Abstract
The ecophysiology of one candidate methanogen class WSA2 (or Arc I) remains largely uncharacterized, despite the long history of research on Euryarchaeota methanogenesis. To expand our understanding of methanogen diversity and evolution, we metagenomically recover eight draft genomes for four WSA2 populations. Taxonomic analyses indicate that WSA2 is a distinct class from other Euryarchaeota. None of genomes harbor pathways for CO2-reducing and aceticlastic methanogenesis, but all possess H2 and CO oxidation and energy conservation through H2-oxidizing electron confurcation and internal H2 cycling. As the only discernible methanogenic outlet, they consistently encode a methylated thiol coenzyme M methyltransferase. Although incomplete, all draft genomes point to the proposition that WSA2 is the first discovered methanogen restricted to methanogenesis through methylated thiol reduction. In addition, the genomes lack pathways for carbon fixation, nitrogen fixation and biosynthesis of many amino acids. Acetate, malonate and propionate may serve as carbon sources. Using methylated thiol reduction, WSA2 may not only bridge the carbon and sulfur cycles in eutrophic methanogenic environments, but also potentially compete with CO2-reducing methanogens and even sulfate reducers. These findings reveal a remarkably unique methanogen ‘Candidatus Methanofastidiosum methylthiophilus' as the first insight into the sixth class of methanogens ‘Candidatus Methanofastidiosa'.
Introduction
As an essential component of the global carbon cycle, methanogenic archaea produce approximately one billion tons of CH4 per year (Thauer et al., 2008). These ‘methanogens', uniquely affiliated to the archaeal phylum Euryarchaeota, facilitate anaerobic conversion of H2, formate, acetate, methyl compounds and simple alcohols to CH4 and CO2. In a variety of anaerobic ecosystems, including both natural and engineered, fermenters, syntrophs and acetogens, metabolically interact with methanogens to accomplish holistic mineralization of organic compounds (Nobu et al., 2015). Methanogens also date back to 3.5 billion years ago based on isotopic signatures of Paleoarchean methane bubbles locked in the Dresser formation (Ueno et al., 2006). Thus, researchers have made continuous efforts to characterize these ecologically, evolutionarily and metabolically important organisms using cultivation-, biochemistry- and genomics-based tools.
Recent cultivation- and metagenomics-based studies on Euryarchaeota methanogens discover novel methanogenic lineages, including permafrost-associated ‘Ca. Methanoflorentaceae' (also known as rice cluster II; Mondav et al., 2014) and gut-associated Thermoplasmata order Methanomassiliicoccales (formerly ‘Ca. Methanoplasmatales' Dridi et al., 2012, Paul et al., 2012; Iino et al., 2013). However, one potentially class-level Euryarchaeota clade thought to be capable of methanogenesis, WSA2 (or Arc 1), (Hugenholtz 2002; Chouari et al., 2005) remains uncharacterized, despite the identification through 16S ribosomal RNA (rRNA) sequencing more than 15 years ago (Dojka et al., 1998). Moreover, members of this clade have been observed in a wide range of natural and engineered environments (for example, freshwater and marine sediments, contaminated groundwater and bioreactors; Dhillon et al., 2005; Cheng et al., 2012; Saito et al., 2015; Wilkins et al., 2015). To fill this gap in our understanding of methanogen phylogeny and WSA2's potential roles in anaerobic biogeochemical cycles, we construct the first WSA2 genomes through metagenomics of methanogenic bioreactors treating wastewater. Using these genomes, we characterize the physiology and methanogenic potential of the sixth class of methanogens and ninth class of Euryarchaeota, ‘Candidatus Methanofastidiosa.'
Materials and methods
Metagenome sequencing, assembly and binning
We collected samples from three anaerobic wastewater treatment samples: one full-scale anaerobic digester (reactor ADurb) and full- and lab-scale bioreactors treating wastewater from purified terephthalate process (Bfssc and U1lsi, respectively). For ADurb and U1lsi, three samples were taken at different time points, October, November and December 2013 and February, April and May 2014, correspondingly. For Bfssc, samples were taken through sampling ports at different depths of the reactor along the sludge bed. Metagenomic DNA was extracted using FastDNA SPIN Kit for Soil kit (MP Biomedicals, Santa Ana, CA, USA). Sequencing libraries were prepared with Kapa Library Preparation kit (Kapa Biosystems, Wilmington, MA, USA) with a genomic DNA fragment size ranging between 300 and 750 bp. These libraries were sequenced on HiSeq2500 with TruSeq SBS Rapid Sequencing kit (Illumina, San Diego, CA, USA), generating paired-end reads up to 165 bp each. The generated reads were trimmed using Trimmomatic v0.30 with a quality cutoff of 30, sliding window of 6 bp and minimum length cutoff of 75 bp (Bolger et al., 2014); digitally normalized and partition using the khmer package (Brown et al., 2012; Pell et al., 2012); and assembled using SPAdes v.3.5.0 (Bankevich et al., 2012). The assembled contigs from three metagenomes corresponding to the same reactor were binned comparatively using MaxBin2.0 (Wu et al., 2014). Genes were then predicted using Prodigal v2.5 (Hyatt et al., 2010) and annotated using Prokka (Seemann, 2014). We manually curated these bins and their annotations as described in our previous metagenomic study (Nobu et al., 2015). For interpretation of physiology and metabolism, WSA2 draft genomes recovered from different samples of the same environment (that is, reactor ADurb time points or Bfssc depths) were collectively analyzed as a pangenome.
Phylogenetic analysis
Genes annotated as small subunit 16S rRNA gene, methyl coenzyme M reductase alpha subunit (McrA) and methylated thiol coenzyme M methyltransferase (Mts) were extracted for phylogenetic analysis. The 16S rRNA genes were aligned against the Greengenes and Silva v123 databases using PYNAST and SINA, respectively (Caporaso et al., 2010; McDonald et al., 2012; Pruesse et al., 2012; Quast et al., 2013). Phylogenetic trees for 16S rRNA genes were constructed in ARB using the neighbor-joining algorithm with 5000 bootstrap replications (Ludwig et al., 2004). The mcrA and mts genes from WSA2 and publically available methanogen genomes were aligned and put into a phylogenetic tree using ClustalW in the MEGA package (Tamura et al., 2007). A representative genome for each Euryarchaeota genus was used to construct a phylogenomic tree including WSA2 genomes using PhyloPhlAn (Segata et al., 2013). To further compare the phylogenetic relationship of these Euryarchaeota, core genomes were predicted for each genus and family with more than one genome publically available by identifying genes with >50% amino acid similarity and >80% alignment coverage across all genomes of that clade using the BLAST+ package (Camacho et al., 2009), based on criteria used in a core genome prediction software (Miele et al., 2011). Core genome overlap between genera and families were determined using the same criteria.
Results and Discussion
Phylogeny of WSA2 based on 16S rRNA, McrA and phylogenomics
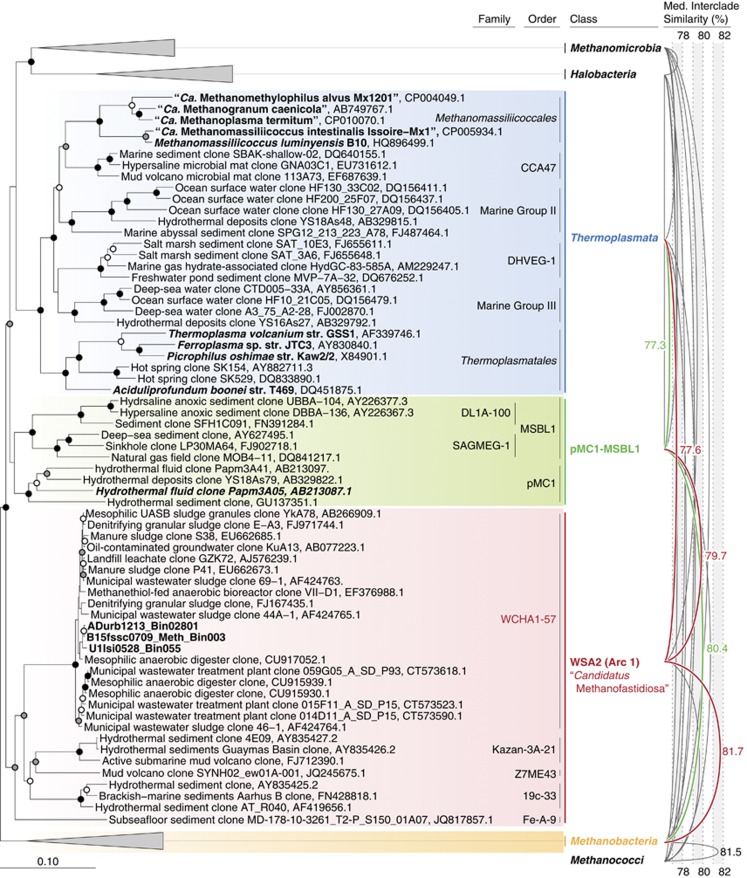
Metagenomics collectively generated genomes for four WSA2 populations from a full-scale anaerobic digester treating a mixture of waste activated sludge and primary sludge in Urbana, IL, USA (population ADurb1213_Bin02801) and lab-scale (U1lsi0214_Bin055 and U1lsi0214_Bin089) and full-scale (B15fssc0709_Meth_Bin003) methanogenic bioreactors treating purified terephthalate wastewater (Supplementary Table 1). For the Urbana anaerobic digester, we sequenced metagenomes in 1-month intervals over 3 months. As for the full-scale reactor, microbial community samples were taken at separate sampling ports along the depth of the reactor sludge bed. Only one high-quality bin was recovered for each WSA2 population identified in the lab-scale reactor. In total, the eight draft genomes reconstructed from seven distinct sequencing efforts range between 1.50 and 1.99 Mb in size and 32.9–35.4% G+C content with estimated completeness of 85.0–92.5% based on 40 universal marker genes (Ciccarelli et al., 2006; Sorek et al., 2007). Draft genomes from the same environment (that is, digester and full-scale reactor) with at least 85% overlap of predicted genes with a 99% similarity cutoff were combined, generating population pangenomes for populations ADurb1213_Bin02801 and B15fssc0709_Meth_Bin003. 16S rRNA gene classification, McrA phylogeny and phylogenomics clearly indicate that these organisms belong to the uncharacterized Euryarchaeota clade WSA2 falling between super-classes Methanomicrobia–Halobacteria–Thermoplasmata and Methanobacteria–Methanococci (Figure 1a and Supplementary Figure 1 and 2). WSA2 is distinct from related Euryarchaeota classes and uncultivated clades based on phylogenetic tree topology with high-bootstrap support, regardless of the reference 16S rRNA alignments used (Greengenes 2013 and Silva v123). Moreover, the median sequence similarities of WSA2 to neighboring classes (76.63–81.7%) are lower than or comparable to the similarity between Methanobacteria and Methanococci (81.5%), confirming that WSA2 is indeed a class-level clade in Euryarchaeota (Figure 1). Based on tree topology and this median sequence similarity criterion, uncultivated Euryarchaeota clades pMC1 and MSBL1 may form another class (tentatively termed pMC1-MSBL1) distinct from WSA2 with phylogenetic radiation between WSA2 and superclass Methanomicrobia–Halobacteria–Thermoplasmata.
Figure 1.
16S rRNA-based phylogeny of WSA2 (red) compared with closely related Euryarchaeota classes and clusters (pMC1 and MSBL1). The tree was constructed using ARB neighbor-joining algorithm and GreenGenes 16S rRNA gene database with 5000 bootstrap replications, sequences at least 1200 bp in length, and Methanobacteria as the outgroup. Bootstrap values >90% (black), 75% (gray) and 50% (white) are indicated. The median sequence similarity of the Euryarchaeota classes and clusters (WSA2, pMC1 and MSBL1) are shown as arcs connecting the cluster labels with horizontal heights indicating similarity (right).
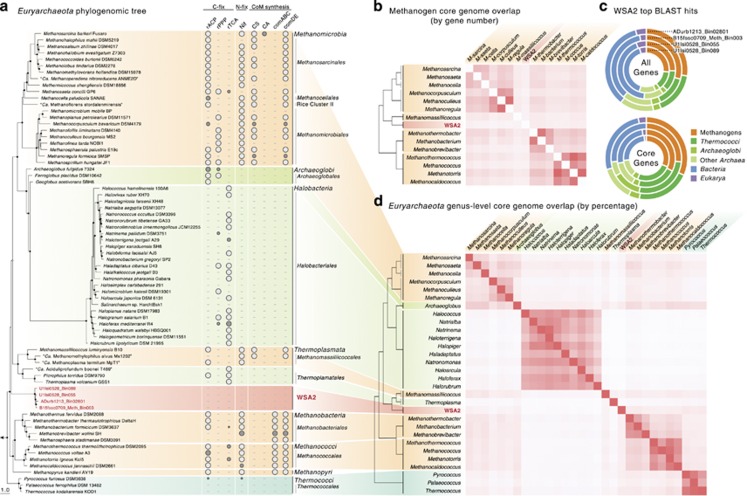
Similarly, genome content also suggests distant relationship with known methanogens and other Euryarchaeota as only 25–28% and 50–58% of genes in each WSA2 genome, respectively, relate to methanogens and Archaea based on top blastp hits (Figure 2c). The remaining Archaea-related genes hit Thermococci (12–16%), Archaeoglobi (3–4%) and other clades outside of Euryarchaeota (11–13%). A notable amount of genes relate to bacterial clades (28–35%) as also previously observed for other Euryarchaeota (Ruepp et al., 2000). However, these WSA2 genes have low average amino acid similarity to genes of known organisms (44–49% Supplementary Figure 3). The core genome of all four WSA2 genomes (defined by >80% amino acid similarity conserved between all genomes) contains 842 genes, which has a similar phylogenetic gene distribution (Figure 2c).
Figure 2.
(a) Euryarchaeota phylogenomics tree of representative genera with Crenarchaeota as the outgroup. Nodes are marked for bootstrap values >75% (filled) and 50% (open). For each genus, one representative species is shown on the tree. Corresponding to each genus, the presence of the gene/pathway in all (white) or only some (gray, size proportional to percentage) of the genomes of that genus are shown. Genes/pathways are shown for carbon fixation (reductive acetyl-CoA pathway—rACP; reductive pentose phosphate pathway—rPPP; and reductive tricarboxylic acid cycle—rTCA), nitrogen fixation (Nif), type I coenzyme M (CoM) synthesis (comABC), type II CoM synthesis (cysteate synthase—CS; cysteate aminotransferase—CA), and final CoM synthesis step (sulfopyruvate decarboxylase—comDE). (b) The overlap of core genomes between methanogenic genera, including WSA2. The intensity of the red color corresponds to the number of overlapping genes. (c) The phylogenetic distribution of WSA2 genes in the entire genome and only core genomes. All four genomes are shown as individual rings. (d) The overlap of core genomes between all Euryarchaeota genera. The intensity of the red color corresponds to the percent overlap of core genes in reference to the genome on the vertical axis.
To further define WSA2's physiology and relationship with other Euryarchaeota beyond this taxonomic analysis, we compare the core genomes of each clade (at genus and class level). Genera of similar phylogeny have pronounced core genome overlap; however, the WSA2 core genome has low overlap with both methanogenic (6–15%) and non-methanogenic (2–15%) Euryarchaeota genera (Figure 2d). Moreover, even though distantly related methanogenic genera in Methanomicrobia, Methanobacteria and Methanococci share many core genes (152–257 genes average), WSA2 shares much fewer genes with these methanogens (69–111 genes average; Figure 2b). The genes shared between WSA2 with Euryarchaeota genera primarily consist of housekeeping genes, including ribosomal proteins, biosynthesis genes and Archaea-specific genes (for example, histone, thermosome and ATP synthase). WSA2 and methanogenic clades all share genes encoding methanogenesis marker proteins and Mcr, responsible for the terminal step of methanogenesis (Supplementary Table 2). Interestingly, WSA2 shares the methylmalonyl-CoA pathway with non-methanogenic genera, which no known methanogen genomes harbor. These findings of Mcr, unique genetic composition and unusual methylmalonyl-CoA pathway suggest that WSA2 may perform methanogenesis and yet have distinct capabilities from typical methanogens.
Uniquely restricted methanogenic metabolism
Even though the WSA2 genomes harbor Mcr, all eight draft genomes from the seven independent-sequencing efforts lack conventional CO2 reduction to CH4 and acetyl-CoA synthase pathway, indicating that neither CO2 nor acetate can serve as substrates. Moreover, while many methanogens missing these pathways respire methanol and methylamines (for example, Methanomethylovorans and Methanosphaera), we could not identify corresponding methyltransferases essential for such metabolism in any of the WSA2 draft genomes. Strikingly, the WSA2 draft genomes consistently encode methylated thiol Mts homologs for funneling C1 compounds into the reductive arm of methanogenesis (Supplementary Table 2). Thus, WSA2 may only be capable of demethylation of methylated thiols for CH4 generation, which would be the first example of such restricted methanogenic catabolism. Moreover, the lack of the methanogenic C1 pathway indicates that WSA2 likely use methylated thiols as electron acceptors rather than donor. Methanol and methylated amine metabolism are observed in methanogens associated with sediments or gut environments as plants produce methanol and methylated amines derive from eukaryotic lipids and osmoregulators; thus, it may be logical that the studied anaerobic digester WSA2 populations lack such metabolism. Perhaps other WSA2 class members associated with marine environments or sediments may possess catabolic capacity beyond methylated thiols.
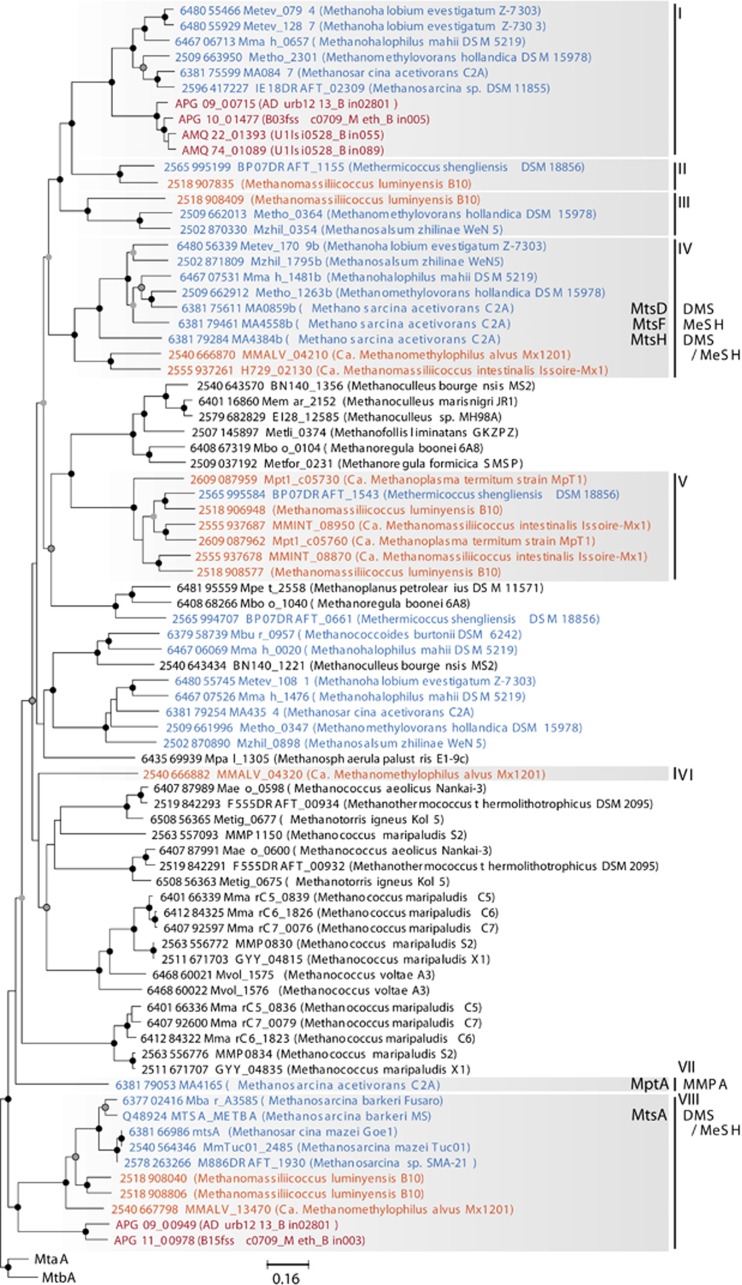
Mts substantially vary in specificity to methylated thiol substrates, such as methanethiol (MeSH), dimethylsulfide (DMS), 3-methylmercaptopropionate (MMPA) and 3-mercaptopropionate (MPA; Tallant and Krzycki, 1997; Fu and Metcalf, 2015). Thus, accurate annotation of WSA2's methanogenic capacity necessitates systemic determination of the specific function of the two Mts homologs conserved among WSA2 genomes. Phylogenetic analysis of all publically available methanogen Mts-related methyltransferase sequences reveals clusters related to (a) methyl-metabolizing Methanosarcinales and Methanomassiliicoccales, (b) non-methyl-metabolizing methanogens or (c) both (Figure 3). We suspect that clusters found in non-methyl-metabolizing methanogens are not catabolic and rather serve physiological roles in methanogens. Clusters related to methyl-metabolizing methanogens were categorized into protein families, of which three have representatives with biochemical or transcriptomic characterization. A recent genetic and transcriptomic study on Methanosarcina acetivorans C2A shows evidence that MtsD, MtsF and MtsH (family IV), respectively, degrade DMS, MeSH and both and MptA (family VII) is critical for MMPA metabolism (Moran et al., 2008; Bose et al., 2009; Oelgeschläger and Rother, 2009; Fu and Metcalf, 2015). M. barkeri MtsA (family VIII) has also been demonstrated to facilitate methyl transfer from DMS, MMPA and MPA, also MeSH to a much lesser extent (LeClerc and Grahame, 1996, Tallant et al., 2001). In WSA2, we identify an Mts fusion protein encoding a methyltransferase and corrinoid protein, both of which relate to family VIII MtsA and MtsB, respectively (Supplementary Figure 4); thus, WSA2 may utilize this MtsA and Mcr to perform methanogenesis through reduction of multi-carbon methylated thiols.
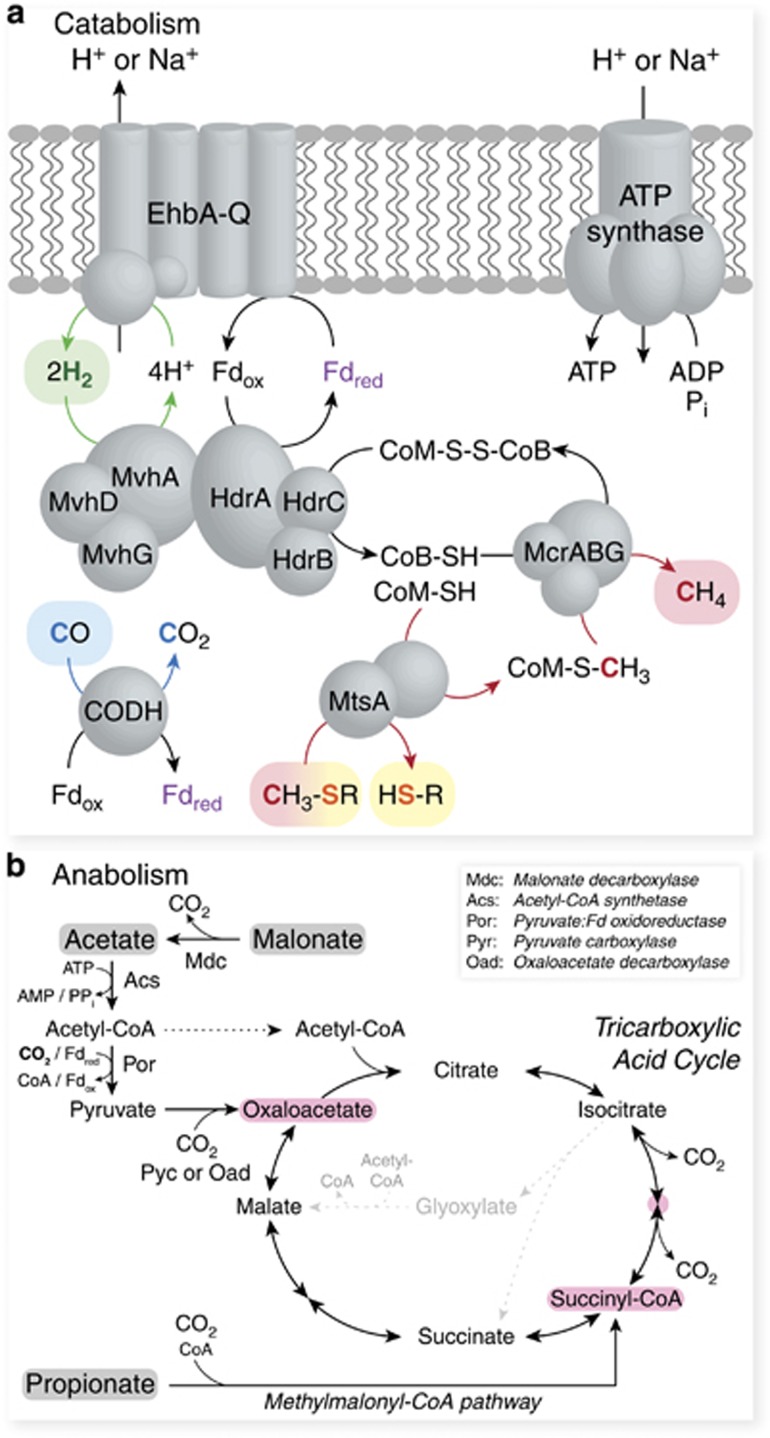
Figure 3.
WSA2 (a) catabolism and (b) anabolism. (a) WSA2 has genes for H2 oxidation through electron-bifurcating hydrogenase (HdrABC–MvhDGA) and H2 cycling by energy-converting hydrogenase (EhbA-Q); CO oxidation by carbon monoxide dehydrogenase (CODH); and methylated thiol reduction and methanogenesis by methylated thiol Coenzyme M methyltransferase corrinoid fusion protein (MtsA) and methyl coenzyme M reductase (McrABG). The proton motive force (or cation gradient) generated by Ehb can support ATP production by ATP synthase. (b) Malonate decarboxylase and acetyl-CoA synthetase can convert malonate and acetate into acetyl-CoA for downstream co-assimilation with CO2 (bolded) through pyruvate:ferredoxin oxidoreductase, pyruvate carboxylase and tricarboxylic acid (TCA) cycle. As identified for other heterotrophic methanogens, WSA2 does not encode the glyoxylate shunt for acetate assimilation (gray with dotted line). The methylmalonyl-CoA pathway can facilitate co-assimilation of propionate and CO2 also into the TCA cycle. WSA2 can use key TCA cycle intermediates (pink) as building blocks for biosynthesis.
Many uncharacterized Mts homologs fall into five other families affiliated with methyl-metabolizing methanogens (I, II, III, V and VI). Although these groups have no experimentally studied representatives, their exclusive affiliation with methyl-degrading methanogens suggests that these Mts homologs relate to methanogenic methyl metabolism. Family I is unlikely to support MeSH, DMS or MMPA metabolism as Methanosarcina acetivorans C2A does not express MA0847 during growth on these substrates (Bose et al., 2009; Fu and Metcalf, 2015). Thus, the metabolic function of family I-related WSA2 Mts-like protein (MtlA) is unclear. Interestingly, MtlA associates with a corrinoid-CoM methyltransferase (MtlB), corrinoid protein (MtlC) and methyltransferase reductive activase (MtlD; Supplementary Table 2). Notably, MtlC is phylogenetically distinct from other known Mts corrinoid proteins (Supplementary Figure 4). Although known Mts can single-handedly catalyze the two-stage methyl transfer from substrate to CoM, methanol and methylamine methyltransferases require two subunits (Tallant et al., 2001); therefore, MtlAB may have substrates and biochemical properties distinct from known Mts. However, the presence of MtlA in other methyl-degrading methanogens warrants further experimental investigation to expand our understanding of diversity in methanogenic methyl metabolism.
To complement the reductive methylated thiol metabolism, the WSA2 genomes also possess H2- and CO-oxidizing pathways for donating reducing power into methanogenesis (Supplementary Table 2). For H2 metabolism, WSA2 can perform (i) electron-bifurcating H2 oxidation using a cytosolic methylviologen-reducing hydrogenase complexed with a heterodisulfide reductase and (ii) proton-pumping H2 generation mediated by a membrane-bound energy-converting hydrogenase (Figure 4). WSA2 may pair these two pathways to accomplish energy-conserving internal H2 cycling similar to Methanosphaera stadtmaniae and Methanomassiliicoccales spp. (Thauer et al., 2008; Borrel et al., 2014) analogous to the Methanosarcina cytochrome-dependent H2 cycling (Kulkarni et al., 2009). Although WSA2 also encodes F420-reducing hydrogenase, none of the draft genomes encode other F420-oxidizing enzymes related to methanogenesis so the physiology function of F420 remains unclear. The WSA2 genomes lack genes encoding cytochromes and methanophenazine synthesis, suggesting that they perform cytochrome-independent methanogenesis like all non-Methanosarcinales methanogens (Thauer et al., 2008).
Figure 4.
Phylogeny of methylated thiol Coenzyme M methyltransferase (Mts) homologs with methanol- (MtaA) and methylamine- (MtbA) specific methlyltransferases as the outgroup. The tree was constructed using Clustalw with default parameters for alignment and neighbor-joining clustering method for tree construction with 5000 bootstrapped replications. Family classifications are assigned to clusters only containing Mts homologs from methyl-metabolizing methanogens (Methanosarcinales—blue, Methanomassiliicoccales—orange) and/or WSA2 (red and bolded). Only three families contain representatives biochemically or transcriptomically characterized in Methanosarcina acetivorans and M. barkeri (bolded). For those Mts, their predicted substrates are noted. Nodes are marked for bootstrap values >90% (black), 75% (gray with outline) and 50% (gray).
Physiology
Archaea are known to utilize a variety of autotrophic carbon fixation pathways (Berg et al., 2010), yet methanogenic Euryarchaeota specifically possess reductive acetyl-CoA pathway, pentose phosphate pathway (rPPP; or Calvin-Benson cycle), and/or tricarboxylic acid cycle (Figure 2a). Methanosarcinales, Methanocellales, ‘Ca. Methanoflorens,' Methanobacteriales, Methanococcales, and Methanopyri members encode reductive acetyl-CoA pathway; Methanomicrobiales members either possess both reductive acetyl-CoA pathway and rPPP or only rPPP; and only several phylogenetically scattered species encode reductive tricarboxylic acid cycle. Interestingly, the Methanomicrobiales members only encoding rPPP all require exogenous organic carbon for growth, suggesting that the rPPP ribulose-1,5-bisphosphate carboxylase/oxygenase in these organisms participates in AMP metabolism rather than carbon fixation (Sato et al., 2007). Likewise, WSA2 may require organic carbon sources for growth (that is, heterotrophic) as they lack any discernable carbon fixation pathway (Figure 2a). In agreement, known methanogens missing such pathways require acetate or complex exogenous nutrients (for example, yeast extract or rumen fluid) for growth (Whitman et al., 1982; Miller and Wolin, 1985; Tanner and Wolfe, 1988; Zellner et al., 1989; Miller and Lin, 2002). In addition, unlike most methanogens, the WSA2 genomes are also deficient in nitrogen fixation genes (that is, Nif) though they do encode non-nitrogen-fixing NifD- and NifH-like genes (NflDH; Figure 2a; Staples et al., 2007). Thus, the draft genomes suggest that WSA2 may be incapable of both autotrophy and nitrogen fixation, a rarity among known methanogens. This genomic analysis also revealed that several gut-associated Methanobacteriales (Methanobrevibater sp. JH1 and Methanosphaera stadtmanae DSM3091) also cannot fix carbon and nitrogen.
In order to determine the carbon source requirement for WSA2, we genomically evaluate their biosynthetic capacity. All WSA2 genomes have genes for gluconeogenesis and tricarboxylic acid (TCA) cycle, but only two genomes encode a complete PPP (Supplementary Table 3). The presence of TCA cycle and malonate decarboxylase suggests that they can utilize acetate as a carbon source and also generate acetate from malonate. They encode a pyruvate:ferredoxin oxidoreductase and pyruvate carboxylase or acetate-CoA assimilation. Similarly, heterotrophic methanogens encode pyruvate:ferredoxin oxidoreductase with either pyruvate carboxylase or oxaloacetate decarboxlyase for acetate assimilation. Unique from other methanogens, WSA2 encode methylmalonyl-CoA pathway that integrate propionate into the TCA cycle. TCA-based propionate assimilation could either split into the oxidative and reductive arms or strictly go through oxidative TCA given acetate or malonate is available as an acetyl-CoA source. Thus, WSA2 may utilize acetate, malonate or propionate with CO2 as carbon sources. However, all genomes lack complete biosynthesis pathways for glycine, homocysteince, homoserine, methionine, proline, threonine and tryptophan and some genomes lack full pathways for isoleucine, leucine, phenylalanine and tyrosine, suggesting severe auxotrophy (Supplementary Table 3). To complement this, they encode amino acid and peptide transporters.
We identify pathways for synthesis of methanogenesis cofactors: Coenzyme B, F420, F430 and cobamide (partial; Supplementary Table 4). The WSA2 genomes also consistently harbor F420 modification genes. Interestingly, they also possess F420 alpha-l-glutamate ligase (CofF) that have only been observed in Methanosarcinales and Methanococcales previously (Li et al., 2003). On the other hand, the WSA2 genomes are missing CoM synthesis pathways. Similarly, several phylogenetically scattered methanogen isolates also lack such genes. All of these methanogens are either dependent on rumen fluid for growth or strongly stimulated by CoM supplementation. Although whether WSA2 and these methanogens possess an unidentified CoM synthesis pathway is unclear, WSA2 is likely adapted to availability of exogenous CoM produced by other methanogens in situ. This suggests that WSA2 can only inhabit ecosystems with other active methanogens. As for methanogens that do encode CoM biosynthesis, Methanomicrobia and Methanomassiliicoccales employ the Methanosarcina-type pathway, while Methanobacteria, Methanococci and Methanopyri employ Methanocaldococcus type (Graupner et al., 2000; Graham et al., 2002, 2009).
Taxonomic classification, core genome comparison, unique methylated thiol-specific methanogenic metabolism and obligate heterotrophy all agree that WSA2 is a novel Euryarchaeota class with distinct features from other methanogens. Their heterotrophic, auxotrophic and ammonia-dependent nature is unique and yet appropriate for the organic-rich anaerobic environments they are often associated with (that is, wastewater treatment sludge and marine sediments) where fermentation and methanogenesis produce the required carbon sources (acetate, propionate and CO2), amino acids, ammonia (Blair and Carter, 1992; Lettinga, 1995) and possibly free CoM. Moreover, fermentative degradation of methionine and methoxylated compounds may yield sufficient DMS to support methanogenesis (Kadota and Ishida, 1972; Zinder and Brock, 1978; Finster et al., 1990; Kiene and Hines, 1995). Thus, we suspect that this novel archaeon is adapted to performing methanogenesis in eutrophic anaerobic environments and bridging the carbon and sulfur cycles. In addition, the existence of a dedicated H2-oxidizing methylated thiol reducing methanogen can significantly change our thermodynamic understanding of anaerobic ecosystems. Typical CO2-reducing methanogenesis (4H2+HCO3−→CH4+3H2O: ΔG°′=−135.56 kJ mol CH4−1) rapidly becomes thermodynamically unfavorable as H2 concentration decreases due to the high stoichiometry of input H2 to generated CH4 (4:1) in contrast with the low stoichiometry (1:1) of WSA2-like methanogenesis (for example, H2+DMS→CH4+CH3SH: ΔG°′=−161.14 kJ mol CH4−1). For example, at 10 Pa H2, CO2 reduction would only yield −18.48 kJ mol CH4−1, while DMS reduction would yied −140.04 kJ mol CH4−1 (50 kPa CH4, 100 μm thiols, 50 mm bicarbonate). In theory, WSA2-like methyl reduction could thrive under low H2 concentrations and maintain H2 lower than CO2-reducing methanogenesis to support H2 producers (that is, syntrophs and fermenters) and also compete against sulfate reducers (4H2+SO42−+H+→HS−+4H2O: ΔG°′=−152.2 kJ mol−1). Thus, WSA2 may have an essential and overlooked ecological role for syntrophy in methanogenic environments (for example, anaerobic digestion) and competitive methanogenesis in sulfate-reducing environments (for example, marine sediments).
Besides these unique features, WSA2 has signatures of cytochrome-independent methanogenesis (heterodisulfide reductase–methylviologen-reducing hydrogenase energy conservation), Methanosarcinales and Methanococcales F420 modification, and Methanosphaera and Methanomassiliicoccales internal H2 cycling. In addition, characterization of WSA2 expands the phylogenetic range of methanogen heterotrophy. In conclusion, we present the first detailed description of the sixth methanogen class WSA2 as a significant step forward in understanding methanogen phylogeny, metabolic diversity and contribution to global biogeochemical cycles with provisional class and species assignment of ‘Candidatus Methanofastidiosa' class. nov. (Me.tha.no.fas.tid.i.o'sa. N.L. pref. methano-, pertaining to methane; N.L. f. adj. fastidiosa, highly critical; referring to the nutritional fastidiousness of the organism, particularly on primary isolation) and ‘Candidatus Methanofastidiosum methylthiophilus' gen. nov. sp. nov. (Me.tha.no.fas.tid.i.o'sum. N.L. pref. methano-, pertaining to methane; N.L. neut. adj. fastidiosum, highly critical; referring to the nutritional fastidiousness of the organism, particularly on primary isolation/me.thyl.thi.o'phi.lus.M.L.n. methyl, the methyl group; thi.o′phi.lus. Gr. n. thion sulfur; Gr. adj. phylos loving; N.L. neut. adj. methylthiophilus methyl- and sulfur- loving).
Acknowledgments
The Roy J Carver Biotechnology Center High-Throughput Sequencing and Genotyping Unit DNA services team provided helpful support for sequencing projects. We thank Professor William W Metcalf at University of Illinois, Urbana-Champaign Department of Microbiology for valuable discussion. We acknowledge the Energy Bioscience Institute for funding through project number OO4J12.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hugler M et al. (2010). Autotrophic carbon fixation in archaea. Nat Rev Microbiol 8: 447–460. [DOI] [PubMed] [Google Scholar]
- Blair NE, Carter WD Jr. (1992). The carbon isotope biogeochemistry of acetate from a methanogenic marine sediment. Geochim Cosmochim Acta 56: 1247–1258. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G, Parisot N, Harris HM, Peyretaillade E, Gaci N, Tottey W et al. (2014). Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics 15: 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose A, Kulkarni G, Metcalf WW. (2009). Regulation of putative methyl-sulphide methyltransferases in Methanosarcina acetivorans C2A. Mol Microbiol 74: 227–238. [DOI] [PubMed] [Google Scholar]
- Brown CT, Howe AC, Zhang Q, Pyrkosz AB, Brom TH. (2012), A reference-free algorithm for computational normalization of shotgun sequencing data arXiv 1203.4802 [q-bio.GN].
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TW, Chang YH, Tang SL, Tseng CH, Chiang PW, Chang KT et al. (2012). Metabolic stratification driven by surface and subsurface interactions in a terrestrial mud volcano. ISME J 6: 2280–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouari R, Le Paslier D, Daegelen P, Ginestet P, Weissenbach J, Sghir A. (2005). Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester. Environ Microbiol 7: 1104–1115. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. (2006). Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287. [DOI] [PubMed] [Google Scholar]
- Dhillon A, Lever M, Lloyd KG, Albert DB, Sogin ML, Teske A. (2005). Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl Environ Microbiol 71: 4592–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dojka MA, Hugenholtz P, Haack SK, Pace NR. (1998). Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol 64: 3869–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. (2012). Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol 62: 1902–1907. [DOI] [PubMed] [Google Scholar]
- Finster K, King GM, Bak F. (1990). Formation of methylmercaptan and dimethylsulfide from methoxylated aromatic compounds in anoxic marine and fresh water sediments. FEMS Microbiol Ecol 74: 295–301. [Google Scholar]
- Fu H, Metcalf WW. (2015). Genetic basis for metabolism of methylated sulfur compounds in Methanosarcina species. J Bacteriol 197: 1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DE, Xu H, White RH. (2002). Identification of coenzyme M biosynthetic phosphosulfolactate synthase: a new family of sulfonate-biosynthesizing enzymes. J Biol Chem 277: 13421–13429. [DOI] [PubMed] [Google Scholar]
- Graham DE, Taylor SM, Wolf RZ, Namboori SC. (2009). Convergent evolution of coenzyme M biosynthesis in the Methanosarcinales: cysteate synthase evolved from an ancestral threonine synthase. Biochem J 424: 467–478. [DOI] [PubMed] [Google Scholar]
- Graupner M, Xu H, White RH. (2000). Identification of the gene encoding sulfopyruvate decarboxylase, an enzyme involved in biosynthesis of coenzyme M. J Bacteriol 182: 4862–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P. (2002). Exploring prokaryotic diversity in the genomic era. Genome Biol 3: reviews0003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T, Tamaki H, Tamazawa S, Ueno Y, Ohkuma M, Suzuki K et al. (2013). Candidatus Methanogranum caenicola: a novel methanogen from the anaerobic digested sludge, and proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a methanogenic lineage of the class Thermoplasmata. Microbes Environ 28: 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota H, Ishida Y. (1972). Production of volatile sulfur compounds by microorganisms. Annu Rev Microbiol 26: 127–138. [DOI] [PubMed] [Google Scholar]
- Kiene RP, Hines ME. (1995). Microbial formation of dimethyl sulfide in anoxic Sphagnum peat. Appl Environ Microbiol 61: 2720–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni G, Kridelbaugh DM, Guss AM, Metcalf WW. (2009). Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri. Proc Natl Acad Sci USA 106: 15915–15920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClerc GM, Grahame DA. (1996). Methylcobamide: coenzyme M methyltransferase isozymes from Methanosarcina barkeri. Physicochemical characterization, cloning, sequence analysis, and heterologous gene expression. J Biol Chem 271: 18725–18731. [DOI] [PubMed] [Google Scholar]
- Lettinga G. (1995). Anaerobic digestion and wastewater treatment systems. Antonie van Leeuwenhoek 67: 3–28. [DOI] [PubMed] [Google Scholar]
- Li H, Xu H, Graham DE, White RH. (2003). Glutathione synthetase homologs encode alpha-L-glutamate ligases for methanogenic coenzyme F420 and tetrahydrosarcinapterin biosyntheses. Proc Natl Acad Sci USA 100: 9785–9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, kumar Yadhu et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele V, Penel S, Duret L. (2011). Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics 12: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TL, Wolin MJ. (1985). Methanosphaera stadtmaniae gen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen. Arch Microbiol 141: 116–122. [DOI] [PubMed] [Google Scholar]
- Miller TL, Lin C. (2002). Description of Methanobrevibacter gottschalkii sp. nov., Methanobrevibacter thaueri sp. nov., Methanobrevibacter woesei sp. nov. and Methanobrevibacter wolinii sp. nov. Int J Syst Evol Microbiol 52: 819–822. [DOI] [PubMed] [Google Scholar]
- Mondav R, Woodcroft BJ, Kim EH, McCalley CK, Hodgkins SB, Crill PM et al. (2014). Discovery of a novel methanogen prevalent in thawing permafrost. Nat Commun 5: 3212. [DOI] [PubMed] [Google Scholar]
- Moran JJ, House CH, Vrentas JM, Freeman KH. (2008). Methyl sulfide production by a novel carbon monoxide metabolism in Methanosarcina acetivorans. Appl Environ Microbiol 74: 540–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobu MK, Narihiro T, Rinke C, Kamagata Y, Tringe SG, Woyke T et al. (2015). Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J 9: 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschläger E, Rother M. (2009). In vivo role of three fused corrinoid/methyl transfer proteins in Methanosarcina acetivorans. Mol Microbiol 72: 1260–1272. [DOI] [PubMed] [Google Scholar]
- Paul K, Nonoh JO, Mikulski L, Brune A. (2012). 'Methanoplasmatales,' Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens. Appl Environ Microbiol 78: 8245–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pell J, Hintze A, Canino-Koning R, Howe A, Tiedje JM, Brown CT. (2012). Scaling metagenome sequence assembly with probabilistic de Bruijn graphs. Proc Natl Acad Sci USA 109: 13272–13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glockner FO. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW et al. (2000). The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407: 508–513. [DOI] [PubMed] [Google Scholar]
- Saito Y, Aoki M, Hatamoto M, Yamaguchi T, Takai K, Imachi H. (2015). Presence of a novel methanogenic archaeal lineage in anaerobic digesters inferred from mcrA and 16S rRNA gene phylogenetic analyses. J Water Envir Tech 13: 279–289. [Google Scholar]
- Sato T, Atomi H, Imanaka T. (2007). Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 315: 1003–1006. [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- Segata N, Bornigen D, Morgan XC, Huttenhower C. (2013). PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4: 2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. (2007). Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318: 1449–1452. [DOI] [PubMed] [Google Scholar]
- Staples CR, Lahiri S, Raymond J, Von Herbulis L, Mukhophadhyay B, Blankenship RE. (2007). Expression and association of group IV nitrogenase NifD and NifH homologs in the non-nitrogen-fixing archaeon Methanocaldococcus jannaschii. J Bacteriol 189: 7392–7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallant TC, Krzycki JA. (1997). Methylthiol:coenzyme M methyltransferase from Methanosarcina barkeri, an enzyme of methanogenesis from dimethylsulfide and methylmercaptopropionate. J Bacteriol 179: 6902–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallant TC, Paul L, Krzycki JA. (2001). The MtsA subunit of the methylthiol:coenzyme M methyltransferase of Methanosarcina barkeri catalyses both half-reactions of corrinoid-dependent dimethylsulfide: coenzyme M methyl transfer. J Biol Chem 276: 4485–4493. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tanner RS, Wolfe RS. (1988). Nutritional requirements of Methanomicrobium mobile. Appl Environ Microbiol 54: 625–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6: 579–591. [DOI] [PubMed] [Google Scholar]
- Ueno Y, Yamada K, Yoshida N, Maruyama S, Isozaki Y. (2006). Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 440: 516–519. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Ankwanda E, Wolfe RS. (1982). Nutrition and carbon metabolism of Methanococcus voltae. J Bacteriol 149: 852–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins D, Lu XY, Shen Z, Chen J, Lee PK. (2015). Pyrosequencing of mcrA and archaeal 16S rRNA genes reveals diversity and substrate preferences of methanogen communities in anaerobic digesters. Appl Environ Microbiol 81: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YW, Tang YH, Tringe SG, Simmons BA, Singer SW. (2014). MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellner G, Stackebrandt E, Messner P, Tindall BJ, Conway de Macario E, Kneifel H et al. (1989). Methanocorpusculaceae fam. nov., represented by Methanocorpusculum parvumMethanocorpusculum sinense spec. nov. and Methanocorpusculum bavaricum spec. nov. Arch Microbiol 151: 381–390. [DOI] [PubMed] [Google Scholar]
- Zinder SH, Brock TD. (1978). Methane, carbon dioxide, and hydrogen sulfide production from the terminal methiol group of methionine by anaerobic lake sediments. Appl Environ Microb 35: 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.