Abstract
MicroRNAs (miRs) play crucial roles in the carcinogenesis and malignant progression of human cancers including nasopharyngeal carcinoma (NPC). In this study, we aimed to investigate the association of serum miR-663 levels with the clinical factors and prognosis of NPC patients. Real-time PCR was performed to examine the amount of miR-663 in serum in NPC patients and healthy controls. Our data showed that the amount of miR-663 in serum was significantly higher in NPC patients than in healthy controls. Moreover, the serum levels of miR-663 were significantly correlated with the grade, lymph node metastasis, and clinical stage of NPC. Furthermore, higher serum miR-663 levels were closely associated with worse 5-year overall survival (OS) and relapse-free survival (RFS) of patients with NPC, and the serum level of miR-663 was found to be an independent predicator for the prognosis of NPC. In addition, after receiving chemoradiotherapy, the serum levels of miR-663 were significantly reduced in NPC patients. In summary, miR-663 was upregulated in the serum of NPC patients, which was downregulated after chemoradiotherapy, and its increased levels were closely associated with malignant progression and poor prognosis in NPC patients. Therefore, the amount of miR-663 in serum may become a potential predicator for the clinical outcome of NPC patients.
1. Introduction
Nasopharyngeal carcinoma (NPC), a head and neck cancer, is most common in Southeast Asia especially in Southern China, and the incidence rate of NPC is approximately 20 cases per 100,000 people in endemic areas [1, 2]. The pathogenesis of NPC involves Epstein-Barr virus (EBV) infection and genetic susceptibility, and chemoradiotherapy is the standard approach for the treatment of NPC [3, 4]. Moreover, the detection of NPC mainly relies on tissue biopsy and cancer imaging, but its early diagnosis is difficult due to its anatomical location [5, 6]. Therefore, it is urgently needed to explore effective biomarkers for early detection of NPC, as well as predication of the prognosis of NPC patients.
MicroRNAs (miRs) are a kind of small noncoding RNAs, containing 22–25 nucleotides [7]. They generally play suppressive role in regulating the gene expression through directly binding to the 3′ untranslational region (UTR) of their target mRNAs, causing translation inhibition or mRNA degradation [7]. In recent years, miRs have been implicated in various cellular biological processes, including cell proliferation, apoptosis, differentiation, metabolism, motility, and tumorigenesis [7–9]. They are involved in the regulation of the expression of many oncogenes or tumors suppressors and thus play promoting or suppressive roles in human cancers [10, 11]. In addition, deregulations of miRs have been found in NPC [12, 13]. Wang et al. reported that miR-429 was downregulated in NPC and played a suppressive role in NPC cell migration and invasion [14]. Mao et al. found that miR-205 could promote the proliferation, migration, and invasion of NPC cells through activation of AKT signaling [15]. MiR-24 was demonstrated to enhance the radiosensitivity of NPC cells by targeting SP1 [16].
MiR-663 has previously been reported to be frequently deregulated in several types of human cancers, indicating that miR-663 may play an important role in carcinogenesis. The expression levels of miR-663 were significantly decreased in gastric cancer [17]. However, it was found to be markedly upregulated in castration-resistant prostate cancer [18]. Recently, it was reported that miR-663 was upregulated in NPC tissues and cell lines, and knockdown of miR-663 suppressed the growth of NPC cells in vitro and in vivo [19]. However, evidence in the serum levels of miR-663 in NPC as well as its clinical significance still remains limited.
In this study, we aimed to investigate the clinical significance of serum miR-663 levels in NPC, as well as its predictive value for prognosis of NPC patients.
2. Materials and Methods
2.1. Ethics Approval
The present study was approved by the Ethics Committee of First People's Hospital of Foshan, Foshan, China. All the participants involved in this study have written the written consent.
2.2. Study Population
A total of 74 cases of serum samples from NPC patients and 27 healthy controls were recruited at the Department of Radiotherapy, Tumor Hospital of First People's Hospital of Foshan. The ages of the NPC patients including 40 male and 34 female were ranged from 28 years to 71 years. The clinical characteristics of NPC patients were summarized in Table 1. Before sample collection, all the participants did not receive any therapy. The serum was isolated from 10 mL of blood by centrifuging at 1,000 ×g at room temperature for 5 min and then at 12,000 ×g at 4°C for 5 min. The samples were stored at −80°C before usage.
Table 1.
Association between serum miR-663 levels and clinicopathological characteristics of patients with nasopharyngeal carcinoma.
| Factors | Number | Low miR-663 (n = 41) | High miR-663 (n = 33) | P value |
|---|---|---|---|---|
| Age | ||||
| <55 | 36 | 20 | 16 | 0.980 |
| ≥55 | 38 | 21 | 17 | |
| Gender | ||||
| Male | 40 | 22 | 18 | 0.939 |
| Female | 34 | 19 | 15 | |
| Grade | ||||
| G1-2 | 41 | 28 | 13 | 0.013 |
| G3 | 33 | 13 | 20 | |
| T stage | ||||
| T1-2 | 39 | 25 | 14 | 0.112 |
| T3-4 | 35 | 16 | 19 | |
| Lymph node metastasis | ||||
| No | 41 | 30 | 11 | 0.001 |
| Yes | 33 | 11 | 22 | |
| Distant metastasis | ||||
| No | 53 | 32 | 21 | 0.172 |
| Yes | 21 | 9 | 12 | |
| Clinical stage | ||||
| I-II | 39 | 29 | 10 | 0.001 |
| III-IV | 35 | 12 | 23 | |
| EBV infection | ||||
| No | 8 | 6 | 2 | 0.239 |
| Yes | 66 | 35 | 31 |
2.3. Real-Time Polymerase Chain Reaction
QIAamp RNA Blood kit (Qiagen, Hilden, Germany) was used to extract the total RNA, according to the manufacturer's instruction. miRNA Reverse Transcription Kit (Life Technologies) was used to convert 1 μg of RNA into cDNA, according to the manufacturer's instruction. Real-time PCR was then performed by using a miRNA Q-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA) on ABI 7500 thermocycler (Applied Biosystems, Carlsbad, CA, USA). The PCR conditions were 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s. Each sample was examined in triplicate and U6 gene was used as an internal reference. The relative expression of miR-663 was analyzed by the 2−ΔΔCt method that normalized to U6 expression.
2.4. Statistical Analysis
Data were expressed as means ± standard deviation (SD). SPSS 20.0 software (SPSS Inc., IL, USA) was used to perform the statistical analysis. Nonparametric t-test was used to analyze the differential expression of serum miR-663 between NPC patients and healthy controls. Chi-square test was used to analyze the association between the serum level of miR-663 and clinicopathological parameters of NPC. Kaplan-Meier analysis with the log-rank test was used to examine the association between the serum level of miR-663 and the 5-year overall survival (OS) and relapse-free survival (RFS). Cox proportional hazard regression model was used to estimate the independent predicators for the prognosis of NPC patients. Paired sample t-test was used to analyze the differential amount of miR-663 in serum between NPC patients before treatment and those after treatment. P value less than 0.05 was considered statistically significant.
3. Results
3.1. The Serum miR-663 Levels Are Higher in NPC Patients Than in Healthy Control
In our study, we firstly examined the levels of miR-663 in serum from NPC and healthy controls using real-time PCR. Our data indicated that the serum miR-663 levels were significantly higher (about 3.5-fold) in NPC patients than in healthy control (P < 0.001, Figure 1).
Figure 1.
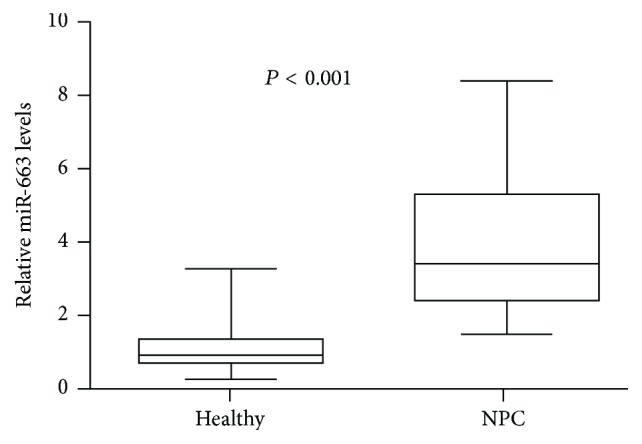
Real-time PCR was used to detect the serum levels of miR-663 in 74 nasopharyngeal carcinoma (NPC) patients and 27 healthy controls. Box-whisker plot (bottom to top) means the minimum, lower quartile, median, upper quartile, and maximum value.
3.2. The Serum miR-663 Levels Are Significantly Associated with Grade, Clinical Stage, and Lymph Node Metastasis in NPC
We further studied the association between the serum miR-663 levels and clinical characteristics in NPC patients. All NPC patients were divided into two groups, high serum levels of miR-663 group and low serum levels of miR-663 group, according to the mean expression of serum miR-663 as a cut-off point. As demonstrated in Table 1, the serum miR-663 levels were not correlated with age (P = 0.980), gender (P = 0.939), T stage (P = 0.112), distant metastasis (P = 0.172), and EBV infection (P = 0.239). However, it was significantly associated with the grade (P = 0.013), clinical stage (P = 0.001), and lymph node metastasis (0.001). Thus, our data suggest that the serum levels of miR-663 may be used as a biomarker for evaluating the malignant progression of NPC.
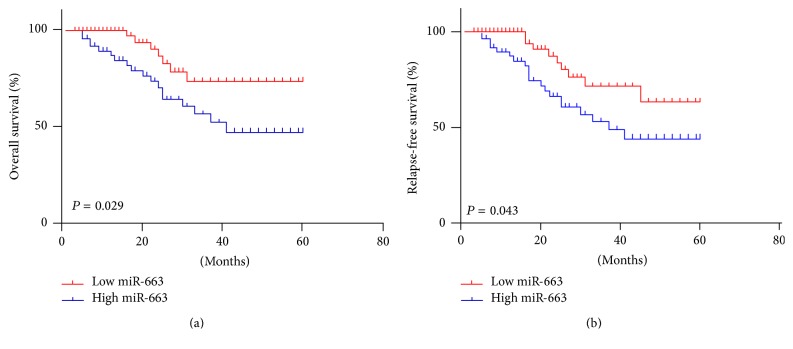
3.3. Higher Serum Level of miR-663 Is Associated with Poor 5-Year Survival Rates in NPC Patients
We further investigated the association between the serum level of miR-663 and the survival rate in NPC patients using the Kaplan-Meier method. We found that the NPC patients with higher serum level of miR-663 showed shorter 5-year overall survival (Figure 2(a), OS, P = 0.029) and relapse-free survival (Figure 2(b), RFS, P = 0.043), when compared to those with lower serum level of miR-663. Therefore, higher serum level of miR-663 is associated with poor prognosis of NPC patients.
Figure 2.
The relationship between serum level of miR-663 and 5-year overall and relapse-free survival rates of nasopharyngeal carcinoma patients.
3.4. The Serum Level of miR-663 Is an Independent Predicator for the Prognosis of NPC Patients
We further investigated the factors that could predicate the prognosis of NPC patients by using the univariate and multivariate analyses. Univariate analysis data indicated that the serum level of miR-663 (P = 0.014), as well as the grade (P = 0.021), lymph node metastasis (P = 0.009), and clinical stage (P = 0.001), was significantly associated with the survival (Table 2). Moreover, as demonstrated in Table 3, the serum level of miR-663 (P = 0.025), lymph node metastasis (P = 0.035), and clinical stage (P = 0.001) were found to be independent factors for predicating the prognosis of NPC patients.
Table 2.
Univariate analysis of prognostic factors of NPC.
| Factors | Hazard ratio | P value |
|---|---|---|
| Age (≥55/<55) | 1.12 | 0.867 |
| Gender (male/female) | 1.1 | 0.882 |
| Grade (G3/G1-2) | 2.73 | 0.021 |
| T stage (T3-4/T1-2) | 1.63 | 0.115 |
| Lymph node metastasis (yes/no) | 3.45 | 0.009 |
| Distant metastasis (yes/no) | 1.61 | 0.124 |
| Clinical stage (III-IV/I-II) | 5.42 | 0.001 |
| EBV infection (yes/no) | 1.48 | 0.343 |
| Serum miR-663 levels (high/low) | 3.88 | 0.014 |
Table 3.
Multivariate analysis of independent prognostic factors of NPC.
| Factors | Hazard ratio | P value |
|---|---|---|
| Grade | 1.73 | 0.077 |
| Lymph node metastasis | 2.43 | 0.035 |
| Clinical stage | 4.32 | 0.001 |
| Serum miR-663 levels | 2.68 | 0.025 |
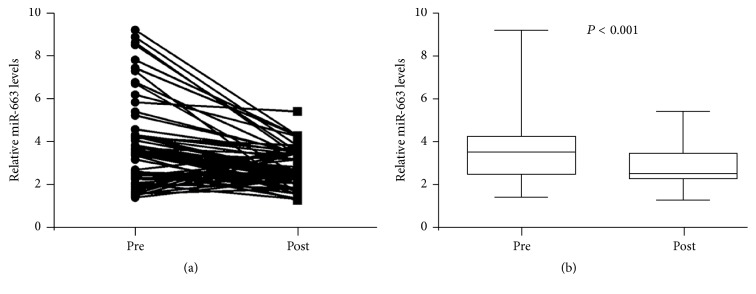
3.5. The Association between Serum Level of miR-663 and Treatment Response
Finally, we compared the serum levels of miR-663 between the NPC patients before treatment and after treatment. As indicated in Figures 3(a) and 3(b), we found that the serum levels of miR-663 were remarkably decreased in NPC patients after receiving the standard chemoradiotherapy (P < 0.001). These data suggest that the serum levels of miR-663 may be used as an important biomarker for monitoring the treatment response.
Figure 3.
(a and b) Real-time PCR was used to detect the serum level of miR-663 in 74 nasopharyngeal carcinoma patients before and after receiving chemoradiotherapy. Box-whisker plot (bottom to top) means the minimum, lower quartile, median, upper quartile, and maximum value. Pre, pretreatment; post, posttreatment.
4. Discussion
The prognosis of patients with stages III and IV NPC was worse when compared with that of patients with stages I and II NPC [20]. Therefore, it is urgently needed to identify novel biomarkers for the early detection of NPC. Recently, the novel biomarkers in the body fluids, such as plasma, serum, saliva, and urine, show promises for the early detection of malignancies including NPC. miRs have been demonstrated to play crucial roles in the development and malignant progression of human cancers. Moreover, accumulating evidences have shown that some serum miRs may become important biomarkers not only for the early detection of human cancers, but also for the evaluation of cancer progression or treatment responses. For instance, the serum expression of miR-199a was significantly decreased in patients with epithelial ovarian cancer, which was significantly associated with the tumor stage, lymph node metastasis, and distal metastasis, suggesting that the downregulation of miR-199a may be a potential indicator for disease progression [21]. Besides, the circulating miR-92a, miR-100, and miR-143 were recently identified as noninvasive biomarkers for bladder cancer diagnosis [22].
MiR-663 has recently been found to generally play a suppressive role in several kinds of human cancers. The expression of miR-663 was significantly lower in pediatric acute myeloid leukemia (AML) cells compared to normal bone marrow due to the hypermethylated promoter, suggesting that downregulation of miR-663 may be involved in the development of AML [23]. Li et al. reported that miR-663 was downregulated in glioblastoma tissues and had inhibitory effects on the proliferation, migration, and invasion of glioblastoma cells via targeting TGF-β1 [24]. Similar findings were also reported that miR-663 acted as a tumor suppressor in glioblastoma by targeting CXCR4 and PIK3CD [25, 26]. Zang et al. indicated that miR-663 could inhibit the tumor growth and invasiveness in pancreatic cancer by directly targeting EEF1A2 [27]. Moreover, treatment with waltonitone in non-small cell lung cancer led to an upregulation of miR-663, which further caused downregulation of bcl-2 and induces cancer cell apoptosis [28]. Overexpression of miR-663 significantly decreased the proliferation, migration, and colony formation of multiple myeloma cells [29]. Besides, other cancer-related factors were also found to be direct targets of miR-663, such as EEF1A2 and HSPG2 [30, 31].
On the contrary, however, several studies also reported that miR-663 played a promoting role in several cancer types [32]. Sand et al. reported that miR-663 was significantly upregulated in cutaneous malignant melanoma compared to benign melanocytic nevi [32]. Liu et al. showed that miR-663 was highly expressed in lung cancer patients and promoted the proliferation of lung cancer cells by directly or indirectly regulating TGFB1, P53, Bax, and Fas [33]. Therefore, it seems that the role of miR-663 is cancer-specific, and miR-663 plays dual roles in different cancer types, probably because it regulates different target genes in different tumor microenvironments. In the present study, we found that the serum miR-663 levels were significantly higher in NPC in patients than in healthy control. Moreover, its expression levels were significantly associated with several important clinicopathological features including grade, lymph node metastasis, and clinical stage, suggesting that the upregulation of serum level of miR-663 may be used as an important biomarker for the malignant progression of NPC. Besides, we further found that NPC patients with higher serum miR-663 levels showed poorer 5-year OS and RFS when compared with those with lower serum miR-663 levels and that serum miR-663 was an independent predicator for the prognosis of NPC. These findings highlight the importance of serum miR-663 in the clinical application in evaluating the prognosis of NPC patients. Consistent with our study, the expression of miR-663 was also found to be increased in NPC tissues and cell lines. Molecular mechanism investigation revealed that miR-663 could promote the proliferation of NPC cells in vitro and in vivo by directly targeted p21(WAF1/CIP1) to promote the cellular G1/S transition [19].
Finally, we found that the serum levels of miR-663 were remarkably decreased in NPC patients after receiving the standard chemoradiotherapy. Recently, miR-774 was also reported to be upregulated in the serum of NPC patients, and its expression level was significantly reduced after the patients had received the standard chemoradiotherapy [34]. Therefore, the serum levels of both miR-663 and miR-774 may be used as important biomarkers for monitoring the treatment response.
5. Conclusion
Our study for the first time demonstrates that the serum expression miR-663 is upregulated in NPC, and high serum miR-663 levels are significantly associated with the malignant progression and poor prognosis in NPC patients. Therefore, the serum level of miR-663 may become a potential predicator for the clinical outcome of NPC patients.
Acknowledgments
This study was supported by Special Fund Project for Technology Innovation of Foshan City (2014AG10003).
Competing Interests
The authors have declared that no competing interests exist.
References
- 1.Chen W. Cancer statistics: updated cancer burden in China. Chinese Journal of Cancer Research. 2015;27, article 1 doi: 10.3978/j.issn.1000-9604.2015.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2016. CA: A Cancer Journal for Clinician. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Yip T. T. C., Ngan R. K. C., Fong A. H. W., Law S. C. K. Application of circulating plasma/serum EBV DNA in the clinical management of nasopharyngeal carcinoma. Oral Oncology. 2014;50(6):527–538. doi: 10.1016/j.oraloncology.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Sze H., Blanchard P., Ng W. T., Pignon J.-P., Lee A. W. M. Chemotherapy for nasopharyngeal carcinoma—current recommendation and controversies. Hematology/Oncology Clinics of North America. 2015;29(6):1107–1122. doi: 10.1016/j.hoc.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Xiao L., Xiao T., Wang Z.-M., Cho W. C. S., Xiao Z.-Q. Biomarker discovery of nasopharyngeal carcinoma by proteomics. Expert Review of Proteomics. 2014;11(2):215–225. doi: 10.1586/14789450.2014.897613. [DOI] [PubMed] [Google Scholar]
- 6.Lee A. W. M., Lin J. C., Ng W. T. Current management of nasopharyngeal cancer. Seminars in Radiation Oncology. 2012;22(3):233–244. doi: 10.1016/j.semradonc.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Fabbri M., Ivan M., Cimmino A., Negrini M., Calin G. A. Regulatory mechanisms of microRNAs involvement in cancer: the strange case of Dr Jekyll and Mr Hyde. Expert Opinion on Biological Therapy. 2007;7(7):1009–1019. doi: 10.1517/14712598.7.7.1009. [DOI] [PubMed] [Google Scholar]
- 9.So A. Y.-L., Zhao J. L., Baltimore D. The Yin and Yang of microRNAs: leukemia and immunity. Immunological Reviews. 2013;253(1):129–145. doi: 10.1111/imr.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng J.-F., Zhuang Y.-Y., Huang F.-T., Zhang S.-N. Noncoding RNAs and pancreatic cancer. World Journal of Gastroenterology. 2016;22(2):801–814. doi: 10.3748/wjg.v22.i2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donzelli S., Cioce M., Muti P., Strano S., Yarden Y., Blandino G. MicroRNAs: non-coding fine tuners of receptor tyrosine kinase signalling in cancer. Seminars in Cell & Developmental Biology. 2016;50:133–142. doi: 10.1016/j.semcdb.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 12.Bruce J. P., Yip K., Bratman S. V., Ito E., Liu F.-F. Nasopharyngeal cancer: molecular landscape. Journal of Clinical Oncology. 2015;33(29):3346–3355. doi: 10.1200/jco.2015.60.7846. [DOI] [PubMed] [Google Scholar]
- 13.Tan G., Tang X., Tang F. The role of microRNAs in nasopharyngeal carcinoma. Tumor Biology. 2014;36(1):69–79. doi: 10.1007/s13277-014-2847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang F., Jiang C., Sun Q., et al. Downregulation of miR-429 and inhibition of cell migration and invasion in nasopharyngeal carcinoma. Molecular Medicine Reports. 2016;13(4):3236–3242. doi: 10.3892/mmr.2016.4940. [DOI] [PubMed] [Google Scholar]
- 15.Mao Y., Wu S., Zhao R., Deng Q. MiR-205 promotes proliferation, migration and invasion of nasopharyngeal carcinoma cells by activation of AKT signalling. Journal of International Medical Research. 2016;44(2):231–240. doi: 10.1177/0300060515576556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang M., Xiao J., Wang J., et al. MiR-24 enhances radiosensitivity in nasopharyngeal carcinoma by targeting SP1. Cancer Medicine. 2016;5(6):1163–1173. doi: 10.1002/cam4.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan J., Hu H., Zhou Z., et al. Tumor-suppressive mir-663 gene induces mitotic catastrophe growth arrest in human gastric cancer cells. Oncology Reports. 2010;24(1):105–112. doi: 10.3892/or_00000834. [DOI] [PubMed] [Google Scholar]
- 18.Jiao L., Deng Z., Xu C., et al. miR-663 induces castration-resistant prostate cancer transformation and predicts clinical recurrence. Journal of Cellular Physiology. 2014;229(7):834–844. doi: 10.1002/jcp.24510. [DOI] [PubMed] [Google Scholar]
- 19.Yi C., Wang Q., Wang L., et al. MiR-663, a microRNA targeting p21WAF1/CIP1, promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene. 2012;31(41):4421–4433. doi: 10.1038/onc.2011.629. [DOI] [PubMed] [Google Scholar]
- 20.Petersson F. Nasopharyngeal carcinoma: a review. Seminars in Diagnostic Pathology. 2015;32(1):54–73. doi: 10.1053/j.semdp.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 21.Zuberi M., Khan I., Gandhi G., Ray P. C., Saxena A. The conglomeration of diagnostic, prognostic and therapeutic potential of serum miR-199a and its association with clinicopathological features in epithelial ovarian cancer. Tumor Biology. 2016;37(8):11259–11266. doi: 10.1007/s13277-016-4993-2. [DOI] [PubMed] [Google Scholar]
- 22.Motawi T. K., Rizk S. M., Ibrahim T. M., Ibrahim I. A. Circulating microRNAs, miR-92a, miR-100 and miR-143, as non-invasive biomarkers for bladder cancer diagnosis. Cell Biochemistry and Function. 2016;34:142–148. doi: 10.1002/cbf.3171. [DOI] [PubMed] [Google Scholar]
- 23.Yan-Fang T., Jian N., Jun L., et al. The promoter of miR-663 is hypermethylated in Chinese pediatric acute myeloid leukemia (AML) BMC Medical Genetics. 2013;14(1, article 74) doi: 10.1186/1471-2350-14-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q., Cheng Q., Chen Z., et al. MicroRNA-663 inhibits the proliferation, migration and invasion of glioblastoma cells via targeting TGF-β1. Oncology Reports. 2016;35(2):1125–1134. doi: 10.3892/or.2015.4432. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y., Chen C., Yu S.-Z., et al. miR-663 suppresses oncogenic function of CXCR4 in glioblastoma. Clinical Cancer Research. 2015;21(17):4004–4013. doi: 10.1158/1078-0432.ccr-14-2807. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y., Chen C., Zhang X., et al. Primate-specific miR-663 functions as a tumor suppressor by targeting PIK3CD and predicts the prognosis of human glioblastoma. Clinical Cancer Research. 2014;20(7):1803–1813. doi: 10.1158/1078-0432.CCR-13-2284. [DOI] [PubMed] [Google Scholar]
- 27.Zang W., Wang Y., Wang T., et al. miR-663 attenuates tumor growth and invasiveness by targeting eEF1A2 in pancreatic cancer. Molecular Cancer. 2015;14, article 37 doi: 10.1186/s12943-015-0315-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Zhang Y., Zhou X., Xu X., et al. Waltonitone induces apoptosis through mir-663-induced Bcl-2 downregulation in non-small cell lung cancer. Tumor Biology. 2015;36(2):871–876. doi: 10.1007/s13277-014-2704-4. [DOI] [PubMed] [Google Scholar]
- 29.Bi C., Chung T.-H., Huang G., et al. Genome-wide pharmacologic unmasking identifies tumor suppressive microRNAs in multiple myeloma. Oncotarget. 2015;6(28):26508–26518. doi: 10.18632/oncotarget.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vislovukh A., Kratassiouk G., Porto E., et al. Proto-oncogenic isoform A2 of eukaryotic translation elongation factor eEF1 is a target of miR-663 and miR-744. British Journal of Cancer. 2013;108(11):2304–2311. doi: 10.1038/bjc.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu H., Li S., Cui X., et al. The overexpression of hypomethylated miR-663 induces chemotherapy resistance in human breast cancer cells by targeting heparin sulfate proteoglycan 2 (HSPG2) The Journal of Biological Chemistry. 2013;288(16):10973–10985. doi: 10.1074/jbc.m112.434340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sand M., Skrygan M., Sand D., et al. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell and Tissue Research. 2013;351(1):85–98. doi: 10.1007/s00441-012-1514-5. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z. Y., Zhang G. L., Wang M. M., Xiong Y. N., Cui H. Q. MicroRNA-663 targets TGFB1 and regulates lung cancer proliferation. Asian Pacific Journal of Cancer Prevention. 2011;12(11):2819–2823. [PubMed] [Google Scholar]
- 34.Yu Q., Zhang F., Du Z., Xiang Y. Up-regulation of serum miR-744 predicts poor prognosis in patients with nasopharyngeal carcinoma. International Journal of Clinical and Experimental Medicine. 2015;8(8):13296–13302. [PMC free article] [PubMed] [Google Scholar]