Abstract
Forced expression of the cytokine-induced large GTPase, human Guanylate-Binding Protein-1 (hGBP-1), in ovarian cancer cell lines increases resistance to paclitaxel. Elevated hGBP-1 RNA in ovarian tumors correlates with shorter recurrence-free survival. In contract, hGBP-1 is part of a gene signature predicting improved prognosis in all subtypes of breast cancers. hGBP-1 does not confer paclitaxel resistance on MCF-7 and TMX2-28 breast cancer cells. Expression of the isotype of the hGBP-1-interacting protein, PIM1, which may contribute to paclitaxel resistance when associated with hGBP-1, is different in breast and ovarian cancer cell lines. Breast cancer cell lines express the 44 kDa isoform of PIM-1, and ovarian cancer cell lines express the 33 kDa isoform.
Keywords: Breast cancer, Paclitaxel, Guanylate-binding protein, Drug resistance
1. Introduction
Almost 1 in every 8 women will get breast cancer in her lifetime [1]. While improvements in early diagnosis and treatment have significantly improved survival rates, over 40,000 women are expected to die of breast cancer in 2016 [1]. Many, if not most, women with advanced disease receive chemotherapy that includes a taxane, usually paclitaxel. Because development of drug resistance still remains a significant problem, understanding how cells become resistant to chemotherapeutic drugs such as paclitaxel could lead to significant improvement in overall survival.
MCF-7 breast cancer cells and OVCAR-8 and SKOV3 ovarian cancer cells were made resistant to paclitaxel to identify a gene signature corresponding to paclitaxel resistance in vitro [2]. One of the genes up-regulated in all three paclitaxel resistant cell lines was the interferon-induced large GTPase, hGBP-1 [2]. Forced expression of hGBP-1 in OVCAR-8, OVCAR-3, and SKOV3 ovarian cancer cells reduces sensitivity to paclitaxel [3,4](Wadi et al., under review). Elevated hGBP-1 also shortens progression-free survival (PFS) in patients with ovarian cancer (Wadi et al., under review). However, hGBP-1 is part of a 5-gene signature correlated with improved PFS for all subclasses of breast cancer [5]. Since paclitaxel is part of the standard treatment of breast cancer patients who receive chemotherapy, the possibility that hGBP-1 promotes paclitaxel resistance is inconsistent with its correlation with improved prognosis.
Human Guanylate-Binding Protein-1 (hGBP-1) is a member of the GBP family of cytokine-induced large GTPases (reviewed in Ref. [6]). Best known for their anti-microbial activities, GBPs are poorly characterized in several tumor types. In glioblastomas, hGBP-1 is induced by EGFR signaling and increases invasion because it is required for EGF-induction of MMP-1 [7]. It also is a predictor of poor prognosis in head and neck cancer and promotes lymph node metastasis in esophageal cancer [8,9]. In addition, increased GBP-1 expression also predicts shorter RFS in ovarian cancer (Wadi et al., in review). An the other hand, hGBP-1 inhibits proliferation and invasion of endothelial cells by down-regulating MMP-1 expression [10,11], thereby inhibiting angiogenesis. Its expression is correlated with improved RFS in both colon and breast cancers [5,12]. Much remains unclear about how hGBP-1 functions and it is still unknown how it contributes to such effects on prognosis in the different tumor types.
hGBP-1 contributes to poor prognosis in ovarian cancer, in part, by contributing to paclitaxel resistance. Here we examine if GBP-1 contributes to paclitaxel resistance in breast cancer cell lines. Forced expression of GBP-1 in MCF-7 breast cancer cells does not decrease sensitive to paclitaxel. Reducing expression of GBP-1 in TMX2-28 cells does not make the cells more sensitive to paclitaxel. Breast cancer lines expressed the 44 kDa isoform of PIM1; ovarian cancer cell lines expressed the 33 kDa isoform. Little is known about functional differences between PIM1 isoforms, but PIM1 is a GBP-1 binding partner and this interaction may contribute to drug resistance. The different in PIM1 isoforms may be responsible for the differences in paclitaxel resistance due to hGBP-1.
2. Materials and methods
2.1. Cells and plasmids
Cells obtained from ATCC (Manassas, VA) were cultured as described [13]. U251 glioblastoma cells and MCF-7 TAX cells were the gift of William Maltese (University of Toledo) [14]. pCMV2(NH) flag-hGBP-1was generated as described (Wadi et al., under review).
MCF-7 Tet-off cells were purchased from Clontech Laboratories, Mountain View, CA. Cells were transfected and stable clones containing either pTRE-hygro (control transfectants) or pTRE-myc-hGBP-1-hygro were screened for inducible expression by Western blotting with anti-myc. Failure to express hGBP-1 in the control cells was confirmed with polyclonal anti-hGBP-1 antisera. Two control clones (8-2c and 9-2c) and two myc-hGBP-1 expressing (4-4 and 7-7) cell lines were used for these experiments described.
2.2. Cytokines, antibiotics, antibodies, paclitaxel
The following reagents were purchased from the indicated sources: anti-Flag monoclonal antibody M2 and rabbit anti-actin (A2066), Sigma-Aldrich (St. Louis, MO); mouse monoclonal anti-βIII tubulin (clone 5G8), Promega Corporation, Madison, WI; mouse anti-PIM1 (sc-374116), Santa Cruz Biotechnology, Dallas, TX; recombinant human interferon gamma (hIFN-γ), PBL Biomedical Laboratories, Piscataway, NJ; hygromycin B solution G-418, puromycin, and tetracycline hydrochloride, Research Products International Corp., Mt. Prospect, IL; and paclitaxel, Calbiochem (cat# 580555).
2.3. Generation of polyclonal anti-hGBP-1 antisera
Rabbit polyclonal antisera against hGBP-1 was generated and immunopurified as described (Wadi et al., under review).
2.4. SDS PAGE and Western blot analysis
Cells were lysed, separated on 8% SDS-PAGE gels, and transferred to PVDF membrane [15]. Secondary antibodies used were horseradish peroxidase-conjugated goat anti-rabbit (1:10,000, Jackson ImmunoResearch, West Grove, PA) and anti-mouse (1:800, Jackson Immunolaboratories).
2.5. Immunofluorescence
Indirect immunofluorescence of tet-regulated MCF-7 cells was performed as previously described [15], except that immunopurified C5391 was used at a dilution of 1:10. For analysis of three antigens, primary antibodies were added together: C5391 (1:50), anti-PIM1 (1:200), and anti-TUBB3 (1:1000). After washing the three secondary antibodies (1:1000 dilution of each) were added together: Alexa 488-conjugated goat anti-rabbit, Alexa 594-conjugated goat anti-mouse IgG1, and Alexa 647-conjugated goat anti-mouse IgG2a. Confocal images were collected using a TCS-SP spectrophotometric multiphoton laser scanning confocal microscope (Leica Microsystems) using 1 µm optical sections.
2.6. SRB assays
Sulforhodamine B (SRB) assay was adapted from Ref. [16]. Cells were plated at 3000 cells per well and in quadruplicate for each drug concentration. After incubation overnight media containing paclitaxel at the indicated concentration was added for 48 h. The contents of plate were precipitated with trichloroacetic acid (TCA) and stained with 0.4% (m/v) Sulforhodamine B (Sigma) solution in 1% (v/v) acetic acid. Wells were washed with 1% (v/v) acetic acid and stained cells were resuspended in 200 µl of 10 mM Tris, pH 10.5. Absorbances at 515 nm, subtracting 620 nm, were read using SpectraMaPlus 384 absorbance micro plate reader. The GI50s from the three SRB assays were averaged, standard deviations were calculated, and significance was determined using a two-tailed T-test after determining variance by F-test in Microsoft Excel.
2.7. Colony assays
Cells were plated into 60-mm plates at 900–1000 cells per plate in triplicate for each drug concentration. The following day the media was replaced with complete DMEM containing appropriate drug concentrations. After 48 h, each plate was washed and received drug-free complete media until individual colonies reached ≥ 50 cells per colony (~7–10 days). Cells were fixed with 100% ice-cold methanol, followed by staining with 1% (m/v) crystal violet solution. Each plate was examined under a dissecting microscope, and all of the colonies with ≥ 50 cells were counted.
2.8. shRNA knockdown of hGBP-1 in TMX2-28 cells
To generate shRNA against hGBP-1, the following forward and reverse oligonucleotides were allowed to anneal and were ligated into BamH1 cut pSIH-H1:
- hGBP-1 shRNA #1
-
forward GATCCGACGAAAGGCATGTACCATACTCGAGTATGG TACATGCCTTTCGTCGTTTTTGreverse AATTCAAAAACGACGAAAGGCATGTACCATACTCGA GTATGGTACATGCCTTTCGTCG
-
- hGBP-1 shRNA #2
-
forward GATCTGAGACGACGAAAGGCATGTACTCGAGTACAT GCCTTTCGTCGTCTCATTTTTGreverse AATTCAAAAATGAGACGACGAAAGCCATGTACTCGA GTACATGCCTTTCGTCGTCTCA
-
- hGBP-1 shRNA #3
-
forward GATCCGGAAATTCTTCCCAAAGAAACTCGAGTTTCTT TGGGAAGAATTTCCGTTTTTGreverse AATTCAAAAACGGAAATTCTTCCCAAAGAAACTCGAG TTTCTTTGGGAAGAATTTCCG
-
- eGFP shRNA #1A
-
forward GATCCTACAACAGCCACAACGTCTATCTTCCTGTCAG AATAGACGTTGTGGCTGTTGTATTTTTGreverse AATTCAAAAATACAACAGCCACAACGTCTATGCTGAC AGGAAGATAGACGTTGTGGCTGTTGTAG
-
- eGFP shRNA #2A
-
forward GATCCCACAAGCTGGAGTACAACTACAACAGCCACT TCCTGTCAGATGGCTGTTGTAGTTGTACTCCAGCTTGTGTTTTTGreverse AATTCAAAAACACAAGCTGGAGTACAACTACAACAG CCATCTGACAGGAAGTGGCTGTTGTAGTTGTACTCCAGCTTGTGG
-
After transfection, cells were characterized by Western blot analysis after IFN-γ treatment to induce hGBP-1.
2.9. Progression-free survival
To determine progression-free survival (PFS) as a function of gene expression, KmPlot was used (kmplot.com/analysis/index.php?p=service&cancer=ovar). The 2015 version of the TCGA database with 1648 ovarian samples was screened for PIM1 expression with the Affymetrix probe of ID number 209193_at and results expressed as median PFS for all stages, histologies, grades, and p53 status of ovarian cancers that underwent optimal debulking. The 2014 version of TCGA database with 4142 breast cancer samples was screened using the same probe against breast cancers of all subtypes, hormone receptor status, grade or treatment.
3. Results and discussion
3.1. MCF-7 cells and paclitaxel resistance
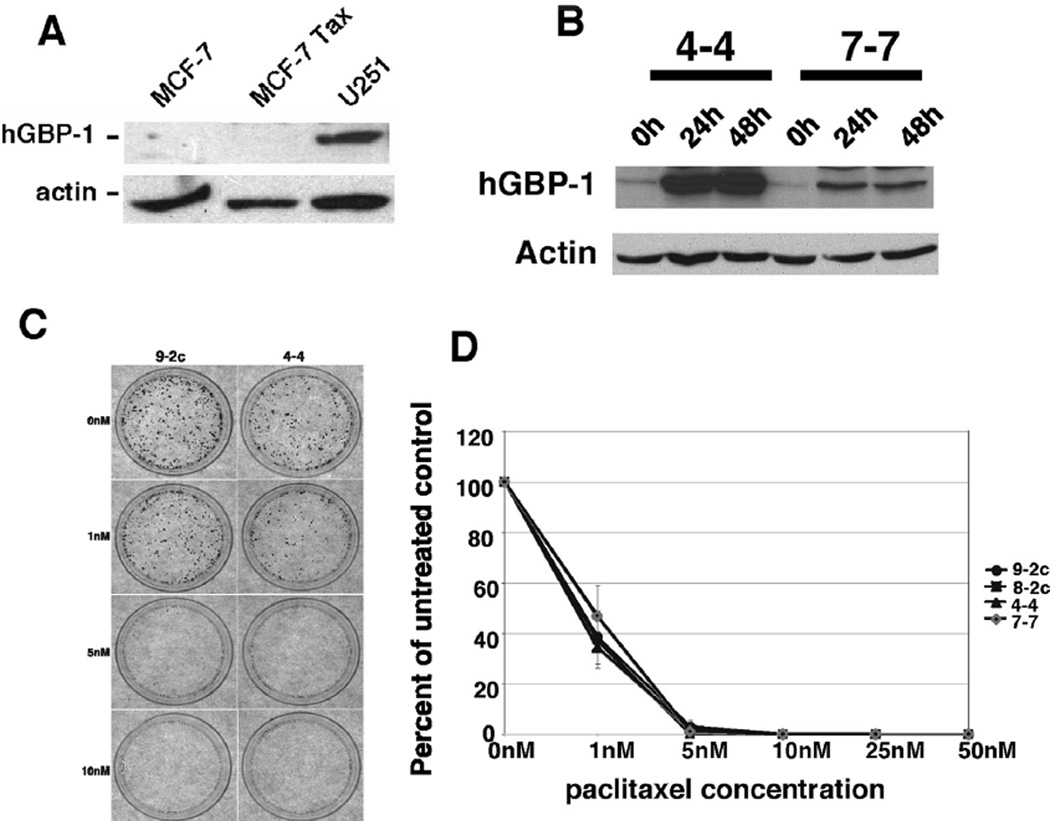
A paclitaxel-resistant version of MCF-7 cells was screened for hGBP-1 expression to determine if hGBP-1 is a prerequisite for paclitaxel (6.64 nM) resistance (Fig. 1A) [14]. Paclitaxel resistant MCF-7 cells did not express hGBP-1. Cells may become resistant to paclitaxel in several ways, so we generated MCF-7 cell lines with tet-regulated expression of hGBP-1. Forced expression of hGBP-1 for 24 or 48 h had no effect on paclitaxel sensitivity (Fig. 1B–D).
Fig. 1.
A. hGBP-1 was not expressed in paclitaxel-sensitive and -resistant MCF-7 cells. U251 glioblastoma cells were used as a positive antibody control. B. Control vector (8-2c and 9-2c) and tetracycline (tet)-regulated MCF-7 cells (4-4 and 7-7) were incubated in the absence of tet for the times indicated and examined for hGBP-1. C. Tet-regulated control and hGBP-1-expressing cells were treated with the concentrations of paclitaxel listed for 48 h. Drug was removed and the cells incubated in media without tet for 7–10 days. Colonies were visualized by staining with crystal violet. Representative photomicrographs are shown. D. The numbers of colonies with 50 or more cells were counted. Results of three experiments performed in triplicate are shown. Results are normalized to the number of colonies generated by untreated control cells. The average number of colonies ± SD are shown (n = 3).
3.2. hGBP-1 expression in breast cancer cell lines
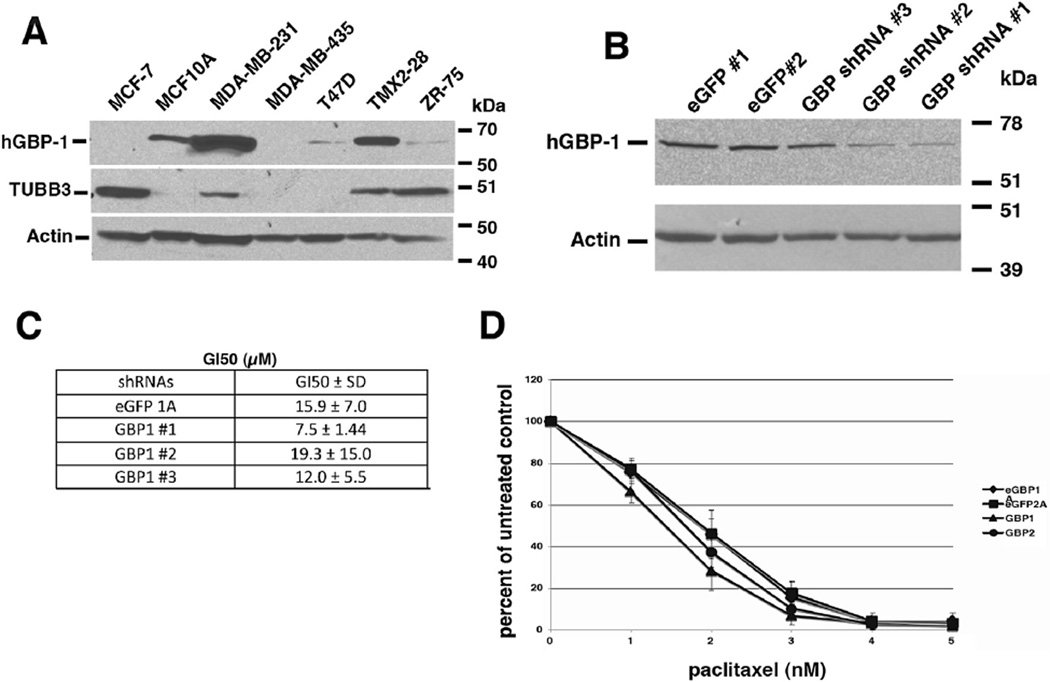
Forced expression of hGBP-1 in MCF-7 cells did not influence paclitaxel-mediated cell death, so we searched for breast cancer cell lines already expressing hGBP-1 for further analysis (Fig. 2A). MCF-7, T47D, and ZR-75 cells are estrogen receptor positive (ER+) and progesterone receptor positive (PR+) invasive ductal carcinoma cells that lack Her2/neu amplification [17–19]. MCF-7 cells had no detectable hGBP-1, while T47D and ZR-75 expressed very low levels (Fig. 2A). MD-MB-435 cells are ER−/PR− but Her2/neu positive. ATCC now lists these cells as melanoma. MDA-MB-231 cells are ER−/PR−/Her2-or triple negative (TN) invasive ductal carcinoma cells [20]. MDA-MB-231 cells robustly express hGBP-1 (Fig. 2A). TMX2-28 is a tamoxifen-resistant variant of MCF-7 cells that are now ER−/PR−/Her2− [21]. These cells also express hGBP-1. MCF10A cells were derived from a mastectomy for benign breast disease and also are ER−/PR−/Her2/neu− [22](Fig. 2A). The cell lines that most robustly expressed hGBP-1 are the triple-negative breast cancers.
Fig. 2.
Triple-negative breast cancer cell lines express hGBP-1. A. Cells were lysed, proteins separated by SDS-PAGE, and analyzed for the expression of hGBP-1, TUBB3, and actin. B. TMX2-28 cells were transfected with two different shRNAs against eGFP (#1 and #2) and three different shRNAs against hGBP-1 (hGBP-1 #1, #2, and #3). Cells were selected in puromycin and analyzed for hGBP-1 expression by Western blot. All three shRNAs against hGBP-1 knocked down hGBP-1 expression but cells containing shRNAs # 1 and 2 were used in the next assays. C. TMX2-28 cells stably expressing the shRNAs were analyzed for paclitaxel sensitivity by SRB assay. D. Colony assays showed that the cells were very sensitive to the drug; the loss of hGBP-1 did not increase sensitivity.
3.3. hGBP-1 does not protect TMX2-28 cells from paclitaxel-mediated killing
To determine if hGBP-1 protects TMX2-28 cells from paclitaxel-induced killing, we knocked down hGBP-1 (Fig. 2B). No differences were observed in the growth inhibition 50 (GI50) for paclitaxel in the presence or absence of hGBP-1 by SRB assay (Fig. 2C). In colony forming assays the differences in GI50s of control and hGBP-1-depleted cell lines were about 1 and 2 nM respectively, differences that are not physiologically significant (Fig. 2D). Thus, hGBP-1 does not reduce sensitivity to paclitaxel in either MCF-7 or TMX2-28 cells.
3.4. Presence and consequences of putative regulators of hGBP-1
In ovarian cancer cell hGBP-1 may be in a three protein complex containing the β-tubulin isoform III, TUBB3, and the serine/threonine kinase, PIM1 and may interact with and activate (TUBB3) [3,4]. Both PIM1 and TUBB3 have been studied independently for their roles in paclitaxel resistance in other tumor types.
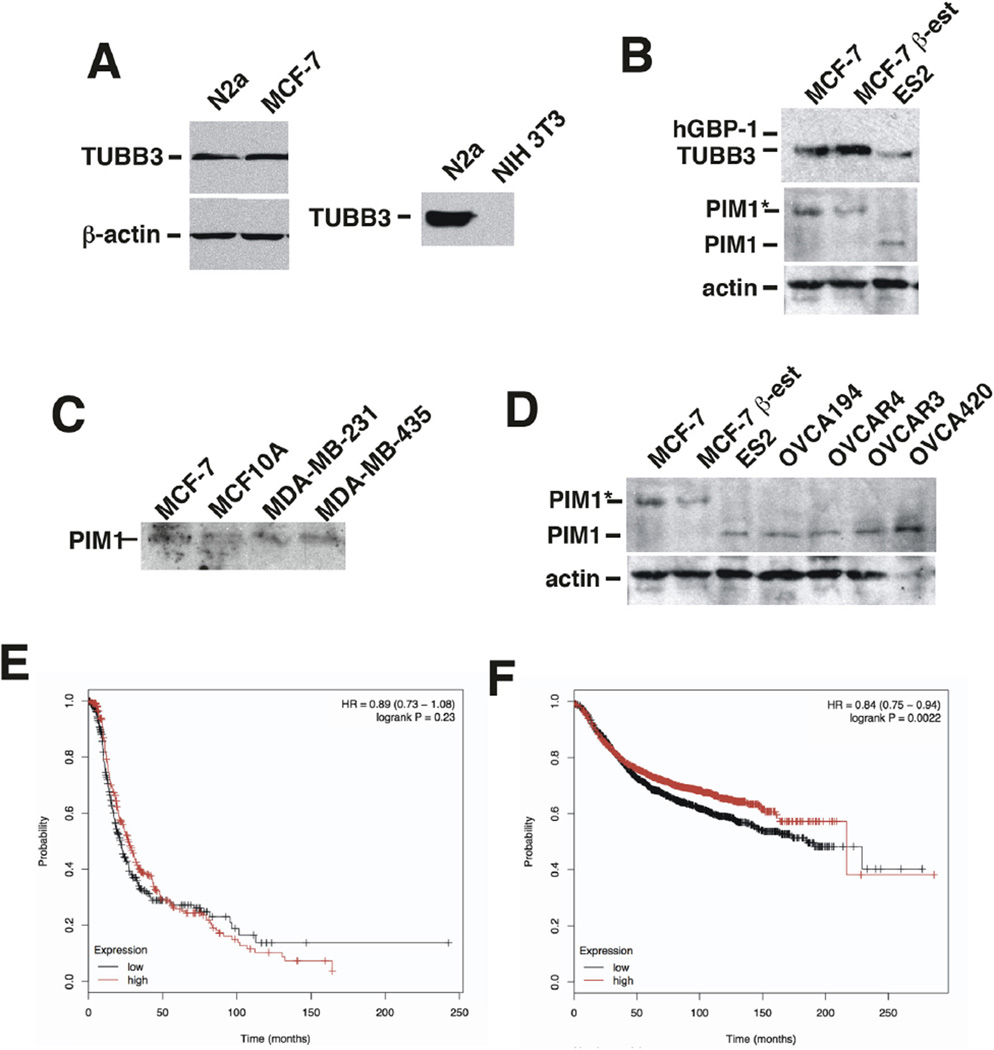
hGBP-1 may behave differently in breast cancer cells is because they lack TUBB3 or PIM1. As expected, N2a cells robustly express TUBB3; NIH 3T3 cells do not (Fig. 3A). MCF-7 cells also express TUBB3 (Fig. 3B). β-estradiol increased the expression of TUBB3 in MCF-7 cells [23]. TUBB3 also was expressed in MDA-MB-231, TMX2-28, and ZR-75 breast cancer cells (Fig. 2A).
Fig. 3.
MCF-7 cells express TUBB3. A. Neuro-2a (N2a) and MCF-7 cell lysates (20 µg) were analyzed for TUBB3 expression by Western blotting. To confirm the specificity of the antibody, NIH 3T3 cell lysates were examined for TUBB3. B. MCF-7 cells express the higher molecular weight isoform of PIM1 (PIM-1L). C. Screen of multiple ovarian cancer cell lines shows that they express the smaller PIM-1 isoform, PIM-1S. E. PFS was determined for patients with ovarian tumors of all grades, stages, histologies, and p53 status that had undergone optimal debulking. The data from 341 tumors with low PIM1 and 340 tumors with high expression are shown. F. PFS was determined from breast cancers of all types, stages, and hormone receptor status. The data from 1778 tumors with low PIM1 expression and 1776 tumors with high expression are shown.
PIM1 has two isoforms, the 44 kDa PIM-1L and the 33 kDa isoform, PIM-1S due to the use of two different translation start sites. PIM-1S lacks a proline-rich domain at the amino terminus and is expected to have different intracellular binding partners than the longer isoform. MCF-7 cells express the 44 kDa isoform of PIM1 (PIM-1L) (Fig. 3B), as did the other breast cancer cells examined (Fig. 3D). Although over-expression of PIM1 may contribute to poor prognosis in tumors by promoting phenotypic changes that include drug resistance, there is little data that addresses isoform specific activities [24–27]. Most studies of PIM1 involved the 33 kDa isoform [27]. All ovarian cancer cell lines we examined expressed the 33 kDa isoform (Fig. 3C).
3.5. PIM1 expression levels predict different PFS in breast and ovarian cancers
To determine if the expression of different isoforms of PIM1 alters disease prognosis, the progression-free survival (PFS) of patients with ovarian tumors was determined for tumors with low or high initial PIM-1 expression [28]. The data used for this analysis were limited to those tumors isolated from patients who underwent optimal debulking surgery. The data included ovarian tumors of all stages, histologies, grades, and p53 status. The PFS of ovarian tumors with elevated levels of PIM-1 is no different than those with low levels of expression of PIM-1 (Fig. 3E). When the same analysis was performed on breast tumors, including all subtypes, hormone receptor statuses, or grade, the tumors with elevated PIM-1 RNA had significantly longer PFS than those with lower levels (Fig. 3F). Thus, the longer isoform of PIM-1 found in breast cancers may be protective. The initial identification of PIM1 as a putative binding partner for hGBP-1 used a protein blot with bacterially expressed hGBP-1 and possible interactions between the two proteins were later examined by protein modeling which most likely used PIM-1S [29]. Because all ovarian cancer cell lines examined expressed PIM-1S (Fig. 3) and expression of hGBP-1 makes ovarian cancer cells resistant to paclitaxel [3,4](Wadi et al., under review), one difference in responses to paclitaxel could be whether or not the express the PIM-1S or PIM-1L.
3.6. Intracellular localization of PIM-1, TUBB3, and hGBP-1 in ovarian and breast cancer cell lines
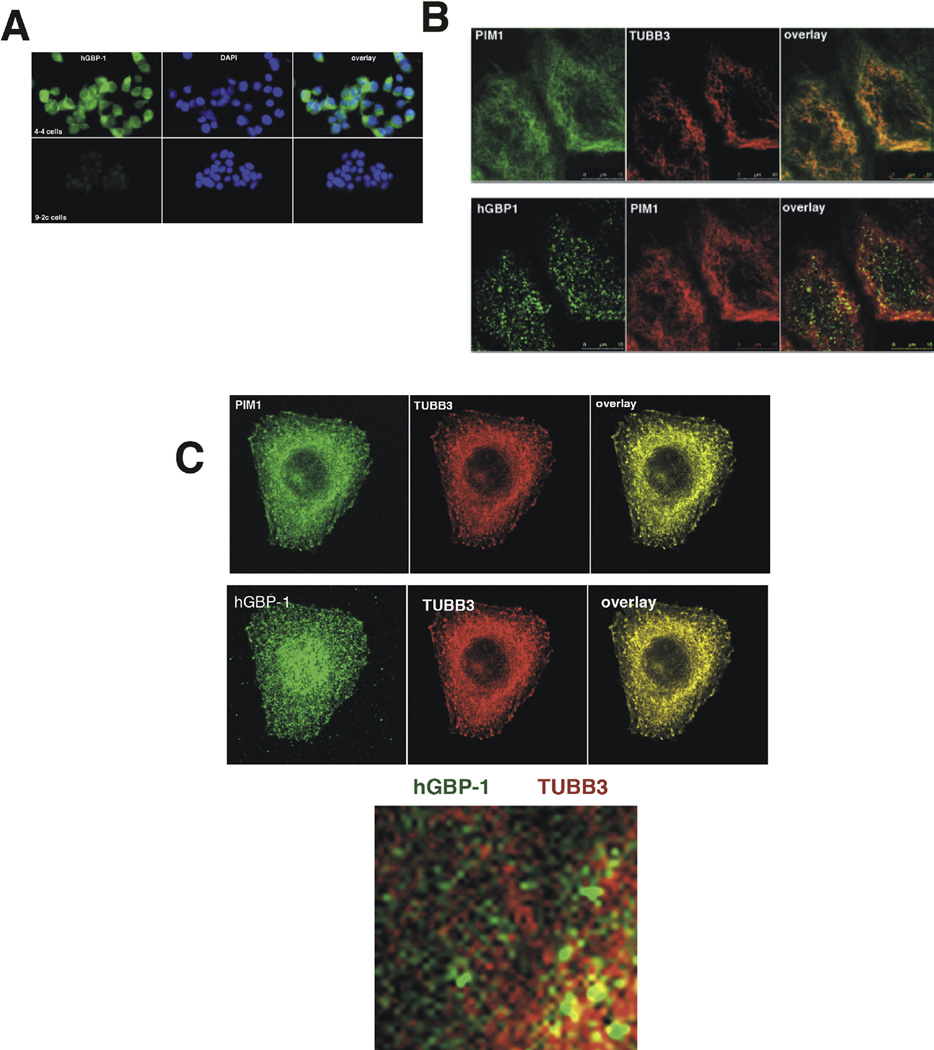
PIM-1L localizes to the plasma membrane and PIM-1S to the nucleus and cytoplasm in prostate cancer cells [27]. To examine the intracellular location of hGBP-1 and whether or not localization is altered in the presence of the PIM1 isoforms, we generated and immunopurified a polyclonal antisera against hGBP-1. In MCF-7 cells with or without hGBP-1, the antisera was specific (Fig. 4A). Triple label indirect immunofluorescence of PIM1, TUBB3, and hGBP-1 was performed in TMX2-28 cells (Fig. 4B). TMX2-28 cells were chosen because they express all three proteins (not shown) and flatten better for imaging than the MCF-7 cells from which they were derived. Although some PIM-1L was detected in the cytosol at increased exposure times, PIM-1L predominantly co-localized with TUBB3 to the microtubules of interphase TMX2-28 cells (Fig. 4B). hGBP-1 localized to the cytosol and to abundant intracellular vesicles. The distinct vesicles containing hGBP-1 are associated exclusively with the microtubules (Fig. 4B). To determine whether differential intracellular localization of the PIM-1S isoform in ovarian cancer cell lines could account for the differences in protection from paclitaxel, OVCA 420 ovarian cancer cells were stained for hGBP-1, PIM1, and TUBB3 (Fig. 4C). Both PIM1 isoforms localize to microtubules. hGBP-1 is located throughout the cytoplasm but is prominently associated with vesicles that exclusively run along the microtubules (Fig. 4C). Based on studies of the murine ortholog, mGBP-2, in NIH 3T3 fibroblasts that do not express TUBB3, this interaction is probably not TUBB3 specific (unpublished). Additionally, hGBP-1 may interact with tubulin in other cell types [30]. This association with microtubules occurs in both the cells where hGBP-1 may contribute to paclitaxel resistance and those in which it does not.
Fig. 4.
A. Immunopurified antiserum was tested on MCF-7 cells ± hGBP-1. After immunopurification, the polyclonal antisera against hGBP-1 only recognized cells expressing hGBP-1. B. TMX2-28 cells were stained for hGBP-1, TUBB3, and PIM1 and analyzed for intracellular distribution by confocal microscopy at 1 µm resolution. These cells express PIM-1L. C. OVCAR 420 cells were analyzed by triple label indirect IF for hGBP-1, TUBB3, and PIM-1 by confocal microscopy with 1 µm sections. OVCAR420 cells express PIM-1L. D. A blow-up of the OVCAR420 cells shows that vesicles containing hGBP-1 associate with microtubules.
hGBP-1 expression has different consequences in ovarian and breast cancer cells. These differences in outcome are accompanied by the expression of different isoforms of the hGBP-1 interacting protein, the proto-oncogene PIM1. Ovarian cancer cell lines express the smaller isoform, PIM-1S and breast cancer cell lines express the larger isoform, PIM-1L.
Acknowledgments
The authors would like to thank Dr. Andrea Kalinoski for her assistance with multiphoton confocal microscopy and Dr. Alan Goodridge for critical reading of the manuscript.
Financial support
These studies were funded by a grant from NIH/NCI (1R21CA132016) to D.J.V.
Abbreviations
- GBP
Guanylate-Binding Protein
- RFS
Recurrence-Free Survival
- SRB
Sulforhodamine B
- Tet
tetracycline.
Footnotes
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2016.08.169.
Conflict of interest
The authors report no conflict of interest.
References
- 1.American Cancer Society. Breast Cancer Facts & Figures 2015–2016. Atlanta, GA: American Cancer Society; 2015. [Google Scholar]
- 2.Duan Z, Lamendola DE, Duan Y, et al. Description of paclitaxel resistance-associated genes in ovarian and breast cancer cell lines. Cancer Chemother. Pharmacol. 2005;55:277–285. doi: 10.1007/s00280-004-0878-y. [DOI] [PubMed] [Google Scholar]
- 3.Duan Z, Foster R, Brakora KA, et al. GBP1 overexpression is associated with a paclitaxel resistance phenotype. Cancer Chemother. Pharmacol. 2005;57:25–33. doi: 10.1007/s00280-005-0026-3. [DOI] [PubMed] [Google Scholar]
- 4.De Donato M, Mariani M, Petrella L, et al. Class III β-tubulin and the cytoskeletal gateway for drug resistance in ovarian cancer. J. Cell Physiol. 2011;227:1034–1041. doi: 10.1002/jcp.22813. [DOI] [PubMed] [Google Scholar]
- 5.Ascierto ML, Idowu MO, Zhao Y, et al. Molecular signatures mostly associated with NK cells are predictive of relapse free survival in breast cancer patients. J. Transl. Med. 2013;11:145. doi: 10.1186/1479-5876-11-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vestal DJ, Jeyaratnam JA. The Guanylate-Binding Proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatases. J. Interferon Cytokine Res. 2011;31:89–97. doi: 10.1089/jir.2010.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Mukasa A, del-Mar Inda M, et al. Guanylate binding protein 1 is a novel effector of EGFR-driven invasion in glioblastoma. J. Exp. Med. 2011;208:2657–2673. doi: 10.1084/jem.20111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Ma G, Jing C, Liu Z. Guanylate-binding protein 1 (GBP1) promotes lymph node metastasis in human esophageal squamous cell carcinoma. Discov. Med. 2016;20:369–378. [PubMed] [Google Scholar]
- 9.Yu C-J, Chang K-P, Chang Y-J, et al. Identification of Guanylate-Binding Protein 1 as a potential oral cancer marker involved in cell invasion using omic-based analysis. J. Proteome Res. 2011;10:3778–3788. doi: 10.1021/pr2004133. [DOI] [PubMed] [Google Scholar]
- 10.Guenzi E, Topolt K, Cornali E, et al. The helical domain of GBP-1 mediates the inhibition of endothelial cell proliferation by inflammatory cytokines. EMBO J. 2001;20:5568–5577. doi: 10.1093/emboj/20.20.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guenzi E, Topolt K, Lubeseder-Martellato C, et al. The guanylate binding protein-1 GTPase controls the invasive and angiogenic capability of endothelial cells through inhibition of MMP-1 expression. EMBO J. 2003;22:3772–3782. doi: 10.1093/emboj/cdg382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britzen-Laurent N, Lipnik K, Ocker M, et al. GBP-1 acts as a tumor suppressor in colorectal cancer cells. Carcinogenesis. 2013;34:153–162. doi: 10.1093/carcin/bgs310. [DOI] [PubMed] [Google Scholar]
- 13.Gorbacheva VY, Lindner D, Sen GC, Vestal DJ. The IFN-induced GTPase, mGBP-2: role in IFN-g induced murine fibroblast proliferation. J. Biol. Chem. 2002;277:6080–6087. doi: 10.1074/jbc.M110542200. [DOI] [PubMed] [Google Scholar]
- 14.Villeneuve DJ, Hembruff SL, Veitch Z, et al. cDNA microarray analysis of isogenic paclitaxel- and doxorubicin-resistant breast tumor cell lines reveals distinct drug-specific genetic signature of resistance. Breast Cancer Res. Treat. 2006;96:17–39. doi: 10.1007/s10549-005-9026-6. [DOI] [PubMed] [Google Scholar]
- 15.Messmer-Blust AF, Balasubramanian S, Gorbacheva VY, et al. The interferon-γ-induced murine Guanylate-Binding Protein-2 (mGBP-2) inhibits Rac activation during cell spreading on fibronectin and after platelet-derived growth factor (PDGF) treatment: role for Phosphatidylinositol 3-Kinase. Mol. Biol. Cell. 2010;15:2514–2528. doi: 10.1091/mbc.E09-04-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voigt W. Sulforhodamine B assay and chemosensitivity. Methods Mol. Med. 2005;110:39–48. doi: 10.1385/1-59259-869-2:039. [DOI] [PubMed] [Google Scholar]
- 17.Soule HD, Vazguez J, Long A, et al. A human cell line from a plerual effusion derived from a breast carcinoma. J. Nat. Cancer Inst. 1973;51:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 18.Keydar I, Chen L, Karby S, et al. Establishment and characterization of a cell line of human breast carcinoma origin. Eur. J. Cancer. 1979;15:659–670. doi: 10.1016/0014-2964(79)90139-7. [DOI] [PubMed] [Google Scholar]
- 19.Engel LW, Yound NA. Human breast carcinoma cells in continuous culture: a review. Cancer Res. 1978;38:4327–4339. [PubMed] [Google Scholar]
- 20.Cailleau R, Young R, Olive M, Reeves WJ. Breast tumor cell lines from pleural effusions. J. Natl. Cancer Inst. 1974;53:661–674. doi: 10.1093/jnci/53.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fasco MJ, Amin A, Pentecost BT, et al. Phenotypic changes in MCF-7 cells during prolonged exposure to tamoxifen. Mol. Cell Endocrinol. 2003;206:33–47. doi: 10.1016/s0303-7207(03)00256-9. [DOI] [PubMed] [Google Scholar]
- 22.Qu Y, Han B, Yu W, et al. Evaluation of MCF10A as a reliable model for normal human mammary epithelial cells. PLoS One. 2015;10:e0131285. doi: 10.1371/journal.pone.0131285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saussede-Aim J, Matera EL, Ferlini C, Dumontet C. Beta3-tubulin is induced by estradiol in human breast carcinoma cells through an estrogen-receptor dependent pathway. Cell Motil. Cytoskelet. 2009;66:378–388. doi: 10.1002/cm.20377. [DOI] [PubMed] [Google Scholar]
- 24.Brault L, Gasser C, Bracher F, et al. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid tumors. Haematologica. 2010;95:1004–1015. doi: 10.3324/haematol.2009.017079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tursynbay Y, Zhang J, Li Z, et al. Pim-1 kinase as cancer drug target: an update (review) Biomed. Rep. 2016;4:140–146. doi: 10.3892/br.2015.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguirre E, Renner O, Narlik-Grassow M, Blanco-Aparicio C. Genetic modeling of PIM proteins in cancer: proviral tagging and cooperation with oncogenes, tumor suppressor genes, and carcinogens. Front. Oncol. 2014;4:1–17. doi: 10.3389/fonc.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Y, Xu K, Dai B, et al. The 44 kDa Pim-1 kinase directly interacts with tyrosine kinase Etk/BMX and protects human prostate cancer cells from apoptosis induced by chemotherapeutic drugs. Oncogene. 2006;25:70–78. doi: 10.1038/sj.onc.1209058. [DOI] [PubMed] [Google Scholar]
- 28.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data of 1287 patients. Endocr.-Relat. Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 29.Persico M, Petrella L, Orteca N, et al. GTP is an allosteric modulator of the interaction between the guanylate-binding protein 1 and the prosurvival kinase PIM1. Eur. J. Med. Chem. 2014;91:132–144. doi: 10.1016/j.ejmech.2014.07.093. [DOI] [PubMed] [Google Scholar]
- 30.Ostler N, Britzen-Laurent N, Leible A, et al. Gamma interferon-induced Guanylate-Binding Protein 1 is a novel actin cytoskeleton remodeling factor. Mol. Cell Biol. 2014;34:196–209. doi: 10.1128/MCB.00664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]