Abstract
Balancer chromosomes are convenient tools used to maintain lethal mutations in heterozygotes. We established a method for engineering new balancers in C. elegans by using the CRISPR/Cas9 system in a non-homologous end-joining mutant. Our studies will make it easier for researchers to maintain lethal mutations and should provide a path for the development of a system that generates rearrangements at specific sites of interest to model and analyse the mechanisms of action of genes.
Genetic balancers (including inversions, translocations and crossover-suppressors) are essential tools to maintain lethal or sterile mutations in heterozygotes. Recombination is suppressed within these chromosomal rearrangements. However, despite efforts to isolate genetic balancers since 19781,2,3,4,5, approximately 15% (map units) of the C. elegans genome has not been covered6 (Supplementary Fig. 1). Because the chromosomal rearrangements generated by gamma-ray and X-ray mutagenesis are random, it is difficult to modify specific chromosomal regions. Here, we used the CRISPR/Cas9 genome editing system to solve this problem. The CRISPR/Cas9 system has enabled genomic engineering of specific DNA sequences and has been successfully applied to the generation of gene knock-outs and knock-ins in humans, rats, mice, zebrafish, flies and nematodes7. Recently, the CRISPR/Cas9 system has been shown to induce inversions and translocations in human cell lines and mouse somatic cells8,9,10. Similarly, inversions up to 57.5 kb have been obtained in the zebrafish germline11. Although a large number of cells can be treated at once for effective CRISPR/Cas9 editing in cell lines, it is more difficult to do so in the germlines of model organisms because of limitations in the ability to introduce genome editing tools. Thus, researchers need an efficient way to engineer the chromosomal structure in multicellular organisms in vivo. In the present study, we established an editing method using the CRISPR/Cas9 system in C. elegans to generate genetic balancers at specific chromosomal sites. The inversions and crossover-suppressors produced were up to 6.7 Mb (~17 cM), lengths 2 orders of magnitude longer than produced in a previous work in the germline of a model organism. To facilitate genomic engineering, we targeted the genome rearrangements in a non-homologous end-joining (NHEJ) mutant background. Our method resulted in a higher proportion of successful rearrangements to generate new balancers. Moreover, we found that the inversion and crossover-suppressor balancers generated in heterozygotes did not result in interchromosomal effects.
Results and Discussion
Experimental design to screen for new balancers on chromosome IV
We designed two sgRNAs (single guide RNA) in the exons of two target genes that result in easily identifiable phenotypes when they are disrupted. We next constructed two targeting vectors that joined the chromosomal breakpoints together, each of which had 2 kb of sequence homologous to each predicted junction point, so that chromosomal rearrangements could be induced by homologous recombination (HR) between the targeted regions and homology vectors (Fig. 1b, Supplementary Fig. 2). A previous study has reported that disabling NHEJ via the RNAi inactivation of the cku-80 gene (a homologue of the human KU80), which acts as a DNA binding protein, significantly improves the HR efficiency in C. elegans12. Therefore, NHEJ disruption may allow for the efficient repair of DSBs by using targeting vectors via HR. One of the NHEJ genes, lig-4 (a human LIG4 homologue), is essential for the final ligation step of the DNA ends. A strain known to contain a disruption in lig-4, tm750, was used in the experimental procedures and is depicted in Fig. 1a.
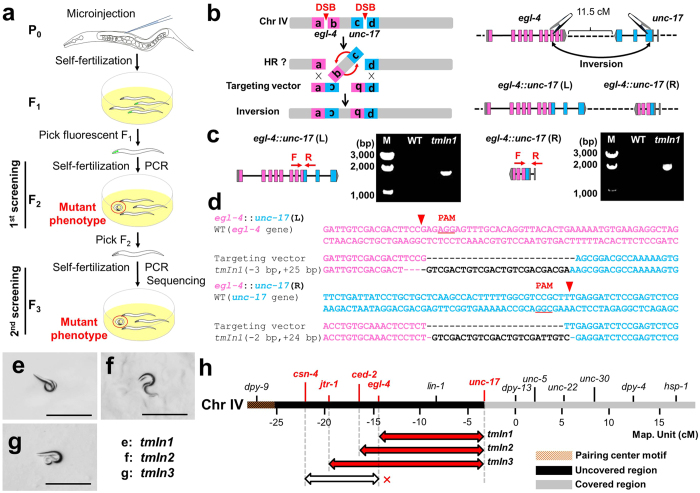
Figure 1. Genetic engineering of new balancers by using the CRISPR/Cas9 system.
(a) Experimental design to screen for inversion balancers. (b) Schematic of the chromosomal rearrangement tmIn1. tmIn1 was created by an inversion between egl-4 and unc-17. (c) PCR amplification of breakpoint junctions in wild-type (WT) and tmIn1 animals. (d) Breakpoint sequence alignments of the targeting vectors and tmIn1 rearrangement. Black bars indicate the cleavage sites. (e) The relative positions of breakpoints on chromosomal balancer IV. The generated balancers are indicated by red double-headed arrows. A white arrow with a cross indicates a failed trial. (f,g,h) Generated balancers showed a recessive larval arrest phenotype. Scale bars represent 100 μm.
We first attempted to generate an inversion balancer on the left arm of chromosome IV (Fig. 1b), which includes part of the largest region of the C. elegans genome that is not covered by current genetic balancers (Supplementary Fig. 1). We co-injected sgRNAs targeting egl-4 and unc-17 with Cas9, the targeting vectors and a Pmyo-2::Venus transgene marker into the gonads of young adult P0 worms. We screened for fluorescent F1 worms that contained larval-arrested F2 progeny caused by unc-17 disruption and confirmed the rearrangements by PCR amplification of both junction points. To examine whether the candidates maintained the chromosomal inversion, F2 worms that laid larval-arrested F3 progeny were further investigated by using both PCR amplification and DNA sequencing. Through these experimental procedures, one chromosomal rearrangement was obtained in each of the 136 F1 worms in the lig-4(tm750) mutant background (Table 1). The rearrangement named tmIn1(IV) exhibited detectable egl-4::unc-17 fused genes at two junction points, as confirmed by PCR amplification of the breakpoints, but the WT did not (Fig. 1c). These breakpoints were verified by DNA sequencing (Fig. 1d). The rearrangement tmIn1(IV) exhibited a recessive larval arrest phenotype (Fig. 1e). Thus, these experimental techniques induced successful chromosomal rearrangements in the germline of a multicellular organism.
Table 1. Summary of experimental efficiencies to generate the genetic balancers IV.
| Balancer name | Cas9 targets | Targeting vector | Distance (cM) | Background genotype | P0 wormsa | F1 wormsb | Phenotype in F2c | F1 PCRd | F2 PCRe | Ratio (%)f |
|---|---|---|---|---|---|---|---|---|---|---|
| tmIn42-44 | egl-4 unc-17 | − | 11.5 | WT (N2) | 146 | 900 | 163 | 4 | 3 | 0.33 |
| tmIn45 | egl-4 unc-17 | − | 11.5 | lig-4 (tm750) | 149 | 723 | 87 | 10 | 1 | 0.13 |
| − | egl-4 unc-17 | + | 11.5 | WT (N2) | 107 | 755 | 102 | 12 | 0 | 0 |
| tmIn1 | egl-4 unc-17 | + | 11.5 | lig-4 (tm750) | 31 | 136 | 40 | 6 | 1 | 0.73 |
| tmIn2 | ced-2 unc-17 | + | 13.5 | lig-4 (tm750) | 48 | 168 | 24 | 2 | 1 | 0.60 |
| tmIn3 | jtr-1 unc-17 | + | 16.4 | lig-4 (tm750) | 46 | 312 | 96 | 25 | 1 | 0.32 |
| − | csn-4 egl-4 | + | 7.6 | lig-4 (tm750) | 39 | 168 | 64 | 4 | 0 | 0 |
aTotal number of injected P0 worms.
bTotal number of fluorescent F1 worms obtained.
cNumber of F1 strains whose progeny showed phenotypes.
dNumber of F1 strains that showed rearrangement-specific PCR bands in the first screening.
eNumber of F2 strains that showed rearrangement-specific PCR bands in the second screening.
fIsolated genetic balancer/total number of fluorescent F1 worms.
We further screened for the generation of large chromosomal balancers on the left arm of chromosome IV. We obtained two additional balancers, tmIn2(IV) and tmIn3(IV), which covered 13.5 cM and 16.4 cM, respectively (Fig. 1h, Table 1, Supplementary Figs 3 and 4). The probability of obtaining inversion strains appeared to decrease as the target size became larger (0.60% and 0.32% for tmIn2 and tmIn3, respectively: Table 1). The tmIn2(IV) and tmIn3(IV) worms exhibited a recessive larval arrest phenotype (Fig. 1f,g). Although target sites between csn-4 and egl-4 covering only 7.6 cM and sgRNA-specific mutations were observed, these chromosomal rearrangements could not be isolated (Fig. 1h, Table 1). One of the target genes (csn-4) was located near pairing centres (PCs), where the chromosome is stabilized by homologue pairing13. The generation frequency decreased at the end of the PC side of the chromosome (Table 1). Heterochromatin is important for maintaining the structural integrity of the genome14. However, tmIn1(IV), tmIn2(IV) and tmIn3(IV) rearrangements were generated on regions of the chromosome arm that are known to contain heterochromatin15 (Table 1). These results suggest that our approach can generate chromosomal rearrangements at specific sites even in heterochromatic regions, whereas these rearrangements were below the generation limit mainly because of the presence of PCs.
Confirmation of the suppression of recombination in tmIn3(IV)
We examined whether tmIn3(IV) could balance a recessive lethal mutation within the inversion interval, as described in Supplementary Fig. 5a. Heterozygous tmIn3(IV) hermaphrodites were mated with heterozygous males carrying a recessive lethal lin-1 mutation (tm5929). After the self-fertilization of F1 worms, the balanced strain lin-1/tmIn3(IV) segregated three phenotypes: WT (lin-1/tmIn3 heterozygotes), lethal (lin-1 homozygotes) and larval arrest (tmIn3 homozygotes) (Supplementary Fig. 5b–g). Thus, the new balancer is a useful tool for maintaining lethal mutations on the left arm of chromosome IV. The segregation of these phenotypes was maintained through more than 20 generations, suggesting that tmIn3 suppresses further recombination of the covered genomic region.
Whole-genome sequence analysis of tmIn3(IV)
To further assess whether the generated balancer strains exhibited rearrangements at unexpected loci, we extracted the genomic DNA from the lin-1(tm5929)/tmIn3(IV) worms and analysed it by whole-genome sequencing (Supplementary Fig. 6, Supplementary Table 1). We observed several structural variants consistent with target-specific rearrangements but no target-independent rearrangements. From these results, we conclude that our methodology can accurately induce target-specific rearrangements.
Isolation of an inversion balancer on chromosome II
Likewise, we sought to generate a chromosomal rearrangement on the left arm of chromosome II, which includes the second-largest region of the C. elegans genome that is not covered by known balancers (Supplementary Fig. 1). We obtained a new balancer named tmIn4(II), which covered 8.6 cM and extensively maintained recessive lethal mutations (Supplementary Figs 7 and 8, Supplementary Table 2). Thus far, we have not been able to generate a balancer near the PCs on chromosome II (Supplementary Fig. 7e, Supplementary Table 2). The results from chromosome IV also indicate that successful chromosomal rearrangements are mainly affected by the distance from the PC region (Fig. 1h).
Genetic engineering of crossover-suppressor and translocation balancers
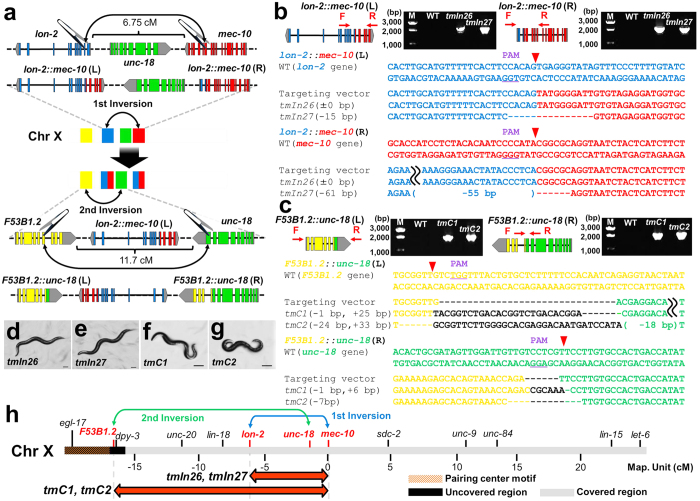
In addition to the generation of inversions, we also engineered two crossover-suppressors, tmC1(X) and tmC2(X), composed of sequential inversions between lon-2 and mec-10, and between F53B1.2 and unc-18 (Fig. 2, Supplementary Table 3). This strategy was used to produce more stable balancers because multiple inversions more effectively prevent recombination6. The crossover suppressors tmC1(X) and tmC2(X) covered most of the left arm of the X chromosome from F53B1.2 to mec-10, covering 17 cM (Fig. 2h).
Figure 2. Genetic engineering of crossover-suppressors by using the CRISPR/Cas9 system.
(a) Schematic of a crossover-suppressor. The Crossover-suppressor was created by the multiple inversions. (b) PCR amplification of the breakpoint junctions in wild-type (WT), tmIn26 and tmIn27 animals. Breakpoint sequence alignments of the targeting vectors and tmIn26 and tmIn27 rearrangements. Black bars indicate the predicted cleavage sites. (c) PCR detection of the breakpoint junctions in WT, tmC1 and tmC2 animals. Breakpoint sequence alignments of targeting vectors and tmC1 and tmC2 rearrangements. (d) The relative positions of breakpoints in the X chromosomal balancers. (e,f) tmIn26 and tmIn27 showed a recessive long phenotype. (g,h) tmC1 and tmC2 showed a recessive uncoordinated phenotype. Scale bars represent 100 μm.
Next, to examine whether our method was also effective between different chromosomes, we generated a chromosomal translocation named tmT3(III;IV) that arose between pal-1(III) and unc-17(IV) (Supplementary Fig. 9). There were no differences in the efficiency of generating inversions or translocations (Table 1, Supplementary Fig. 9d; Fisher’s test, P > 0.05). Thus, the results demonstrate that these experimental methodologies successfully provided a systematic approach to target chromosomal rearrangements at specific sites. Because the DNA repair pathways are highly conserved across species, our methodology may provide a universal approach for engineering chromosomal rearrangements.
Generation of transgene-integrated strains by using the CRISPR/Cas9 system
To facilitate balancer chromosome usage, we developed a technique using the CRISPR/Cas9 system that produced multi-copy fluorescent gene integration in tmC1(X) from extrachromosomal arrays16 (Supplementary Fig. 10a). We first generated an extrachromosomal line tmC1;tmEx4487 that expressed Pmyo-2::Venus together with Punc-18::unc-18 (which rescues unc-18 disruption) and a dpy-3 genome sequence as the sgRNA target. We co-injected the dpy-3 genome sequence-specific sgRNA with a Cas9 expression vector and a Pdpy-7::DsRed transgene marker into the gonads of tmC1;tmEx4487 worms. We isolated F1 worms with Venus and DsRed expression and screened F2 progeny for dumpy (Dpy) phenotypes and Venus expression. The breakpoints were examined by PCR amplification (Supplementary Fig. 10b). Venus fluorescent Dpy worms that carried the balancer chromosome and harboured the Pmyo-2::Venus transgene were isolated as tmC1[F53B1.2 lon-2 unc-18 mec-10 Pmyo-2::Venus Punc-18::unc-18] (Supplementary Fig. 10c,d).
Examination of the repair mechanisms that generate rearrangements
During the course of isolation of genetic balancers, we found that only a portion of the phenotype-positive lines yielded PCR-positive alleles; in the case of tmIn1, only 6 of the 40 phenotype-positive lines were PCR-positive (Table 1). This finding implies that DSBs are often repaired without inversion or with structural changes that are unable to be amplified by PCR, thus suggesting that breakpoints are often repaired by a mechanism other than HR. To determine whether the targeting vectors and lig-4 mutant background were truly necessary, we injected the genome-editing and marker plasmids without targeting vectors in the gonads of WT and lig-4 worms. In the absence of the targeting vectors, we still obtained inversions (in 0.33% of WT and 0.13% of lig-4(tm750) offspring), but the probability was decreased compared with that observed in the lig-4(tm750) background injected with targeting vectors (0.73%). Recent studies have shown that the CRISPR/Cas9-induced DSB repair of germ cells in C. elegans is often mediated by polymerase theta-mediated end-joining (TMEJ)17. These observations suggest that in the lig-4(tm750) mutant background, the targeting vectors may be required for HR, but TMEJ may also induce rearrangements. A previous report has also identified the generation of inversions that depended on the LIG4 gene in human cells without repair templates18, suggesting that NHEJ may also be involved in the process.
Upon closer inspection of the repaired regions in the rearrangements in 9 strains obtained from the lig-4 mutant background by using targeting vectors, we found that only one strain (tmIn26) contained complete copies of the targeting vector sequences at both breakpoints (Supplementary Table 4). Another strain tmT3 contained one complete copy at a breakpoint but contained an indel at another breakpoint. The other 7 strains had indels at both breakpoints. Thus, of 18 breakpoints, 3 appeared to be repaired by HR, whereas 15 were repaired by TMEJ. This phenomenon suggests that each breakpoint is repaired by either system stochastically.
In the case of 4 inversion strains without targeting vectors, we found that all the breakpoints contained some indels of the genome sequence (Supplementary Table 4 and Supplementary Fig. 11). This result suggests that TMEJ (for these 4 strains) or NHEJ (except for the case of tmIn45), might be used to repair the breakpoints.
It should also be noted that we were unable to obtain any inversions in the wild-type background by using targeting vectors (0/755 F1 animals). Although it is expected that all three repair mechanisms (HR using targeting vectors, NHEJ and TMEJ) can repair breakpoints, we could not find any evidence for successful rearrangements among the 102 phenotype-positive candidates (Table 1). The probability of successful rearrangements appeared even lower than that in the wild-type background without targeting vectors. Although the mechanisms for this phenomenon remain unclear, we speculate that the introduction of targeting vectors could mobilize NHEJ, thus quickly resulting in the repair of breakpoints without inversion19,20.
Conclusion
In summary, our strategy systematically generated chromosomal inversion, translocation and crossover-suppressor balancers at specific sites. These new balancers covered 8% of the C. elegans genome, remaining 7% of the 15% of the genome that was previously uncovered by balancers. It should be noted that our crossover-suppressor lines containing a fluorescent marker are ideal for the analysis of lethal mutations. Many of the common balancer lines used by the C. elegans research community have translocations and thus suffer from aneuploidy, which is inconvenient for phenotypic analyses6. In contrast, inversion and crossover-suppressor balancer lines have structural variations within their own chromosomes, are free from aneuploidy and are more straightforward to use for the examination of mutant phenotypes. Unfortunately, the crossover-suppressors used to date in the field have complex chromosomal structural changes. Our strategy using CRISPR/Cas9 resulted in minimal additional chromosomal changes. Our crossover-suppressors with double inversions covered a larger genomic region than did simple inversion balancers. Finally, we were able to introduce locus-specific fluorescent markers into these crossover-suppressor lines16.
Methods
Nematode strains
Caenorhabditis elegans wild-type strain Bristol N2 was used in this study. Lines carrying lig-4(tm750), lin-1(tm5929), and mlt-7(tm1794) mutations were obtained previously21. Nematodes were grown by using standard genetic protocols22.
Plasmid construction
We used site-directed mutagenesis to insert the guide sequences into a Peft-3::Cas9 + sgRNA dual expression vector (pDD162, Addgene plasmid 47549, Cambridge, MA). We designed G(N)19–25NGG specific sgRNA sequences as previously described23 (Supplementary Table 5). The sgRNA sequences were designed to target the exons of genes with easily identifiable loss-of-function phenotypes, such as uncoordinated (Unc), dumpy (Dpy), long (Lon), or lethal (Let). The Cas9-sgRNA plasmids were made by using a Clontech In-Fusion PCR Cloning Kit (Clontech Laboratories, Palo Alto, CA) as previously described24.
Targeting vectors were constructed by inserting 2 kb of homologous sequences for each target site into pBluescript KS(+) by using a Clontech In-Fusion PCR Cloning Kit (Clontech Laboratories) as previously described24 (Supplementary Table 5). We designed targeting vectors to join two DNA sequences so that junction is the centre of predicted cleavage sites which are located within 3 bp of PAM (promoter adjacent motif) sequences25. For example, in the case for the tmIn1, the left targeting vector contained a chimeric fusion of 1 kb upstream sequence from the putative cleavage site in the egl-4 gene and the reverse-directed 1 kb upstream sequence of the unc-17 gene from the predicted cleavage site. The right targeting vector is composed of a chimeric fusion of the reverse-directed 1 kb downstream sequence from the putative cleavage site in the egl-4 gene and 1 kb downstream sequence of the unc-17 gene from the predicted cleavage site. These target vectors used together, can cause a inversion.
A Cas9 integration-site dpy-3 genome fragment containing approximately 500 bp of sequence homologous to the target site was inserted into pPD95.79, using EcoRI and BamHI sites as previously described16. Plasmids for the transgenic markers Pmyo-2::Venus and Pdpy-7::DsRed were generated as previously described26.
DNA microinjection
Plasmids were prepared for injection using Qiagen’s Midi Plasmid Purification Kit (QIAGEN, Hilden, Germany). The targeting vectors were linearized from purified plasmids by PCR amplification and were purified using Illustra GFX PCR DNA and a Gel Band Purification Kit (GE Healthcare, Little Chalfont, UK). To generate new balancers, the following concentrations of injection mix were used: 45 ng/μl Cas9-sgRNA #1 dual expression vector, 45 ng/μl Cas9-sgRNA #2 dual expression vector, 40 ng/μl targeting vector (left side), 40 ng/μl targeting vector (right side) and 30 ng/μl Pmyo-2::Venus transgene marker. To generate the integrated strain, the following concentrations of injection mix were used: 100 ng/μl Cas9-sgRNA dual expression vector and 40 ng/μl Pdpy-7::DsRed transgene marker. The injection mix was centrifuged for 3 min at 15,000 × g at 4 °C in Ultrafree-MC filter devices (Millipore, Massachusetts, MA). The injection mix was injected into the germ lines of adult hermaphrodite worms by using standard methods as previously described26. Importantly, the total Cas9-sgRNA plasmid concentration of the injection mix should be lower than 100 ng/μl. When the Cas9-sgRNA concentration exceeded 100 ng/μl, the F1 progeny were sterile.
Screening for the generation of new balancers using the CRISPR/Cas9 system
To screen for new genetic balancers, injected P0 worms were grown on NGM plates at 20 °C for three days. We picked fluorescent F1 worms to individual plates at 20 °C (for example, Table 1 F1 worms). First screening: after three days, we selected plates which contained phenotype-positive F2 worms (for example, Table 1 phenotype-positive worms). By this way, we chose the F1 worms whose genome was cut by Cas9 at the target sites. We then picked F1 worms to lysis buffer (500 μg/ml proteinase K, 100 mM NaCl, 50 mM Tris, 20 mM EDTA, and 1% SDS) and confirmed by nested-PCR amplification with primers (Supplementary Table 5), whose sequences are not included in the targeting vectors (for example, Table 1 F1 PCR). Second screening: To examine whether the rearrangements occurred in the germline of the animals and they were heritable, we then picked F2 animals and performed the same PCR as above (for example, Table 1 F2 PCR). We isolated positive bands and determined and aligned the sequences of both breakpoints. After we identified strains with heritable rearrangements, we singled their F2 progeny to individual plates at 20 °C and cultured them for three days and confirmed the presence of the phenotype-positive F3 in the plates.
Test for balancer chromosome
The tmIn3(IV) rearrangement was chosen to examine whether it could balance a recessive lethal mutation. Heterozygous tmIn3/+ hermaphrodites were mated with heterozygous lin-1/+ males. The F1 progeny from each cross plate were transferred to individual plates at 20 °C for three days. After self-fertilization, the lin-1/tmIn3 hermaphrodites produced offspring that segregated into three genotypes, lin-1/tmIn3, tmIn3/tmIn3 and lin-1/lin-1, and were distinguishable according to their phenotypes.
The tmIn4(II) rearrangement was also examined to determine whether it could balance a recessive lethal mutation. Heterozygous tmIn4/+ hermaphrodites were mated with heterozygous mlt-7/+ males. The F1 progeny from each cross plate were transferred to individual plates at 20 °C for three days. After self-fertilization, mlt-7/tmIn4 hermaphrodites produced offspring segregating into three genotypes, mlt-7/tmIn4, tmIn4/tmIn4 and mlt-7/mlt-7, which were distinguishable according to their phenotypes.
Whole-genome sequencing
Genomic DNA was extracted from starved worms. Fragmentation of the genome into approximately 140 bp segments and preparation of genomic libraries were performed using automated Library Builder system (Thermo Fisher Scientific). Then, sequence templates were synthesised from the prepared libraries using the Ion Chef system, and the templates were sequenced by Ion Proton (Thermo Fisher Scientific, Massachusetts, MA) according to standard protocols (https://ioncommunity.thermofisher.com/docs/DOC-8775).
Detection of structural variants
Raw sequencing reads were primarily mapped to the reference sequence by using TMAP software (https://github.com/iontorrent/TMAP). The reference sequence was prepared by adding sequences of Pmyo-2::Venus, pDD162 (Addgene plasmid 47549) and pBlueScript II KS(+) to the C. elegans genome sequence (ftp://ftp.wormbase.org/pub/wormbase/species/c_elegans/sequence/genomic). After primary mapping, we calculated the mean value for all read lengths. The product of the average read length times the number of reads was divided by the length of the reference sequence. The result was defined as the coverage (Supplementary Table 1). Then, the genomic rearrangements were detected by following processes.
From the primary mapping results, we obtained clipped reads, which contained both mapped and unmapped sequences (Supplemental Fig. 6a, solid and broken lines). We selected unmapped sequences that were longer than 20 bp and extracted all the continuous 16-base sequences from the unmapped reads and their complementary sequences as queries for the following realignment. From the reference sequence, the regions that perfectly matched the queries were searched by using the Aho-Corasick algorithm27. Then, the unmapped sequences were compared and aligned to the neighbouring sequences of each matched regions using the Smith–Waterman algorithm28. For the algorithm, the values used for matching, mismatching and gap score were +2, −1 and −2, respectively. Through this alignment, we detected the most homologous regions for each unmapped sequence. If there was more than one candidate for the most homologous region of an unmapped sequence, we selected the one that was nearest to the mapped region of the original clipped read.
As a result, we obtained split reads (SR) whose 5′- and 3′-regions were mapped to different sites of the reference29. Next, the split reads were classified into the following 5 categories: deletion-, insertion-, inversion-, translocation- and translocational inversion-type SR. When the 3′-region of an SR was aligned downstream or upstream of the site where the 5′-region of the read was mapped, the SR was defined as a deletion- or insertion-type SR. Otherwise, when the 5′-region of an SR was aligned to the reverse strand of the 3′-region of the read in the same linkage group, the SR was defined as an inversion-type SR. If the 5′- and 3′-regions of an SR were aligned to different linkage groups, the read was defined as a translocation-type SR. If an SR was determined to be both translocation and inversion-type, the SR was defined as a translocational inversion-type SR (Supplementary Fig. 6a, middle panel). After classification, we eliminated SRs that were also detected in control data. If the number of deletion-type SRs that contained a common gap between the 5′- and 3′-regions was greater than 2, we defined the region as a deletion candidate. Additionally, we investigated the combination of two types of SRs to detect complicated variant candidates. When combined deletion- and insertion-type SRs were located on both sides of a region, the region was defined as an insertion candidate. Similarly, when two inversion-type SRs were located on both sides of a region, the region was defined as a local inversion or inverted insertion candidate. The translocated insertion and inverted translocational insertion candidates were also defined using two translocation- and translocational inversion-type SRs. If there were gaps near the border of a variant region, the variant was also defined as a deletion candidate (Supplementary Fig. 6a, lower panel). To improve the reliability, complicated variants were removed when fewer than ten reads contained common variant regions.
We also counted the number of reads covering each base of the reference sequence as the depth of sequence. Regions in which the depth values were greater than one were defined as mapped regions (Supplementary Table 1). In mapped regions, the depth values were divided by the coverage value, and the quotient was defined as the normalized depth (ND) (Supplementary Fig. 6b, left panel). Then, the ratio of the ND between the balanced strains and tmIn3 was calculated as the depth ratio (DR) value (Supplementary Fig. 6b, right panel). A low DR value meant that the copy number of the base was lower than that in the control, thus suggesting that the base was deleted in the balanced strain. Finally, we evaluated the variant candidates investigated by SR analysis using DR values. When the DR value of a deleted region was higher than 0.75, the variant was removed. Furthermore, when the DR value of insertion variants was greater than 1.75 or 2.5, the variants were defined as duplications or multiplications, respectively.
Generation of the tmC1;tmEx4487 transgenic line
To generate tmC1;tmEx4487 transgenic worms, 20 ng/μl Cas9 integration-site dpy-3 genome fragment, 160 ng/μl Pmyo-2::Venus and 20 ng/μl Punc-18::unc-18 (unc-18 rescue construct) were co-injected into tmC1 worms by using standard methods as previously described30.
Generation of integrated strains by using the CRISPR/Cas9 system
Integration of extrachromosomal arrays into a balancer line was performed as previously described16. To screen for integrated strains, we first removed the lig-4 (tm750) background, and the injected P0 tmC1;tmEx4487 worms were grown on NGM plates at 20 °C for four days. After self-fertilization, we picked F1 worms with Venus and DsRed fluorescence and transferred them to individual plates, where they were incubated at 20 °C for four days. If their F2 progeny carried integrated Pmyo-2::Venus constructs in tmC1, Dpy progeny would express Venus in the pharynx. In contrast, in F2 progeny carrying only tmEx4487, Dpy progeny would not express Venus. To confirm integration, F2 Dpy animals were transferred to individual plates and grown at 20 °C for four days. After self-fertilization, if the F3 Dpy progeny carried the desired integration, all Dpy progeny would express Venus.
Additional Information
How to cite this article: Iwata, S. et al. Engineering new balancer chromosomes in C. elegans via CRISPR/Cas9. Sci. Rep. 6, 33840; doi: 10.1038/srep33840 (2016).
Supplementary Material
Acknowledgments
We thank our laboratory members for their helpful discussions and suggestions.
Footnotes
Author Contributions S.I. designed and performed the experiments, and drafted the manuscript. S.Y. participated in the design of the studies. Y.S. performed the computational analysis of whole-genome sequence. S.Y., Y.S. and S.H. helped to write the manuscript. S.M. conceived and coordinated the studies, and corrected the manuscript.
References
- Herman R. K. Crossover suppressors and balanced recessive lethal in Caenorhabditis elegans. Genetics 88, 49–65 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth R. E. & Baillie D. L. The genetic analysis of a reciprocal translocation, eT1 (III; V), in Caenorhabditis elegans. Genetics 99, 415–428 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor A. & Deàk P. The isolation and genetic analysis of a Caenorhabditis elegans translocation (szT1) strain bearing an X-chromosome balancer. J Genet 64, 143–157 (1985). [Google Scholar]
- Zetka M. C. & Rose A. M. The meiotic behavior of an inversion in Caenorhabditis elegans. Genetics 131, 321–332 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley M. & Riddle D. LG II balancer chromosomes in Caenorhabditis elegans: mT1 (II; III) and the mIn1 set of dominantly and recessively marked inversions. Mol Genet Genomics 266, 385–395 (2001). [DOI] [PubMed] [Google Scholar]
- Edgley M. K., Baillie D. L. & Rose A. M. Genetic balancers. The C. elegans Research Community, WormBook WormBook, doi: /10.1895/wormbook.1.89.1, http://www.wormbook.org (Apr 6, 2006) (2006). [DOI] [PMC free article] [PubMed]
- Sander J. D. & Joung J. K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32, 347–355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P. S. & Matthew M. Targeted genomic rearrangements using CRISPR/Cas technology. Nature Commun 5, 3728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco R. B. et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell reports 9, 1219–1227 (2014). [DOI] [PubMed] [Google Scholar]
- Maddalo D. et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516, 423–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao An. et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res 41, e141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. D. Rapid and precise engineering of the C. elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199, 363–377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M. et al. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat Cell Biol 11, 934–942 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J. C. & Gary H. K. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet 5, e1000435 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res 21, 227–236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshina S., Suehiro Y., Kage-Nakadai E. & Mitani S. Locus-specific integration of extrachromosomal transgenes in C. elegans using the CRISPR/Cas9 system. Biochem Biophys Rep 5, 70–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schendel R. et al. Polymerase Theta is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nat Commun 6, 7394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biol 16, 111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarz A. et al. Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am J Cancer Res 2, 249–268 (2012). [PMC free article] [PubMed] [Google Scholar]
- Abdisalaam S. et al. Scanning fluorescence correlation spectroscopy techniques to quantify the kinetics of DNA double strand break repair proteins after γ-irradiation and bleomycin treatment Nucleic Acids Res 42, e5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K. & Mitani S. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem Biophys Res Commun 269, 64–69 (2000). [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E. et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10, 741–743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Cai G., Hall E. O. & Freeman G. J. In-FusionTM assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques 43, 354–359 (2007). [DOI] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A. & Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K., Yoshina S., Inoue H. & Mitani S. An efficient transgenic system by TA cloning vectors and RNAi for C. elegans. Biochem Biophys Res Commun 349, 1345–1350 (2006). [DOI] [PubMed] [Google Scholar]
- Aho A. V. & Corasick M. J. Efficient string matching: an aid to bibliographic search. Commun. ACM. 18, 333–340 (1975). [Google Scholar]
- Smith T. F. & Waterman M. S. Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197 (1981). [DOI] [PubMed] [Google Scholar]
- Tattini L., D’Aurizio R. & Magi A. Detection of genomic structural variants from next-generation sequencing data. Front. Bioeng. Biotechnol. 3, 92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. & Fire A. DNA transformation. Methods Cell Biol 48, 451–482 (1995). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.