Abstract
The bacteria in the midgut of Anopheles stephensi adult females from laboratory colonies were studied by sequencing the V4 region of 16S rRNA genes, with respect to three experimental factors: stable or cured Wolbachia infection; sugar or blood diet; and age. Proteobacteria and Bacteroidetes dominated the community [>90% of operational taxonomic units (OTUs)]; most taxa were in the classes Flavobacteriia, Gammaproteobacteria, and Alphaproteobacteria, and were assigned to Elizabethkingia (46.9%), Asaia (6.4%) and Pseudomonas (6.0%), or unclassified Enterobacteriaceae (37.2%). Bacterial communities were similar between Wolbachia-cured and Wolbachia-infected mosquito lines, indicating that the gut microbiota were not dysregulated in the presence of Wolbachia. The proportion of Enterobacteriaceae was higher in mosquitoes fed a blood meal compared to those provided a sugar meal. Collectively, the bacterial community had a similar structure in older Wolbachia-infected mosquitoes 8 days after the blood meal, as in younger Wolbachia-infected mosquitoes before a blood meal, except that older mosquitoes had a higher proportion of Enterobacteriaceae and lower proportion of Elizabethkingia. Consistent presence of certain predominant bacteria (Elizabethkingia, Asaia, Pseudomonas, and Enterobacteriaceae) suggests they would be useful for paratransgenesis to control malaria infection, particularly when coupled to a Wolbachia-based intervention strategy.
Keywords: Anopheles, Wolbachia, malaria vectors, paratransgenesis
Introduction
Strategies for controlling mosquito-vectored pathogens based on infection with the symbiotic bacterium Wolbachia include vector population replacement and population suppression (Sinkins, 2004; Werren et al., 2008; Iturbe-Ormaetxe et al., 2011; Bourtzis et al., 2014). Although many mosquito species are naturally infected with various strains of Wolbachia, artificially but stably induced Wolbachia infection in mosquito host species not naturally having Wolbachia infection was originally established by embryonic injection followed by trans-generational spread (Dobson et al., 1999; Sinkins, 2004; Bourtzis et al., 2014). Certain combinations of Wolbachia strains and mosquito species suppress propagation and transmission of dengue viruses and malaria parasites in mosquito hosts (Bourtzis et al., 2014). However, in nature the host range of Wolbachia does not include some of the most important vectors of human pathogens, such as Anopheles stephensi, A. gambiae, and Aedes aegypti (Hilgenboecker et al., 2008). This host range has been expanded through forced infections by inoculations into mosquito embryos, resulting in stable infection across subsequent generations in A. aegypti and A. stephensi with little fitness cost (Xi et al., 2005; Bian et al., 2013; Joshi et al., 2014). A recent study found strongly negative interactions between the commensal gut microflora in mosquitoes and propensity of the mosquitoes to harbor Wolbachia infection after intrathoracic inoculation, which might explain why natural infections of Wolbachia in some mosquito species are not found in nature (Hughes et al., 2014). Hughes et al. (2014) reported that Wolbachia vertical transmission was achievable in Anopheles species only when the gut microbiota (especially, Asaia bacteria) were perturbed (i.e., eliminated) with antibiotics. Furthermore, Rossi et al. (2015) observed that Wolbachia failed to colonize the female reproductive organs of anophelines, due to presence of Asaia infection in those tissues (Rossi et al., 2015). The conclusion of both studies is that commensal microbiota impede Wolbachia vertical transmission in mosquitoes (Hughes et al., 2014; Rossi et al., 2015). However, what remains to be determined is whether the commensal microbiota of mosquitoes that have been stably infected trans-generationally with Wolbachia by embryonic injection differ from that of Wolbachia-uninfected mosquitoes of the same species and held under the same conditions. Based on the results of studies reviewed here (Hughes et al., 2014; Rossi et al., 2015), one would predict that stably infected mosquitoes must differ in their microbiota due to the inhibitory effects of Asaia and other gut symbionts on Wolbachia infection. Because stably infected mosquitoes are those most likely to be used in attempts to control vector populations or pathogen transmission, the interactions between stable Wolbachia infection and gut microbiota clearly become important.
After the wAlbB Wolbachia strain was successfully transferred from A. albopictus into A. stephensi by embryonic microinjection, this strain of Wolbachia spread rapidly into a laboratory population of A. stephensi by vertical transmission over the course of several generations without any manipulation of gut microbiota (Bian et al., 2013). Further, wAlbB infection conferred partial resistance to development of the malaria parasite Plasmodium falciparum in A. stephensi (Bian et al., 2013), and significantly up-regulated the mosquitoes’ innate immune response, including generation of reactive oxygen species from midgut epithelial cells (Pan et al., 2012). One way to increase the inhibitory effect of Wolbachia on virus or parasite development in mosquito hosts is to increase the intensity of Wolbachia infection (Correa and Ballard, 2014). Another strategy is to couple paratransgenesis of gut microbiota with a Wolbachia-based vector population replacement/suppression strategy. The latter concept is promising because certain symbionts such as Asaia and Pantoea agglomerans, when engineered to express molecules having anti-malaria parasite properties, result in reduced malaria parasite load in host mosquitoes (Wang et al., 2012; Bongio and Lampe, 2015). However, the innate immune response triggered by Wolbachia infection is non-specific and it may affect the commensal microflora (Pan et al., 2012). Thus, it remains unclear if introduced or genetically modified bacteria will persist in Wolbachia-infected mosquitoes due to elevated innate immune response and putative incompatibility between Wolbachia and gut microbiota. If Wolbachia wAlbB infection with its non-specific immune effects in A. stephensi, and engineered gut microbiota with specific anti-parasite effects, are found to be compatible; then a synergistic, dual strategy could be developed to suppress malaria parasite or virus development in mosquitoes. In this study, and as a step toward determining the extent of this compatibility, we quantified the diversity of the bacterial community in stably Wolbachia-infected and Wolbachia-cured A. stephensi mosquitoes. Moreover, because of the dynamics of microbiota associated with key life history events in mosquitoes, we further compared bacterial communities when sugar or meals were provided, and when the mosquitoes were young or aged.
Materials and Methods
Ethics Statement
All procedures involving vertebrate animals were in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and were conducted under protocol 03/14-036-00, approved by the Michigan State University Institutional Animal Care and Use Committee.
Insects
Mosquito colonization procedures are described elsewhere (Chen et al., 2015). Briefly, adult mosquitoes were held in cages at 27°C and 85% humidity under a light/dark 12:12-h photoperiod without dawn/dusk transitions, and were provided a 10% sucrose solution ad libitum by cotton wick. To initiate egg development, bovine blood with sodium heparin (Hemostat Lab, Dixon, CA, USA) was fed to adult mosquitoes via an artificial membrane feeder. Two days after the blood meal, oviposition substrates (consisting of filter paper moistened with water in a Petri dish) were placed in the cages. Eggs were transferred to plastic containers with distilled water for hatching. First Bite (Kyorin, Himeji, Japan) and Tetramin tropical fish food flakes (Tetra, Blacksburg, VA, USA) were provided ad libitum to first instar larval mosquitoes. Thereafter, pet food (Purina Cat Chow; Nestleé) was given once per day ad libitum.
The stable wAlbB infection in the A. stephensi isofemale line (designated LB1) and the aposymbiotic A. stephensi line (wAlbB cured, designated LBT) were previously established (Bian et al., 2013). Aposymbiotic line LBT was derived from the LB1 strain after treatment with tetracycline (Bian et al., 2013). Approximately 4 years and 21 generations passed since that treatment was done, before this study was conducted. The presence or absence of Wolbachia in LB1 or LBT was confirmed by PCR of DNA extractions from abdomens using primers wsp81F (TGGTCC AATAAGTGATGAAGAAAC) and wsp691R (AAAAATTAAACGCTACTCCA). No Wolbachia infection was found in LBT individuals but all LB1 individuals were positive across the experimental groups (Supplementary Figure S1). Females of each strain were sampled 1 week after adult emergence, designated here as “LB1-SM” and “LBT-SM” given that they had sugar meals but no blood meals yet. Other females were provided a blood meal at 7 days after adult emergence by allowing them to feed on anesthetized BALB/c mice for 20 min. These mosquitoes were sampled 24 h after the blood meal, and were designated “LB1-BM” and “LBT-BM,” respectively. Finally, other blood fed females of the LB1 and LBT strains were sampled after an interval of 1 week following the blood meal as “aged” mosquito samples (15 days after adult emergence and 8 days after the blood meal), during which time they were provisioned 10% sugar solution. These mosquitoes were designated “LB1-PBM” and “LBT-PBM,” respectively.
DNA Extraction, Library Construction, and 16S rRNA Sequencing
All DNA extractions were performed in a laminar flow biosafety cabinet to avoid contamination. Prior to dissection, mosquitoes were disinfected externally with 70% ethanol (three rinses), followed by a rinse with sterile water. Midguts were dissected with sterile forceps, and transferred to 200 μl of sterile phosphate-buffered saline (PBS) buffer. Midguts from six individuals were homogenized by crushing with a sterile pestle in the same tube as a single sample. The volume was re-suspended in 200 μl of lysis buffer and DNA extracted with the DNeasy Blood & Tissue Kit (Qiagen) following the manufacturer’s recommendations. Six biological replicates (total 36 midguts) were used for each of the six experimental treatments; a total of 216 mosquitoes were used in this study. The DNA concentration was measured using QubitTM dsDNA HS Assay Kits and the DNA integrity was tested by PCR using primers 63F (CAGGCCTAACACATGCAAGTC) and 1387R (CGGAACATGTGWGGCGGG). Amplicon tagging and sequencing were conducted at the Research Technology Support Facility (RTSF) at Michigan State University. DNA was amplified by using the primers 515f (GTG CCA GCM GCC GCG GTA A) and 806r (NNN NNN GGA CTA CHV GGG TWT CTA AT), which targets the V4 region of bacterial 16S rDNA. The reverse primer contains a 6-bp error-correcting barcode unique to each sample. The purified amplicons were pooled, loaded on an Illumina MiSeq flow cell, and sequenced in a 2 × 250 bp paired end format using a 500 cycle v2 reagent cartridge. Base calling was done by Illumina Real Time Analysis (RTA) v1.18.54 and output of the RTA was demultiplexed and converted to FastQ format by Illumina Bcl2fastq v1.8.4.
DNA Sequence Analysis
Sequencing reads from the FastQ format were processed and analyzed using the Mothur package1 v.1.37.0. After denoising procedures by PyroNoise, Uchime, preclustering, good quality sequences (>250 bp) without detectable sequencing errors or chimeras and removing rare operational taxonomic units (OTUs; 5 or fewer) were used for assigning OTUs using an average neighbor algorithm (97% similarity cutoff). OTUs were classified at the genus level using the Bayesian method. When there were no references for the interested bacteria in the database used by Mothur, we retrieved the representative sequences for specific OTUs and submitted the sequences to GenBank (NCBI) for a BLAST search for purposes of classification. Data were further analyzed by first trimming sequence quality with different cutoffs using standard filtering tools (Schloss et al., 2009). Rarefaction curves were built to estimate sample coverage. Non-parametric estimates of richness were made using the Chao index and Abundance Coverage Estimator (cut-off threshold, 97%). Within-sample diversity (or α-diversity) was calculated using Simpson and Shannon diversity indices. Welch’s t-test was used to compare diversity indexes between Wolbachia-infected and Wolbachia-cured mosquitoes with different diets and at different rearing ages. Community composition (or β-diversity) was compared among treatments using Bray–Curtis dissimilarities. Differences in community composition among treatments were tested using permutational multivariate analysis of variance (PERMANOVA). Richness and α-diversity analyses were done in mothur. β-diversity analyses were done in R (v. 3.2.0). Bray–Curtis dissimilarity matrices were created using the vegdist() function (method = “bray”) and PERMANOVA tests (999 permutations) were done using the adonis() function in the vegan package [v. 2.3-2 (Oksanen et al., 2015)]. Principal coordinate analyses (PCoA) ordinations were created using the pcoa() function in the ape package [v. 3.4 (Paradis et al., 2004)].
Nucleotide Sequence Accession Number
The raw sequences of this study have been deposited in the Sequence Read Archive (accession number: SRP072068).
Results
Characteristics of Midgut Bacteria Community Libraries
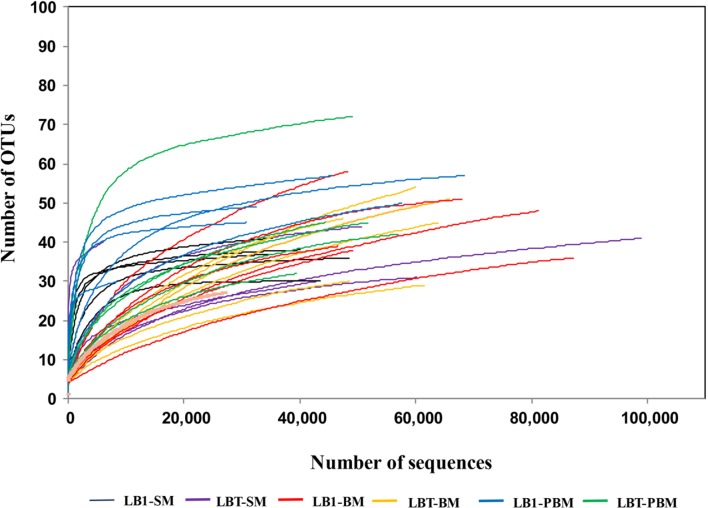
Approximately 2,130,826 sequence reads were successfully retained. After removal of short (<250 bp), possibly chimeras and rare OTUs (5 or fewer, see Supplementary Table S1), a total of 1,704,538 good quality sequences were available for further analysis (Supplementary Table S2). These sequences were assigned to 394 OTUs (Supplementary Table S1). Sequence number for each sample was normalized to the minimal readings (6,056) by randomly subsampling to minimize the biases generated by sequencing depth (Supplementary Table S2). The average Good’s coverage was 99.9% (Supplementary Table S2). The rarefaction curves (Figure 1) showed that the samples from some of these mosquito collections did not reach saturation to an asymptote, indicating that additional rare bacterial taxa likely occur in them, but all deflected as they rose with sampling effort in a characteristic concave function (Figure 1).
FIGURE 1.
Rarefaction analysis of bacterial 16S rRNA gene libraries from Wolbachia-infected and Wolbachia-cured A. stephensi samples that were sugar fed, blood fed, and post-blood fed. Operational taxonomic units (OTUs) were grouped with a 97% similarity. LB1-SM, Wolbachia-infected mosquitoes with sugar meal; LBT-SM, Wolbachia-cured mosquitoes with sugar meal; LB1-BM, Wolbachia-infected mosquitoes with blood meal; LBT-BM, Wolbachia-cured mosquitoes with blood meal; LB1-PBM, post-blood fed Wolbachia-infected mosquitoes; LBT-PBM, post-blood fed Wolbachia-cured mosquitoes.
Effects of Wolbachia Infection, Diet, and Age on Composition of the Gut Microbiota
Operational taxonomic units were assigned to over 16 bacterial phyla and to unclassified bacteria (Supplementary Table S3). Single OTUs assigned to the classes Flavobacteriia, Gammaproteobacteria, and Alphaproteobacteria were most frequent, together accounting for more than 90% of the sequences, and were dominant in experimental treatments (Figure 2A). Sequences of bacteria in the classes Actinobacteria and Betaproteobacteria were also detected, but they were not uniformly represented in every sample. Some bacteria (2% of the total community) could not be assigned to a certain rank (unclassified), suggesting the existence of previously uncharacterized taxa. However, OTUs assigned to other classes were not abundant (i.e., <1%) and/or were absent from some samples or groups. The top 10 OTUs at the genus level comprised 99.2% of the total sequences (Figure 2B). The predominant OTUs were represented by Elizabethkingia (46.9%), unclassified Enterobacteriaceae (37.2%), Asaia (6.4%) and Pseudomonas (6.0%).
FIGURE 2.
Gut bacterial composition at class level and the relative abundance of the top 10 genera in Wolbachia-infected and Wolbachia-cured mosquito samples by dietary treatment. (A) Taxonomic classification of bacterial reads retrieved from the Wolbachia-infected and Wolbachia-cured mosquitoes. (B) Relative abundance of 10 most abundant OTUs (97% similarity). Unclassified Enterobacteriaceae or Micrococcaceae represent the reads that were assigned to the family Enterobacteraceae or Micrococcaceae, but could not be assigned to a genus. Wol+, Wolbachia-infected mosquito; Wol-, Wolbachia-cured mosquito.
There was no significant difference in bacterial class abundance between Wolbachia-infected (LB1-SM) and Wolbachia-cured mosquitoes (LBT-SM) (Welch’s t-test) (Figure 2A). At the genus level, the most abundant OTU in the midgut of A. stephensi LBT (Wolbachia-cured) was Elizabethkingia, accounting for 65% of the community (Figure 2B). A similar proportion of Elizabethkingia was found in A. stephensi LB1 (69%), indicating that Wolbachia infection did not significantly alter its relative abundance (p > 0.05). Next to Elizabethkingia, unclassified Enterobacteriaceae were the second most abundant OTU in sugar-fed young Anopheles mosquitoes (LB1-SM and LBT-SM), which accounted for between 8 and 15% of the OTUs (Figure 2B). The number of Enterobacteriaceae OTUs was not statistically different between LB1-SM and LBT-SM (p > 0.05; Figure 2B). Asaia OTUs ranged in abundance from 5 to 8% of the total bacterial community (Figure 2B) in LB1-SM and LBT-SM, respectively, indicating that the colonization and persistence of Asaia in A. stephensi midguts occurred in a stable Wolbachia infection mosquito line as well as in A. stephensi without Wolbachia infection (p > 0.05; Figure 2B). Similar to Asaia, Pseudomonas OTUs were equally associated with both mosquito lines (p > 0.05; Figure 2B). As expected, the relative abundance of Wolbachia was 8.6% of the total bacterial OTUs in LB1-SM while it was nearly absent (<0.01% of OTUs) in LBT-BM (Figure 2B).
At 24 h after a blood meal, the relative abundance of Elizabethkingia decreased from 65 to 19% in LBT (p < 0.05) and decreased from 69 to 34% in LB1 mosquitoes, respectively (p < 0.05). By contrast, after a blood meal the Enterobacteriaceae OTUs increased 7.1 and 4.4-fold in LB1-BM and LBT-BM within 24 h (p < 0.05), respectively. Compared to sugar meals, the relative abundance of Asaia or Pseudomonas was not significantly changed by blood feeding (p > 0.05; Figure 2B). The relative abundance of Wolbachia in the midgut decreased to 0.18% of the total taxa in 24 h after blood meals (Figure 2B).
In mosquitoes at age 15 days (that is, 8 days post-blood meal), the frequency of Elizabethkingia OTUs represented 42% of the bacterial community in guts of Wolbachia-infected mosquitoes (LB1-PBM), showing that relative abundance of these OTUs in 15 days old LB1-PBM was significantly different from 7 days old LBT-SM mosquitoes (p < 0.05; Figure 2B). However, there was no statistical difference between LBT-SM and LBT-PBM mosquitoes (p > 0.05; Figure 2B). The relative abundance of Enterobacteriaceae OTUs in 15 days old LBT and LB1 mosquitoes was 2.2 and 3.4-fold higher than that in young LBT and LB1 mosquitoes (p < 0.05), respectively. The abundance of Asaia was consistent and not affected by age (p > 0.05), and accounted for 8.9 and 6.5% of bacterial community in older Wolbachia-infected and Wolbachia-cured mosquitoes, respectively. The abundance of Pseudomonas in older, Wolbachia-infected mosquitoes (LBT-PBM) was 2.6-fold higher than in young LBT mosquitoes (p < 0.05), while it remained stable between young and old, Wolbachia-cured mosquitoes (p > 0.05; Figure 2B). The relative abundance of Wolbachia OTUs in LB1-PBM reached up to 7.4% of total bacterial community level, which was comparable to that in LB1-SM (p > 0.05; Figure 2B).
Alpha and Beta Bacterial Diversity in Wolbachia-Infected and Wolbachia-Cured Mosquitoes of Different Diets and Ages
To analyze differences in alpha diversity, we compared diversity (Shannon index) and richness (Chao1). There was no significant difference in Shannon diversity between LB1-SM and LBT-SM or LB1-BM and LBT-BM, showing that intracellular Wolbachia infection did not affect microbial diversity in young mosquito midguts (Table 1; Supplementary Figure S2). However, the Shannon index was different between LB1-PBM and LBT-PBM samples (p < 0.05), indicating that Wolbachia infection only changed gut microbial diversity in older mosquito. After blood feeding, the Shannon diversity in LB1-BM was not significantly different from that in LB1-SM (p > 0.05) and there was no significant difference between LBT-BM and LBT-SM (p > 0.05) (Table 1; Supplementary Figure S2). Thirdly, microbial diversity was significantly different between older mosquitoes (LBT-PBM) and young mosquitoes (LBT-SM) with Wolbachia infection (p < 0.05). However, in non-Wolbachia infection mosquitoes, the microbial diversity was similar between older mosquitoes (LB1-PBM) and young mosquitoes (LB1-SM) (p < 0.05; Table 1; Supplementary Figure S2). For community richness comparisons (Chao1), no significant differences were observed between Wolbachia-infected mosquitoes (LB1-SM, LB1-BM, or LB1-PBM) and Wolbachia-cured ones (LBT-SM, LBT-BM, or LBT-PBM) (p > 0.05; Supplementary Figure S2). However, blood-fed mosquitoes with Wolbachia infection had a significantly lower number of estimated OTUs (Chao1) than sugar-fed ones with Wolbachia infection (i.e., LB1-BM vs. LB1-SM, p < 0.05) while there was no significant difference between blood-fed and sugar-fed mosquitoes without Wolbachia infection (LBT-BM vs. LBT-SM, p > 0.05) as shown in Supplementary Figure S2, indicating that the blood meal decreased richness in Wolbachia infected mosquitoes. However, the richness (Chao1) in the guts of older mosquitoes (LB1-PBM or LBT-PBM) was not significantly different from that in of younger ones (LB1-BM or LBT-BM) after blooding feeding (p > 0.05; Supplementary Figure S2).
Table 1.
Richness and diversity estimation of the 16S rRNA gene libraries.
| Sample | Cutoffs | Simpson | Shannon | Chao | Ace |
|---|---|---|---|---|---|
| LBT-SM | 0.03 | 0.58 ± 0.23 | 0.99 ± 0.71 | 32.15 ± 14.16 | 41.24 ± 14.35 |
| LB1-SM | 0.03 | 0.50 ± 0.01 | 1.13 ± 0.23 | 39.91 ± 10.31 | 41.23 ± 10.07 |
| LBT-BM | 0.03 | 0.57 ± 0.12 | 0.78 ± 0.19 | 39.81 ± 32.14 | 22.07 ± 25.94 |
| LB1-BM | 0.03 | 0.50 ± 0.11 | 0.92 ± 0.16 | 25.96 ± 11.81 | 40.49 ± 24.43 |
| LBT-PBM | 0.03 | 0.41 ± 0.04 | 1.12 ± 0.06 | 39.23 ± 13.51 | 64.59 ± 24.09 |
| LB1-PBM | 0.03 | 0.33 ± 0.07 | 1.56 ± 0.31 | 36.76 ± 10.51 | 37.21 ± 9.64 |
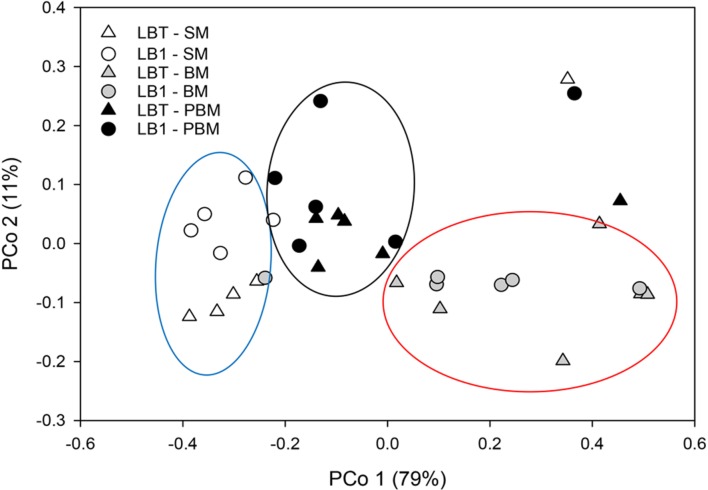
For beta diversity analysis, we examined the relationships in gut microbiota between different diet, age and Wolbachia infection status by using principal coordinate analyses (PCoA) based on Bray–Curtis distance (Figure 3). The first two components captured 90% of the variance among samples. Community composition differed among all three diet types (PERMANOVA; F = 10.93, p < 0.05, Table 2). Bacterial communities from the mosquitoes fed on sugar separated from those fed on blood and those PBM along PC1, which explained 79% of the variation (Table 3). However, community composition was similar between Wolbachia-infected and Wolbachia-cured groups across all diet treatments (PERMANOVA; F = 2.86, p > 0.05, Table 2).
FIGURE 3.
Principal coordinate analysis of bacterial the community structure using Bray–Curtis distances. Symbols represent one mosquito collection (six pooled mosquito guts). Distances between symbols on the ordination plot reflect relative dissimilarities in community structures.
Table 2.
Pseudo F table of PERMANOVA analysis based on Bray–Curtis dissimilarities.
| Source of variance | Sum of squares | Degrees of freedom | Mean square | F | R2 | p∗ |
|---|---|---|---|---|---|---|
| Wolbachia | 0.186 | 1 | 0.186 | 2.861 | 0.053 | 0.086 |
| Diet | 1.418 | 2 | 0.709 | 10.931 | 0.404 | 0.001 |
| Wolbachia∗Diet | 0.088 | 2 | 0.044 | 0.681 | 0.025 | 0.549 |
| Residuals | 1.816 | 28 | 0.065 | 0.518 | ||
| Total | 3.509 | 33 | 1 | |||
∗OTUs with 5 or fewer sequences were excluded. Significant p-values (≤0.05) are bolded.
Table 3.
Pseudo F table of pairwise comparisons of diet types using PERMANOVA.
| Pairwise comparison | Source of variance | Sum of squares | Degrees of freedom | Mean square | F | R2 | p∗ |
|---|---|---|---|---|---|---|---|
| SM vs. BM | Diet | 1.314 | 1 | 1.314 | 19.660 | 0.496 | 0.001 |
| Residuals | 1.337 | 20 | 0.067 | 0.504 | |||
| Total | 2.652 | 21 | 1 | ||||
| SM vs. PBM | Diet | 0.308 | 1 | 0.308 | 4.261 | 0.176 | 0.021 |
| Residuals | 1.444 | 20 | 0.072 | 0.824 | |||
| Total | 1.752 | 21 | 1 | ||||
| BM vs. PBM | Diet | 0.522 | 1 | 0.522 | 8.208 | 0.272 | 0.004 |
| Residuals | 1.399 | 22 | 0.064 | 0.728 | |||
| Total | 1.921 | 23 | 1 | ||||
∗OTUs with 5 or fewer sequences were excluded. Significant p-values (≤0.05) are bolded.
Discussion
The bacteria (i.e., microbiota) associated with mosquitoes have significant roles throughout the mosquito lifecycle, in providing energy and nutrients, and regulating development, fecundity, and immune responses (Sinkins, 2004; Chouaia et al., 2012; Pan et al., 2012; Coon et al., 2014). These symbiotic bacteria find shelter and nutrients within this environment and could properly be referred to as commensals for these beneficial reasons, even if the beneficial associations of the bacteria to mosquitoes are not entirely clear. Stable, intracellular Wolbachia infection in A. stephensi (LB1) was achieved previously through embryonic inoculation and multi-generational, vertical passage (Bian et al., 2013). It was done without perturbation of the natural microbiota (as shown here) and without excessive mortality after blood meals (Bian et al., 2013; Joshi et al., 2014). More recently and by contrast, vertical persistence of Wolbachia infection in A. stephensi was achieved only when symbiotic bacteria were perturbed by antibiotics (Hughes et al., 2014). In that same study, a combination of Wolbachia and Asaia caused mosquito mortality after a blood meal, a finding suggestive of a physiologic incompatibility between Wolbachia infection and presence of a constitutive microflora when the stressors of the blood meal arrive (Hughes et al., 2014). Indeed, if such an incompatibility exists, it would obviate the possibility of a dual, anti-malaria parasite strategy based on immune-mediating effects of Wolbachia infection and paratransgenesis of select members of that microflora. This possibility, in part, motivated the research presented here.
The similarity of the gut bacterial community structure, as measured by composition of taxa as well as by α- and β-diversity, between Wolbachia-infected LB1 and Wolbachia-cured LBT mosquitoes indicates that there were no significant Wolbachia-mediated effects on the structure of the commensal bacterial community; and that the bacterial assembly was resilient to any effects, such as innate immune effects, that the presence of Wolbachia might impose. Therefore, coexistence of these gut bacteria with Wolbachia was not problematic in the stable Wolbachia-infected Anopheles progeny. These findings contrast with other studies demonstrating very strongly negative effects of combination of gut bacteria and Wolbachia on host fitness (high mortality rate after a blood meal), particularly due to Asaia bacteria (Hughes et al., 2014). One explanation for the difference is in the way the Wolbachia infection was originally introduced into the mosquitoes: embryonic microinjection followed by stable, trans-generational perpetuation of infection (Bian et al., 2013); or intrathoracic inoculation without perpetuation (Hughes et al., 2014). The latter likely led to a larger and more acute dose of the bacteria, than one inherited in the germ line. However, the detailed mechanisms need to be further investigated.
The blood meal profoundly affects bacterial community composition in the midgut of several mosquito species, including A. gambiae, A. stephensi, and A. coluzzii (Wang et al., 2011; Gimonneau et al., 2014; Tchioffo et al., 2016). The main influencing factors included dramatic changes in mosquito gut temperature and appearance of oxidative stress (Luckhart et al., 1998; Peterson and Luckhart, 2006; Benoit et al., 2011). Especially, the ingested red blood cells from animals carry an enormous amount of hemoglobin, which causes a massive release of heme in the midgut, thus leading to a dramatic change in gut conditions (Wang et al., 2011; Tchioffo et al., 2016). Blood meals induce midgut epithelia to produce nitric oxide which is also a source for free radicals (Luckhart et al., 1998). Hematophagous mosquitoes exhibit detoxification mechanisms in response to these free radicals, but the role of commensal bacteria in the gut in that same process and how those bacteria survive them are not known (Peterson and Luckhart, 2006; Benoit et al., 2011). After blood meals, Enterobacteriaceae dramatically increased, which was consistent with those reported in A. gambiae (Wang et al., 2011). Wang et al. (2011) found several stress response systems existing in the Enterobacteriaceae genomes. Possibly, this group of bacteria interacts with the mosquito host in response to the oxidative stressors present in the blood meal bolus (Wang et al., 2011). Therefore, blood-induced mortality in transiently Wolbachia-infected Anopheles mosquito could be caused by a combinative effect, that is, innate immune response elicited by Wolbachia infection and by dysregulated microbiota (Luckhart et al., 1998; Pan et al., 2012; Hughes et al., 2014). Both of them caused an unusual high level of active radicals in midgut, which can cause destructive damage to mosquitoes (Luckhart et al., 1998). Although there was a dramatic change in microbial structure after blood meals (compared to sugar meals), the core microbial compositions and community structure between Wolbachia-infected and Wolbachia-cured mosquitoes were rather similar (Figure 3).
When switching diet from blood to sugar meals and aging the insects, we found that the microbial community structure was different between the young (before blood meal) and aged mosquitoes (post-blood meal) in both mosquito lines. However, the relative abundance of transiently increased bacteria such as Enterobacteriaceae decreased when blood digestion was complete; the dominant Elizabethkingia abundance at age 15 days, and after the blood meal, was similar to before the blood meal (Figure 2B). The dominant bacterial community patterns we observed here were similar to those reported in A. gambiae (Wang et al., 2011). Collectively, Asaia, Elizabethkingia, and Enterobacteriaceae were present and predominant in aged, blood-fed, Wolbachia-infected A. stephensi, providing the possibility to devise an efficient malaria control reagent based on a combination of effects of Wolbachia and effects of paratransgenic gut microbiota (Federici et al., 2003; Favia et al., 2008; Cirimotich et al., 2011; Boissière et al., 2012; Capone et al., 2013; Bongio and Lampe, 2015; Chen et al., 2015).
Previous studies demonstrated that Wolbachia infected the midgut in A. stephensi and A. gambiae mosquitoes though its distribution level was much lower than that in the fat body (Bian et al., 2013). After a blood meal, the dramatic decrease of Wolbachia OTUs in LB1 gut was consistent with a previous observation by Hughes et al. (2014). Several factors including heme release, ROS level change, and interactions with gut bacteria contributed to this change (Pan et al., 2012; Bian et al., 2013; Hughes et al., 2014). However, after removing gut bacteria by antibiotic treatment (especially Asaia), Wolbachia persisted at a level similar to that non-blood fed mosquitoes, indicating gut microbiota suppressed Wolbachia level after a blood meal (Hughes et al., 2014).
Operational taxonomic units of Asaia (up to 8.9% of the total taxa) were consistently detected in Wolbachia-infected A. stephensi guts here. The relative abundance of Asaia in LBT or LB1 mosquitoes was similar regardless of Wolbachia infection, diet switching and rearing ages. This observation differs from the study conducted by Hughes et al. (2014) where Asaia concentration was much higher in blood fed mosquitoes than in non-blood fed ones. The initial assembly of the gut microbiota might account for this discrepancy: Asaia, Pseudomonas, and unclassified Gammaproteobacteria were the most predominant bacteria (Hughes et al., 2014). Asaia is a common commensal of several mosquito species such as Aedes, Anopheles, and Culex (Favia et al., 2008; Rossi et al., 2015). Its physiological roles in mosquitoes have been suggested to be nutrient scavenging and regulation of larval development (Chouaia et al., 2012). Recent studies showed antagonism between Asaia and Wolbachia in the reproduction organs (such as ovary; Rossi et al., 2015). Further, antagonistic interactions between Wolbachia and the gut microbiota, particularly Asaia, resulted in significant fitness costs in the mosquito host (Hughes et al., 2014). Our observation here showed that Asaia was stably maintained in both LB1 and LBT guts, which was consistent with Rossi et al. (Rossi et al., 2015).
The relative abundance of Elizabethkingia OTUs accounted for more than 60% of the total microbial taxa, highlighting their importance in Anopheles mosquitoes. Presumably, they contribute to metabolism of sugars in the gut environment, as evidenced by the SusC/SusD-like polysaccharide transport system(s) and glycosidases in their genome (Kukutla et al., 2014). Supplementation of Elizabethkingia cells in the sugar meal for A. stephensi led to approximately 50% more egg production compared to those without supplementation, suggesting that Elizabethkingia contributed to animal erythrocyte lysis and thus increased host’s fecundity in mosquitoes (S. Chen, unpublished). Elizabethkingia was frequently detected in water, sediments and insects including field caught, semi-natural reared, and insectary reared mosquitoes, showing that it adapted to diverse ecological niches in nature, but it is commonly found in Anopheles midguts (Chen et al., 2015). Pyrosequencing analysis showed that Elizabethkingia OTUs were more abundant in larval A. gambiae mosquitoes than in the aquatic medium in which they were being reared, but were present in the latter suggesting a common environmental source (Wang et al., 2011). Furthermore, Elizabethkingia anophelis and its nearly identical taxon, Elizabethkingia meningoseptica, were among the most predominant bacteria in adult A. gambiae and A. stephensi (Wang et al., 2011; Akhouayri et al., 2013; Ngwa et al., 2013).
Author Contributions
SC and EW wrote the manuscript. DJ and SC performed the experiments. SC, JZ, ZX, BN, and EW performed the data analysis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer HS and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
This project was funded by NIH grant R37 AI21884.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01485
References
- Akhouayri I. G., Habtewold T., Christophides G. K. (2013). Melanotic pathology and vertical transmission of the gut commensal Elizabethkingia meningoseptica in the major malaria vector Anopheles gambiae. PLoS ONE 8:e77619 10.1371/journal.pone.0077619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit J., Lopez-Martinez G., Patrick K., Phillips Z., Krause T., Denlinger D. (2011). Drinking a hot blood meal elicits a protective heat shock response in mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 108 8026–8029. 10.1073/pnas.1105195108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian G., Joshi D., Dong Y., Lu P., Zhou G., Pan X. (2013). Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science 340 748–751. 10.1126/science.1236192 [DOI] [PubMed] [Google Scholar]
- Boissière A., Tchioffo M. T., Bachar D., Abate L., Marie A., Nsango S. E., et al. (2012). Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 8:e1002742 10.1371/journal.ppat.1002742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongio N. J., Lampe D. J. (2015). Inhibition of Plasmodium berghei development in mosquitoes by effector proteins secreted from Asaia sp. Bacteria using a novel native secretion signal. PLoS ONE 10:e0143541 10.1371/journal.pone.0143541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis K., Dobson S. L., Xi Z., Rasgon J. L., Calvitti M., Moreira L. A., et al. (2014). Harnessing mosquito–Wolbachia symbiosis for vector and disease control. Acta Trop. 132(Suppl.), S150–S163. 10.1016/j.actatropica.2013.11.004 [DOI] [PubMed] [Google Scholar]
- Capone A., Ricci I., Damiani C., Mosca M., Rossi P., Scuppa P., et al. (2013). Interactions between Asaia, Plasmodium and Anopheles: new insights into mosquito symbiosis and implications in malaria symbiotic control. Parasit. Vectors 6 1–13. 10.1186/1756-3305-6-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Bagdasarian M., Walker E. D. (2015). Elizabethkingia anophelis: molecular manipulation and interactions with mosquito hosts. Appl. Environ. Microbiol. 81 2233–2243. 10.1128/AEM.03733-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouaia B., Rossi P., Epis S., Mosca M., Ricci I., Damiani C., et al. (2012). Delayed larval development in Anopheles mosquitoes deprived of Asaia bacterial symbionts. BMC Microbiol. 12(Suppl. 1):S2 10.1186/1471-2180-1112-S1181-S1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirimotich C. M., Dong Y., Clayton A. M., Sandiford S. L., Souza-Neto J. A., Mulenga M., et al. (2011). Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332 855–858. 10.1126/science.1201618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon K. L., Vogel K. J., Brown M. R., Strand M. R. (2014). Mosquitoes rely on their gut microbiota for development. Mol. Ecol. 23 2727–2739. 10.1111/mec.12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa C. C., Ballard J. W. O. (2014). What can symbiont titres tell us about co-evolution of Wolbachia and their host? J. Invertebr. Pathol. 118 20–27. 10.1016/j.jip.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Dobson S., Bourtzis K., Braig H., Jones B., Zhou W., Rousset F., et al. (1999). Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochem. Mol. Biol. 29 153–160. 10.1016/S0965-1748(98)00119-2 [DOI] [PubMed] [Google Scholar]
- Favia G., Ricci I., Marzorati M., Negri I., Alma A., Sacchi L. (2008). Bacteria of the genus Asaia: a potential weapon against malaria. Adv. Exp. Med. Biol. 627 49–59. 10.1007/978-0-387-78225-6_4 [DOI] [PubMed] [Google Scholar]
- Federici B. A., Park H.-W., Bideshi D. K., Wirth M. C., Johnson J. J. (2003). Recombinant bacteria for mosquito control. J. Exp. Biol. 206 3877–3885. 10.1242/jeb.00643 [DOI] [PubMed] [Google Scholar]
- Gimonneau G., Tchioffo M. T., Abate L., Boissière A., Awono-Ambéné P. H., Nsango S. E., et al. (2014). Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect. Genet. Evol. 28 715–724. 10.1016/j.meegid.2014.09.029 [DOI] [PubMed] [Google Scholar]
- Hilgenboecker K., Hammerstein P., Schlattmann P., Telschow A., Werren J. H. (2008). How many species are infected with Wolbachia?–a statistical analysis of current data. FEMS Microbiol. Lett. 281 215–220. 10.1111/j.1574-6968.2008.01110.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes G. L., Dodson B. L., Johnson R. M., Murdock C. C., Tsuijimoto H., Suzuki Y. (2014). Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 111 12498–12503. 10.1073/pnas.1408888111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I., Walker T., O’ Neill S. L. (2011). Wolbachia and the biological control of mosquito-borne disease. EMBO Rep. 12 508–518. 10.1038/embor.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi D., Mc Fadden M. J., Bevins D., Zhang F., Xi Z. (2014). Wolbachia strain wAlbB confers both fitness costs and benefit on Anopheles stephensi. Parasit. Vector 7:336 10.1186/1756-3305-7-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukutla P., Lindberg B. G., Pei D., Rayl M., Yu W., Steritz M., et al. (2014). Insights from the genome annotation of Elizabethkingia anophelis from the malaria vector Anopheles gambiae. PLoS ONE 9:e97715 10.1371/journal.pone.0097715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckhart S., Vodovotz Y., Cui L., Rosenberg R. (1998). The mosquito Anopheles stephensi limits malaria parasite development with inducible synthesis of nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 95 5700–5705. 10.1073/pnas.95.10.5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngwa C. J., Glöckner V., Abdelmohsen U. R., Scheuermayer M., Fischer R., Hentschel U., et al. (2013). 16S rRNA gene-based identification of Elizabethkingia meningoseptica (Flavobacteriales: Flavobacteriaceae) as a dominant midgut bacterium of the Asian malaria vector Anopheles stephensi (Diphteria: Culicidae) with antimicrobial activities. J. Med. Entomol. 50 404–414. 10.1603/ME12180 [DOI] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’Hara R. B. (2015). Vegan: Community Ecology. Available at: http://cran.r-project.org/package=vegan [Google Scholar]
- Pan X., Zhou G., Wu J., Bian G., Lu P., Raikhel A., et al. (2012). Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 109 E23–E31. 10.1073/pnas.1116932108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20 289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Peterson T. M. L., Luckhart S. (2006). A mosquito 2-Cys peroxiredoxin protects against nitrosative and oxidative stresses associated with malaria parasite infection. Free Radic. Biol. Med. 40 1067–1082. 10.1016/j.freeradbiomed.2005.10.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi P., Ricci I., Cappelli A., Damiani C., Ulissi U., Mancini M. V., et al. (2015). Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasit. Vector 8 1–10. 10.1186/s13071-015-0888-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkins S. P. (2004). Wolbachia and cytoplasmic incompatibility in mosquitoes. Insect Biochem. Mol. Biol. 34 723–729. 10.1016/j.ibmb.2004.03.025 [DOI] [PubMed] [Google Scholar]
- Tchioffo M. T., Boissière A., Abate L., Nsango S. E., Bayibéki A. N., Awono-Ambéné P. H., et al. (2016). Dynamics of bacterial community composition in the malaria mosquito’s epithelia. Front. Microbiol. 6:1500 10.3389/fmicb.2015.01500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Ghosh A. K., Bongio N., Stebbings K. A., Lampe D. J., Jacobs-Lorena M. (2012). Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 109 12734–12739. 10.1073/pnas.1204158109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Gilbreath T. M., Kukutla P., Yan G., Xu J. (2011). Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS ONE 6:e24767 10.1371/journal.pone.0024767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Baldo L., Clark M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6 741–751. 10.1038/nrmicro1969 [DOI] [PubMed] [Google Scholar]
- Xi Z., Khoo C. C., Dobson S. L. (2005). Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science 310 326–328. 10.1126/science.1117607 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.