Abstract
The mating system transition in polyploid Brassica napus (AACC) from out-crossing to selfing is a typical trait to differentiate it from their diploid progenitors. Elucidating the mechanism of mating system transition has profound consequences for understanding the speciation and evolution in B. napus. Functional complementation experiment has shown that the insertion of 3.6 kb into the promoter of self-incompatibility male determining gene, BnSP11-1 leads to its loss of function in B. napus. The inserted fragment was found to be a non-autonomous Helitron transposon. Further analysis showed that the inserted 3.6 kb non-autonomous Helitron transposon was widely distributed in B. napus accessions which contain the S haplotype BnS-1. Through promoter deletion analysis, an enhancer and a putative cis-regulatory element (TTCTA) that were required for spatio-temporal specific expression of BnSP11-1 were identified, and both might be disrupted by the insertion of Helitron transposon. We suggested that the insertion of Helitron transposons in the promoter of BnSP11-1 gene had altered the mating system and might facilitated the speciation of B. napus. Our findings have profound consequences for understanding the self-compatibility in B. napus as well as for the trait variations during evolutionary process of plant polyploidization.
Polyploidization drives speciation and diversification in flowering plants. Its impact on plant genomes has primarily been thought of as ‘genomic shock’ accompanied by rapid and extensive genomic and epigenomic changes1,2,3,4. In this process, duplicated genes (whole genome duplication, WGD) can be lost, retained or maintained as duplicates, often undergoing subfunctionalization and neofunctionalization5,6. As a result, polyploids often show different phenotypic traits than their presumed progenitors with respect to morphology, ecology, cytology, and physiology; these new phenotypes may contribute to speciation, adaption to the environment and enhancement of the utility of polyploids for agriculture7,8,9,10. The molecular mechanisms underlying the evolution of these novel phenotypes remain largely unknown.
In Brassica, mating system transition is particularly well suited to studying the molecular mechanism of phenotypic trait variations in polyploids. The three basic diploid species B. rapa (AA, 2n = 20), B. oleracea (CC, 2n = 18) and B. nigra (BB, 2n = 16) are self-incompatible, but all three cultivated allotetraploids, B. napus (AACC, 2n = 38), B. carinata (BBCC, 2n = 34) and B. juncea (AABB, 2n = 36) are self-compatible, indicating that the mating system has evolved from outcrossing to self-fertilization (selfing) in association with polyploid formation. In many flowering plants, loss of self-incompatibility (SI) caused the transition of mating systems from outcrossing to self-fertilization, with a significant impact on the evolution of these species11. Selfing can increase homozygosity and cause inbreeding depression in offspring, however it also confers advantages such as reproductive assurance when pollinators or mates are scarce and higher efficiency of gamete transmission than out-crossing12,13,14,15. The molecular mechanism of mating system transition has also been a major focus in evolutionary biology.
Self-incompatibility in the Brassicaceae is controlled sporophytically by the multi-allelic S locus (i.e., pollen SI phenotype is determined by the diploid genotype of the pollen-producing parent)16. An S locus consists mainly of two genes SP11/SCR (S-locus protein 11/S-locus cysteine rich protein) and SRK (S-locus receptor kinase) determining recognition specificity in pollen17,18 and stigma19,20, respectively. The S locus is also called the ‘S haplotype’ as the S-locus genes are transmitted to progeny as one unit21. S haplotypes of Brassica can be divided into two classes. Class-II haplotypes are generally recessive to class-I haplotypes in pollen, but they are co-dominant in the stigma19. When pollen and stigma carry the same S haplotype of Brassica, self-incompatible interaction occurs, leading to the arrest of ‘self’ pollen at the stigma surface22. Any mutations in genes involved in female specificity, male specificity or downstream signaling pathways could cause the loss of SI. Recent studies in wild Brassicaceae species demonstrated that the evolution of self-compatibility (SC) tends to be driven by mutations in the male rather than the female components11, such as in Arabidopsis thaliana, Leavenworthia alabamica, Capsella rubella, Arabidopsis kamchatica and others21,23,24,25,26. However, in domesticated Brassica species, female self-compatible mutations are more frequent11, such as in B. napus (mutations of SRK27), B. rapa (mutations of SRK28 and MLPK, M-locusprotein kinase29) and B. oleracea (mutation of SRK30). As the determinant of SI recognition specificity in pollen, SP11/SCR gene expression was tightly regulated and coordinated with stamen development to confer a successful self-incompatible reaction. In B. rapa, class II BrSP11-60 was found to be expressed mainly in the anther tapetum, while transcripts of class I SP11/SCR genes were detected clearly both early in anther tapetum development and late in pollen development by RNA gel blot analysis18,22,31,32,33. The class I SP11/SCR protein product was present in the tapetum and pollen, but late in anther development the SP11/SCR protein was mainly localized in the pollen coat34. Also, the dominance relationship between SI alleles (SP11/SCR gene) was proposed to be responsible for SC in the polyploid species26,27,35,36.
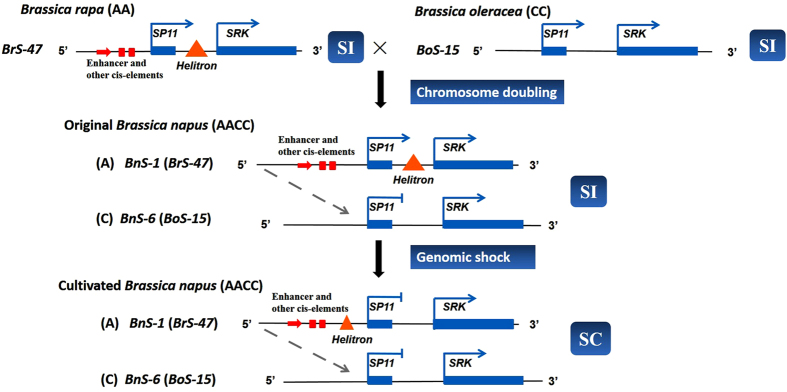
Cultivated B. napus is a self-compatible species, although it carries S haplotypes. In B. napus cultivar ‘Westar’, a dominant S haplotype BnS-1 derived from B. rapa haplotype BrS-47 on the A genome and a recessive S haplotype BnS-6 derived from the B. oleracea haplotype BoS-15 on the C genome were identified, with both SP11/SCR genes having lost their function27. An insertion of a DNA element of 3606 bp in the promoter region of BnSP11-1 gene on the A genome was responsible for the SC of ‘Westar’27,37. But how the insertion was generated and affected the evolution of B. napus remains unknown. In this study, by analyzing the 3.6 kb fragment inserted in the promoter of BnSP11-1, a non-autonomous Helitron transposon was identified. Further analysis showed that this Helitron transposon did not appear in B. rapa (BrS-47) but was widely distributed in B. napus (BnS-1), which indicated that it moved into the promoter of BnSP11-1 gene after formation of the polyploid species B. napus. By promoter deletion analysis, we found the insertion had disrupted the enhancer sequence and other cis-regulatory elements required for the spatio-temporal specific expression of BnSP11-1. We propose that the movement of the Helitron type transposon caused the transition of mating system, with a significant impact on the origin and evolution of B. napus. Our results yield insight into the complex mechanisms of both loss of self-incompatibility in B. napus and phenotypic trait variation in polyploid plants.
Results
Validation of the role of the inserted fragment in BnSP11-1 gene
To confirm the role of the inserted 3606 bp element in BnSP11-1 for the SC of ‘Westar’ proposed by Okamoto et al.27 and Tochigi et al.37, the SP11/SCR gene in BrS-47 (BnS-1 was derived from BrS-47) was used to complement the function of BnSP11-1 in ‘Westar’ (Additional Information: Figure S1A). RT-PCR analysis showed that BrSP11-47 transcripts can be detected in mature buds of ten transgenic plants and all of them were self-incompatible to varying degrees (Additional Information: Figure S1C). T1 progeny plants were also self-incompatible and the trait co-segregated with the introduced DNA. Transgenic line ‘W-3’ showed a higher level of BrSP11-47 transcripts and stronger self-incompatibility (setting only several seeds by self-pollination) than other lines, so it was used for further analysis (Additional Information: Figure S1B,C). Pollination assays showed that when the pollen of ‘Westar’ was applied to the stigma of ‘W-3’, compatible interaction occurs with many pollen tubes penetrating the stigma and resulting in normal pod set. When the pollen of ‘W-3’ was applied to the stigma of ‘Westar’, self-incompatible reaction occurs, no pollen tubes were observed, and the pods set few seeds (Fig. 1). These results showed that BnSRK-1 had normal function and the non-functional BnSP11-1 gene (with the 3606 bp insertion) on the A genome was responsible for the SC of ‘Westar’ in B. napus.
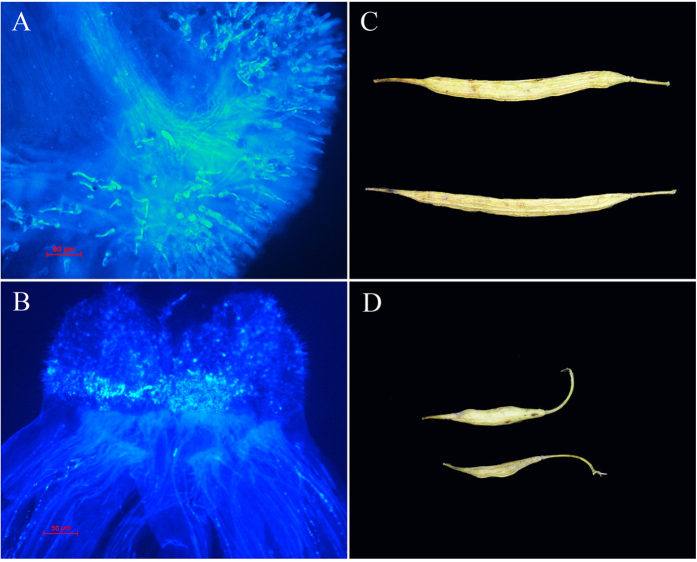
Figure 1. Pollination assays of the transgenic line ‘W-3’ and the wild type ‘Westar’.
(A) ‘Westar’ pollen placed on the stigma of ‘W-3’ shows compatible interaction, with many pollen tubes penetrating the stigma; (B)‘W-3’ pollen placed on the stigma of ‘Westar’ shows self-incompatible reaction, with no pollen tubes observed; (C) Pods set many seeds in the compatible pollination; (D) Pods set few seeds in the incompatible pollination. Bars = 50 μm in (A,B).
Helitron like transposon identification in the promoter of the BnSP11-1 gene and evolutionary analysis
The 3606 bp DNA element inserted in the promoter of the BnSP11-1 gene (Genbank accession AB270773) was analyzed to explore how it was generated and affected the evolution of B. napus. The inserted fragment showed no sequence similarity to known transposable elements. However, manually it was found to contain all the structural characteristics of a novel family of putative rolling circle transposable elements in eukaryotes, termed Helitron38. Like other Helitrons reported, the 3606 bp element was inserted precisely between the nucleotides 5′-A and T-3′, and did not cause duplication of the insertion site sequence. Furthermore, this fragment starts with 5′-TC, ends with 3′-CTAG, and is accompanied by a predicted small hairpin structure near the 3′ end (Fig. 2). As it lacks sequences similar to DNA helicase and RPA-like proteins which are necessary for being autonomous, we concluded that the insertion in BnSP11-1 is a non-autonomous Helitron type transposable element.
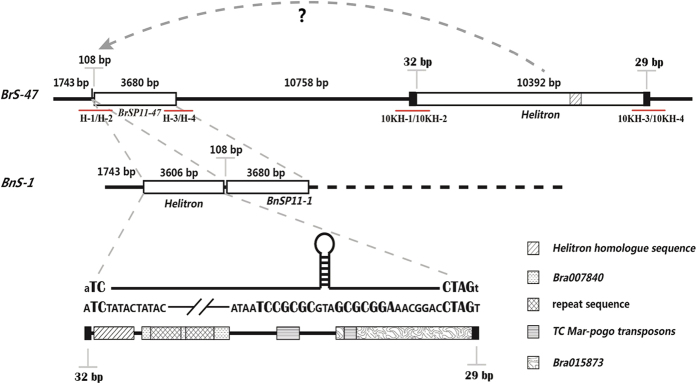
Figure 2. Two non-autonomous Helitron type transposable elements inserted independently in the S haplotypes BrS-47 and BnS-1.
The Helitron in BrS-47 lies downstream of the SP11/SCR gene but is absent from BnS-1. The Helitron in BnS-1 lies in the promoter region of the SP11/SCR gene. Both Helitrons shared similar boundaries, small hairpin structures, and embedded (captured) sequences.
Plant Helitrons often capture gene fragments during their movement38. By BLASTN analysis of the inserted element in BRAD (Brassica database, http://brassicadb.org/brad/), we found that positions 542 to 1333 bp showed almost 90% similarity to Bra007840 and positions 2292 to 3606 bp were similar to Bra015873. Interestingly, positions 114 to 474 bp showed 85% similarity to a BAC sequence (Genbank accession AB180899.1) that contains the SP11/SCR gene of S haplotype BrS-47. As S haplotype BnS-1 was derived from BrS-47, the BAC sequence was further analyzed. To our surprise, another Helitron type transposable element with a length of 10393 bp was identified downstream of the BrSP11/SCR-47 gene (Fig. 2). These two Helitron type transposable elements showed sequence similarity at the termini (32 bp at the 5′ terminal and 29 bp at the 3′ terminal), shared the same predicted small hairpin structure near the 3′ end, and even captured a similar small gene fragment (positions 114 to 474 bp of the 3606 bp fragment) during their movement (Fig. 2).
To detect the distribution of these two Helitron transposable elements in B. napus, 123 inbred lines were collected and analyzed. Three primer combinations: H-1/H-2 and H-3/H-4 flanking the 5′ and 3′ end of the Helitron transposable element (Fig. 2), and SpeS1-5/SpeS1-6 which can specifically amplify the BnSP11-1 intron were designed (Additional Information: Table S2). All three pairs of primers showed amplification in 85 lines but not in the remaining 38 lines (Additional Information: Table S3), indicating that the Helitron transposable element inserted in BnSP11-1 was widely distributed in B. napus having the S haplotype BnS-1, and moved into the promoter after formation of B. napus. In addition, two primer combinations: 10KH-1/10KH-2 and 10KH -3/10KH-4 flanking the 5′ and 3′ end of the 10393 bp Helitron (Fig. 2; Additional Information: Tables S1 and S2, Figure S3) did not show amplification in any of the 123 B. napus inbreds (Additional Information: Table S3), indicating that it moved away from downstream of BrSP11/SCR-47 gene after the formation of B. napus.
Disruption of cis-regulatory elements in the promoter of BnSP11-1
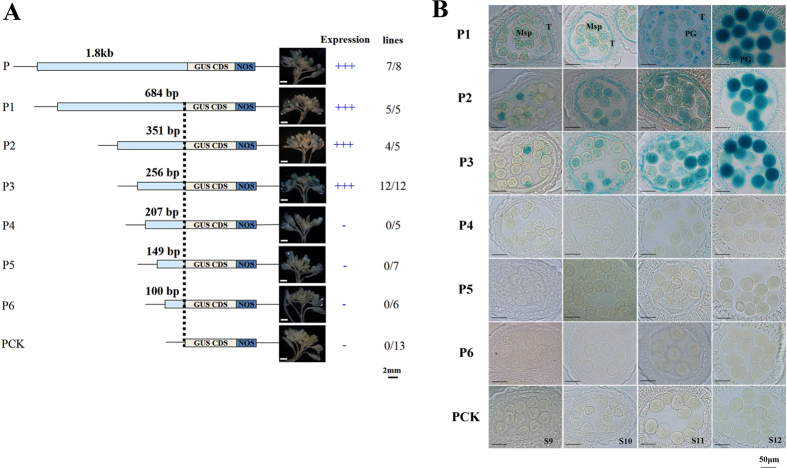
Overlapping PCR technology was used to isolate the promoter of the BnSP11-1 gene that did not contain the Helitron. A fragment of 1851 bp 5′-upstream of the translation initiation site was obtained and used to drive GUS (β-glucuronidase) gene expression in Arabidopsis (P-GUS). GUS staining results showed that the promoter was functional (Additional Information: Figure S2). To identify and characterize cis-regulatory elements involved in promoter strength and specificity, promoter deletion analysis was conducted. Six 5′-deletion promoter fragments linked to GUS were introduced into Arabidopsis (P1-GUS to P6-GUS, Fig. 3A). GUS staining can be detected in older buds in P1-GUS to P3-GUS but not in P4-GUS to P6-GUS under a stereo microscope (Fig. 3A). By semi-thin section analysis, no obvious difference in GUS staining (in the tapetum, microspores and mature pollen from stage 9 to stage 12) was observed in P1-GUS (−681 bp), P2-GUS (−351 bp) and P3-GUS (−256 bp) transgenic plants (Fig. 3B). However, GUS staining cannot be detected in P4-GUS (−207 bp), P5-GUS (−149 bp), P6-GUS (−100 bp) fusions and the control construct PCK at any stage of anther development (Fig. 3B). It indicated that the region from −256 bp (P3-GUS) to −207 bp (P4-GUS) contained cis-regulatory elements responsible for the spatial and temporal expression patterns of the BnSP11-1 gene.
Figure 3. BnSP11-1 promoter deletion analysis.
(A) Promoter deletion constructs (P1-GUS to P6-GUS), the control construct PCK and summary of GUS staining results. Numbers indicate positions relative to the BnSP11-1 translation start site. Photographs were taken after 16 h GUS incubation. Relative expression of each promoter construct in stamen is represented by +(positive) or −(negative). ‘Lines’ column indicates the number of individual transformants displaying stamen GUS activity over total number of transformants analyzed; Bars = 2 mm. (B) Representative GUS staining results for semi-thin section from stage 9 to stage 12 of lines transgenic for the indicated BnSP11-1 promoter deletion constructs and the control construct PCK. T, tapetum; Ms, microsporocyte; Tds, tetrads; Msp, microspore; PG, pollen grain; Bars = 50 μm.
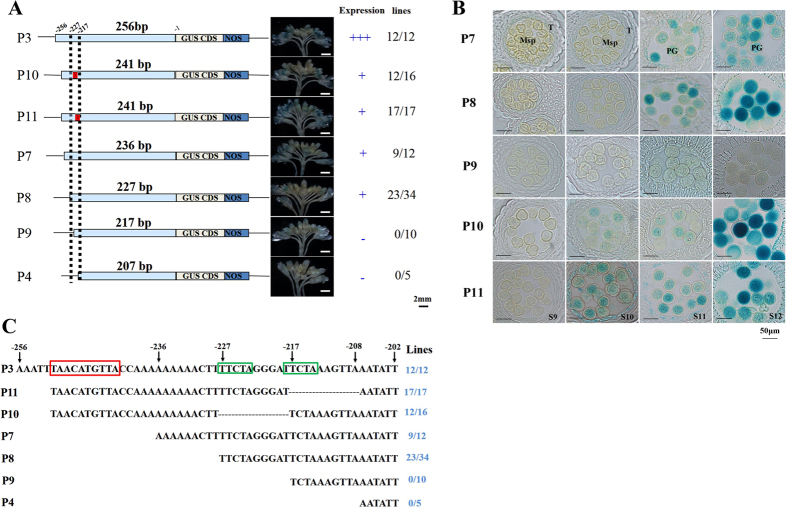
Further, P7-GUS to P9-GUS were constructed based on the region from −256 bp (P3-GUS) to −207 bp (P4-GUS) (Fig. 4A). GUS staining was detected in P7-GUS and P8-GUS, but not in P9-GUS (Fig. 4A), showing that the region near −227 bp to −217 bp has cis-regulatory elements responsible for the spatial and temporal expression of BnSP11-1. Compared with P3-GUS, P7-GUS and P8-GUS initiated gene expression at later stages: GUS staining was detected weakly at stage 9 and 10, but strongly at stage 11 and 12 (Fig. 4A,B). Therefore, the 20 bp sequence from −256 to −237 bp was proposed to determine early stage expression strength of BnSP11-1. A putative 10 bp palindromic sequence (TAACTAGTTA) was identified, which was considered an enhancer (Fig. 4C).
Figure 4. Further deletion analysis of the BnSP11-1 promoter.
(A) Promoter deletion constructs (P7-GUS, P8-GUS, P9-GUS, P10-GUS and P11-GUS) and representative GUS staining results. Numbers indicate positions relative to the BnSP11-1 translation start site. Photographs were taken after 16 h GUS incubation. Relative expression of each construct in stamen is represented by +(positive) or −(negative). ‘Lines’ indicate numbers of individual transformants displaying stamen GUS activity over total number of transformants analyzed; Bars = 2 mm. (B) Representative GUS staining results for semithin sections from stages 9 to 12 in transgenic lines of P7, P8, P9, P10 and P11. T, tapetum; Msp, microspore; PG, pollen grain; Bars = 50 μm. (C) Summary of BnSP11-1 promoter deletion analysis results. By sequence alignment of the deleted promoter fragments, a palindromic sequence (TAACTAGTTA, red box) and a putative cis-element (TTCTA, green box) responsible for the spatial and temporal expression patterns of BnSP11-1 were identified.
To identify the cis-regulatory elements exactly, P10-GUS and P11-GUS were constructed (Fig. 4A). Unexpectedly, P10-GUS and P11-GUS had almost the same staining patterns as P7-GUS and P8-GUS (Fig. 4A,B). We speculated that both the deleted regions in P10-GUS (−227 bp to −218 bp) and P11-GUS (−217 bp to −208 bp) were necessary for early stage BnSP11-1 expression. Also, they played redundant roles in pollen grain expression at late stages. By analyzing the sequences in P10-GUS, P11-GUS and P9-GUS, a putative cis-element (TTCTA) located in both deleted regions was identified (Fig. 4C). We also concluded that the Helitron type transposable element inserted 108 bp upstream of the translation initiation site has disrupted enhancer sequences and other cis-regulatory elements responsible for the normal expression of BnSP11-1.
Discussion
Unlike their progenitors B. rapa (AA) and B. oleracea (CC) that are self-incompatible and mainly used as vegetables, cultivated allotetraploid B. napus (AACC) genotypes are self-compatible and mainly grown for seed harvest. The mating system transition from SI to SC has therefore been a key event which may have contributed to B. napus speciation, increasing its adaptation to the environment and its utility for agriculture. In this study, we found the mating system transition from SI to SC in B. napus was caused by a Helitron-like transposon that inserted in the promoter of BnSP11-1 gene. The inserted Helitron-like transposon was widely distributed in B. napus containing the S haplotype BnS-1. By promoter deletion analysis we found the insertion of the Helitron-like transposon had disrupted the enhancer sequence and other cis-regulatory elements responsible for the normal expression of BnSP11-1 gene.
TEs (transposable elements) are one of the major components of plant genomes and also one of the major drivers of plant genome evolution39,40. Interspecific hybridization, accompanied by ‘genomic shock’, has been proposed to induce bursts of transposition attributable to the interaction of merged genomes, as first proposed by McClintock1. Beyond their considerable contribution to genome structure, they also influence gene expression2,41,42,43. Helitrons, as eukaryotic DNA transposable elements, are predicted to amplify by a rolling-circle mechanism and constitute about 2% of the A. thaliana genome38. In maize, most Helitrons are non-automonous elements with truncated pseudogenes and/or mobile elements that are considered to be responsible for rich in transpecific genetic diversity and the loss of function of some nuclear genes44,45,46,47. In B. rapa, an insertion of a Helitron was reported to be responsible for the yellow seed trait of cultivar yellow sarson48. Both the DNA elements inserted independently in the promoter region of the BnSP11-1 gene and downstream of the BrSP11-47 gene were non-autonomous Helitron transposons, which contain no transposase gene(s) but some truncated pseudogenes. The BnSP11-1 and BrSP11-47 associated Helitrons showed sequence similarity at the termini (32 bp at the 5′ terminal and 29 bp at the 3′ terminal), the same predicted small hairpin structure near the 3′ end and even a similar captured gene fragment (position 114 to 474 of the 3606 bp fragment) (Fig. 2). As the transposition mechanism of Helitrons is unclear, we could not extend our hypothesis to propose that the BnSP11-1 associated Helitron might be resulted from the movement of the BrSP11-47-associated one during B. napus speciation and evolution. By analyzing 123 inbred lines of B. napus, we found that all 85 lines which contain the S haplotype BnS-1 also had the 3606 bp Helitron transposon inserted in the promoter of BnSP11-1, but none of them have the Helitron inserted downstream of BrSP11-47 (Additional Information: Table S3). As a result, the movement of the Helitron transposon caused the mating system transition from SI to SC of the polyploid species B. napus (Fig. 1). Our results provided evidence that transposons played a key role for the evolution of polyploid species.
Mutations disabling male specificity (SP11/SCR gene) of the SI system are expected to be more strongly selected in wild species as mutant pollen grains are more easily transmitted to the offspring than mutant ovules during the pollination process, contrasting with the prevalent mutations disabling female specificity in domesticated species11,49,50,51,52. However, in the present investigation, Helitron transposon insertion in BnSP11-1 gene (male self-compatible mutation) was responsible for SC in ‘Westar’. SP11/SCR expression was tightly regulated and was under the control of a gene regulatory network involved in anther development. Shiba et al.34 suggested that the sporophytic and gametophytic expression patterns of the SP11/SCR gene are controlled by different cis-regulatory elements. The intact BnSP11-1 gene promoter can drive GUS gene expression in anther tapetum and microspores at stage 9, reaching maximum expression at stage 12, when anthers contain tricellular pollen grains and appear bilocular after degeneration and breakage of the septum below the stomium53 (Additional Information: Figure S2). In B. rapa, the highly conserved 192-bp upstream region was sufficient to drive the unique expression of BnSP11-934. However, we observed delayed GUS expression in tapetum and pollen in P7-GUS (−236 bp) and P8-GUS (−227 bp) with detailed semi-thin section analysis (Fig. 4B). Further, we identified a putative 10 bp palindromic sequence (TAACTAGTTA) considered an enhancer, and a putative cis-element (TTCTA), that played redundant roles in pollen grain expression at late stages (Fig. 4C). Revealing the molecular mechanism of SP11/SCR gene expression and regulation might provide a foundation for mating system transition research in the Brassicaceae species.
There are two S haplotypes that determined the SI reaction located on A and C genome, respectively in B. napus, both of them might contribute to the mating system transition. Here we report that Helitron-like transposon insertion on A genome was responsible for mating system transition in ‘Westar’ (Figs 1 and 2). While on C genome, expression of the recessive BnSP11-6 gene cannot be detected, though the DNA sequence was intact, which might be suppressed by the dominant nonfunctional BnSP11-1 gene on A genome27. In the Brassicaceae, complex dominance interactions among S-haplotypes have been reported54,55,56, and the underlying molecular mechanism have been partially revealed in B. rapa31,32,57. If the reported mechanism of dominance relationship between SI alleles in B. rapa contributed to the mating system transition in allotetraploid B. napus needs to be explored. In allotetraploid A. kamchatica, the degradation of male components was responsible for the loss of SI, both homeologous copies of the SP11/SCR gene ought to have lost their function by interspecific crosses analysis and also the dominance interactions of the SI alleles may be involved26. Zhai et al.36 proposed that besides the mutation of SP11/SCR genes and dominance relationship between SI alleles, other factor(s) independent of the S locus are involved in the SC of some accessions in B. napus. Although, the precise mechanism of dominance relationship between SI alleles is still unclear, and it was proposed to be responsible for the SC in the polyploid species26,27,35,36.
Based on those results, a speculative model for the origin and speciation of B. napus was proposed (Fig. 5): After the formation of the original B. napus plants by inter-specific hybridization between B. rapa and B. oleracea, the expression of the recessive SP11/SCR gene located on the B. oleracea-derived C genome was suppressed by an unknown mechanism. However, the SP11/SCR and SRK genes on the B. rapa-derived A genome were expressed normally, therefore the original B. napus plants were inferred to be self-incompatible. The movement of a Helitron transposon disrupted the enhancer sequence and other cis-regulatory elements responsible for the normal expression of BnSP11-1 gene, conferring SC and permitting newly formed B. napus plants to produce seeds. SC shows particular advantages when out-crossing mates are scarce50,51, such as they may be for a newly formed polyploidy. So, we suggested that the movement of Helitron transposons and the dominance interactions between the S alleles might coordinately contribute to the origin and speciation of B. napus by changing the mating system from cross pollination to self-pollination. Our findings provide insight into the self-compatibility of B. napus as well as other trait variations associated with the evolutionary process of plant polyploidization.
Figure 5. A proposed model for the origin and formation of B. napus.
In the original B. napus plants, BrS-47 was dominant over BoS-15 in pollen, which may suppress the expression of recessive SP11/SCR gene located on the C genome. As the SP11/SCR and SRK genes located on the A genome can be expressed normally, the original B. napus plants were inferred to be self-incompatible. The genome of the newly formed amphidiploid plants was unstable, with transposable elements playing a pivotal role in providing variation for genome reorganization. A Helitron transposon disrupted cis-regulatory elements responsible for the normal expression of BnSP11-1 and conferred.
Methods
Plant Materials and Growth Conditions
The wild type self-compatible B. napus line ‘Westar’, its transgenic plants and B. rapa line ‘9-117’ with S haplotype BrS-47 were grown in the greenhouse with a 16/8 h day/night photoperiod and day/night temperatures of 22 °C/15 °C. A. thaliana plants (ecotype Columbia) were grown at 22 °C, 16/8 h light/dark in the greenhouse.
Investigation of SI phenotype
Self-incompatibility phenotype was measured as follows58: when three to five flowers were set on the major inflorescence, the major inflorescence and two or three secondary ramifications were bagged for self-pollination after removing the apical buds artificially. Every two days, bags were slipped gently in order to assure enough self-pollination. Self-seeds were produced by bud-pollination. About two weeks later, bags were removed to allow the seeds to develop. After seedpods were mature, the number of seeds produced was counted, and self-compatibility index (SCI) was calculated as the ratio of number of seeds to number of flowers59. Plants with SCI ≥ 2 were referred as self-compatible and plants with SCI < 2 were considered as self-incompatible58.
Promoter Region of BnSP11-1 and Promoter Deletion Constructs
Overlapping PCR technology was used to isolate the clean promoter fragment (no Helitron type transposable element contained) of the BnSP11-1 gene from ‘Westar’. Primer combination BnS1PRO-3/BnS1PRO-4 (5′-ACGCGTCGACAGCTTCACTCTTGGACTGTC-3′/5′-TAACAATCATTATAAATACATATCCAACAGAAGTTGCGTA-3′) was used to amplify the 5′- flanking region of the Helitron type transposable element and primer combination BnS1PRO-5/BnS1PRO-2 (5′-TACGCAACTTCTGTTGGATATGTATTTATAATGATTGTTA-3′/5′-TCCCCCGGGGATTCAGAAAAGTGATAAAAGATTC-3′) was used to amplify the 3′-flanking region of the Helitron type transposable element. SalI and SmaI restriction sites were added to the 5′ and 3′ ends of the primers BnS1PRO-3 and BnS1PRO-2 respectively. A fragment of 1851 bp of the 5′-upstream region of translation initiation site was obtained. A cassette containing the GUS coding region followed by the nopaline synthase polyadenylation signal from pBI101 (CLONTECH, Palo Alto, CA) was subcloned into the binary vector pCAMBIA 230060 with restriction enzymes Hind III and EcoRI to construct promoter-GUS fusions. The amplified fragments were subcloned into the modified binary vector pCAMBIA 230060 to yield the 1851-bp SP11-1 promoter-GUS construct. Based on the resultant promoter sequence of BnSP11-1, a series of deletion constructs (P1-GUS to P11-GUS) were generated. BnS1PRO-2 (5′-TCCCCCGGGGATTCAGAAAAGTGATAAAAGATTC-3′) which contains a SmaI restriction site to the 3′ end was used as the antisense primer to amplify the deletion promoter fragment for all deletion constructs. Sense primers for each deletion construct were listed as follows: P1-GUS: (5′-ACGCGTCGACGACACACCATCACCACTTCTTT-3′), P2-GUS: (5′-ACGCGTCGACCTTTTAGACCTCCTTAATAGCCTG-3′), P3-GUS: (5′-ACGCGTCGACAAATTTAACATGTTACCAAAAAAA-3′), P4-GUS: (5′-ACGCGTCGACAATATTTGGACCCGTTAATCTC-3′), P5-GUS: (5′-ACGCGTCGACTTTAGTTAAAAAATCTGTTTTACG-3′), P6-GUS: (5′-ACGCGTCGACTAATGATTGTTAACAAGGAAAC-3′), P7-GUS: (5′-ACGCGTCGACAAAAAAACTTTTCTAGGGATTCT-3′), P8-GUS: (5′-ACGCGTCGACTTCTAGGGATTCTAAAGTTAAATA-3′), P9-GUS: (5′-ACGCGTCGACTCTAAAGTTAAATATTTGGACC-3′), P10-GUS: (5′-ACGCGTCGACTAACATGTTACCAAAAAAAAACTTTCTAAAGTTAAATATTTGGACCCG-3′) and P11-GUS: (5′-ACGCGTCGACTAACATGTTACCAAAAAAAAACTTTTCTAGGGATAATATTTGGACCCGTTAATCTCGTTG-3′). All the sense primers contain SalI restriction site at the 5′ end. All the amplified deletion promoter fragments were subcloned independently into the modified binary vector pCAMBIA 230060 to yield the P1-GUS to P11-GUS constructs. The modified binary vector pCAMBIA 230060 was used as the negative control.
Vector Construction of BrSP11-47 gene and Plant Transformation
Overlapping PCR technology was used to clone the promoter sequence and the coding sequence (CDS) of BrSP11-47 (GenBank accession no. AB180899) from a B. rapa line ‘9-117’ with homozygous BrS-47. Primer combination S1E1/S1E2 (5′-TCCCCCGGGTACGACCTGCTGATATTCTCC-3′/5′-ATCAGATTAGCTTCCACTTCTTGAATATGACCTGAAACG-3′) was used to amplify the promoter region and the first exon and primer combination S1E3/S1E4 (5′-CGTTTCAGGTCATATTCAAGAAGTGGAAGCTAATCTGAT-3′/5′-GGGTTACCCTAACACAATTTACATACACAAGAATAA-3′) was used to amplify the second exon of BrSP11-47. SmaI and BstEII restriction sites were added to the 5′ and 3′ ends of the primers S1E-1 and S1E-4 respectively. Finally, a fragment of 2345 bp containing the promoter region and the CDS of BrSP11-47 was obtained. This fragment was then subcloned into the binary vector pCAMBIA230160 to yield the 2301-1 + 4 construct.
The construct 2301-1 + 4 was introduced into A. tumefaciens GV3101 host cells. Plant transformation was carried out following the method of Dun et al.61. The transformed plants with roots were subsequently transplanted in experimental plots from which T1 seeds were harvested. DNA from transgenic plants was analyzed by PCR, combining the primers S1E1 (5′-TCCCCCGGGTACGACCTGCTGATATTCTCC-3′) and PC2301R (5′-GCAACAGGATTCAATCTTAAGAA-3′) designed from the sequence of the nopaline synthase polyadenylation signal present in the vector to verify the presence of the SP11 transgene.
Pollination Assay
Floral buds of the B. napus plants were emasculated one day before anthesis to avoid pollen contamination. Pollination was performed the next day. Some pollinated pistils were left to set seeds. The rest were cut at the peduncle 16 hours after pollination, fixed for 2 h in ethanol: acetic acid (3:1), softened in 1 N NaOH at 60 °C for 1.5 h, and stained with 0.01% (w/v) decolorized aniline blue for 2.5 h in 2% (w/v) K3PO4. Pistils were gently squashed onto a microscopic slide glass by placing the cover glass over the pistils. Samples were examined under a fluorescence microscope (Ax 10, Zeiss).
GUS assay
All promoter-GUS constructs were introduced into Arabidopsis wild-type plants (ecotype Columbia) by Agrobacterium-mediated transformation. GUS activity was visualized by staining different stage flowers in the T3 generation of homozygous transgenic lines, overnight in X-Gluc solution62, and then tissues were cleared in 75% (v/v) ethanol. Treated flower buds were observed and photographed under a stereomicroscope, then embedded in Technovit 7100 resin (HeraeusKulzer, http://www.heraeus.com/) as described previously by Zhu et al.63. Afterwards, transverse sections of the anthers approximately 12 μm thick were cut from the embedded blocks using a Leica Ultracut R ultra-microtome (Leica). The sections were photographed under a microscope and the anther development stages were determined53.
RT-PCR and qRT-PCR
Total RNA was extracted using a plant mini RNeasy kit (Qiagen). Five micrograms of RNA was DNase-treated using a DNA-free kit (Ambion, http://www.ambion.com). First-strand cDNA synthesis was performed using a SuperScript kit (Gibco BRL, http://www.invitrogen.com). The reverse transcription products were used as templates for PCR to examine the expression of BrSP11-47 in transgenic plants. Real-time RT-PCR was also performed using a Bio-Rad IQ5 with SYBR Green detection (http://www.bio-rad.com/). Primer combination RT-SCR1-L/RT-SCR1-R (5′-TGTTTCATATTCATCGTTTCAGG-3′/5′-CTCTTGTCCATACCCTTCGAATA-3′) was used for RT-PCR analysis. Primer combination RT-SCR1-1/RT-SCR1-2 (5′-GCTAATCTGATGAATCCGTGCG-3′/5′-TTTGTGCATTCGCAACGTGG-3′) was used for Real-time RT-PCR analysis. Actin (Gene-Bank accession no.: AF111812) was amplified with RAC1-P3/RAC1-P4 and used as an internal control to normalize transcript levels for all the expression analyses.
Additional Information
How to cite this article: Gao, C. et al. Helitron-like transposons contributed to the mating system transition from out-crossing to self-fertilizing in polyploid Brassica napus L. Sci. Rep. 6, 33785; doi: 10.1038/srep33785 (2016).
Supplementary Material
Acknowledgments
We are thankful to Dr. Mayank Gautam for thoroughly and thoughtfully editing through the English version of the manuscript. This work was funded by China Postdoctoral Science Foundation (2014M552055; 2015T80816), National Natural Science Foundation of China (Project 31571706).
Footnotes
Author Contributions C.B.G. and G.L.Z. conceived and performed experiments and wrote the manuscript; C.Z.M. conceived strategies, designed experiments and edited the manuscript. W.Z., T.Z., Z.Q.L. and Y.Y. participated in the promoter deletion analysis. M.W., Y.Y., Z.Q.D. and Y.L. participated in the DNA methylation analysis. B.L. and J.J.L. helped to edit the manuscript. J.X.S., J.X.T. and T.D.F. provided comments on entire study and manuscript. All authors read and approved the manuscript.
References
- McClintock B. The significance of responses of the genome to challenge. Science 226, 792–801 (1984). [DOI] [PubMed] [Google Scholar]
- Kashkush K., Feldman M. & Levy A. A. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33, 102–106 (2003). [DOI] [PubMed] [Google Scholar]
- Ha M. et al. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA 106, 17835–17840 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraitshtein Z., Yaakov B., Khasdan V. & Kashkush K. Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 186, 801–812 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. & Conery J. S. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000). [DOI] [PubMed] [Google Scholar]
- Adams K. L. & Wendel J. F. Polyploidy and genome evolution in plants. Curr. Opin.Plant Biol. 8, 135–141 (2005). [DOI] [PubMed] [Google Scholar]
- Levin D. A. Polyploidy and novelty in flowering plants. Am. Nat. 122, 1–25 (1983). [Google Scholar]
- Ramsey J. & Schemske D. W. Neopolyploidy in flowering plants. Ann. Rev. Ecol. Syst. 33, 589–639 (2002). [Google Scholar]
- Gaeta R. T., Pires J. C., Iniguez-Luy F., Leon E. & Osborn T. C. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19, 3403–3417 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H. & Willis J. H. Plant speciation. Science 317, 910–914 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K. K. & Tsuchimatsu T. Evolution of selfing: recurrent patterns in molecular adaptation. Annu. Rev. Ecol. Evol. Syst. 46, 593–622 (2015). [Google Scholar]
- Darwin C. R. The effects of cross and self fertilisation in the vegetable kingdom Ch. 9 (John Murray, London, 1876). [Google Scholar]
- Charlesworth D. & Charlesworth B. Inbreeding depression and its evolutionary consequences. Ann. Rev. Ecol. Syst. 18, 237–268 (1987). [Google Scholar]
- Goodwillie C., Kalisz S. & Eckert C. G. The evolutionary enigma of mixed mating systems in plants: occurrence, theoretical explanations, and empirical evidence. Annu. Rev. Ecol. Evol. Syst. 36, 47–79 (2005). [Google Scholar]
- Busch J. W. & Delph L. F. The relative importance of reproductive assurance and automatic selection as hypotheses for the evolution of self-fertilization. Ann. Bot. 109, 553–562 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. J. Self-incompatibility systems in angiosperms.(III). Cruciferae. Heredity, 9, 52–68 (1955). [Google Scholar]
- Schopfer C. R., Nasrallah M. E. & Nasrallah J. B. The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700 (1999). [DOI] [PubMed] [Google Scholar]
- Takayama S. et al. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 97, 1920–1925 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J. C., Howlett B., Boyes D. C., Nasrallah M. E. & Nasrallah J. B. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA 88, 8816–8820 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki T. et al. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403, 913–916 (2000). [DOI] [PubMed] [Google Scholar]
- Vekemans X., Poux C., Goubet P. M. & Castric V. The evolution of selfing from outcrossing ancestors in Brassicaceae: what have we learned from variation at the S-locus? J. Evolutionary Biol. 27, 1372–1385 (2014). [DOI] [PubMed] [Google Scholar]
- Schopfer C. R. & Nasrallah J. B. Self-incompatibility. Prospects for a novel putative peptide-signaling molecule. Plant Physiol. 124, 935–940 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. L. et al. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl. Acad. Sci. USA 106, 5246–5251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimatsu T. et al. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464, 1342–1346 (2010). [DOI] [PubMed] [Google Scholar]
- Busch J. W., Joly S. & Schoen D. J. Demographic signatures accompanying the evolution of selfing in Leavenworthia alabamica. Mol. Biol. Evol. 28, 1717–1729 (2011). [DOI] [PubMed] [Google Scholar]
- Tsuchimatsu T., Kaiser P., Yew C. L., Bachelier J. B. & Shimizu K. K. Recent loss of self-incompatibility by degradation of the male component in allotetraploid Arabidopsis kamchatica. PLoS Genetics 8, e1002838 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S. et al. Self-compatibility in Brassica napus is caused by independent mutations in S-locus genes. Plant J. 50, 391–400 (2007). [DOI] [PubMed] [Google Scholar]
- Fujimoto R., Sugimura T., Fukai E. & Nishio T. Suppression of gene expression of a recessive SP11/SCR allele by an untranscribed SP11/SCR allele in Brassica self-incompatibility. Plant.Mol. Biol. 61, 577–587 (2006). [DOI] [PubMed] [Google Scholar]
- Murase K. et al. A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303, 1516–1519 (2004). [DOI] [PubMed] [Google Scholar]
- Nasrallah J. B., Rundle S. J. & Nasrallah M. E. Genetic evidence for the requirement of the Brassica S-locus receptor kinase gene in the self-incompatibility response. Plant J. 5(3), 373–384 (1994). [Google Scholar]
- Shiba H. et al. The dominance of alleles controlling self-incompatibility in Brassica pollen is regulated at the RNA level. Plant Cell 14, 491–504 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba H. et al. Dominance relationships between self-incompatibility alleles controlled by DNA methylation. Nat. Genet. 38, 297–299 (2006). [DOI] [PubMed] [Google Scholar]
- Tarutani Y. et al.Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 466, 983–986 (2010). [DOI] [PubMed] [Google Scholar]
- Shiba H. et al. A pollen coat protein, SP11/SCR, determines the pollen S-specificity in the self-Incompatibility of Brassica species. Plant Physiol. 125, 2095–2103 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah J. B., Liu P., Sherman-Broyles S., Schmidt R. & Nasrallah M. E. Epigenetic mechanisms for breakdown of self-incompatibility in interspecific hybrids. Genetics 175, 1965–1973 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W. et al. Gene expression and genetic analysis reveal diverse causes of recessive self-compatibility in Brassica napus L. BMC Genomics 15, 1037 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochigi T., Udagawa H., Li F., Kitashiba H. & Nishio T. The self-compatibility mechanism in Brassica napus L. is applicable to F1 hybrid breeding. Theor. Appl. Genet. 123, 475–482 (2011). [DOI] [PubMed] [Google Scholar]
- Kapitonov V. V. & Jurka J. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 98, 8714–8719 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H. H. Mobile elements: drivers of genome evolution. Science 303, 1626–1632 (2004). [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 15, 621–627 (2005). [DOI] [PubMed] [Google Scholar]
- Wessler S. R. Phenotypic diversity mediated by the maize transposable elements Ac and Spm. Science 242, 399–405 (1988). [DOI] [PubMed] [Google Scholar]
- Naito K. et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461, 1130–1134 (2009). [DOI] [PubMed] [Google Scholar]
- Studer A., Zhao Q., Ross-Ibarra J. & Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat.Genet. 43, 1160–1163 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal S. K. The Maize Genome Contains a Helitron Insertion. Plant Cell 15, 381–391 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Gallavotti A., Stryker G. A., Schmidt R. J. & Lal S. K. A novel class of Helitron-related transposable elements in maize contain portions of multiple pseudogenes. Plant.Mol. Biol. 57, 115–127 (2005). [DOI] [PubMed] [Google Scholar]
- Lai J., Li Y., Messing J. & Dooner H. K. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc. Natl. Acad. Sci. USA 102, 9068–9073 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M. et al. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat.Genet. 37, 997–1002 (2005). [DOI] [PubMed] [Google Scholar]
- Li X. et al. A large insertion in bHLH transcription factor BrTT8 resulting in yellow seed coat in Brassica rapa. PloS One. 7, e44145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama M. K., Zhang Y. & Newbigin E. On the origin of self-incompatibility haplotypes: transition through self-compatible intermediates. Genetics 157, 1805–1817 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch J. W. & Schoen D. J. The evolution of self-incompatibility when mates are limiting. Trends Plant Sci. 13, 128–136 (2008). [DOI] [PubMed] [Google Scholar]
- Tsuchimatsu T. & Shimizu K. K. Effects of pollen availability and the mutation bias on the fixation of mutations disabling the male specificity of self-incompatibility. J. Evolutionary Biol. 26, 2221–2232 (2013). [DOI] [PubMed] [Google Scholar]
- Gervais C. E., Castric V., Ressayre A. & Billiard S. Origin and diversification dynamics of self-incompatibility haplotypes. Genetics 188, 625–636 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P. M. et al. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322 (1999). [Google Scholar]
- Hatakeyama K., Takasaki T., Watanabe M. & Hinata K. Molecular characterization of S locus genes, SLG and SRK, in a pollen-recessive self-incompatibility haplotype of Brassica rapa L. Genetics 149, 1587–1597 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson K. Non-linear dominance relationships between S alleles. Heredity 21, 345–362 (1966). [Google Scholar]
- Kakizaki T. et al. Linear dominance relationship among four class-II S haplotypes in pollen is determined by the expression of SP11 in Brassica self-incompatibility. Plant Cell Physiol. 44, 70–75 (2003). [DOI] [PubMed] [Google Scholar]
- Tarutani Y. et al. Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 466, 983–986 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Development of SCAR markers linked to self-incompatibility in Brassica napus L. Mol.Breed. 21, 305–315 (2008). [Google Scholar]
- Yang G. et al. Genetic analysis of four self-incompatible lines in Brassica napus. Plant. Breed. 120, 57–61 (2001). [Google Scholar]
- Hajdukiewicz P., Svab Z. & Maliga P. The small, versatilep PZP family of Agrobacterium binary vectors for plant transformation. Plant. Mol. Biol. 25, 989–994 (1994). [DOI] [PubMed] [Google Scholar]
- Dun X. et al. BnaC. Tic40, a plastid inner membrane translocon originating from Brassica oleracea, is essential for tapetal function and microspore development in Brassica napus. Plant J. 68, 532–545 (2011). [DOI] [PubMed] [Google Scholar]
- Willemsen V., Wolkenfelt H., de Vrieze G., Weisbeek P. & Scheres B. The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125, 521–531 (1998). [DOI] [PubMed] [Google Scholar]
- Zhu Y. et al. A separation defect of tapetum cells and microspore mother cells results in male sterility in Brassica napus: the role of abscisic acid in early anther development. Plant. Mol. Biol. 72, 111–123 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.