Abstract
BACKGROUND
Neuraxial anesthesia and epidural steroid injection techniques require precise anatomical targeting to ensure successful and safe analgesia. Previous studies suggest that only some of the tissues encountered during these procedures can be identified by spectroscopic methods, and no previous study has investigated the use of Raman, diffuse reflectance, and fluorescence spectroscopies. The authors hypothesized that real-time needle-tip spectroscopy may aid epidural needle placement and tested the ability of spectroscopy to distinguish each of the tissues in the path of neuraxial needles.
METHODS
For comparison of detection methods, the spectra of individual, dissected ex vivo paravertebral and neuraxial porcine tissues were collected using Raman spectroscopy (RS), diffuse reflectance spectroscopy (DRS), and fluorescence spectroscopy (FS). Real-time spectral guidance was tested using a 2 mm inner diameter fiber optic probe-in-needle device. Raman spectra were collected during the needle’s passage through intact paravertebral and neuraxial porcine tissue and analyzed afterward. The RS tissue signatures were verified as mapping to individual tissue layers using histochemical staining and widefield microscopy.
RESULTS
Raman spectroscopy revealed a unique spectrum for all ex vivo paravertebral and neuraxial tissue layers; DRS and FS spectra were not distinct for all tissues. Moreover, when accounting for the expected order of tissues, real-time Raman spectra recorded during needle insertion also permitted identification of each paravertebral and neuraxial porcine tissue.
CONCLUSIONS
This study demonstrates Raman spectroscopy can distinguish the tissues encountered during epidural needle insertion. This technology may prove useful during needle placement by providing evidence of its anatomical localization.
Introduction
Injection of pharmacologic agents into the epidural space is a common technique to control obstetric, perioperative, and radicular pain. Generally regarded as a safe procedure, minor complications including wrong tissue injection and dural puncture do occur. Fortunately, serious side effects including stroke and spinal cord injury are uncommon, but are catastrophic when they transpire.1-5 The loss of resistance (LOR) technique is the most common method of locating the epidural space, but suffers from a lack of specificity.6 Numerous techniques have been proposed to improve localization of the epidural space, including the use of ultrasound, loss of resistance assistance devices, fluoroscopy, and optical devices.7-22 However, these methods lack the resolution to identify each tissue in the path of an epidural needle.
Epidural anesthesia (EA) provides superior obstetric and postoperative analgesia compared to parenteral opioids, has a favorable risk/benefit profile, and may reduce morbidity and mortality in some patient populations.23-28 Patient anatomical characteristics including increased body mass index,29,30 congenital spinal malformations,31-34 advanced age,35 and previous spine surgery36-39 can make localization of the epidural space difficult. While the accidental dural puncture rate most commonly reported is 1-2%, it has been reported to be as high as 6%.10,40-44 Additionally, the failure rate for epidural catheter analgesia is as high as 22%. 6,10,43-45 In some clinical situations, up to half of these failures may be due to an inability to accurately locate the epidural space.41,44 As a result of the rare, but serious, complications during epidural corticosteroid injection (ESI), the U.S. Food and Drug Administration convened a panel of experts to determine practices to decrease their incidence.5
As each tissue has different proportions of molecules with unique spectral properties, optical spectroscopy provides biochemical tissue information and a method to distinguish different tissues from one another. Optical spectroscopy involves delivering light onto tissue and collecting the back scattered light. The technology is based on intrinsic tissue signal without exogenous injection or treatment, raising minimal safety concerns when translating this modality for clinical purposes. Multi-modal spectroscopy (MMS), the combination of diffuse reflectance, fluorescence, and Raman spectroscopies, provides complementary tissue information.46 Diffuse reflectance spectroscopy (DRS) is based on white light absorption. Molecules with distinctive diffuse reflectance spectra include oxy-/deoxy-hemoglobin, β-carotene, and melanin. Fluorescence spectroscopy (FS) is based on ultra-violet (UV) induced fluorescence,47 and collagen, elastin, keratin, NADH, and flavin adenine dinucleotide generate strong UV-induced fluorescence signals. Raman spectroscopy (RS) is based on inelastic light scattering; tissues with greater amounts of proteins, lipids, and collagen have unique Raman spectra. To date, no study has utilized DRS, FS, and RS to characterize each tissue encountered during neuraxial anesthesia and ESI techniques.
We, therefore, conducted our current study to evaluate whether some combination of DRS, FS, and RS differentiates each tissue from the skin to the spinal cord along the trajectory of epidural needles. Our hypothesis is that these spectroscopic techniques can identify and differentiate each tissue during probe-in-needle insertion into ex vivo porcine neuraxial and paravertebral tissues.
Methods
Study Design
This study was approved by the Institutional Animal Care and Use Committee of Massachusetts General Hospital, Boston, MA, U.S. (Protocol #: 2014N000209). The first part of this study was performed in dissected, individual, ex vivo porcine neuraxial and paravertebral tissues. DRS, FS, and RS data was collected from epidermis/dermis, adipose, skeletal muscle, supraspinous/intraspinous ligament, ligamentum flavum, epidural adipose, dura mater, and spinal cord. The second part of the study was performed in ex vivo porcine neuraxial and paravertebral tissue corresponding to the lumbar and lower thoracic spinal column excised en bloc. Data was collected as a RS probe-in-needle was inserted in a stepwise fashion from the dermis to the spinal cord. Tissues were subsequently identified by a neuropathologist.
Part 1: Characterization of spectral signature using dissected tissues
Animal tissue was obtained from 40 – 50 kilogram, female, Yorkshire swine euthanized with 100 milligrams per kilogram of pentobarbital given intravenously. The neuraxial and paravertebral tissues, including the spinal column from the lumbar to lower thoracic vertebrae, were excised en bloc and refrigerated at 4°C for less than 24 hours before use or frozen at −80°C within three hours of excision and thawed at 4°C for 24 hours before scanning. Tissue stored by both methods was dissected and scanned to determine if meaningful signal differences were detected. Key tissue planes were dissected and excised. After spectroscopy scans, each tissue sample was fixed using 4 % formalin in phosphate-buffered saline and embedded in paraffin. Subsequently, 4 µm-thick sections were dewaxed and stained with hematoxylin and eosin before examination by a neuropathologist.
Diffuse reflectance and fluorescence spectra were acquired by custom-built tissue scanners.48 A broad-band laser driven light source (EQ-99-FC-S LDLS, Energetiq Technology, Woburn, MA, U.S.) was the excitation source for DRS. A 355nm UV laser (SNV-40F-000, Teem Photonics, Meylan, France) was the excitation source for FS. Both used custom-built optical fiber probes (one excitation, six collection fibers) to deliver the broad band light and collect the back-scattered light. XY scanning was performed by a motorized stage (MS-6000-XY, Applied Scientific Instrumentation, Eugene, OR, U.S.). Back-scattered reflectance light was delivered to a miniature spectrometer (HR-2000+, Ocean Optics, Dunedin, FL, U.S.). LabView (National Instruments, Austin, TX, U.S.) and MATLAB software (Mathworks, Natick, MA, U.S.) controlled the systems. Dissected tissues were placed on top of a quartz plate. From each tissue, 20 diffuse reflectance spectra were acquired from a 5 mm2 area. 15 mW of power was delivered to the sample with a 10 millisecond integration time. For fluorescence, 20 spectra were acquired from identical tissue locations as the DRS spectra to guarantee the same probe-sample distance. 3 mW of power was delivered to the sample with a 100 millisecond integration time. No correction was made for photobleaching, and no filters were used for fluorescence measurements as full spectrum information is recorded from the spectrophotometer.
Raman spectra were acquired by a custom-built near infrared Raman microscopy as described previously (Supplemental Figure 1).49 A 785nm wavelength Ti: Sapphire laser was used as an excitation source; XY tissue scanning was performed by galvanometer mirrors. A 1.2 numerical aperture water immersion objective lens with 60× magnification was used to focus the laser light onto the sample and collect the back-scattered light. Back-scattered Raman light from the sample passed through two dichroic mirrors, was collected by a multi-mode fiber, delivered to the spectrograph, and detected by a thermoelectric-cooled, back-illuminated and deep depleted charge-coupled device. LabView software (National Instruments), a 16-bit data acquisition board (PCI-6251, National Instruments), and MATLAB software (Mathworks) were used to control the system, and acquire the data. Dissected tissues were placed on top of quartz coverslips (043210-KJ, Alfa Aesar, Haverhill, MA, U.S.). 25 spectra were acquired from a 1444 µm2 area to account for tissue inhomogeneities. 60 mW of power was delivered to the sample with a 5 second integration time.
Part 2: Real-time characterization of RS in situ
Raman spectra were acquired by a multimodal spectroscopy system using a 830 nm diode laser to generate Raman signal from tissue.46 The laser light is filtered and coupled to an optical fiber Raman probe that delivers light to, and collects light from, a sample. The 2 mm outer diameter Raman probe50 consists of a single excitation fiber surrounded by 15 collection fibers; specialized filters and a ball lens are at the distal probe tip. Return light is passed through a spectrograph and dispersed onto a charge-coupled device detector. Tissue was acquired as described in Part 1: Characterization of Spectral Signature Using Dissected Tissues. A 2 mm inner diameter, custom-build, stainless steel needle with a short, cutting bevel (Supplemental Figure 2) and stylette were inserted into the tissue and advanced at 1–2 mm intervals. After each 1–2 mm movement, the stylette was removed, the RS probe was inserted, and ten RS spectra were acquired with 60 mW of power and a 1 second integration time. The stylette was reinserted between steps. The needle was advanced along trajectories that correspond to the midline and paramedian approaches to epidural space localization. The needle depth was measured and recorded for each distinct spectrum. After insertion of the probe-in-needle to the posterior longitudinal ligament, the needle was left in place. Dissection then occurred along the length of the needle, and tissue was removed at the depth of each distinct spectrum.
Spectrum Analysis
Diffuse Reflectance Spectroscopy
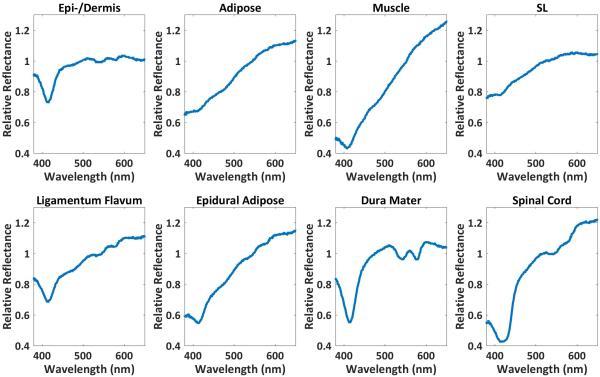
In order to compensate for the spectrum from the broadband source, the raw reflectance spectrum from each tissue was divided by the reflectance spectrum acquired from a 20% white reflectance standard (Spectralon, Labsphere, NH, U.S.).48 For each tissue type, a mean spectrum was calculated from ten of the 20 tissue spectra (Figure 1). For each remaining test spectrum, eight Pearson correlation coefficients were calculated between the remaining spectrum and eight mean spectra using MATLAB. A Pearson correlation coefficient is defined as where N is the number of scalar observations for each variable, μA and σA are the mean and SD of A, respectively, and μB and σB are the mean and SD of B, respectively. The correlation coefficient for each test spectrum and mean spectrum pair was calculated using the test spectrum and mean spectrum as variables A and B, respectively. The resultant correlation coefficients represent the degree of similarity between each test tissue spectrum and the mean spectrum of eight tissue types, with higher correlation coefficients signifying greater similarity. Each test spectrum was thus classified to the tissue type with the highest correlation coefficient.
Figure 1.
For each tissue type, a mean spectrum was calculated from ten of the 20 tissue spectra acquired from a 5 mm2 area. Some of the reflectance spectra from dissected tissues can clearly be differentiated from one another while the spectra from others have subtle or no differences. SL, supraspinous/intraspinous ligament.
Fluorescence Spectroscopy
The FS data were analyzed in the same manner as that of DRS (Figure 2).
Figure 2.
For each tissue type, a mean spectrum was calculated from ten of the 20 tissue spectra acquired from identical tissue locations as the DRS spectra to guarantee the same probe-sample distance. Some of the fluorescence spectra from dissected tissues can clearly be differentiated from one another while the spectra from others have subtle or no differences. SL, supraspinous/intraspinous ligament.
Raman Spectroscopy
For quantitative analysis, the acquired Raman spectra from the tissue specimens were decomposed as a summation of basis components, albumin, actin, collagen, triolein, and phosphatidylcholine. Each acquired spectrum was represented by five fitting coefficients, representing the relative amounts of basis components. Albumin and actin were selected to represent plasma and cytoskeleton proteins. Collagen was selected as it is the main component of connective tissues. Triolein and phosphatidylcholine were selected as non-membrane and membrane lipids respectively. The acquired spectra from these agents were normalized and used as the basis spectra for spectrum analysis (Figure 3) and represented as a vector. Wavenumbers (Raman shift) represent the energy shift between excitation photons and scattered photons from a sample. Typically, wavenumbers between 600 cm−1 and 1800 cm−1 are used due to good molecular specificity. For biochemical decomposition of the tissue, the ordinary least squares fitting method was used (Figure 4). To avoid data overfitting, the fitting process was carefully monitored by restricting the fitting coefficient to non-negative values. Acquired Raman spectra were normalized and decomposed as a summation of basis components as previously described.51-53
Figure 3.
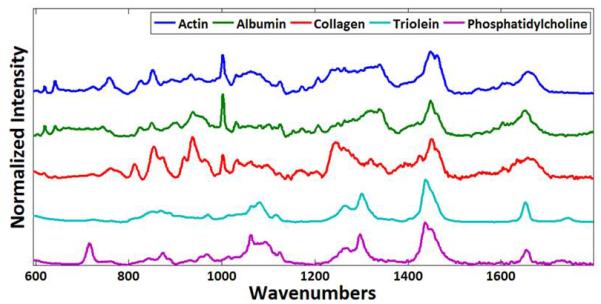
The five basis components albumin, actin, collagen, triolein, and phosphatidylcholine have unique Raman spectra.
Figure 4.
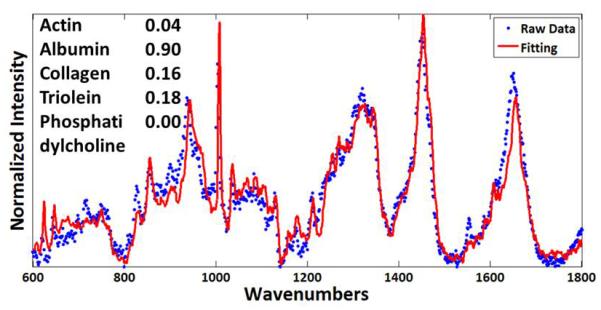
Fitting coefficients determined using basis spectra represent the relative amount of each basis component. A representative fitting result from the skeletal muscle is shown.
A decision algorithm was developed based on the five fitting coefficients. Ten Raman spectra from each of the eight tissues were used to develop the decision algorithm and 15 spectra were used for testing the algorithm.
Machine Learning Classification Model
As variation was evident in fitting coefficients between dissected and non-dissected tissue, a machine learning classification model was used to enable autonomous identification of real-time acquired RS spectra. The use of a machine learning classification model allows for robust identification of tissue type that can algorithmically compensate for the variation seen in fitting coefficients between dissected and non-dissected tissue. Acquired spectra were separated into a training data set and a testing data set; 10 spectra from each dissected tissue type were used for training, while 80 spectra from each tissue acquired during probe-in-needle insertion were used to test the tissue identification accuracy of the algorithm. The five fitting coefficients, representing the relative amounts of albumin, actin, collagen, triolein, and phosphatidylcholine in each spectra, were used as a 5-dimensional vector representation of each spectrum. The classification model was programmed in Python based on a Gaussian Mixture Model algorithm, a type of machine learning algorithm that clusters data points using maximum-likelihood estimation.54 The model was trained using the Raman spectra fitting coefficients to create data clusters for each tissue type in five-dimensional vector space and then classified test spectra by identifying the most probable cluster to the fitting coefficient vector of the test spectrum (Supplemental Figure 3).
Statistics
MATLAB software (Mathworks, Natick, MA, U.S.) was used to analyze the diffuse reflectance, fluorescence, and Raman spectra data from dissected tissues and to calculate correlation coefficients. The fitting coefficient of each basis component represents the “relative” amount of a basis component within each tissue since each has a different Raman cross section. For example, the epidermis/dermis has a fitting coefficient of 0.09 for actin and 0.84 for collagen. This does not indicate that the collagen content is more than nine times the actin content as collagen and actin have different Raman cross sections. However, the 0.22 (actin) and 0.64 (collagen) fitting coefficients within the ligamentum flavum indicate that the actin content within the ligamentum flavum is 144% greater than within epidermis/dermis and the collagen content is 24% less.
A RS decision algorithm (Supplemental Figure 4) was developed based on the five fitting coefficients for classification of the dissected tissue. The RS decision algorithm was empirically designed, and verified as described. Ten Raman spectra (40%) from each of the eight tissues were used to develop the decision algorithm, and 15 spectra (60%) were used for testing the algorithm. First, the tissue spectra were divided into two groups based on the collagen content. The high collagen tissues (> 0.5) are further categorized based on the relative ratios of collagen and total protein (actin + albumin). Tissues are further subdivided based on actin content.
The low collagen tissues (< 0.5) are further categorized based on the total protein content (> 0.5 and < 0.5). Low collagen, low protein content tissues are differentiated from one another using the relative ratio of triolein and phosphatidylcholine.
Results
Tissue refrigerated at 4°C for less than 24 hours before use or frozen at −80°C within several hours of excision and thawed at 4°C for 24 hours before scanning did not have meaningful differences in DRS, FS, and RS signal.
Part 1: Characterization of spectral signature using dissected tissues
Diffuse Reflectance Spectroscopy
The normalized reflectance spectra from each tissue (Figure 1) show visible differences as previously reported.20 Tissue prediction results using DRS spectra are summarized in Table 1. The overall prediction accuracy of DRS is 89%.55 The sensitivity for accurate tissue prediction using DRS is less than 90% for two tissues (LF, 60%; epidural adipose, 80%), while the specificity is greater than 90% for all tissues.
Table 1.
Tissue prediction by diffuse reflectance spetroscopy, fluorescence spetroscopy, and a fitting coefficients-based decision algorithm for Raman spectroscopy.
|
How often each of the 10 (DRS, FS) or 15 (RS) spectra from histologically confirmed dissected
tissue were correctly identified, and when incorrectly identified, what they were labeled as. |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Epi- /dermis |
Adipose | Muscle | SL | LF | EA | DM | SC | Sensitivity (%) (95% CI) |
Specificity (%) (95% CI) |
|
| Diffuse Reflectance (Accuracy = 89%) | ||||||||||
|
Dissected tissue identfied by
histopathology |
||||||||||
| epidermis/dermis | 10 | 100 (66-100) |
97 (89-99) |
|||||||
| Adipose | 9 | 1 | 90 (54-99) |
97 (89-99) |
||||||
| Muscle | 9 | 1 | 90 (54-99) |
100 (93-100) |
||||||
| SL | 1 | 9 | 90 (54-99) |
95 (87-98) |
||||||
| Ligamentum Flavum | 1 | 3 | 6 | 60 (27-86) |
98 (91-99) |
|||||
| Epidural Adipose | 2 | 8 | 80 (44-96) |
98 (91-99) |
||||||
| Dura Mater | 10 | 100 (65-100) |
100 (93-100) |
|||||||
| Spinal Cord | 10 | 100 (65-100) |
100 (93-100) |
|||||||
| Fluorescence (Accuracy = 81%) | ||||||||||
| epidermis/dermis | 10 | 100 (65-100) |
84 (73-91) |
|||||||
| Adipose | 10 | 100 (65-100) |
100 (93-100) |
|||||||
| Muscle | 9 | 1 | 90 (54-99) |
100 (93-100) |
||||||
| SL | 6 | 4 | 40 (13-72) |
97 (89-99) |
||||||
| Ligamentum Flavum | 5 | 2 | 2 | 1 | 20 (3-55) |
100 (93-100) |
||||
| Epidural Adipose | 10 | 100 (65-100) |
100 (93-100) |
|||||||
| Dura Mater | 10 | 100 (65-100) |
98 (91-99) |
|||||||
| Spinal Cord | 10 | 100 (65-100) |
99 (91-99) |
|||||||
| Raman (Accuracy = 100%) | ||||||||||
| epidermis/dermis | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 (74-100) |
100 (95-100) |
| Adipose | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 100 (74-100) |
100 (95-100) |
| Muscle | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 100 (74-100) |
100 (95-100) |
| SL | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 100 (74-100) |
100 (95-100) |
| Ligamentum Flavum | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 100 (74-100) |
100 (95-100) |
| Epidural Adipose | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 100 (74-100) |
100 (95-100) |
| Dura Mater | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 100 (74-100) |
100 (95-100) |
| Spinal Cord | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 100 (74-100) |
100 (95-100) |
In the leftmost column are the dissected tissues as identified by histopathology. The next eight columns hold the number of the 10 spectra (DRS, FS) or 15 spectra (RS) from each tissue assigned as each of the eight tissues based on the one with the highest correlation coefficient (DRS, FS) or decison algorithm (RS). This table reports how often each of the 10-15 spectra from histologically confirmed dissected tissue were correctly identified by the three spectroscopic methods and when incorrectly identified, what they were labeled as. DM, dura mater; EA, epidural adipose; LF, ligamentum flavum; SL, supraspinous/intraspinous ligament; SC, spinal cord.
Fluorescence Spectroscopy
Strong fluorescence signals are generated from collagen in connective tissues. Therefore, tissues containing greater amounts of collagen (skin, supraspinous/intraspinous ligament, ligamentum flavum, dura mater) have a higher signal (Figure 2). Adipose, muscle, and spinal cord do not generate strong fluorescence signals. FS cannot effectively quantitate signal strength due to photobleaching and is not sensitive enough to accurately differentiate low collagen content tissues. Generated fluorescence signals are distorted by the optical absorption and scattering properties of overlying tissue. Therefore, acquired spectral shapes are different from pure fluorescence spectra. Tissue prediction results using FS spectra are summarized in Table 1. The overall prediction accuracy of FS is 81%. Poor distinction is made between the collagen-rich layers (skin, supraspinous/intraspinous ligament, ligamentum flavum). The sensitivity for accurate tissue prediction using FS is less than 90% for two tissues (SL, 40%; LF, 20%), while the specificity is greater than 90% for all tissues except epidermis/dermis (84%).
Raman Spectroscopy
Unlike the previous DRS and FS prediction methods, where tissue prediction is based on the similarity between acquired and averaged, representative spectra, the RS tissue prediction method is developed based on a combination of molecular fitting coefficients. Figure 4 demonstrates the fitting result for the Raman spectrum from the skeletal muscle. The fitting spectrum (red line) is similar to the raw data (blue dots) and the five fitting coefficients provide the chemical composition of the measured tissue.56
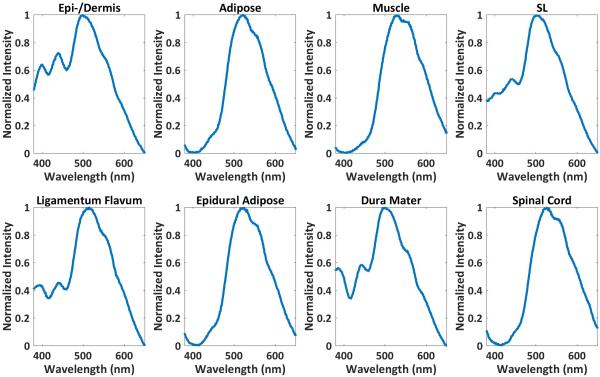
Normalized averaged Raman spectra from the eight dissected tissues are shown in Figure 5. The red highlighted Raman band at 939 cm−1 indicates a collagen specific Raman signature while the yellow Raman band at 1450 cm−1 represent a lipid specific Raman signal. Strong collagen signals are found in the spectra of epidermis/dermis, supraspinous/intraspinous ligament, ligamentum flavum, and dura mater. Strong lipid signals are found in adipose, epidural adipose, and spinal cord.
Figure 5.
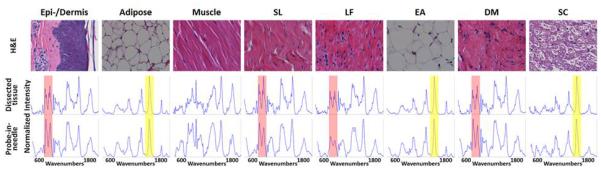
For each tissue type, a mean spectrum was calculated from 10 of the 25 tissue spectra acquired from a 1444-μm2 area. Raman spectra contain information on chemical composition. Thus, the Raman spectra from each tissue can be differentiated. Spectra acquired from dissected (middle) and probe-in-needle experiments (bottom row) for the same tissues are similar. Top, Hematoxylin and eosin (H&E)–stained tissue (40× magnification). The red (939 cm−1) and yellow (1450 cm−1) bands indicate collagen- and lipid-specific Raman signatures. DM, dura mater; EA, epidural adipose; LF, ligamentum flavum; SC, spinal cord; SL, supraspinous/intraspinous ligament.
Quantitative analysis results are shown in Figure 6 and Table 2. Figure 6 demonstrates the differences in basis component composition between each of the eight tissues dissected and scanned by RS.
Figure 6.
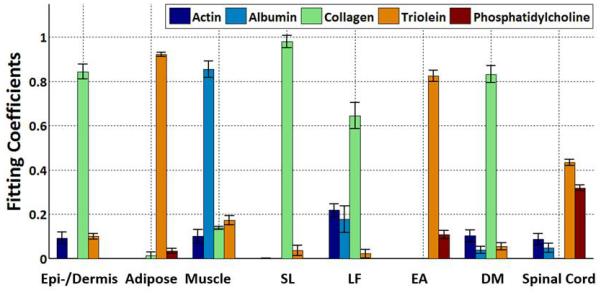
Raman spectra from dissected tissues experiments were decomposed into the linear superposition of the five basis spectra. Each tissue layer is defined by five fitting coefficients. Every tissue layer from epidermis/dermis to the spinal cord can be clearly differentiated based on the fitting coefficients. DM, dura mater; EA, epidural adipose; LF, ligamentum flavum; SL, supraspinous/intraspinous ligament.
Table 2.
The fitting coefficients of the basis Raman spectra identify a unique combination for each dissected tissue layer.
| Albumin | Actin | Collagen | Triolein | PC | |
|---|---|---|---|---|---|
| epidermis/dermis | 0.09±0.03 | 0.00±0.00 | 0.84±0.03 | 0.10±0.01 | 0.00±0.00 |
| Adipose | 0.00±0.00 | 0.00±0.00 | 0.01±0.02 | 0.92±0.01 | 0.03±0.01 |
| Muscle | 0.10±0.03 | 0.85±0.04 | 0.14±0.01 | 0.17±0.02 | 0.00±0.00 |
| SL | 0.00±0.00 | 0.00±0.00 | 0.98±0.03 | 0.04±0.02 | 0.00±0.00 |
| Ligamentum Flavum | 0.22±0.03 | 0.18±0.06 | 0.64±0.06 | 0.02±0.02 | 0.00±0.00 |
| Epidural Adipose | 0.00±0.00 | 0.00±0.00 | 0.00±0.00 | 0.82±0.03 | 0.11±0.02 |
| Dura Mater | 0.10±0.03 | 0.04±0.02 | 0.83±0.04 | 0.05±0.02 | 0.00±0.00 |
| Spinal Cord | 0.09±0.03 | 0.05±0.02 | 0.00±0.00 | 0.43±0.01 | 0.32±0.01 |
Values are represented as mean fitting coefficient ± SD. PC, phosphatidylcholine; SL, supraspinous/intraspinous ligament.
Results of the RS decision algorithm can be seen in Supplemental Figure 4. The high collagen group (> 0.5) includes epidermis/dermis, supraspinous/intraspinous ligament, ligamentum flavum, and dura mater. These tissues have low lipid content and are further categorized based on the relative ratios of collagen and total protein (actin + albumin). supraspinous/intraspinous ligament (SL) has a ratio > 100. Ligamentum flavum has a ratio of < 3.6. epidermis/dermis and dura mater have collagen/total protein ratios between 3.6 and 100 and can be differentiated based on their actin content. 3.6 and 100 were found to be the optimal collagen/total protein ratios for differentiating the high collagen tissues from one another.
The low collagen group (< 0.5) includes skeletal muscle, adipose, epidural adipose, and spinal cord. Skeletal muscle can be differentiated due to its high actin and albumin content (total protein > 0.5). The remaining three tissues (adipose, epidural adipose, spinal cord; total protein < 0.5) are differentiated from one another using the relative ratio of triolein and phosphatidylcholine. Adipose has a ratio of > 20; epidural adipose has a ratio between 5 and 20; and spinal cord has a ratio of < 5. 5 and 20 were found to be the optimal triolein/phosphatidylcholine ratios for differentiating the low collagen, low total protein tissues from one another. Table 1 shows the tissue prediction results. All eight tissue layers can be differentiated.
Table 1 shows the RS tissue prediction results. All eight tissue layers can be differentiated from one another. The overall prediction accuracy of RS is 100%. The sensitivity for accurate tissue prediction using RS is 100% for all tissues, while the specificity is 100% for all tissues.
The tissues identified during dissection, and subsequently scanned by DRS, FS, and RS, were confirmed by histology. All tissues underwent H & E staining and were independently assessed by a neuropathologist who confirmed the tissue types (Figure 5).
Part 2: Real-time characterization of RS in situ
Normalized, averaged Raman spectra acquired from in situ probe insertion are shown in Figure 5. Overall the spectra have a qualitatively smoother profile than those from dissected tissues due to different spectral resolutions (4 cm−1 for dissected tissue measurements vs. 16 cm−1 for probe-in-needle measurements) of the two instruments. Nonetheless, the RS spectrum of each tissue is similar to those acquired from the individually dissected tissues. The acquired Raman spectra were processed in the same fashion as spectra from dissected tissues (Figure 7). The fitting coefficients were similar to those from the dissected tissues except for those of supraspinous/intraspinous ligament and ligamentum flavum. Both tissues have higher lipid content than the dissected tissues. Adipose, removed during dissection studies, is actually interspersed with the supraspinous/intraspinous ligament and immediately superficial and adjacent to ligamentum flavum in swine. Thus, as the RS probe samples a volume of tissue within 1 mm of the probe tip, some adipose is sampled along with these thin ligaments.
Figure 7.
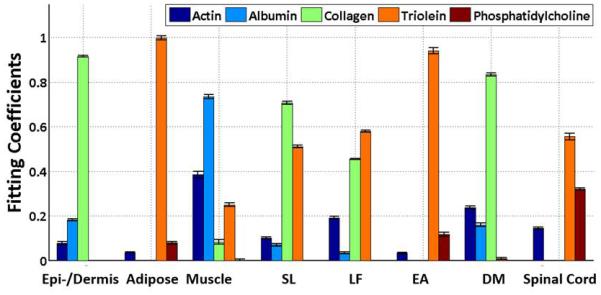
Raman spectra from probe-in-needle experiments were decomposed into the linear superposition of the five basis spectra. Each tissue layer is defined by five fitting coefficients. Every tissue layer from the epidermis/dermis to the spinal cord can be clearly differentiated based on the fitting coefficients. DM, dura mater; EA, epidural adipose; LF, ligamentum flavum; SL, supraspinous/intraspinous ligament.
With the training spectra presented to the machine learning classification model in random order, the algorithm has 85.5% accuracy across the entire test data set. Specifically, the model produced 100% accuracy of identification of 7 out of 8 issue types, including epidermis/dermis, adipose, skeletal muscle, ligamentum flavum, epidural adipose, dura mater, and spinal cord. Because of similarity of supraspinous/intraspinous ligament (SL) spectra to ligamentum flavum (LF) spectra, the current model cannot be used to differentiate SL and LF with spectra classified in random order; real-time classification, accounting for the expected order of transition between layers enables differentiation between all eight tissue types.
Discussion
While obstetric epidural analgesia, perioperative epidural analgesia, and epidural injection of corticosteroids are useful techniques for acute and chronic pain, both minor and serious complications do occur. Thus, a technique to aid with epidural needle placement may be beneficial if it can identify each tissue in the path of the needle, potentially preventing wrong site needle insertion and drug injection.
Our studies show that Raman spectroscopy identifies each tissue in the path of an epidural needle in an ex vivo porcine model. Characterization of the DRS, FS, and RS spectral signatures showed that the Raman spectrum from each tissue is unique. Further investigation in which a RS probe was inserted in situ corroborated these results. RS may be useful to safely and effectively aid epidural needle placement during steroid injection and catheter placement procedures.
Other methods have been used to aid with epidural needle placement. However, these methods lack resolution and are unable to distinguish between each tissue encountered at the epidural needle tip during insertion. Ultrasound is increasingly used to aid neuraxial anesthetic placement. A recent meta-analysis concluded that neuraxial ultrasound more accurately identifies the lumbar intervertebral space over palpation, accurately predicts the distance from the skin to the epidural space, and improves efficacy.57 However, with some practitioners, the actual depth from the skin to the intended space may differ by as much as 15 mm from that estimated by ultrasound.7-9,21 Significant training and experience is necessary before one can properly utilize ultrasound for assistance with epidural needle and catheter placement. In addition, the acquisition and maintenance of the equipment is costly. As bone is not penetrated by ultrasound, it is not possible to acquire a comprehensive view of the epidural space but the practitioner is limited to small windows. Real-time use of ultrasound during epidural needle advancement is typically not possible, and ultrasound is most often used to assess the location and depth of neuraxial structures before blind needle advancement.11
Fluoroscopy can help guide the needle to, and confirm placement of the needle tip in the epidural space.14,16,22 However, its use is labor and time intensive, requires additional equipment and often manpower, and exposes the patient and medical personnel to radiation.
Despite the relatively low specificity of this technique, a number of LOR assistance devices have also been developed and studied, including the Episure Autodetect syringe (Indigo Orb, Inc., Irvine, CA, USA) ,19 Epidrum (Exmoor Innovations Ltd, Taunton, UK),58 and Compuflo (Milestone Scientific, Livingston, NJ) injection pump.59 A pilot study found fewer epidural catheter failures when the Episure Autodetect syringe was used for the placement compared to a glass syringe.12 Similarly, use of the Epidrum device led to a lower epidural failure rate and less time to identify epidural space compared to use of the traditional loss of resistance syringe.13
Previously, reflectance spectroscopy was shown to distinguish between the ligamentum flavum and dura mater as well as aid with epidural space localization.15,20 Another study using visible and near-infrared spectroscopy found that upon entry of the epidural needle into the epidural space an increase in the lipid content of the tissue was detected. 18 No data from either technique were published on the other tissues that are encountered during epidural needle insertion. As midline ligamentum flavum is absent up to 74% of the time in the cervical and high thoracic region,60 it is important to identify all tissue layers encountered to prevent injury and improve accuracy. Moreover, reflection or image based methods will identify two areas of the same tissue as separate tissue types if the areas have different tissue densities. Additionally, diffuse reflectance spectroscopy is sensitive to tissue inhomogeneities, probe-sample distance, and pressure variation.61 Fluorescence spectroscopy only differentiates tissues with high collagen content from those with low collagen content. Thus, neither method is ideal to accurately and precisely distinguish each tissue in the path of an epidural needle. Conversely, Raman spectroscopy is specific to the chemical composition of the tissue. Areas of different density within the same tissue will change the Raman intensity, but the spectral shape will not be altered. Furthermore, the sampling space of Raman spectroscopy extends 1 mm from the tip of the probe, allowing identification of tissue before the tip of the needle enters it.
In this study, Raman spectroscopy was found to have greater prediction accuracy in identifying and differentiating tissues than DRS and FS. Furthermore, the Raman spectrum of each tissue was found to be unique and can be identified based on the relative amount of albumin, actin, collagen, triolein, and phosphatidylcholine. This identification held for the Raman spectra collected during in situ probe in needle insertion as well. Neither DRS nor FS was able to distinguish between all tissues of interest. Additionally, the machine learning classification model developed based on Raman spectra demonstrates the capability for autonomous tissue identification that can be used for real-time classification of tissues.
There are potential limitations to use of this technology during epidural needle placement. Raman spectroscopy differentiated each dissected tissue but did not clearly distinguish between supraspinous/intraspinous ligament and ligamentum flavum during probe-in-needle insertion secondary to adipose interspersed within the undissected tissue. Although all tissues can be differentiated during real-time probe-in-needle insertion if the expected order of transition between layers is taken into account, future collection and use of a large training data set from in situ measurements are likely to allow differentiation of these two tissues with use of the machine learning classification model. Additionally, a smaller Raman spectroscopy probe, necessary for clinical purposes and currently being built in our laboratory, will sample a smaller tissue volume at the tip of the needle and is less likely to include information from multiple adjacent tissues. While RS can identify tissue at the tip of the needle during neuraxial procedures, it cannot be used to guide the needle trajectory. It may be beneficial in the future to use RS for needle tip tissue identification along with US or fluoroscopy to aid with needle trajectory. Our current analysis model includes the five basis components as outlined in Spectrum Analysis under Raman Spectroscopy. If other compounds that strongly absorb Raman signal are present in these tissues, such as heme from minor bleeding during needle insertion or calcified ligaments in elderly patients, detection accuracy may be affected. The solution to this potential issue is to include the new component such as hemoglobin and/or calcium salts into the basis model. For example, 12 basis components are used for breast cancer diagnosis.62 Intravenous medications and electrolyte changes will not affect RS signal since their concentration is much lower than the major compounds responsible for Raman spectra within tissues. Raman signal depends on temperature, but this is extremely minor effect from the small temperature changes in biologic tissues. Patient movement may affect the tissue density and RS signal strength but the spectral shape, and hence prediction accuracy, will not be affected.
It remains to be seen whether or not use of this technology along with the traditional LOR technique results in a change in procedure time, number of needle passes, procedure success, or complication rates. Future studies include miniaturization of the RS probe for use in an epidural needle, RS characterization of neuraxial and paravertebral tissues in a live animal model and in human cadaveric tissue in order to support the application of this technology for use in a human trial.
In conclusion, we have described a novel method, using Raman spectroscopy, to identify each tissue at the tip of neuraxial needles. RS can potentially be used along with current techniques for epidural space localization. It remains to be seen if use of this technology decreases epidural needle complication rates.
Supplementary Material
Brief Summary Statement: Raman spectroscopy differentiates every tissue from the skin to the spinal cord in an ex vivo porcine model. This novel spectroscopic technique may be adapted to aid epidural needle placement.
Acknowledgements
The authors thank the staff of the Knight Surgical Research Lab at the Massachusetts General Hospital (Boston, Massachusetts), especially Dr. Michael Duggan, D.V.M., and John Beagle, B.S., for their time and expertise as well as the generous donation of swine tissue. Luis Galindo, B.S., a senior engineer within the Massachusetts Institute of Technology Laser Biomedical Research Center (Cambridge, Massachusetts), helped perform probe-in-needle experiments.
Funding: This work is supported by National Science Foundation, Arlington, Virginia (1549613). TAA acknowledges support in the form of research time from Dr. Jeanine Wiener-Kronish, M.D., Anesthetist-in-Chief, Massachusetts General Hospital Department of Anesthesia, Critical Care and Pain Medicine (Boston, MA, U.S.). JWK, RRD, and PTCS acknowledge support from NIH 9P41EB015871-28 and the Massachusettes Institute of Technology SkolTech initiative (Cambridge, MA, U.S.), and Samsung Advanced Institute of Technology (Seoul, South Korea). PTCS further acknowledge support from DP3-DK101024 01, 1-U01-NS090438-01, 1-R01-EY017656-06A1, 1R21NS091982-01, 1-R01-HL121386-01A1, 1U01CA202177-01, the Singapore-Massachusettes Institute of Technology Alliance 2 (Cambridge, MA, U.S.), the Biosym IRG of Singapore-Massachusettes Institute of Technology Alliance Research and Technology Center (Cambridge, MA, U.S.), Massachusettes Institute of Technology SkolTech initiative (Cambridge, MA, U.S.), Hamamatsu Corporation (Hamamatsu City, Japan), the Koch Institute for Integrative Cancer Research Bridge Initiative (Cambridge, MA, U.S.), and Connecticut Children's Medical Center Hartford, CT, U.S.).
TAA, JWK, RRD, and PTCS are named as authors on patents related to several multimodal spectroscopy medical devices designed for needle tip tissue identification.
Footnotes
Conflicts of Interest: TAA, JWK, RRD, and PTCS have equity interest in a for profit company (Medisight Corporation, Cambridge, MA 02141) designed to commercialize multi-modal spectroscopy for needle tip tissue identification. All other authors have no conflicts of interest.
IRB: Permission was obtained from the MGH Institutional Animal Care and Use Committee (Protocol 2014N000209).
References
- 1.Bicket MC, Chakravarthy K, Chang D, Cohen SP. Epidural steroid injections: an updated review on recent trends in safety and complications. Pain Manag. 2015;5:129–46. doi: 10.2217/pmt.14.53. [DOI] [PubMed] [Google Scholar]
- 2.Brull R, McCartney CJ, Chan VW, El-Beheiry H. Neurological complications after regional anesthesia: contemporary estimates of risk. Anesth Analg. 2007;104:965–74. doi: 10.1213/01.ane.0000258740.17193.ec. [DOI] [PubMed] [Google Scholar]
- 3.Dahlgren N, Tornebrandt K. Neurological complications after anaesthesia. A follow-up of 18,000 spinal and epidural anaesthetics performed over three years. Acta Anaesthesiol Scand. 1995;39:872–80. doi: 10.1111/j.1399-6576.1995.tb04190.x. [DOI] [PubMed] [Google Scholar]
- 4.Kang XH, Bao FP, Xiong XX, Li M, Jin TT, Shao J, Zhu SM. Major complications of epidural anesthesia: a prospective study of 5083 cases at a single hospital. Acta Anaesthesiol Scand. 2014;58:858–66. doi: 10.1111/aas.12360. [DOI] [PubMed] [Google Scholar]
- 5.Rathmell JP, Benzon HT, Dreyfuss P, Huntoon M, Wallace M, Baker R, Riew KD, Rosenquist RW, Aprill C, Rost NS, Buvanendran A, Kreiner DS, Bogduk N, Fourney DR, Fraifeld E, Horn S, Stone J, Vorenkamp K, Lawler G, Summers J, Kloth D, O'Brien D, Jr., Tutton S. Safeguards to prevent neurologic complications after epidural steroid injections: consensus opinions from a multidisciplinary working group and national organizations. Anesthesiology. 2015;122:974–84. doi: 10.1097/ALN.0000000000000614. [DOI] [PubMed] [Google Scholar]
- 6.Tran DQ, Gonzalez AP, Bernucci F, Finlayson RJ. Confirmation of loss-of-resistance for epidural analgesia. Reg Anesth Pain Med. 2015;40:166–73. doi: 10.1097/AAP.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 7.Balki M, Lee Y, Halpern S, Carvalho JC. Ultrasound imaging of the lumbar spine in the transverse plane: the correlation between estimated and actual depth to the epidural space in obese parturients. Anesth Analg. 2009;108:1876–81. doi: 10.1213/ane.0b013e3181a323f6. [DOI] [PubMed] [Google Scholar]
- 8.Chin KJ, Karmakar MK, Peng P. Ultrasonography of the adult thoracic and lumbar spine for central neuraxial blockade. Anesthesiology. 2011;114:1459–85. doi: 10.1097/ALN.0b013e318210f9f8. [DOI] [PubMed] [Google Scholar]
- 9.Chin KJ, Perlas A, Singh M, Arzola C, Prasad A, Chan V, Brull R. An ultrasound-assisted approach facilitates spinal anesthesia for total joint arthroplasty. Can J Anaesth. 2009;56:643–50. doi: 10.1007/s12630-009-9132-8. [DOI] [PubMed] [Google Scholar]
- 10.Eappen S, Blinn A, Segal S. Incidence of epidural catheter replacement in parturients: a retrospective chart review. Int J Obstet Anesth. 1998;7:220–5. doi: 10.1016/s0959-289x(98)80042-3. [DOI] [PubMed] [Google Scholar]
- 11.Grau T. The evaluation of ultrasound imaging for neuraxial anesthesia. Canadian Journal of Anesthesia. 2003;50:R30–37. [Google Scholar]
- 12.Habib AS, George RB, Allen TK, Olufolabi AJ. A pilot study to compare the Episure Autodetect syringe with the glass syringe for identification of the epidural space in parturients. Anesth Analg. 2008;106:541–3. doi: 10.1213/ane.0b013e3181606c0a. [DOI] [PubMed] [Google Scholar]
- 13.Kim SW, Kim YM, Kim SH, Chung MH, Choi YR, Choi EM. Comparison of loss of resistance technique between Epidrum(R) and conventional method for identifying the epidural space. Korean J Anesthesiol. 2012;62:322–6. doi: 10.4097/kjae.2012.62.4.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SY, Kim YY, Kim AR. Incidence of intravascular insertion in thoracic epidural catheterization by using real time fluoroscopy. Korean J Anesthesiol. 2012;62:251–5. doi: 10.4097/kjae.2012.62.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin SP, Mandell MS, Chang Y, Chen PT, Tsou MY, Chan KH, Ting CK. Discriminant analysis for anaesthetic decision-making: an intelligent recognition system for epidural needle insertion. Br J Anaesth. 2012;108:302–7. doi: 10.1093/bja/aer369. [DOI] [PubMed] [Google Scholar]
- 16.Mehta M, Salmon N. Extradural block. Confirmation of the injection site by X-ray monitoring. Anaesthesia. 1985;40:1009–12. doi: 10.1111/j.1365-2044.1985.tb10558.x. [DOI] [PubMed] [Google Scholar]
- 17.Perlas A, Chaparro LE, Chin KJ. Lumbar Neuraxial Ultrasound for Spinal and Epidural Anesthesia: A Systematic Review and Meta-Analysis. Reg Anesth Pain Med. 2016;41:251–60. doi: 10.1097/AAP.0000000000000184. [DOI] [PubMed] [Google Scholar]
- 18.Rathmell JP, Desjardins AE, van der Voort M, Hendriks BH, Nachabe R, Roggeveen S, Babic D, Soderman M, Brynolf M, Holmstrom B. Identification of the epidural space with optical spectroscopy: an in vivo swine study. Anesthesiology. 2010;113:1406–18. doi: 10.1097/ALN.0b013e3181fcee47. [DOI] [PubMed] [Google Scholar]
- 19.Riley ET, Carvalho B. The Episure syringe: a novel loss of resistance syringe for locating the epidural space. Anesth Analg. 2007;105:1164–6. doi: 10.1213/01.ane.0000281935.78144.82. [DOI] [PubMed] [Google Scholar]
- 20.Ting CK, Tsou MY, Chen PT, Chang KY, Mandell MS, Chan KH, Chang Y. A new technique to assist epidural needle placement: fiberoptic-guided insertion using two wavelengths. Anesthesiology. 2010;112:1128–35. doi: 10.1097/ALN.0b013e3181d3d958. [DOI] [PubMed] [Google Scholar]
- 21.Tran D, Kamani AA, Al-Attas E, Lessoway VA, Massey S, Rohling RN. Single-operator real-time ultrasound-guidance to aim and insert a lumbar epidural needle. Can J Anaesth. 2010;57:313–21. doi: 10.1007/s12630-009-9252-1. [DOI] [PubMed] [Google Scholar]
- 22.Uchino T, Iwasaka H, Kudo K, Takatani J, Mizutani A, Miura M, Noguchi T. Use of imaging agent to determine postoperative indwelling epidural catheter position. Korean J Pain. 2010;23:247–53. doi: 10.3344/kjp.2010.23.4.247. HS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Block BM, Liu SS, Rowlingson AJ, Cowan AR, Cowan JA, Jr., Wu CL. Efficacy of postoperative epidural analgesia: a meta-analysis. JAMA. 2003;290:2455–63. doi: 10.1001/jama.290.18.2455. [DOI] [PubMed] [Google Scholar]
- 24.Guay J, Choi P, Suresh S, Albert N, Kopp S, Pace NL. Neuraxial blockade for the prevention of postoperative mortality and major morbidity: an overview of Cochrane systematic reviews. Cochrane Database Syst Rev. 2014;1:CD010108. doi: 10.1002/14651858.CD010108.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halpern SH, Muir H, Breen TW, Campbell DC, Barrett J, Liston R, Blanchard JW. A multicenter randomized controlled trial comparing patient-controlled epidural with intravenous analgesia for pain relief in labor. Anesth Analg. 2004;99:1532–8. doi: 10.1213/01.ANE.0000136850.08972.07. [DOI] [PubMed] [Google Scholar]
- 26.Hausman MS, Jr., Jewell ES, Engoren M. Regional versus general anesthesia in surgical patients with chronic obstructive pulmonary disease: Does avoiding general anesthesia reduce the risk of postoperative complications? Anesth Analg. 2014;120:1405–12. doi: 10.1213/ANE.0000000000000574. [DOI] [PubMed] [Google Scholar]
- 27.Popping DM, Elia N, Van Aken HK, Marret E, Schug SA, Kranke P, Wenk M, Tramer MR. Impact of epidural analgesia on mortality and morbidity after surgery: systematic review and meta-analysis of randomized controlled trials. Ann Surg. 2014;259:1056–67. doi: 10.1097/SLA.0000000000000237. [DOI] [PubMed] [Google Scholar]
- 28.Wu CL, Cohen SR, Richman JM, Rowlingson AJ, Courpas GE, Cheung K, Lin EE, Liu SS. Efficacy of postoperative patient-controlled and continuous infusion epidural analgesia versus intravenous patient-controlled analgesia with opioids: a meta-analysis. Anesthesiology. 2005;103:1079–88. doi: 10.1097/00000542-200511000-00023. [DOI] [PubMed] [Google Scholar]
- 29.Hood DD, Dewan DM. Anesthetic and obstetric outcome in morbidly obese parturients. Anesthesiology. 1993;79:1210–8. doi: 10.1097/00000542-199312000-00011. [DOI] [PubMed] [Google Scholar]
- 30.Tonidandel A, Booth J, D'Angelo R, Harris L, Tonidandel S. Anesthetic and obstetric outcomes in morbidly obese parturients: a 20-year follow-up retrospective cohort study. Int J Obstet Anesth. 2014;23:357–64. doi: 10.1016/j.ijoa.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Kavanagh T, Jee R, Kilpatrick N, Douglas J. Elective cesarean delivery in a parturient with Klippel-Feil syndrome. Int J Obstet Anesth. 2013;22:343–8. doi: 10.1016/j.ijoa.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Ko JY, Leffert LR. Clinical implications of neuraxial anesthesia in the parturient with scoliosis. Anesth Analg. 2009;109:1930–4. doi: 10.1213/ANE.0b013e3181bc3584. [DOI] [PubMed] [Google Scholar]
- 33.O'Connor PJ, Moysa GL, Finucane BT. Thoracic epidural anesthesia for bilateral reduction mammoplasty in a patient with Klippel-Feil syndrome. Anesth Analg. 2001;92:514–6. doi: 10.1097/00000539-200102000-00043. [DOI] [PubMed] [Google Scholar]
- 34.Smith KA, Ray AP. Epidural anesthesia for repeat cesarean delivery in a parturient with Klippel-Feil syndrome. J Anaesthesiol Clin Pharmacol. 2011;27:377–9. doi: 10.4103/0970-9185.83686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer-Bender A, Kern A, Pollwein B, Crispin A, Lang PM. Incidence and predictors of immediate complications following perioperative non-obstetric epidural punctures. BMC Anesthesiol. 2012;12:31–7. doi: 10.1186/1471-2253-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crosby ET, Halpern SH. Obstetric epidural anaesthesia in patients with Harrington instrumentation. Can J Anaesth. 1989;36:693–6. doi: 10.1007/BF03005423. [DOI] [PubMed] [Google Scholar]
- 37.Daley MD, Rolbin SH, Hew EM, Morningstar BA, Stewart JA. Epidural anesthesia for obstetrics after spinal surgery. Reg Anesth. 1990;15:280–4. [PubMed] [Google Scholar]
- 38.Feldstein G, Ramanathan S. Obstetrical lumbar epidural anesthesia in patients with previous posterior spinal fusion for kyphoscoliosis. Anesth Analg. 1985;64:83–5. [PubMed] [Google Scholar]
- 39.Hubbert CH. Epidural anesthesia in patients with spinal fusion. Anesth Analg. 1985;64:843. doi: 10.1213/00000539-198508000-00019. [DOI] [PubMed] [Google Scholar]
- 40.Apfel CC, Saxena A, Cakmakkaya OS, Gaiser R, George E, Radke O. Prevention of postdural puncture headache after accidental dural puncture: a quantitative systematic review. Br J Anaesth. 2010;105:255–63. doi: 10.1093/bja/aeq191. [DOI] [PubMed] [Google Scholar]
- 41.Berger CW, Crosby ET, Grodecki W. North American survey of the management of dural puncture occurring during labour epidural analgesia. Can J Anaesth. 1998;45:110–4. doi: 10.1007/BF03013247. [DOI] [PubMed] [Google Scholar]
- 42.Choi PT, Galinski SE, Takeuchi L, Lucas S, Tamayo C, Jadad AR. PDPH is a common complication of neuraxial blockade in parturients: a meta-analysis of obstetrical studies. Can J Anaesth. 2003;50:460–9. doi: 10.1007/BF03021057. [DOI] [PubMed] [Google Scholar]
- 43.Pan PH, Bogard TD, Owen MD. Incidence and characteristics of failures in obstetric neuraxial analgesia and anesthesia: a retrospective analysis of 19,259 deliveries. Int J Obstet Anesth. 2004;13:227–33. doi: 10.1016/j.ijoa.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 44.Stacey RG, Gass CW. Epidural failure rate. Int J Obstet Anesth. 1999;8:294–5. doi: 10.1016/s0959-289x(99)80118-6. [DOI] [PubMed] [Google Scholar]
- 45.Desai A, Alemayehu H, Weesner KA, St Peter SD. Impact of epidural failures on the results of a prospective, randomized trial. Eur J Pediatr Surg. 2016;26:160–3. doi: 10.1055/s-0034-1396414. [DOI] [PubMed] [Google Scholar]
- 46.Scepanovic OR, Volynskaya Z, Kong CR, Galindo LH, Dasari RR, Feld MS. A multimodal spectroscopy system for real-time disease diagnosis. Rev Sci Instrum. 2009;80:0431031–9. doi: 10.1063/1.3117832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller MG, Georgakoudi I, Zhang Q, Wu J, Feld MS. Intrinsic fluorescence spectroscopy in turbid media: disentangling effects of scattering and absorption. Appl Opt. 2001;40:4633–46. doi: 10.1364/ao.40.004633. [DOI] [PubMed] [Google Scholar]
- 48.Lue N, Kang JW, Yu CC, Barman I, Dingari NC, Feld MS, Dasari RR, Fitzmaurice M. Portable optical fiber probe-based spectroscopic scanner for rapid cancer diagnosis: a new tool for intraoperative margin assessment. PLoS One. 2012;7:e3088701–10. doi: 10.1371/journal.pone.0030887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang JW, Lue N, Kong CR, Barman I, Dingari NC, Goldfless SJ, Niles JC, Dasari RR, Feld MS. Combined confocal Raman and quantitative phase microscopy system for biomedical diagnosis. Biomed Opt Express. 2011;2:2484–92. doi: 10.1364/BOE.2.002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Motz JT, Hunter M, Galindo LH, Gardecki JA, Kramer JR, Dasari RR, Feld MS. Optical fiber probe for biomedical Raman spectroscopy. Appl Opt. 2004;43:542–54. doi: 10.1364/ao.43.000542. [DOI] [PubMed] [Google Scholar]
- 51.Buschman HP, Motz JT, Deinum G, Romer TJ, Fitzmaurice M, Kramer JR, van der Laarse A, Bruschke AV, Feld MS. Diagnosis of human coronary atherosclerosis by morphology-based Raman spectroscopy. Cardiovasc Pathol. 2001;10:59–68. doi: 10.1016/s1054-8807(01)00063-1. [DOI] [PubMed] [Google Scholar]
- 52.Shafer-Peltier KE, Haka AS, Motz JT, Fitzmaurice M, Dasari RR, Feld MS. Model-based biological Raman spectral imaging. J Cell Biochem Suppl. 2002;39:125–37. doi: 10.1002/jcb.10418. [DOI] [PubMed] [Google Scholar]
- 53.Kang JW, Nguyen FT, Lue N, Dasari RR, Heller DA. Measuring uptake dynamics of multiple identifiable carbon nanotube species via high-speed confocal Raman imaging of live cells. Nano Lett. 2012;12:6170–4. doi: 10.1021/nl302991y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dempster AP, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society. Series B (Methodological) 1977;39:1–38. LN. [Google Scholar]
- 55.Griner PF, Mayewski RJ, Mushlin AI, Greenland P. Selection and interpretation of diagnostic tests and procedures. Principles and applications. Ann Intern Med. 1981;94:557–92. [PubMed] [Google Scholar]
- 56.Shafer-Peltier KE, Fitzmaurice M, Crowe J, Myles J, Dasari RR, Feld MS. Raman microspectroscopic model of human breast tissue: implications for breast cancer diagnosis in vivo. Journal of Raman Spectroscopy. 2002;33:552–63. HA. [Google Scholar]
- 57.Perlas A, Chaparro LE, Chin KJ. Lumbar neuraxial ultrasound for spinal and epidural anesthesia: A systematic review and meta-analysis. Reg Anesth Pain Med. 2016;41:251–60. doi: 10.1097/AAP.0000000000000184. [DOI] [PubMed] [Google Scholar]
- 58.Sawada A, Kii N, Yoshikawa Y, Yamakage M. Epidrum((R)): a new device to identify the epidural space with an epidural Tuohy needle. J Anesth. 2012;26:292–5. doi: 10.1007/s00540-011-1278-1. [DOI] [PubMed] [Google Scholar]
- 59.Ghelber O, Gebhard RE, Vora S, Hagberg CA, Szmuk P. Identification of the epidural space using pressure measurement with the compuflo injection pump--a pilot study. Reg Anesth Pain Med. 2008;33:346–52. doi: 10.1016/j.rapm.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 60.Lirk P, Kolbitsch C, Putz G, Colvin J, Colvin HP, Lorenz I, Keller C, Kirchmair L, Rieder J, Moriggl B. Cervical and high thoracic ligamentum flavum frequently fails to fuse in the midline. Anesthesiology. 2003;99:1387–90. doi: 10.1097/00000542-200312000-00023. [DOI] [PubMed] [Google Scholar]
- 61.Lim L, Nichols B, Rajaram N, Tunnell JW. Probe pressure effects on human skin diffuse reflectance and fluorescence spectroscopy measurements. J Biomed Opt. 2011;16:0110121–9. doi: 10.1117/1.3525288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barman I, Dingari NC, Saha A, McGee S, Galindo LH, Liu W, Plecha D, Klein N, Dasari RR, Fitzmaurice M. Application of Raman spectroscopy to identify microcalcifications and underlying breast lesions at stereotactic core needle biopsy. Cancer Res. 2013;73:3206–15. doi: 10.1158/0008-5472.CAN-12-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.