Abstract
Clinical outcome and mutations of 96 core-binding factor acute myeloid leukemia (AML) patients 18–60 years old were examined. Complete remission (CR) after induction was 94.6%. There was no significant difference in CR, leukemia-free-survival (LFS) and overall survival (OS) between t(8;21) (N=67) and inv(16) patients (N=29). Univariate analysis showed hematopoietic stem cell transplantation at CR1 as the only clinical parameter associated with superior LFS. Next-generation sequencing based on a myeloid gene panel was performed in 72 patients. Mutations in genes involved in cell signaling were associated with inferior LFS and OS, whereas those in genes involved in DNA methylation were associated with inferior LFS. KIT activation loop (AL) mutations occurred in 25 patients, and were associated with inferior LFS (P=0.003) and OS (P=0.001). TET2 mutations occurred in 8 patients, and were associated with significantly shorter LFS (P=0.015) but not OS. Patients negative for KIT-AL and TET2 mutations (N=41) had significantly better LFS (P<0.001) and OS (P=0.012) than those positive for both or either mutation. Multivariate analysis showed that KIT-AL and TET2 mutations were associated with inferior LFS, whereas age ⩾40 years and marrow blast ⩾70% were associated with inferior OS. These observations provide new insights that may guide better treatment for this AML subtype.
Introduction
Acute myeloid leukemia (AML) is a group of heterogeneous diseases with distinct clinicopathologic, cytogenetic and genetic characteristics. Conventional therapeutic approaches include induction and consolidation chemotherapy. Patients at high risk of relapse receive allogeneic hematopoietic stem cell transplantation (HSCT) as post-remission therapy. AMLs with translocation involving core-binding factors (CBF) including t(8;21)(q22;q22) and inv(16)(p13;q22) or t(16;16)(p13;q22) constitute a distinct clinicopathologic subtype as defined by the World Health Organization (WHO), and occur in 15–20% of adult patients.1, 2 In general, CBF-AML has a superior outcome compared with other cytogenetic subtypes, with a long-term overall survival (OS) of 40–60%.3, 4, 5, 6, 7 However, their clinical outcome is highly heterogeneous. Mutations of type III receptor tyrosine kinase KIT in the activation loop at exon 17 (D816, N822) and the extracellular domain at exon 8, which are rare in other AMLs, occurred in 12–46% cases of t(8;21) and 9–53% of cases of inv(16).8, 9, 10, 11, 12, 13, 14, 15 These mutations may induce ligand-independent KIT activation and are generally associated with a higher risk of relapse and poorer prognosis. The impacts of other gene mutations frequently identified in AML, including RAS and FLT3, on outcome in these AML are less well defined. With the advent of next-generation sequencing (NGS), the genomic landscape of AML can be examined in detail. In particular, The Cancer Genome Atlas (TCGA) examined the genomic profile of 200 de novo AML and provided important genomic information of this disease.16 However, the number of CBF-AML in TCGA was limited.
In this study, we examined the clinical outcome of a consecutive cohort of patients with CBF-AML who were treated with a uniform approach and examined their mutation spectrum using NGS. The aim was to define the clinicopathologic characteristics and identify novel gene mutations of prognostic values for the design of better therapeutic strategies in this AML subtype.
Materials and methods
Patients
Consecutive patients aged 18–60 years, diagnosed with t(8;21)(q22;q22) or inv(16)(p13;q22) CBF-AML in six regional hospitals in Hong Kong between January 2003 and December 2015, were retrospectively analyzed. The diagnoses were confirmed by fluorescence in situ hybridization or reverse transcription-PCR for RUNX1/RUNX1T1 and CBFB/MYH11 fusions. All patient records were retrieved and independently reviewed by two investigators. Clinicopathologic features, including age, gender, presenting white cell counts, marrow blast percentage, additional cytogenetic abnormalities, induction and consolidation regimens, HSCT and the source of HSC, were analyzed. The study was approved by the institutional review boards of Hospital Authority (HKU/HA HKW UW14-430; KC/KE-15-0039/ER-3; KW/EX-15-052/85-05; NTWC/CREC/15013) and research ethics committee of Hong Kong Sanatorium & Hospital (REC-2015-02).
Treatments
Specific chemotherapy regimens are shown in Supplementary Table S1. Induction chemotherapy comprised daunorubicin and cytarabine. Reassessment bone marrow was performed between days 21 and 28 after induction. Consolidation comprised four courses of high-dose cytarabine. Before 2012, two courses of daunorubicin and etoposide were given as consolidation before high-dose cytarabine. Remission and relapse were defined by standard criteria. Salvage chemotherapy included ICE (idarubicin, cytarabine, etoposide), MAC (mitoxantrone, cytarabine), FLAG (fludarabine, cytarabine, granulocyte-colony stimulating factor) and CLARA (clofarabine, cytarabine). Patients who achieved second complete remission (CR2) received the same salvage chemotherapy as consolidation until HSCT or leukemic progression (Supplementary Table S2).
Allogeneic hematopoietic stem cell transplantation
Indications of HSCT at CR1 for eligible patients included two or more inductions to achieve CR1, additional chromosomal abnormalities and presence of KIT mutations. HSCT was recommended for all eligible patients in CR2. Allogeneic HSCT was performed in Queen Mary Hospital. Bu-Cy (busulfan; cyclophosphamide) was employed as myeloablative conditioning, and Flu-Bu (fludarabine, busulfan) and Cy-TBI (total body irradiation) for non-myeloablative conditioning. Patients received standard antimicrobial and graft versus host disease prophylaxis as described previously.17 Patients who relapsed after HSCT received one of the salvage regimens aforementioned followed by infusion of mobilized peripheral blood HSC from the original donors.18
Next-generation sequencing
Diagnostic bone marrow samples were collected. DNA was extracted using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) and analyzed by MiSeq NGS with the TruSight Myeloid sequencing panel (Illumina, San Diego, CA, USA). The panel targeted 54 genes covering full coding sequence of 15 genes and exonic hot spot for 39 genes (Supplementary Table S3). Workflows of MiSeq sequencing library preparation, variant calling and annotation as well as detection of FLT3 internal tandem duplication by ITDseek has been previously described.19 Complex insertions and deletions were detected by an in-house designed algorithm INDELseek on a Cray XC30 supercomputer (Cray Inc., Seattle, WA, USA).
Orthogonal validation of detected variants
Mutations with variant allele frequency (VAF) <20% were confirmed by one of the following methods according to the specific gene mutations involved. These included microfluidic PCR using Access Array 48.48 (Fluidigm, South San Francisco, CA, USA) and a different primer panel followed by MiSeq NGS, bi-directional Sanger sequencing or PCR fragment analysis by capillary electrophoresis using ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, CA, USA). Only confirmed variants were analyzed in this study.
Survival and statistical analyses
Leukemia-free survival (LFS) was defined as the time between first complete remission (CR1) to first relapse. Unless otherwise specified, LFS was censored at the date of the last follow-up or death. Overall survival (OS) was defined as the time between diagnosis and death or the date of the last follow up. Treatment-related mortality was defined as death within 30 days of the last chemotherapy or HSCT. Numerical data were compared using Mann–Whitney U-test for nonparametric and Student's t-test for parametric parameters. Categorical data were compared using χ2 test. Different thresholds for age (increment of 10 years, from 20 to 50 years), white cell counts (increments of 10 × 109/l, from 10 to 100 × 109/l) and marrow blasts percentage (increment of 10%, from 30 to 90%) were tested to identify the optimal cutoffs that best defined LFS and OS. Parameters that fulfilled the proportional hazard assumption with a P-value<0.1 in univariate analyses were further evaluated by multivariate analysis with the Cox proportional hazards model. Survival curves were constructed using the Kaplan–Meier method and compared by the log-rank test. All analyses were performed using SPSS (IBM Corp., Armonk, NY, USA). A P-value of 0.05 was considered statistically significant.
Results
Clinicopathologic characteristics and induction chemotherapy
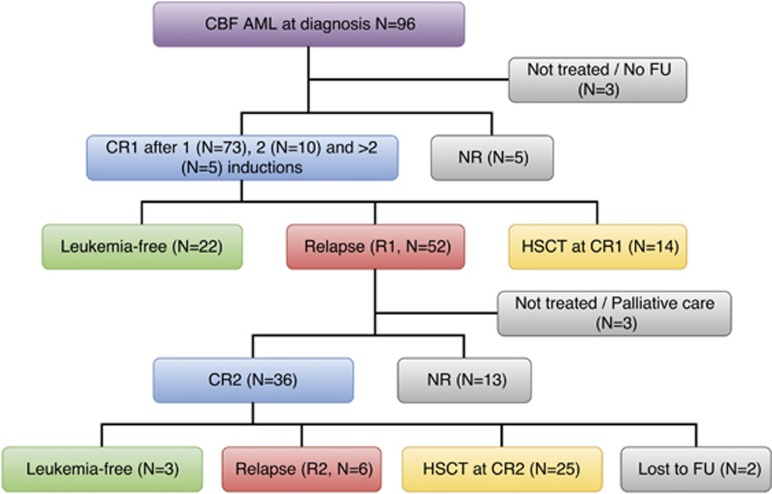
During the study period, 96 patients with CBF-AML were diagnosed. Of these patients, 91 received a standard 7-day regimen of cytarabine (100 mg/m2/day) and 3-day regimen of daunorubicin (50 mg/m2, N=72; 90 mg/m2, N=18; not specified, N=1). One patient received 5:2, and one patient received MAC. Three patients did not receive chemotherapy (Figure 1 and Supplementary Table S2). All 96 patients were included in the survival analysis.
Figure 1.
Treatment outcome of 96 CBF-AML patients in this study. Outcome of HSCT has been described in the text. FU, follow-up; NR, nonremission; R1, first relapse; R2, second relapse.
Treatment outcome
Out of 93 patients, 88 (94.6%) achieved CR1 after one (N=73), two (N=10), three (N=4) and four (N=1) courses of induction. Five patients died of refractory leukemia. There was no significant difference in CR rates between AML with t(8;21) and inv(16) (95.5% versus 92.6%, P=0.579), and they had similar LFS and OS (Supplementary Figure S1). Fourteen patients received HSCT at CR1, of whom 8 had remained in remission after a median follow-up of 83.8 (44.6–151.3) months. For 74 patients not receiving HSCT at CR1, 22 had remained in remission after a median follow-up of 27.1(4.4–121.6) months. A total of 52 patients had relapsed at a median of 8.9 (1–25.6) months from CR1. Of these patients, 49 received reinduction chemotherapy including MAC (N=22), FLAG (N=9), ICE (N=7), 7:3 (N=4), CLARA (N=2) and others (N=5). Thirty-six patients (73.5%) achieved CR2. Of these patients, 25 underwent allogeneic HSCT at CR2, of whom 13 (52%) had remained in remission after a median follow-up of 67.0 (26.5–137.5) months, 10 (40%) had relapsed at a median of 4.9 (1.6–37.8) months post HSCT and 2 had died of transplant-related mortality. For the other 11 CR2 patients not receiving HSCT, 6 had relapsed at a median of 6.6 (5.4–7.8) months from CR2; 3 had remained in remission 52.6 (2.5–107.4) months from CR2; and 2 had been lost to follow-up (Table 1).
Table 1. Clinicopathologic characteristics of 96 patients with CBF-AML.
| Features | Number (%) |
|---|---|
| Gender | |
| Male | 53 (55.2%) |
| Female | 43(44.8%) |
| Age (median, range) (years) | 41 (18–60) |
| Presenting WCC (median, range) (× 109/l) | 16.4 (1.6–396.6) |
| BM blast % (median, range) | 50 (20–100) |
| CBF-AML | |
| t(8;21) | 67 (69.8%) |
| inv(16) | 29 (30.2%) |
| Other cytogenetic abnormalities | |
| Sole | 31 (32.3%) |
| Additionala | 65 (67.7%) |
| Induction to achieve CR1b | |
| One | 73 (83.0%) |
| >One | 15 (17.0%) |
| First salvage chemotherapy | |
| ICE | 1 |
| FLAG | 1 |
| MAC | 7 |
| Second course of 7+3 | 6 |
| HSCT | 42 (43.8%) |
| Status at HSCT | |
| CR1 | 14 |
| CR2 | 25 |
| >CR2 | 2 |
| R1 | 1 |
| Source of HSC | |
| Sibling | 24 |
| Matched unrelated donor | 18 |
| No HSCT | 54 (56.3%) |
Abbreviations: AML, acute myeloid leukemia; BM, bone marrow; CBF, core-binding factor; CR1, first complete remission; CR2, second complete remission; FLAG, fludarabine, cytarabine, granulocyte-colony stimulating factor; HSCT, hematopoietic stem cell transplantation; ICE, idarubicin, cytarabine, etoposide; MAC, mitoxantrone, cytarabine; R1, first relapse; WCC, white cell count; 7:3, cytarabine, daunorubicin.
Additional chromosomal abnormalities included trisomy-X (n=10), trisomy-Y (n=24) and others (n=31).
A total of 88 patients achieved CR1.
Mutational analysis
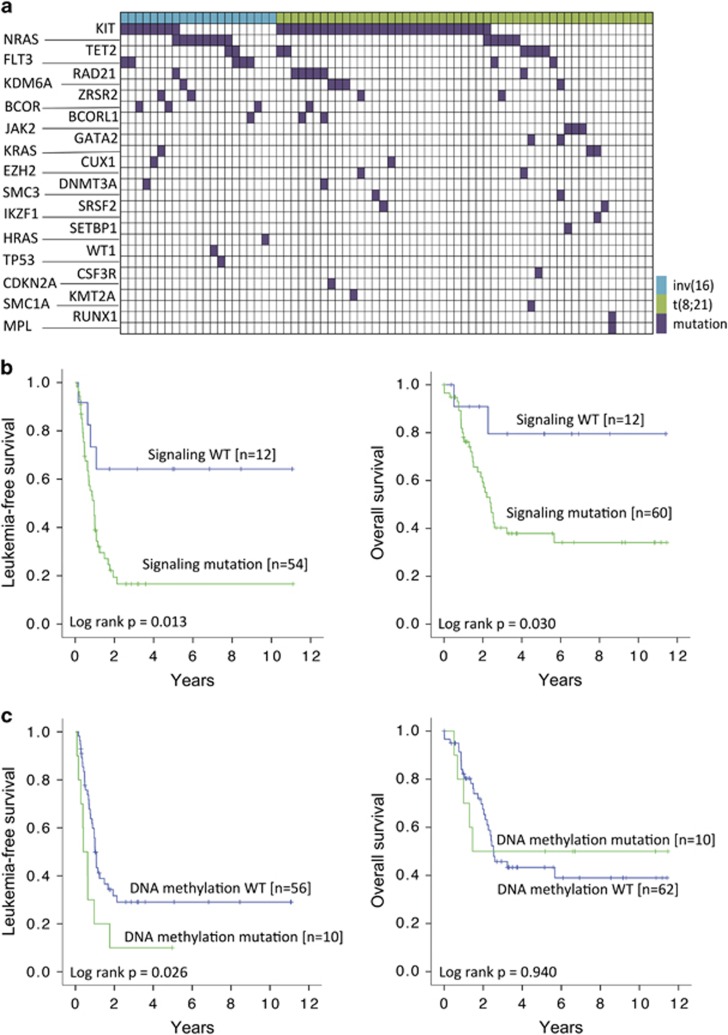
Genomic studies based on NGS of a myeloid gene panel were carried out in 72 patients, of whom 70 had received induction chemotherapy. When genes were categorized into functional groups, the most common mutations were those involved in cell signaling (N=60), chromatin modification (N=15), DNA methylation (N=10), cohesin complex (N=10), RNA splicing (N=6), tumor suppression (N=5) and transcription (N=5) (Supplementary Table S4). Six patients had no mutations and 36 patients showed two or more mutations. The most common recurrent mutations occurred in KIT (N=37, 51.4%), RAS (HRAS=1; KRAS=3; NRAS=13; overall: 23.6%) and TET2 (N=8, 11.1%). Other recurrent mutations included FLT3 (N=7, 9.7%) and RAD21 (N=7, 9.7%) (Figure 2a and Supplementary Figure S2A). The nature of these mutations is shown in Supplementary Table S5.
Figure 2.
Mutational spectrum and its impact on 72 CBF-AML patients. (a) Each column represented data from a single CBF-AML patient. Genetic mutation is colored in purple, t(8;21) in green and inv(16) in blue. (b) Survival impacts of mutations of genes involved in cell signaling. (c) Survival impacts of mutations of genes involved in DNA methylation.
Functional group analysis
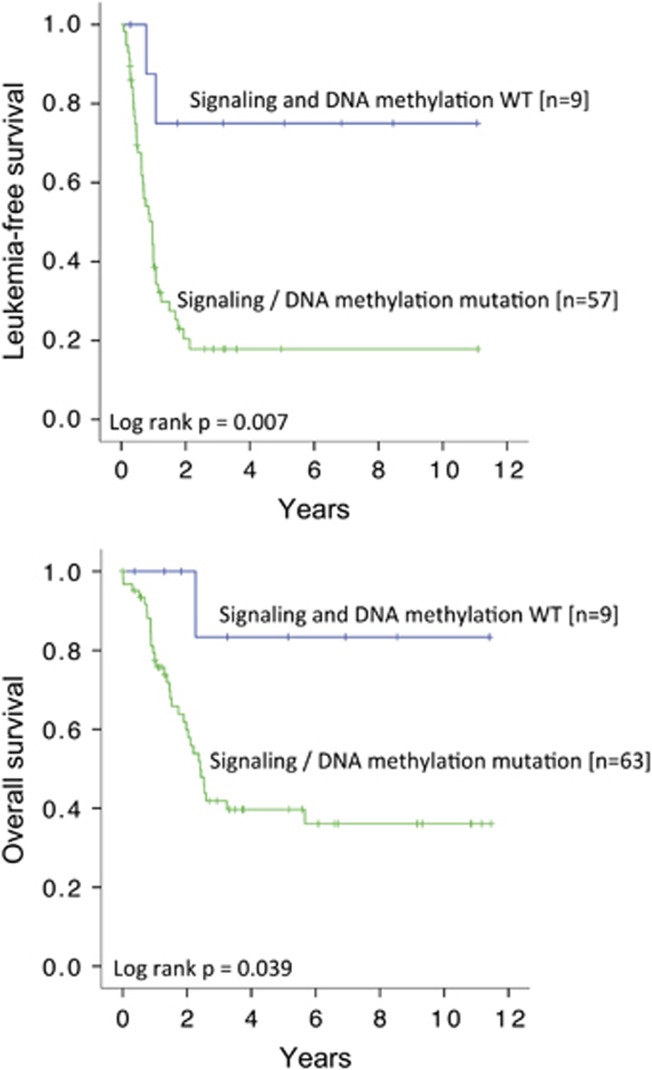
To overview the mutation spectrum and to provide mechanistic insights into the pathogenesis of CBF-AML, we evaluated the impact of gene mutations, categorized into specific functional groups, on LFS and OS. Gene mutations involved in molecular pathways of cell signaling were associated with inferior LFS and OS (Figure 2b). Gene mutations involved in DNA methylation were associated with inferior LFS but not OS (Figure 2c). Mutations in other functional groups have no significant impact on either LFS or OS. Importantly, patients negative for gene mutations in both signaling and DNA methylation, compared with those positive for either or both mutations, had significantly superior 10-year LFS (75.0% versus 17.9%, P=0.007) and 10-year OS (83.3% versus 36.1%, P=0.039) (Figure 3).
Figure 3.
Impacts of gene mutations in cell signaling and DNA methylation on LFS and OS.
KIT mutations
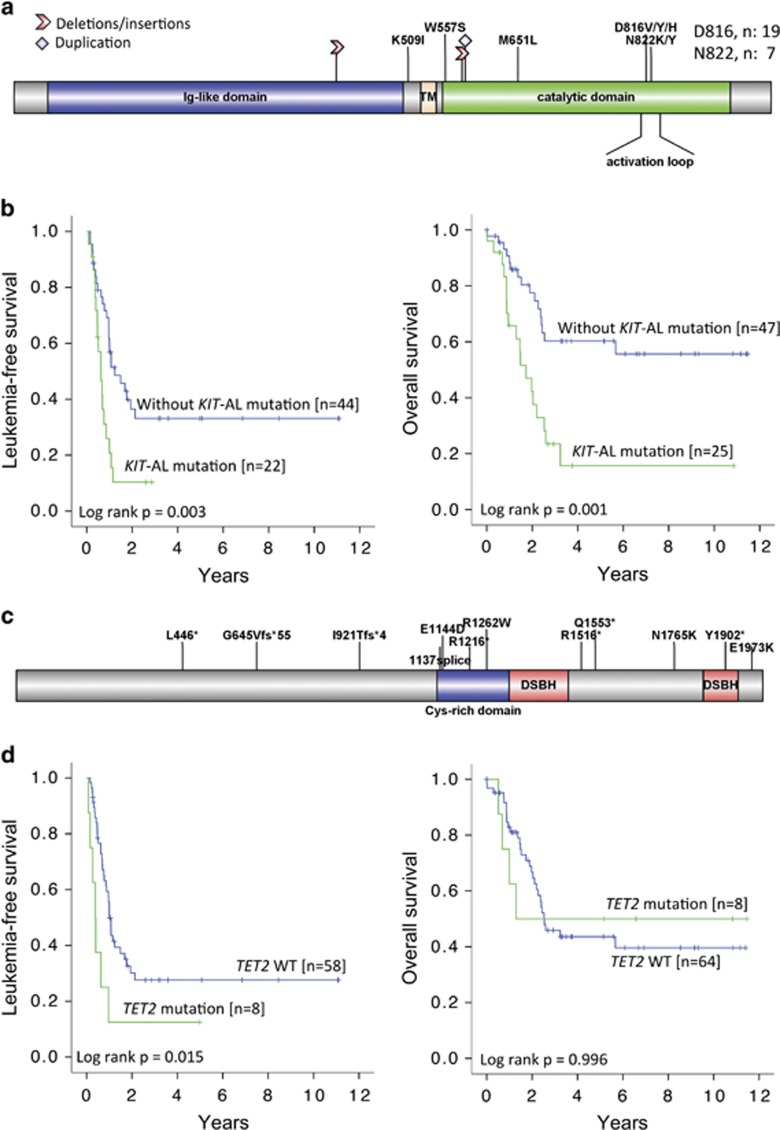
KIT mutations were identified in 29 patients with t(8;21) and 8 patients with inv(16). Mutation locations are shown in Figure 4a, being most prevalent in exon 17 (N=26, D816 and N822 occurring at the activation loop (AL) of the tyrosine kinase domain) and exon 8 (N=13), with others scattered in exons 9, 10, 11 and 12. Seven patients had two or more mutations. AL mutations, when compared with non-AL mutations, were associated with significantly higher median VAF (42% versus 12%, P=0.001), comparable median presenting white cell counts (11.7 × 109 versus 20 × 109/l, P=0.100) and CR rates (88% versus 97.8%, P=0.136), but inferior LFS (P= 0.022) and OS (P=0.003) (Supplementary Figure S2B). When patients with non-AL mutations were compared with patients without the mutations, LFS and OS were similar (Supplementary Figure S2C). Therefore, patients with AL mutations, when compared with patients having non-AL or no mutations, had significantly inferior LFS (P=0.003) and OS (P=0.001) (Figure 4b).
Figure 4.
KIT and TET2 mutations in CBF-AML. (a) Mutations in KIT were concentrated at the AL in exon 17 as shown. Other mutations were scattered in different exons. (b) Kaplan–Meier analyses showing that KIT-AL mutations were associated with inferior LFS and OS. (c) Mutations in TET2 were scattered throughout the coding sequence. (d) Kaplan–Meier analyses showing that TET2 mutations were associated with inferior LFS but not OS (right panel).
TET2 mutations
TET2 mutations, being distributed throughout the coding sequence, were found in eight patients (Figure 4c). All but one patients had VAF >40% (median: 43%, range: 16–53%), and three patients had two or more mutations. Patients with mutations, when compared with patients without mutations, showed a significantly shorter LFS but comparable OS (Figure 4d).
Other mutations
Mutations in RAS (median VAF: 33%, range: 10–77%), FLT3 (median VAF: 5%, range: 1–53%) and RAD21 (median VAF: 25%, range 16–46%) did not affect LFS or OS (Supplementary Figures S3A–C).
KIT-AL and TET2 double mutations
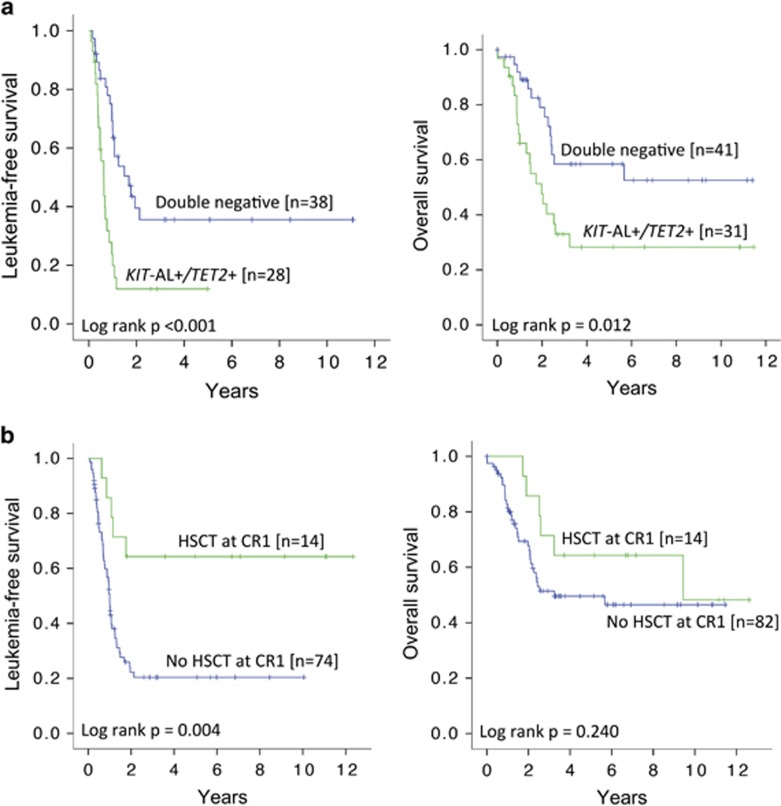
As KIT-AL and TET2 mutations were associated with an inferior outcome, the impact of combined KIT-AL and TET2 mutations was examined. Patients were divided into two groups. Group 1 comprised patients negative for KIT-AL mutations (including wild-type KIT and non-AL KIT mutations) and TET2 mutations (double negative, N=41). Group 2 comprised patients with either one or both of KIT-AL and TET2 mutations (N=31). Group 1 compared with group 2 patients had similar CR rates (97.4% versus 90.3%, P=0.20), but significantly superior 10-year LFS (35.6% versus 11.9%, P<0.001) and 10-year OS (52.6% versus 28.3%, P=0.012) (Figure 5a).
Figure 5.
Impacts of KIT-AL and TET2 mutations and HSCT at CR1. (a) Kaplan–Meier analyses showing that patients who were negative for both KIT-AL and TET2 mutations had superior LFS and OS compared with those who were positive for either or both of these mutations. (b) Kaplan–Meier analyses showing that patients receiving HSCT at CR1 were associated with superior LFS but not OS.
Impact of allogeneic HSCT
Patients undergoing HSCT in CR1, when compared with those without, had significantly better LFS (P=0.004) but similar OS (Figure 5b). The impact of HSCT in CR1 for each genetic subgroup was not evaluated because of the small patient number in each category. HSCT from HLA-identical sibling donors or matched unrelated donors showed comparable LFS and OS (Supplementary Figure S4).
Prognostic factors
Univariate analysis showed that HSCT at CR1 was significantly associated with better LFS. Mutations in cell signaling and DNA methylation as functional groups and KIT-AL and TET2 mutations as individual genes were significantly associated with inferior LFS (Table 2). Age ⩾40 years, presenting white blood cell count ⩾100 × 109/l and marrow blasts ⩾70% were significantly associated with inferior OS. Mutations in cell signaling and KIT-AL were also associated with an inferior OS. Multivariate analysis showed that KIT-AL and TET2 mutations were significant adverse factors for LFS, whereas age ⩾40 years and marrow blast ⩾70% were adverse factors for OS. An adverse indicator for OS in univariate analysis notwithstanding, KIT-AL mutation did not fulfill the proportional hazard assumption and was excluded from multivariate analysis.
Table 2. Univariate and multivariate analysis of leukemia-free survival (LFS) and overall survival (OS).
|
LFS |
OS |
|||||||
|---|---|---|---|---|---|---|---|---|
| P-value | Hazard ratio |
95% CI |
P-value | Hazard ratio |
95% CI |
|||
| Lower | Upper | Lower | Upper | |||||
| Univariate analysis | ||||||||
| Male gender | 0.977 | 0.99 | 0.59 | 1.67 | 0.448 | 1.27 | 0.68 | 2.38 |
| Age ⩾40 years | 0.807 | 1.07 | 0.63 | 1.80 | 0.013 | 2.31 | 1.20 | 4.48 |
| WCC ⩾100 × 109/l | 0.557 | 1.54 | 0.37 | 6.42 | 0.010 | 5.04 | 1.46 | 17.37 |
| BM blasts ⩾70% | 0.762 | 0.91 | 0.48 | 1.72 | 0.010 | 2.43 | 1.23 | 4.79 |
| >1 induction to CR1 | 0.584 | 0.82 | 0.40 | 1.68 | 0.299 | 1.50 | 0.70 | 3.21 |
| HSCT at CR1 | 0.006 | 0.28 | 0.11 | 0.70 | 0.246 | 0.60 | 0.25 | 1.43 |
| Mutations in signaling | 0.020 | 3.42 | 1.22 | 9.63 | 0.047 | 4.26 | 1.02 | 17.80 |
| Mutations in methylation | 0.030 | 2.26 | 1.08 | 4.72 | 0.940 | 0.96 | 0.37 | 2.50 |
| KIT-AL | 0.004 | 2.46 | 1.33 | 4.55 | 0.001 | 3.09 | 1.56 | 6.12 |
| TET2 | 0.019 | 2.64 | 1.17 | 5.97 | 0.996 | 1.00 | 0.35 | 2.85 |
| RAS | 0.785 | 0.91 | 0.45 | 1.84 | 0.804 | 0.90 | 0.41 | 2.00 |
| FLT3 | 0.664 | 0.80 | 0.28 | 2.23 | 0.966 | 0.98 | 0.30 | 3.19 |
| RAD21 | 0.284 | 0.57 | 0.20 | 1.60 | 0.406 | 0.61 | 0.18 | 1.98 |
| KIT-ALNeg and TET2Neg | 0.001 | 0.35 | 0.19 | 0.65 | 0.015 | 0.43 | 0.22 | 0.85 |
| Multivariate analysis | ||||||||
| Age ⩾40 years | – | – | – | – | 0.013 | 2.36 | 1.19 | 4.65 |
| WCC ⩾100 × 109/l | – | – | – | – | 0.109 | 2.90 | 0.79 | 10.65 |
| BM blasts ⩾70% | – | – | – | – | 0.036 | 2.16 | 1.05 | 4.44 |
| KIT-AL | 0.001 | 2.84 | 1.50 | 5.36 | – | – | – | – |
| TET2 | 0.004 | 3.43 | 1.48 | 7.96 | – | – | – | – |
Abbreviations: AL, activation loop; BM, bone marrow; CI, confidence interval; CR1, first complete remission; HSCT, hematopoietic stem cell transplantation; WCC, white cell count.
KIT-ALNeg and TET2Neg was not entered into multivariate analysis, as it would mutually exclude KIT-AL and TET2. Parameters showing statistical significance are highlighted in bold.
Discussion
Patients with CBF-AML are clinically heterogeneous despite similar cytogenetic aberrations, implying that secondary mutations might be pathogenetically important. In fact, mutations involving KIT, FLT3 and RAS were commonly reported in CBF-AML.8, 10, 11, 20 Knock-in mouse models of RUNX1/RUNX1T1 or CBFB/MYH11 induced a preleukemia hematopoietic state and required additional mutations for the development of AML,21, 22 supporting the proposition of a ‘second-hit' leukemogenic model. With NGS, a recent study recruiting both adult and pediatric patients from two clinical trials has identified mutations in cell signaling, cohesion complex and chromatin modification, being associated with higher risk of relapse.14
In this study, we examined the mutation spectrum in CBF-AML and the clinicopathologic features of an exclusively adult patient population who were treated with a uniform algorithm. Mutations of genes involved in cell signaling were associated with significantly inferior LFS and OS, whereas those in DNA methylation were associated with a significantly inferior LFS. Remarkably, patients without any mutations in genes involved in cell signaling and DNA methylation had long-term LFS and OS of ∼80%. The identity of genes in these categories was diverse, with KIT and TET2 being the most common. Other functional groups had no impact on clinical outcome. These results underscore the genetic heterogeneity in CBF-AML, and provide clinical evidence for the importance of concomitant mutations in these leukemias.
In this study, KIT mutations were identified in >50% of tested patients, with KIT-AL mutations in 35% of them. The prevalence was comparable with those reported based on targeted sequencing.9, 10 Importantly, only KIT-AL mutations but not other mutations were associated with inferior LFS and OS. Overexpression of KIT-AL mutant has been shown to cooperate with RUNX1-RUNX1T1 or CBFB-MYH11 to induce AML in mice.21, 22 However, KIT mutations are known to be unstable during disease evolution, with nearly 50% of cases losing or changing their mutations at relapse.10 The mechanistic link between KIT-AL mutations and inferior prognosis in CBF-AML would have to be further elucidated.
We also showed for the first time that TET2 mutations were an adverse prognostic factor in CBF-AML. Earlier studies examining TET2 mutations with bidirectional Sanger sequencing or 454-based NGS failed to detect any TET2 mutations in CBF-AML.23, 24 Of AML cases examined in TCGA, mutation in TET2 was shown in only 1 of 7 cases with t(8;21), and none of 11 cases with inv(16).16 A more recent study based on RNA-sequencing has shown TET2 mutations in 3 of 20 cases with t(8;21), but none in 28 cases with inv(16).15 In this study, we have shown a similar frequency (6/51) of TET2 mutations in t(8;21) and an apparently higher frequency (2/21) in inv(16). The scattering of mutations throughout the coding sequence and the lack of hot spot mutations supported the proposition that they were loss-of-function mutations. In all patients but one, TET2 mutations had VAF >40%, suggesting that the TET2-mutant clone was predominant and occurred in a heterozygous state. TET2 functions as a dioxygenase that converts 5 methylcytosine to 5 hydroxymethylcytosine, with a pivotal role in DNA demethylation.25 In mice, loss of TET2 has been shown to induce genome-wide enhancer methylation that in a CBF-AML model collaborates to induce an aggressive leukemia.26, 27 Further studies are needed to validate the very poor prognostic impact and define the pathogenetic/mechanistic basis of concomitant KIT-AL and TET2 mutations.
In our CBF-AML patients not receiving allogeneic HSCT in CR1, the poor long-term LFS of only 20–30% (Supplementary Figure S5) was comparable with the results from an earlier report on t(8;21) AML from our center28 and other reports from Asia,29, 30 but inferior to results reported from larger international studies.31, 32 It is unclear whether the inferior results of chemotherapy in our patients were because of a different gene mutation spectrum, the small size of this cohort or a difference in treatment regimens. However, the long-term OS was similar to that reported previously,5 supporting the notion that relapsed CBF-AML remained sensitive to salvage chemotherapy and patients could still be rescued by allogeneic HSCT.
Because relapsed patients are still salvageable, it remains uncertain whether frontline allogeneic HSCT is needed for CBF-AML. Accordingly, we showed an improved LFS but not OS in our patients receiving allogeneic HSCT in CR1, consistent with results from other studies employing frontline HSCT in CBF-AML, either because of institute policy or guided by minimal residual leukemia.29, 30, 33 With improvement in supportive care and advent of reduced-intensity, the indications and potential benefit of upfront HSCT for selected CBF-AML patients, particularly those with unfavorable gene mutation profiles, should be explored.
Our observations have important clinical implications. Patients without KIT-AL and TET2 mutations (‘double-negative' patients) have favorable outcome, suggesting this to be a distinct genetic subgroup that may be curable with chemotherapy. In fact, patients without any mutations involved in cell signaling (which included KIT-AL mutations) and DNA methylation (which included TET2 mutations) had an even more favorable LFS and OS. With the decreasing cost of NGS, a more extensive examination of genetic mutations at diagnosis may become a plausible strategy for prognostication. Furthermore, minimal residual disease monitoring has been shown to predict treatment outcome and provide guidance to HSCT indication in CBF-AML.33, 34, 35, 36, 37 Treatment algorithms incorporating mutational profiles and minimal residual disease monitoring should be tested to define whether better risk stratification may be achieved in this AML subtype.
Acknowledgments
This work was supported by Collaborative Research Fund (CityU9/CRF/13G), Blood Cancer Fund (200007147) and SK Yee Medical Foundation (2151209). AYHL received an endowment from the Li Shu Fan Medical Foundation.
Author contributions
CYC and GMKL designed research, analyzed data, performed statistical analysis and wrote the manuscript. CHA designed research, performed NGS and analyzed data. TLC and ESKM designed research and analyzed data. JPYS, HG, AKWL, RL, SFY, HKKL, HSYL, JSML, THL, CKL and SYL recruited and treated the patients. KFW, LLPS, CSPT, CCS and HWWW collected the samples and analyzed data. YLK and AYHL recruited and treated the patients, designed research, analyzed data and wrote the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Ustun C, Marcucci G. Emerging diagnostic and therapeutic approaches in core binding factor acute myeloid leukaemia. Curr Opin Hematol 2015; 22: 85–91. [DOI] [PubMed] [Google Scholar]
- Sinha C, Cunningham LC, Liu PP. Core binding factor acute myeloid leukemia: new prognostic categories and therapeutic opportunities. Semin Hematol 2015; 52: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenk RF, Benner A, Krauter J, Buchner T, Sauerland C, Ehninger G et al. Individual patient data-based meta-analysis of patients aged 16 to 60 years with core binding factor acute myeloid leukemia: a survey of the German Acute Myeloid Leukemia Intergroup. J Clin Oncol 2004; 22: 3741–3750. [DOI] [PubMed] [Google Scholar]
- Marcucci G, Mrozek K, Ruppert AS, Maharry K, Kolitz JE, Moore JO et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. J Clin Oncol 2005; 23: 5705–5717. [DOI] [PubMed] [Google Scholar]
- Appelbaum FR, Kopecky KJ, Tallman MS, Slovak ML, Gundacker HM, Kim HT et al. The clinical spectrum of adult acute myeloid leukaemia associated with core binding factor translocations. Br J Haematol 2006; 135: 165–173. [DOI] [PubMed] [Google Scholar]
- Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010; 116: 354–365. [DOI] [PubMed] [Google Scholar]
- Solh M, Yohe S, Weisdorf D, Ustun C. Core-binding factor acute myeloid leukemia: heterogeneity, monitoring, and therapy. Am J Hematol 2014; 89: 1121–1131. [DOI] [PubMed] [Google Scholar]
- Boissel N, Leroy H, Brethon B, Philippe N, de Botton S, Auvrignon A et al. Incidence and prognostic impact of c-Kit, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML). Leukemia 2006; 20: 965–970. [DOI] [PubMed] [Google Scholar]
- Cairoli R, Beghini A, Grillo G, Nadali G, Elice F, Ripamonti CB et al. Prognostic impact of c-KIT mutations in core binding factor leukemias: an Italian retrospective study. Blood 2006; 107: 3463–3468. [DOI] [PubMed] [Google Scholar]
- Allen C, Hills RK, Lamb K, Evans C, Tinsley S, Sellar R et al. The importance of relative mutant level for evaluating impact on outcome of KIT, FLT3 and CBL mutations in core-binding factor acute myeloid leukemia. Leukemia 2013; 27: 1891–1901. [DOI] [PubMed] [Google Scholar]
- Paschka P, Du J, Schlenk RF, Gaidzik VI, Bullinger L, Corbacioglu A et al. Secondary genetic lesions in acute myeloid leukemia with inv(16) or t(16;16): a study of the German-Austrian AML Study Group (AMLSG). Blood 2013; 121: 170–177. [DOI] [PubMed] [Google Scholar]
- Qin YZ, Zhu HH, Jiang Q, Jiang H, Zhang LP, Xu LP et al. Prevalence and prognostic significance of c-KIT mutations in core binding factor acute myeloid leukemia: a comprehensive large-scale study from a single Chinese center. Leuk Res 2014; 38: 1435–1440. [DOI] [PubMed] [Google Scholar]
- Chen W, Xie H, Wang H, Chen L, Sun Y, Chen Z et al. Prognostic significance of kit mutations in core-binding factor acute myeloid leukemia: a systematic review and meta-analysis. PLoS One 2016; 11: e0146614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duployez N, Marceau-Renaut A, Boissel N, Petit A, Bucci M, Geffroy S et al. Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood 2016; 127: 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavallee VP, Lemieux S, Boucher G, Gendron P, Boivin I, Armstrong RN et al. RNA-sequencing analysis of core binding factor AML identifies recurrent ZBTB7A mutations and defines RUNX1-CBFA2T3 fusion signature. Blood 2016; 127: 2498–2501. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AY, Suen CK, Lie AK, Liang RH, Yuen KY, Kwong YL. Quantification of polyoma BK viruria in hemorrhagic cystitis complicating bone marrow transplantation. Blood 2001; 98: 1971–1978. [DOI] [PubMed] [Google Scholar]
- Leung AY, Tse E, Hwang YY, Chan TS, Gill H, Chim CS et al. Primary treatment of leukemia relapses after allogeneic hematopoietic stem cell transplantation with reduced-intensity conditioning second transplantation from the original donor. Am J Hematol 2013; 88: 485–491. [DOI] [PubMed] [Google Scholar]
- Au CH, Wa A, Ho DN, Chan TL, Ma ES. Clinical evaluation of panel testing by next-generation sequencing (NGS) for gene mutations in myeloid neoplasms. Diagn Pathol 2016; 11: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opatz S, Polzer H, Herold T, Konstandin NP, Ksienzyk B, Zellmeier E et al. Exome sequencing identifies recurring FLT3 N676K mutations in core-binding factor leukemia. Blood 2013; 122: 1761–1769. [DOI] [PubMed] [Google Scholar]
- Wang YY, Zhao LJ, Wu CF, Liu P, Shi L, Liang Y et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci USA 2011; 108: 2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Melenhorst JJ, Alemu L, Kirby M, Anderson S, Kench M et al. KIT with D816 mutations cooperates with CBFB-MYH11 for leukemogenesis in mice. Blood 2012; 119: 1511–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann S, Alpermann T, Grossmann V, Kowarsch A, Nadarajah N, Eder C et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia 2012; 26: 934–942. [DOI] [PubMed] [Google Scholar]
- Aslanyan MG, Kroeze LI, Langemeijer SM, Koorenhof-Scheele TN, Massop M, van Hoogen P et al. Clinical and biological impact of TET2 mutations and expression in younger adult AML patients treated within the EORTC/GIMEMA AML-12 clinical trial. Ann Hematol 2014; 93: 1401–1412. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011; 333: 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev 2015; 29: 910–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatlen MA, Arora K, Vacic V, Grabowska EA, Liao W, Riley-Gillis B et al. Integrative genetic analysis of mouse and human AML identifies cooperating disease alleles. J Exp Med 2016; 213: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SK, Au WY, Kwong YL, Lam CK, Liang RH, Chan LC. Hematological features and treatment outcome in acute myeloid leukemia with t(8;21). Hematol Oncol 1997; 15: 93–103. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Kim HJ, Kim JW, Jeon YW, Shin SH, Lee SE et al. Identification of molecular and cytogenetic risk factors for unfavorable core-binding factor-positive adult AML with post-remission treatment outcome analysis including transplantation. Bone Marrow Transplant 2014; 49: 1466–1474. [DOI] [PubMed] [Google Scholar]
- Qin YZ, Xu LP, Chen H, Jiang Q, Wang Y, Jiang H et al. Allogeneic stem cell transplant may improve the outcome of adult patients with inv(16) acute myeloid leukemia in first complete remission with poor molecular responses to chemotherapy. Leuk Lymphoma 2015; 56: 3116–3123. [DOI] [PubMed] [Google Scholar]
- Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood 2002; 100: 4325–4336. [DOI] [PubMed] [Google Scholar]
- Nguyen S, Leblanc T, Fenaux P, Witz F, Blaise D, Pigneux A et al. A white blood cell index as the main prognostic factor in t(8;21) acute myeloid leukemia (AML): a survey of 161 cases from the French AML Intergroup. Blood 2002; 99: 3517–3523. [DOI] [PubMed] [Google Scholar]
- Zhu HH, Zhang XH, Qin YZ, Liu DH, Jiang H, Chen H et al. MRD-directed risk stratification treatment may improve outcomes of t(8;21) AML in the first complete remission: results from the AML05 multicenter trial. Blood 2013; 121: 4056–4062. [DOI] [PubMed] [Google Scholar]
- Yin JA, O'Brien MA, Hills RK, Daly SB, Wheatley K, Burnett AK. Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: results of the United Kingdom MRC AML-15 trial. Blood 2012; 120: 2826–2835. [DOI] [PubMed] [Google Scholar]
- Jourdan E, Boissel N, Chevret S, Delabesse E, Renneville A, Cornillet P et al. Prospective evaluation of gene mutations and minimal residual disease in patients with core binding factor acute myeloid leukemia. Blood 2013; 121: 2213–2223. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu DP, Liu QF, Qin YZ, Wang JB, Xu LP et al. In adults with t(8;21)AML, posttransplant RUNX1/RUNX1T1-based MRD monitoring, rather than c-KIT mutations, allows further risk stratification. Blood 2014; 124: 1880–1886. [DOI] [PubMed] [Google Scholar]
- Willekens C, Blanchet O, Renneville A, Cornillet-Lefebvre P, Pautas C, Guieze R et al. Prospective long-term minimal residual disease monitoring using RQ-PCR in RUNX1-RUNX1T1-positive acute myeloid leukemia: results of the French CBF-2006 trial. Haematologica 2016; 101: 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.