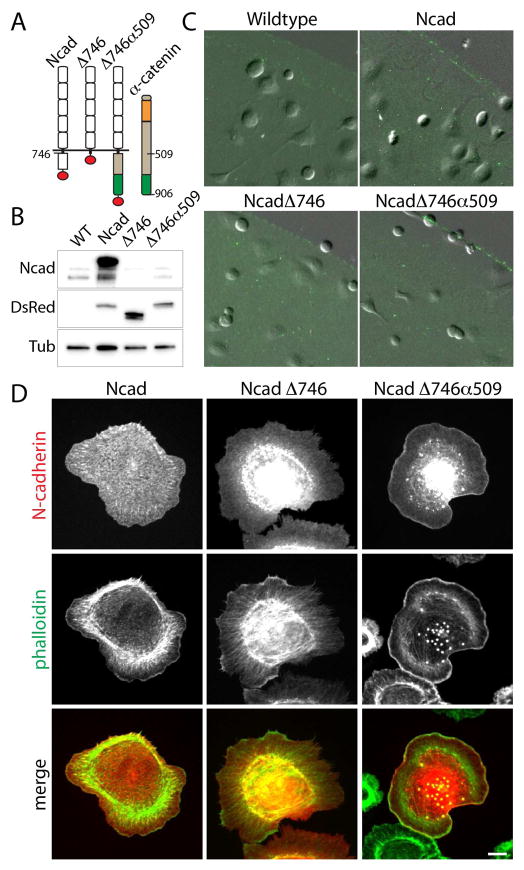
Figure 3. N-cadherin mutants expressed in HGF-treated, transformed MDCK epithelial cells.
(A) Schematic of N-cadherin mutants. Three N-cadherin constructs were used in this study: the full-length (Ncad), the cytoplasmic deleted (NcadΔ746), the chimera of the extracellular domain of N-cadherin and the C-terminus of α-catenin (NcadΔ746α509). The green segment of α-catenin represents the actin-binding domain. All constructs were tagged with tandem dimer DsRed tomato (red circle). (B) Western blot of N-cadherin expressing cells using the cytoplasmic domain specific N-cadherin antibody (Ncad), DsRed antibody (DsR), and tubulin antibody (Tub). (C) Cell adhesion to N-cadherin coated elastomer. All cells adhere to N-cadherin coated surface (marked by the fluorescent particles) but not on non-coated elastomer. (D) N-cadherin (red) and phalloidin (green) distribution of N-cadherin expressing cells plated on purified N-cadherin coated surface. The full-length N-cadherin formed puncta along cell peripheral and otherwise localized on plasma membrane. The cytoplasmic deletion (NcadΔ746) or the addition of actin binding domain of α-catenin (NcadΔ746α509) prevented efficient trafficking of the mutant N-cadherins to plasma membrane and the significant portion of proteins accumulated in Golgi. While NcadΔ746 localized uniformly on the membrane, NcadΔ746α509 accumulated at lamellipodia, most evident in the highest expressers. Scale bar 10 μm.
The actin cytoskeletal network is a critical requirement of traction force generation. On the N-cadherin coated surface, the full-length N-cadherin clustered around the tips of actin bundles similar to focal adhesion like structures (Figure 1B and 3D). In the absence of the cytoplasmic domain of N-cadherin, the NcadΔ746 mutant appeared uniformly on the plasma membrane of adhered cells, and the actin cytoskeleton emanated from the center of cells without distinct focus and bundling (Figure 3D). Interestingly, the chimeric NcadΔ746α509 mutant localized to the edge of lamellipodia where intense actin accumulation was also observed (Figure 3D). These observations were particularly pronounced for the highest expressers of the chimeric N-cadherin. Both the cytoplasmic domain mutants accumulated in the Golgi (Figure 3D), suggesting that these mutants are inefficiently trasported through the secretion pathway. Despite the mutations, these cells adhered to a similar extent to the wildtype cells (Figure 3C), suggesting that minimal dependency of the N-cadherin cytoplasmic sequence for the N-cadherin mediated adhesion. However, the cytoplasmic sequence had a significant impact in the organization of actin network while adhering to the N-cadherin-coated substrates.
Traction force analysis of N-cadherin mutant expressing cells
While all the full-length N-cadherin and N-cadherin mutant expressing cells adhered to the N-cadherin coated substrate (Figure 3C and 3D), their traction force generation was not the same. On purified N-cadherin coated substrates, traction stress of the full-length N-cadherin expressing cells (Figure 4A Ncad) was greater than that of wildtype transformed cells (only expresses endogenous N-cadherin) on N-cadherin coated surfaces (Figure 1C). The lower traction stress generated by the wildtype cells relative to the full-length N-cadherin expressing cells may be due to the lower level of total N-cadherin expressed in the wildtype cells (Figure 3B).
On purified N-cadherin coated substrate, the NcadΔ746 mutant expressing cells exerted a lower level of traction stress than the full-length N-cadherin expressing cells (Figure 4A), but traction stress exerted by the NcadΔ746 mutant expressing cells was not insignificant. In contrast, the chimeric NcadΔ746α509 mutant expressing cells exerted a similar level of traction stress as the full-length N-cadherin expressing cells (Figure 4A). The distinct actin organization of the full-length from the chimeric N-cadherin expressing cells (Figure 3D) suggest that, while the full-length and chimeric N-cadherin expressing cells both exerted similar magnitude of traction forces, the underlying mechanism of force-generation may be different.
The difference in the magnitudes of traction stress may be due to the difference in the intrinsic ability of cells to generate contractile force. To test this hypothesis, the elastomer substrate was coated with fibronectin to analyze traction force generation via the integrin complex. On fibronectin-coated substrates, the full-length or the NcadΔ746 mutant expressing cells exerted the similar levels of traction stress, whereas the chimeric N-cadherin expressing cells exerted a slightly higher level of traction stress than others (Figure 4B). These data suggest that the NcadΔ746 mutant expressing cells do have an ability to generate traction force, and that the reduced traction stress on the N-cadherin-coated surface is likely due to the absence of the N-cadherin cytoplasmic domain.
Discussion
N-cadherin is often up-regulated in aggressive cancers, and N-cadherin mediated cell-cell adhesion may play a role in the progression of cancers [29, 30]. Therefore, the regulation of N-cadherin-mediated cell-cell adhesion and traction force transmission is likely to be critical for understanding how cancer cells invade and metastasize. In our study, N-cadherin mediated junctions exert traction stress in the order of 100 Pa (Figure 2D and 4A). This is consistent with previous studies using normal, E-cadherin expressing MDCK cells and polyacrylamide gel substrate coated with purified E-cadherin [31–33]. While these studies (including this study) measured forces exerted to the artificial cadherin coated substrates, these data serve as a first estimate of forces between cadherins junctions to promote 3D collective cell migration.
The mechanism by which the cytoplasmic domain deleted N-cadherin expressing cells exert significant traction stress remains unresolved. One possibility is that other cadherins expressed in these cells (e.g. K-cadherin or cadherin 11, but not E-cadherin as a minimal level of E-cadherins is expressed in these cells) [24] are interacting with purified N-cadherins on the substrate. However, this is unlikely as the removal of endogenous N-cadherin completely eliminated cell-cell adhesion [24], suggesting that N-cadherin as the predominant cadherin for cell-cell adhesion in these cells. Alternatively, the cytoplasmic domain deleted N-cadherin may be interacting with other trans-membrane proteins that indirectly link the mutant N-cadherin to the actin cytoskeleton. Our attempts to isolate such binding partners using immuno-precipitation analysis has not be successful, suggesting that such interactions, if they exist, are likely to be weak or transient interactions that are not preserved in the assay.
Interestingly, on the purified N-cadherin-coated substrate, the actin cytoskeleton of the cytoplasmic domain deleted N-cadherin expressing cells are more dispersed than well-defined actin bundles formed in the full-length N-cadherin expressing cells (Figure 3D). While it is unclear as to how this dispersed actin network is generating forces, this unique actin organization may be the source of the lower traction stress observed (Figure 4A). Further analysis is needed to resolve the mechanism by which these actin networks transmit traction force via N-cadherin junctions. Our analysis demonstrates that the cytoplasmic domain deleted N-cadherin expressing cells exert significant traction forces, and this could represent an alternative mode of force-transmission without the cytoplasmic domain of N-cadherin.
Supplementary Material
Highlights.
N-cadherin mediated traction force analysis of HGF-transformed MDCK cells.
The assembly of N-cadherin puncta requires the cytoplasmic domain of N-cadherin.
Loss of N-cadherin cytoplasmic domain reduces, but does not eliminate, traction.
Acknowledgments
We thank Dr. Kent Leach (University of California Davis) for the use of the Discovery HR2 Hybrid rheometer. This work was supported by NIH EUREKA GM094798, UC Davis Bridge Fund, NIH R03 EB021636 and NSF 1562095 (all to SY).
Abbreviations
- MDCK
Madin-Darby canine kidney
- HGF
Hepatocyte Growth Factor
- EMT
Epithelial-to-Mesenchymal Transition
- N
neural
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 2.Scarpa E, Mayor R. Collective cell migration in development. J Cell Biol. 2016;212:143–155. doi: 10.1083/jcb.201508047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins C, Nelson WJ. Running with neighbors: coordinating cell migration and cell-cell adhesion. Current opinion in cell biology. 2015;36:62–70. doi: 10.1016/j.ceb.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das T, Safferling K, Rausch S, Grabe N, Boehm H, Spatz JP. A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nature cell biology. 2015;17:276–287. doi: 10.1038/ncb3115. [DOI] [PubMed] [Google Scholar]
- 5.Li L, Hartley R, Reiss B, Sun Y, Pu J, Wu D, Lin F, Hoang T, Yamada S, Jiang J, Zhao M. E-cadherin plays an essential role in collective directional migration of large epithelial sheets. Cellular and molecular life sciences: CMLS. 2012;69:2779–2789. doi: 10.1007/s00018-012-0951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, Ladoux B. Force mapping in epithelial cell migration. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reffay M, Parrini MC, Cochet-Escartin O, Ladoux B, Buguin A, Coscoy S, Amblard F, Camonis J, Silberzan P. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nature cell biology. 2014;16:217–223. doi: 10.1038/ncb2917. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi N, Mizutani T, Kawabata K, Haga H. Leader cells regulate collective cell migration via Rac activation in the downstream signaling of integrin beta1 and PI3K. Scientific reports. 2015;5:7656. doi: 10.1038/srep07656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kollimada SA, Kulkarni AH, Ravan A, Gundiah N. Advancing Edge Speeds of Epithelial Monolayers Depend on Their Initial Confining Geometry. PloS one. 2016;11:e0153471. doi: 10.1371/journal.pone.0153471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zehnder SM, Suaris M, Bellaire MM, Angelini TE. Cell Volume Fluctuations in MDCK Monolayers. Biophys J. 2015;108:247–250. doi: 10.1016/j.bpj.2014.11.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poujade M, Grasland-Mongrain E, Hertzog A, Jouanneau J, Chavrier P, Ladoux B, Buguin A, Silberzan P. Collective migration of an epithelial monolayer in response to a model wound. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haga H, Irahara C, Kobayashi R, Nakagaki T, Kawabata K. Collective movement of epithelial cells on a collagen gel substrate. Biophys J. 2005;88:2250–2256. doi: 10.1529/biophysj.104.047654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tambe DT, Hardin CC, Angelini TE, Rajendran K, Park CY, Serra-Picamal X, Zhou EH, Zaman MH, Butler JP, Weitz DA, Fredberg JJ, Trepat X. Collective cell guidance by cooperative intercellular forces. Nature materials. 2011;10:469–475. doi: 10.1038/nmat3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angelini TE, Hannezo E, Trepat X, Marquez M, Fredberg JJ, Weitz DA. Glass-like dynamics of collective cell migration. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4714–4719. doi: 10.1073/pnas.1010059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaritsky A, Welf ES, Tseng YY, Rabadan MA, Serra-Picamal X, Trepat X, Danuser G. Seeds of Locally Aligned Motion and Stress Coordinate a Collective Cell Migration. Biophys J. 2015;109:2492–2500. doi: 10.1016/j.bpj.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen DJ, Nelson WJ, Maharbiz MM. Galvanotactic control of collective cell migration in epithelial monolayers. Nature materials. 2014;13:409–417. doi: 10.1038/nmat3891. [DOI] [PubMed] [Google Scholar]
- 17.Shih W, Yamada S. N-cadherin as a key regulator of collective cell migration in a 3D environment. Cell adhesion & migration. 2012;6:513–517. doi: 10.4161/cam.21766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Rooij J, Kerstens A, Danuser G, Schwartz MA, Waterman-Storer CM. Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J Cell Biol. 2005;171:153–164. doi: 10.1083/jcb.200506152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sperry RB, Bishop NH, Bramwell JJ, Brodeur MN, Carter MJ, Fowler BT, Lewis ZB, Maxfield SD, Staley DM, Vellinga RM, Hansen MD. Zyxin controls migration in epithelial-mesenchymal transition by mediating actin-membrane linkages at cell-cell junctions. Journal of cellular physiology. 2010;222:612–624. doi: 10.1002/jcp.21977. [DOI] [PubMed] [Google Scholar]
- 20.Hoj JP, Davis JA, Fullmer KE, Morrell DJ, Saguibo NE, Schuler JT, Tuttle KJ, Hansen MD. Cellular contractility changes are sufficient to drive epithelial scattering. Exp Cell Res. 2014;326:187–200. doi: 10.1016/j.yexcr.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Montesano R, Matsumoto K, Nakamura T, Orci L. Identification of a fibroblast-derived epithelial morphogen as hepatocyte growth factor. Cell. 1991;67:901–908. doi: 10.1016/0092-8674(91)90363-4. [DOI] [PubMed] [Google Scholar]
- 22.O’Brien LE, Tang K, Kats ES, Schutz-Geschwender A, Lipschutz JH, Mostov KE. ERK and MMPs sequentially regulate distinct stages of epithelial tubule development. Developmental cell. 2004;7:21–32. doi: 10.1016/j.devcel.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Leroy P, Mostov KE. Slug is required for cell survival during partial epithelial-mesenchymal transition of HGF-induced tubulogenesis. Molecular biology of the cell. 2007;18:1943–1952. doi: 10.1091/mbc.E06-09-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shih W, Yamada S. N-cadherin-mediated cell-cell adhesion promotes cell migration in a three-dimensional matrix. J Cell Sci. 2012;125:3661–3670. doi: 10.1242/jcs.103861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drees F, Reilein A, Nelson WJ. Cell-adhesion assays: fabrication of an E-cadherin substratum and isolation of lateral and Basal membrane patches. Methods Mol Biol. 2005;294:303–320. doi: 10.1385/1-59259-860-9:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mertz AF, Che Y, Banerjee S, Goldstein JM, Rosowski KA, Revilla SF, Niessen CM, Marchetti MC, Dufresne ER, Horsley V. Cadherin-based intercellular adhesions organize epithelial cell-matrix traction forces. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:842–847. doi: 10.1073/pnas.1217279110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Style RW, Boltyanskiy R, German GK, Hyland C, MacMinn CW, Mertz AF, Wilen LA, Xu Y, Dufresne ER. Traction force microscopy in physics and biology. Soft matter. 2014;10:4047–4055. doi: 10.1039/c4sm00264d. [DOI] [PubMed] [Google Scholar]
- 28.Martiel JL, Leal A, Kurzawa L, Balland M, Wang I, Vignaud T, Tseng Q, Thery M. Measurement of cell traction forces with ImageJ. Methods in cell biology. 2015;125:269–287. doi: 10.1016/bs.mcb.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121:727–735. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 30.Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- 31.Tabdili H, Langer M, Shi Q, Poh YC, Wang N, Leckband D. Cadherin-dependent mechanotransduction depends on ligand identity but not affinity. J Cell Sci. 2012;125:4362–4371. doi: 10.1242/jcs.105775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maruthamuthu V, Gardel ML. Protrusive activity guides changes in cell-cell tension during epithelial cell scattering. Biophys J. 2014;107:555–563. doi: 10.1016/j.bpj.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barry AK, Tabdili H, Muhamed I, Wu J, Shashikanth N, Gomez GA, Yap AS, Gottardi CJ, de Rooij J, Wang N, Leckband DE. alpha-catenin cytomechanics--role in cadherin-dependent adhesion and mechanotransduction. J Cell Sci. 2014;127:1779–1791. doi: 10.1242/jcs.139014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.