Abstract
Bolidomonas is a genus of picoplanktonic flagellated algae that is closely related to diatoms. Triparma laevis, a species belonging to the Parmales, which are small cells with a siliceous covering, has been shown to form a monophyletic group with Bolidomonas. We isolated several novel strains of Bolidophyceae that have permitted further exploration of the diversity of this group using nuclear, plastidial and mitochondrial genes. The resulting phylogenetic data led us to formally emend the taxonomy of this group to include the Parmales within the Bolidophyceae, to combine Bolidomonas within Triparma and to define a novel species, Triparma eleuthera sp. nov. The global distribution of Bolidophyceae was then assessed using environmental sequences available in public databases, as well as a large 18S rRNA V9 metabarcode data set from the Tara Oceans expedition. Bolidophyceans appear ubiquitous throughout the sampled oceans but always constitute a minor component of the phytoplankton community, corresponding to at most ~4% of the metabarcodes from photosynthetic groups (excluding dinoflagellates). They are ~10 times more abundant in the small size fraction (0.8–5 μm) than in larger size fractions. T. eleuthera sp. nov. constitutes the most abundant and most widespread operational taxonomic unit (OTU) followed by T. pacifica, T. mediterranea and the T. laevis clade. The T. mediterranea OTU is characteristic of Mediterranean Sea surface waters and the T. laevis clade OTU is most prevalent in colder waters, in particular off Antarctica.
Introduction
The heterokont genus Bolidomonas (class Bolidophyceae) contains flagellated picophytoplankton cells (Guillou et al., 1999a). Two species, Bolidomonas pacifica and B. mediterranea, were initially described, differing in the angle of insertion between flagella, in swimming patterns and in 18S rRNA gene sequences (Guillou et al., 1999a). A variety, B. pacifica var. eleuthera, was later proposed (but not formally described) based on both cultures and environmental sequences (Guillou et al., 1999b). Analyses of photosynthetic pigments as well as nuclear 18S rRNA and plastid RubisCO large subunit (rbcL) sequences (Guillou et al., 1999a; Daugbjerg and Guillou, 2001) demonstrated the sister relationship between Bolidomonas and diatoms, even though Bolidomonas possesses two flagella and lacks the siliceous frustule characteristic of diatoms. The Bolidophyceae were thus proposed to be an intermediate group between diatoms and all other Heterokontophyta (Guillou et al., 1999a). Bolidophyceae culture and environmental sequences have been predominantly reported from oligotrophic waters in the Pacific Ocean and Mediterranean at low concentrations (Guillou et al., 1999b).
The Parmales is a group of pico-sized marine protists with cells surrounded by silica plates (Booth and Marchant, 1987), first observed by microscopy in oceanic samples (Iwai and Nishida, 1976). Originally, they were thought to be resting stages of silicified loricate choanoflagellates (Silver et al., 1980), but the discovery of a chloroplast in Parmales cells revealed that they are phytoplankton (Marchant and McEldowney, 1986). The taxonomy of the Parmales is based on morphological features that can only be observed with scanning electron microscopy (SEM). Three genera have been described: the family Pentalaminaceae contains one genus, Pentalamina, with one species, and the family Triparmaceae has two genera, Tetraparma (four species) and Triparma (five species, four subspecies and four forma). Pentalamina has two circular shield plates of equal size, a larger ventral plate and two triradiate girdle plates. Tetraparma has three shield plates of equal size, a smaller ventral plate, a triradiate dorsal plate and three girdle plates. Triparma has three shield plates of equal size, a larger ventral plate, a triradiate dorsal plate and three girdle plates (Booth and Marchant, 1987, 1988; Kosman et al., 1993; Bravo-Sierra and Hernández-Becerril, 2003; Konno and Jordan, 2007; Konno et al., 2007). However, morphological variations are observed in field samples suggesting phenotypic plasticity in response to environmental conditions (Konno et al., 2007). Although they are most abundant in polar and subarctic waters (Booth et al., 1980; Nishida, 1986; Taniguchi et al., 1995; Komuro et al., 2005; Ichinomiya et al., 2013; Ichinomiya and Kuwata, 2015), parmaleans have a worldwide distribution including tropical waters (Silver et al., 1980; Kosman et al., 1993; Bravo-Sierra and Hernández-Becerril, 2003).
Typical heterokont features led Booth and Marchant (1987) to establish the Parmales, a new microalgal order tentatively belonging to the class Chrysophyceae. Recently, a culture strain of the Parmales species Triparma laevis was isolated from the western North Pacific off Japan (Ichinomiya et al., 2011). Transmission electron microscope observations showed the typical ultrastructure of photosynthetic heterokonts, with two endoplasmic reticulate membranes surrounding the chloroplast, a girdle and two to three thylakoid lamellae, as well as a mitochondrion with tubular cristae. Molecular phylogenetic analyses based on 18S rRNA and rbcL gene sequences revealed that this species did not group with the Chrysophyceae, but rather with the Bolidophyceae (Ichinomiya et al., 2011).
The discovery of the close relationship between the Bolidophyceae and the Parmales is highly relevant for elucidating the evolutionary history of diatoms (Guillou, 2011). Diatoms constitute the most ecologically successful phytoplankton group accounting for ~20% of primary production in modern oceans with an estimated diversity of 200 000 species (Armbrust, 2009). To date, however, phylogenetic analyses have relied on a single strain of Parmales and a few strains of Bolidomonas, this coverage being insufficient to determine the relationships between species within this key evolutionary lineage. In addition, only limited information is available from SEM surveys of field samples and from environmental clone libraries concerning the oceanic distribution and relative abundance of Bolidomonas and Parmales species.
In recent years, the massive sequencing of metabarcodes, that is, small regions of marker genes such as the V4 or V9 regions of the 18S rRNA gene (Pawlowski et al., 2012; Logares et al., 2014), has provided in-depth information on the structure of eukaryotic microbial communities. Analyses of samples from the Tara Oceans expedition (de Vargas et al., 2015) and the Ocean Sampling Day (Kopf et al., 2015) are providing new opportunities to map the distribution of specific taxonomic groups across world oceans.
In this study, we first used nuclear, plastid and mitochondrial gene sequences to analyze the phylogenetic relationships of several new silicified Parmales and flagellated Bolidomonas strains isolated from different oceanic regions as well as related environmental sequences. We then assessed the distribution of the major Bolidophyceae taxa across the world oceans using the V9 rRNA metabarcoding data set obtained in the frame of the Tara Oceans expedition.
Materials and methods
Culture isolation and maintenance
Silicified Parmales strains were isolated from seawater samples collected from the Oyashio region of the western North Pacific in July 2009 during a cruise of the RV Wakatakamaru of the Tohoku National Fisheries Research Institute. T. laevis f. longispina and T. strigata were isolated from a sample collected at 50 m at St. A3 (42°30′N, 145°00′E) and T. aff. verrucosa from a sample collected at 50 m at St. A7 (41°30′N, 145°30′E), the light intensity at these depths corresponding to <0.5% of surface light intensity. Strains were isolated by serial dilution combined with cell labeling by the fluorescent dye PDMPO (2-(4-pyridyl)-5-([4-(2-dimethylaminoethylaminocarbamoyl)-methoxy]phenyl)oxazole) (LysoSensor Yellow/Blue DND-160; Molecular Probe, Eugene, OR, USA) as described previously (Ichinomiya et al., 2011). Strains were subsequently maintained in the f/2 medium (Guillard and Ryther, 1962) at 5 °C under 30 μmol photons m−2 s−1 (14:10 L:D photoperiod). All new silicified strains were deposited to the NIES (National Institute for Environmental Studies) culture collection (http://mcc.nies.go.jp; Table 1). Three novel flagellated Bolidophyceae strains were isolated from North Sea, Atlantic Ocean and Mediterranean Sea samples by serial dilution and maintained in the K/2 medium at the temperature specified in Table 1. These strains were deposited in the Roscoff Culture Collection (RCC; http://www.roscoff-culture-collection.org; Table 1). Strains previously isolated were obtained either from the NIES or RCC collections (Table 1).
Table 1. Origin and culture conditions of Bolidophyceae strains.
| ID | Species | Previous genus assignment | Strain name | Other name | Ocean origin | Depth of isolation (m) | Growth temperature (°C) | 18S rRNA | ITS rRNA | 16S rRNA | rbcL | nadH1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NIES-2565 | Triparma laevis f. inornata | Triparma | TOY-0807 | Western Pacific Ocean | 80 | 5 | AB546639 | This study | LN735526 | AB546640 | This study | |
| NIES-3699 | Triparma laevis f. longispina | TLLOY-0907 | Western Pacific Ocean | 50 | 5 | This study | This study | LN735530 | This study | This study | ||
| NIES-3701 | Triparma strigata | TSOY-0907 | Western Pacific Ocean | 50 | 5 | This study | This study | LN735531 | This study | This study | ||
| NIES-3700 | Triparma aff. verrucosa | TVOY-0907 | Western Pacific Ocean | 50 | 5 | This study | This study | LN735532 | This study | This study | ||
| RCC205 | Triparma pacifica | Bolidomonas | OLI 31 SE3 | CCMP1866, NIES2682 | Equatorial Pacific Ocean | 15 | 20 | HQ912557, AF123595 | This study | LN735325 | AF372696 | |
| RCC206 | Triparma pacifica | Bolidomonas | OLI 31 SC-A | Equatorial Pacific Ocean | 15 | 20 | This study | This study | LN735329 | This study | ||
| RCC208 | Triparma eleuthera | Bolidomonas | OLI 120 SD-A | Equatorial Pacific Ocean | 5 | 20 | AF167154 | This study | LN735332 | AF333978 | This study | |
| RCC210 | Triparma eleuthera | Bolidomonas | OLI 46 SE-A | Equatorial Pacific Ocean | 15 | 20 | AF167153 | This study | LN735333 | This study | This study | |
| RCC212 | Triparma eleuthera | Bolidomonas | OLI 41 SA-A | Equatorial Pacific Ocean | 15 | 20 | AF167155 | This study | LN735334 | AF333979 | This study | |
| RCC216 | Triparma pacifica | Bolidomonas | OLI 94 SCH | Equatorial Pacific Ocean | 5 | 20 | This study | This study | LN735335 | This study | ||
| RCC238a | Triparma mediterranea | Bolidomonas | MIN 129–20m Aa | MIN B11E5, CCMP1867, NIES-2681 | Mediterranean Sea | 20 | 20 | AF123596, HQ710555 | AY702144 | AF333977 | ||
| RCC239 | Triparma mediterranea | Bolidomonas | MIN 129–20m Ab | MIN B11E7 | Mediterranean Sea | 20 | 20 | This study | This study | LN735367 | This study | |
| RCC1657 | Triparma sp. | MICROVIR 3K_0 | NIES-3369 | North Sea | 10 | 15 | KF422625 | This study | LN735273 | This study | ||
| RCC1730 | Triparma eleuthera | SOLAS_F | NIES-3370 | Atlantic Ocean | 5 | 20 | This study | This study | LN735286 | This study | This study | |
| RCC2347 | Triparma eleuthera | BOUM117–22 | NIES-3371 | Mediterranean Sea | 25 | 22 | KF422629 | This study | LN735356 | This study | This study |
Abbreviations: ITS, Internal Transcribed Spacer; NIES, National Institute of Environmental Sciences Collection; RCC, Roscoff Culture Collection.
Strains are available from the RCC (http://www.roscoff-culture-collection.org/) or the NIES culture collection (http://mcc.nies.go.jp/). Lines in bold correspond to novel strains.
This strain has been contaminated and is not available anymore from the RCC collection.
SEM observations
Silicified cultures were filtered through a polycarbonate membrane filter (0.6 μm pore size; Advantech, Tokyo, Japan) and air-dried at room temperature after rinsing with distilled water to remove salts. The filter was coated with Pt/Pd and examined by SEM (JSM-6390LV; JEOL, Tokyo, Japan).
Flagellated cultures were fixed with a solution of glutaraldehyde at 2.5% containing a 0.1 m sodium cacodylate buffer (pH 7.2) with 0.25 m sucrose. The fixed cells were placed on a polylysine-coated SEM glass plate and kept overnight at 4 °C. The glass plate was rinsed two times with the cacodylate–sucrose buffer, and postfixed with 1% OSO4 in cacodylate buffer for 1 h at 4 °C. Another double rinse with cacodylate buffer was carried out before dehydration in a graded ethanol series (30%, 50%, 75%, 90%, 95% and 100%). The 100% ethanol was then substituted two times with 100% of t-butyl alcohol. After 1 h at 4 °C, the glass plate was dried in a freeze drying device JEOL JFD-310 (JEOL). Before observation, the glass plate was coated with osmium in a vacuum evaporator coater HPC-1S (Vacuum Device Ltd, Ibaraki, Japan) and then examined by SEM (Hitachi S-4800, Tokyo, Japan).
Genomic DNA extraction and PCR amplification
Cells were harvested in exponential growth phase and concentrated by centrifugation. Total nucleic acids were extracted using the Nucleospin Plant II Kit (Macherey-Nagel, Düren, Germany) for RCC strains and the FastDNA SPIN Kit (MP Biomedicals, Solon, OH, USA) for NIES strains. All primer sequences and PCR conditions are listed in Supplementary Table S1. The nearly full-length nuclear 18S rRNA gene was PCR amplified using eukaryotic primers (Lepère et al., 2011). The nuclear region containing the internal transcribed spacer (ITS) 1 and 2, as well as the 5.8S rRNA gene were obtained by PCR amplification using universal primers (White et al., 1990). The plastid-encoded rbcL gene was amplified using primers Parma_rbcLF1 and rbcSR. Parma_rbcLF1 was designed to amplify the nearly full-length rbcL gene (ca. 1700 bp) of Bolidophyceae (Supplementary Table S1). The mitochondrion-encoded NADH dehydrogenase subunit 1 (nad1) gene was amplified using nested primer sets designed for Bolidophyceae (but which also amplify a bacterial nad1-like gene). All PCR amplifications were conducted using Phusion high-fidelity DNA polymerase (Finnzymes, Thermo Fisher Scientific, Waltham, MA, USA) in a 20 μl reaction volume with 0.25 μm of each primer. The PCR products were purified with the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) and sent directly for sequencing either to the Roscoff Genomer service or to the Macrogen Company (Seoul, South Korea). Sequences were deposited to GenBank under the following accession numbers: KR998385–KR998423 and KU160636.
Environmental sequences
A reference data set of near-complete sequences of genes coding for nuclear 18S rRNA, plastid 16S rRNA gene, rbcL, nad1 and ITS (ITS1+5.8S rDNA+ITS2) sequences from Bolidophyceae was compiled. In particular, we retrieved all sequences assigned as ‘Bolidophyceae' in the PR2 database for nuclear 18S rRNA (Guillou et al., 2013) and in the PhytoRef database for plastid 16S rRNA (Decelle et al., 2015). This data set was used to identify similar environmental sequences in the GenBank (release 206.0; February 2015) and CAMERA (http://camera.calit2.net/—note that this service no longer exists but the data are available on iMicrobe at http://data.imicrobe.us/) databases using MegaBLAST and BLAST (e-value e−10). For 18S rRNA, only Genbank sequences longer than 800 bp were kept for analysis (Supplementary Table S2). For CAMERA, only data retrieved from the ‘all 454 reads' database-matched Bolidophyceae sequences were used.
The nuclear 18S and plastid 16S rRNA envi-ronmental sequences were manually checked for chimeras. Two subsequences of 300 bp were extracted from the 5′ and 3′ ends, respectively, and assigned to a taxonomic group based on BLAST searches against GenBank or PhytoRef. Sequences that presented conflicting taxonomic assignation between the two parts were examined in detail to confirm their chimerical nature and not considered further (Supplementary Table S2).
Nuclear 18S rRNA sequences were clustered at 99% similarity with CD-HIT-EST (Li and Godzik, 2006) to obtain OTUs. OTUs with representative sequences on the 18S rRNA phylogenetic tree based on full-length sequences (see below) were annotated following the clades described (Supplementary Table S2).
Phylogenetic analysis
All culture and environmental sequences were aligned with MAFFT using the E-INS-i algorithm (Katoh et al., 2002). Gaps were treated as missing characters. CAMERA reads were excluded from phylogenetic analysis as they represented different regions of a given gene. Only 18S rRNA sequences longer than 1500 bp were used for phylogenetic reconstruction. Phylogenetic analyses were performed with three different methods: maximum-likelihood (ML), distance (neighbor-joining) and Bayesian analyses. The TrN+I+G, K2+G, GTR+G, T92+G and TN93+G+I models were selected for the 18S rDNA, 16S rDNA, rbcL, ITS and nad1 sequence data, respectively, using Akaike information criterion in Modeltest 2.1.4 (Posada, 2008). Neighbor-joining and ML analyses were performed using MEGA 6.0 (Tamura et al., 2013) and PhyML 3.0 (Guindon et al., 2010) with Subtree Pruning and Regrafting tree topology search operations and approximate likelihood ratio test with Shimodaira–Hasegawa-like procedure (Guindon et al., 2010), respectively. Markov chain Monte Carlo iterations were conducted for 1 000 000 generations sampling every 100 generations with burning length 1 00 000 using MrBayes 3.2.2 (Ronquist and Huelsenbeck, 2003). MAFFT and MrBayes programs were run within Geneious 7.1.7 (Kearse et al., 2012). For the definition of environmental clades, we followed the criteria of Guillou et al. (2008): a clade has to contain two or more environmental sequences obtained from different locations and/or samples in a given location and has to have strong phylogenetic support.
Tara Oceans 18S rDNA metabarcodes
The Tara Oceans project sampled oceanic waters over the course of a two and a half year expedition (Pesant et al., 2015). Here, we used the primary eukaryotic metabarcoding data set, consisting of >530 million quality-checked eukaryotic V9 rDNA reads obtained from 293 size-fractionated plankton communities sampled at 47 stations (Supplementary Figure S1), two depths (surface and deep chlorophyll maximum (DCM)) and four size fractions (0.8–5, 5–20, 20–180 and 180–2000 μm). V9 rDNA metabarcodes (129 bp on average) were sequenced with Illumina HiSeq (Genoscope, Evry, France) at a typical depth of 2 million reads per sample (Supplementary Table S3). Detailed information on sampling and analysis is provided in Pesant et al. (2015) and de Vargas et al. (2015), respectively
Metabarcode sequences for each data set were clustered into OTUs using the SWARM software (Mahé et al., 2014). Each OTU corresponds to a set of sequences differing by one base at most. For the present analysis, we only considered OTUs that contained at least 10 reads and that had a BLAST similarity higher than 80% to an 18S rRNA sequence from GenBank. OTUs were assigned to a taxonomic group based on ggsearch against an annotated eukaryotic V9 database (de Vargas et al., 2015) derived from the PR2 database (Guillou et al., 2013), which provides an eight-level taxonomic hierarchy (kingdom, superdivision, division, class, order, family, genus, species). Taxa were regrouped at the class level and we extracted the 36 OTUs that were classified as Bolidophyceae-and-relatives (Supplementary Table S4). We compared the contribution of Bolidophyceae metabarcodes to the total number of ‘photosynthetic' metabarcodes. Groups considered as photosynthetic included Chlorophyta, Rhodophyta, Cryptophyta, Haptophyta, Ochrophyta (i.e., photosynthetic heterokonts such as diatoms), but not dinoflagellates for which the differentiation of photosynthetic vs heterotrophic species is highly complex. Tara Oceans data have been deposited to the International Nucleotide Sequence Database Collaboration and are accessible at http://doi.pangaea.de/10.1594/PANGAEA.843017 and 843022. Analyses, statistics and graphics of the Tara Oceans data were performed with the R software using the following libraries: ggplot2, mapproj, maps, mapdata, reshape2, scales and gridExtra (R Core Team, 2015).
Results
Characterization of novel strains
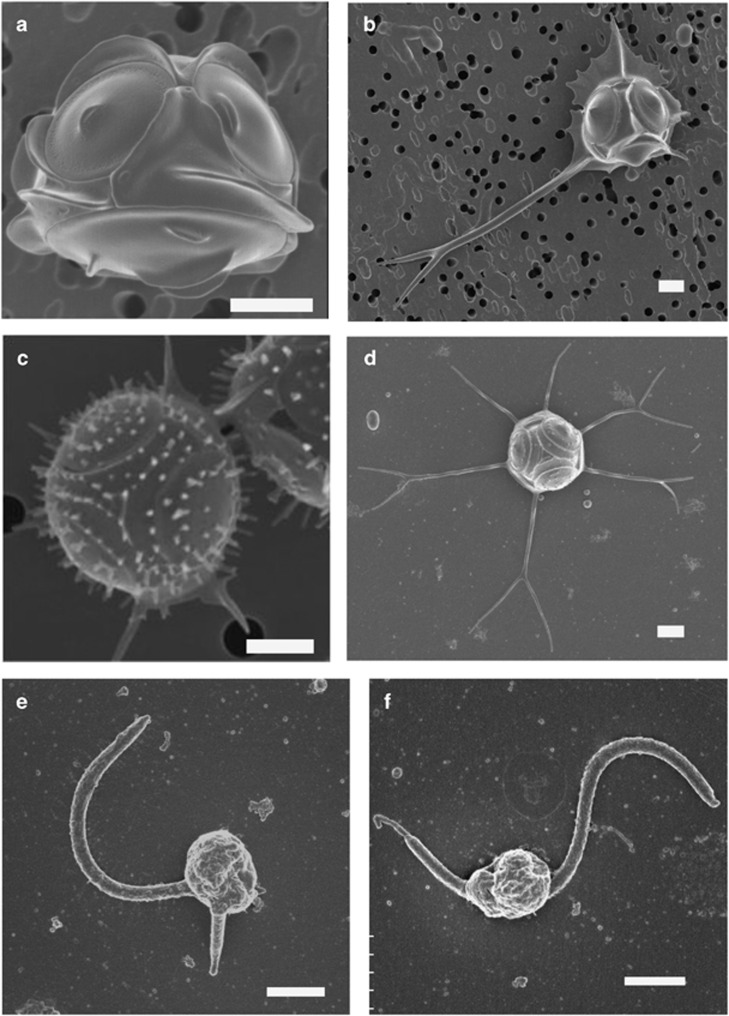
The silicified Parmales strain initially isolated (NIES-2565) and the three new strains (NIES-3699, NIES-3700 and NIES-3701) have cells with a diameter in the range 2.4–3.6 μm (Figures 1a–d), surrounded by three shield plates, one dorsal plate, one ventral plate and three girdle plates, demonstrating that they all belong to the genus Triparma (Booth and Marchant, 1987). Strains NIES-2565 and NIES-3699 possess three girdle plates with a single wing without any surface ornamentation (Figures 1a and b), corresponding to the description of T. laevis (Ichinomiya et al., 2011). More precisely, in NIES-2565 each girdle plate has a rounded wing at the margin without spines (Figure 1a), which is characteristic of the inornata form of T. laevis (Konno et al., 2007). In contrast, for NIES-3699, each wing on the girdle plate is irregular at the margin and has a buttressed spine (Figure 1b). One of the spines is very long (up to 14 μm) and bifurcated at the end, thus being typical of the longispina form of T. laevis (Konno et al., 2007). All plates in cells of NIES-3701 are covered by straight or forked tubular processes (Figure 1c). Girdle plates do not have a wing, but have two straight spines. These features are typical of the species T. strigata (Booth and Marchant, 1987; Kosman et al., 1993). The morphological features of the fourth strain, NIES-3700 (Figure 1d), have never been reported before. This strain shares features with both T. strigata and T. verrucosa. It lacks central area structures on shield and ventral plates like T. strigata, but no triradiate keel is found. It possesses two long girdle-plate spines like T. verrucosa, but lacks dense radiating papillae characteristic of this latter species (Kosman et al., 1993). For these reasons, this strain is hereafter identified as T. aff. verrucosa.
Figure 1.
SEM of the novel strains used in this study. (a) T. laevis f. inornata NIES-2565, (b) T. laevis f. longispina NIES-3699, (c) T. strigata NIES-3701, (d) T. aff. verucossa NIES-3700, (e) T. eleuthera sp. nov. RCC2347 and (f) Triparma sp. RCC1657. Scale bar=1 μm.
In contrast to the significant morphological variation observed between silicified strains, all new flagellated strains isolated from the Atlantic Ocean, North Sea and Mediterranean Sea (Table 1) possess the typical minimalist morphology of the genus Bolidomonas with very small cells possessing two flagella of unequal length and morphology (Figures 1e and f).
Phylogenetic analysis
Full or partial DNA sequences were obtained for Triparma and Bolidomonas strains for the genes coding for nuclear 18S rRNA and ITS, plastid 16S rRNA and rbcL and mitochondrial nad1 (Table 1). We also included environmental sequences related to the Bolidophyceae lineage (Supplementary Table S2) in all phylogenetic analyses.
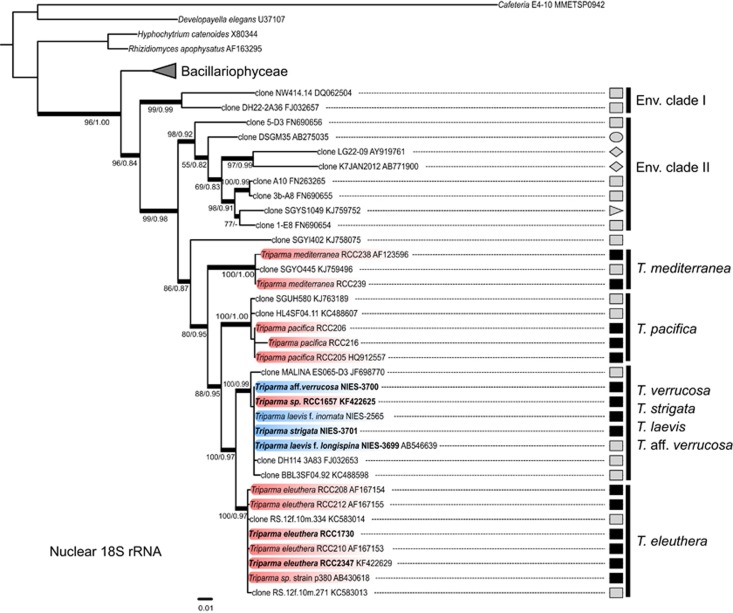
In agreement with the work of Ichinomyia et al. (2011), the phylogeny based on full-length nuclear 18S rRNA gene sequences clearly demonstrates that the genus Bolidomonas (Guillou et al., 1999a) is polyphyletic and that the order Parmales falls within the class Bolidophyceae (Figure 2). This prompted us to emend several taxonomic levels (see Taxonomic Appendix) and in particular to combine the genus Bolidomonas into Triparma following the anteriority rule of the International Code of Botanical Nomenclature. Remarkably, the full-length nuclear 18S rRNA gene sequences from the strains of T. laevis f. inornata, T. laevis f. longispina, T. strigata, T. aff. verrucosa and from the flagellated strain RCC1657 were very similar, sharing between 99.9% and 100% identity (Figure 2) despite obvious morphological differences (Figure 1). These sequences, together with the environmental sequence MALINA ES065-D3 from the Arctic, formed a well-supported clade (Figure 2) that we hereafter call for convenience the ‘T. laevis clade'. The T. laevis clade was sister to the clade of T. eleuthera, a novel species defined here for a previously invalid taxon, B. pacifica var. eleuthera (Guillou et al., 1999b), which contains RCC strains and two environmental sequences from the Red Sea. These two clades clustered with the T. pacifica clade, which included two environmental sequences from the Atlantic and Pacific Oceans. Finally, T. mediterranea formed the most basal group together with one environmental sequence from the Gulf Stream.
Figure 2.
ML tree inferred from nuclear 18S rRNA sequences belonging to the Bolidophyceae lineage. Only bootstrap and SH-like support values higher than 50 and 0.7, respectively, are shown. Nodes supported by Bayesian posterior probabilities over 0.95 are shown by thick branches. Sequences in bold correspond to novel strains. Sequences in blue and red correspond to non-motile silicified and motile strains, respectively. Symbols represent the origin of the sequences: environmental samples (gray), cultures (black), euphotic zone (square), deep-sea (right-pointing triangle), fish microbiome (left-pointing triangle), sediments (circle) and freshwater (diamond).
A number of environmental 18S rRNA gene sequences retrieved from PR2/GenBank according to their affiliation with Bolidophyceae did not group with Triparma sequences and formed two distinct clades (env. I and II) basal to the Bolidophyceae. These sequences originated from a range of environments including freshwater and marine sediments (Figure 2). The monophyly of these clades with the Bolidophyceae was supported by neighbor-joining bootstrap, ML and Bayesian analyses.
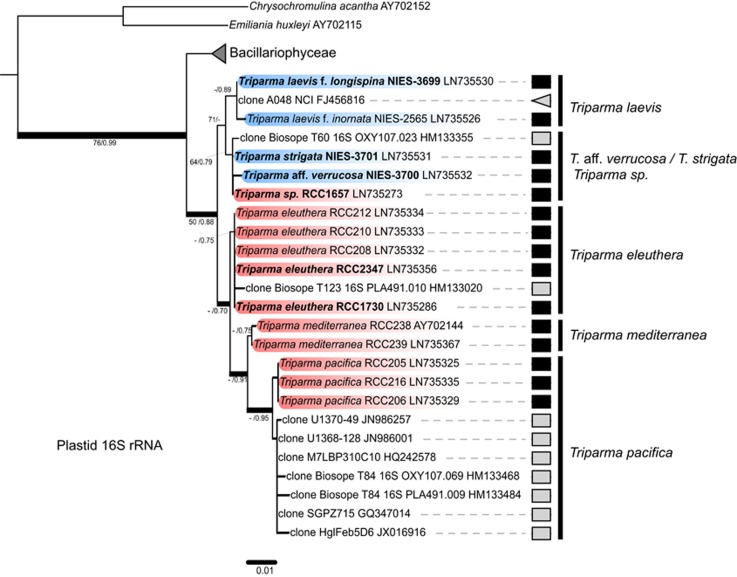
In contrast to the situation for the 18S rRNA gene, all other molecular markers (plastid 16S rRNA (Figure 3), plastid rbcL (Supplementary Figure S4), ITS rRNA (Supplementary Figure S5), mitochondrial nad1 (Supplementary Figure S6)) revealed the presence of two distinct subclades within the silicified Triparma species. One subclade contained the two forms of T. laevis (f. inornata, and longispina) and the other included T. strigata, T. aff. verrucosa and the flagellated strain RCC1657. The subclades were well supported in the rbcL and ITS trees but not in the plastid 16S rRNA tree (Figure 3). The close relationship between silicified Triparma and flagellated T. eleuthera was observed in all of our ML and Bayesian analyses, except for the plastid 16S rRNA gene for which the former group was allied to a clade formed by the three flagellated species T. eleuthera, T. mediterranea and T. pacifica (Figure 3).
Figure 3.
ML tree inferred from plastid 16S rRNA gene nucleotide sequences belonging to the Bolidophyceae lineage. Legend as in Figure 2.
Oceanic distribution from sequences available in public databases
To determine the oceanic distribution of Bolidophyceae, we first searched for similar sequences in GenBank and in PR2 (Guillou et al., 2013) as well as PhytoRef (Decelle et al., 2015), which contain annotated eukaryotic nuclear 18S and plastid 16S rRNA sequences, respectively, as well as CAMERA, which contains large-scale environmental metagenomic data sets (Supplementary Table S2). Only environmental sequences from nuclear 18S and plastid 16S rRNA genes were retrieved from these databases. The absence of rbcL, ITS or nad1 sequences in GenBank is explained by the fact that these genes are typically not used as molecular markers in environmental diversity studies. The data retrieved from CAMERA consisted of 135 reads that were reduced to 32 reads closely related to Bolidophyceae after careful checking by alignment with the full sequences used for the phylogenetic analysis and exclusion of reads without information on sample location.
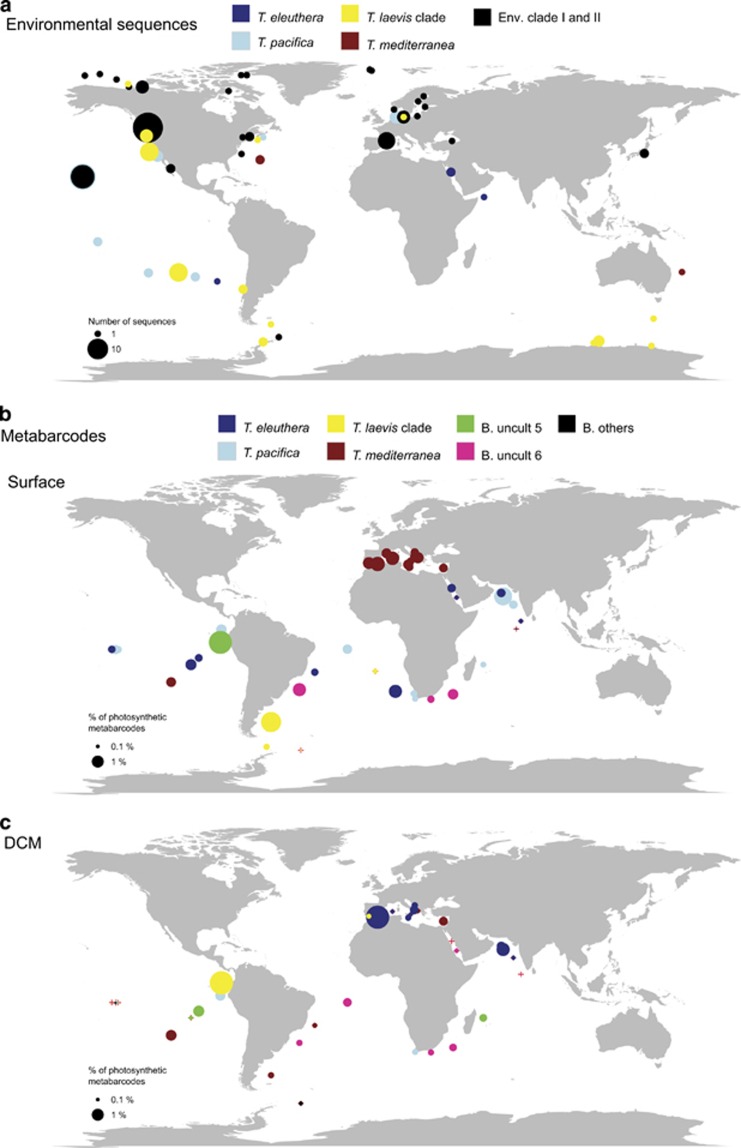
After clustering, 50% of the environmental 18S rRNA gene sequences could not be assigned to a specific clade (Supplementary Table S2). Sequences related to T. pacifica were the most abundant followed by those from the T. laevis clade (Supplementary Figure S3). T. pacifica sequences originated mostly from the Pacific Ocean and T. eleuthera from the Red Sea and Pacific Ocean, whereas the T. laevis clade was characteristic of the Southern Ocean and was also found near the Pacific coast (Figure 4a). A large number of sequences from Arctic or sub-Arctic locations, as well as from the Baltic Sea, were short and difficult to align and could not be affiliated to the four major Bolidophyceae clades.
Figure 4.
Oceanic distribution of Bolidophyceae. (a) Based on available environmental sequences from GenBank and CAMERA databases (see Supplementary Table S2). (b) Based on Tara Oceans V9 metabarcodes from the 0.8 to 5 μm plankton size fraction from surface waters. (c) Idem but at the DCM. Color indicates for subpanel a, the taxonomic affiliation of the sequences (Supplementary Table S2) and for subpanels b and c, the dominant OTU at each station (OTUs 7–36 have been regrouped in the category ‘B. others'). Surface of the circle is proportional to the number of sequences (a) or to the total contribution of Bolidophyceae OTUs to sequences from photosynthetic groups (b and c). Red crosses correspond to the location of the Tara Oceans stations where no Bolidophyceae metabarcodes have been detected.
Oceanic distribution based on Tara Oceans 18S rRNA V9 metabarcodes
Thirty-six Tara Oceans 18S V9 rDNA OTUs were classified as Bolidophyceae, representing a total of 105 113 reads (Supplementary Table S4). Their contribution to metabarcodes affiliated to photosynthetic groups (excluding dinoflagellates) reached a maximum of 3.8% and was slightly higher, on average, in surface waters compared with at the DCM (Table 2). Bolidophyceae were present in the 0.8–5 μm plankton size fraction at virtually all stations. In contrast, they were absent at more than a third of the stations in the larger size fractions and their mean contribution decreased ~5-fold compared with the 0.8–5 μm fraction (Table 2), confirming that this group is essentially pico-/nanoplanktonic in size.
Table 2. Bolidophyceae sequences as a function of size fraction and depth in the Tara Oceans V9 data set.
| Fraction (μm) | Depth | No. of samples | % of samples with Bolido sequences |
Bolido/photo (%) |
|
|---|---|---|---|---|---|
| Mean | Max | ||||
| 0.8–5 | SUR | 40 | 100 | 0.73 | 3.68 |
| 0.8–5 | DCM | 33 | 97 | 0.52 | 3.78 |
| 5–20 | SUR | 41 | 68 | 0.15 | 0.88 |
| 5–20 | DCM | 33 | 58 | 0.10 | 0.89 |
| 20–180 | SUR | 42 | 45 | 0.08 | 1.00 |
| 20–180 | DCM | 28 | 43 | 0.04 | 0.47 |
| 180–2000 | SUR | 45 | 47 | 0.23 | 3.34 |
| 180–2000 | DCM | 31 | 55 | 0.10 | 1.09 |
Abbreviations: DCM, deep chlorophyll maximum; SUR, surface.
Number of samples analyzed, percentage of samples with Bolidophyceae sequences, mean and maximum contribution of Bolidophyceae to photosynthetic taxa.
In surface waters, Bolidophyceae in the 0.8–5 μm size fraction displayed the highest contribution to photosynthetic groups in the Mediterranean Sea and in regions relatively close to the coast (but not strictly coastal since Tara Oceans only sampled offshore pelagic waters) of the Indian, Atlantic and Pacific Oceans. In contrast, at the DCM their contribution appeared significant at only two stations, one near Gibraltar and the second in the equatorial Pacific Ocean (Figure 4c).
The six most abundant OTUs contributed 86% of total Bolidophyceae sequences in the four size fractions considered (Supplementary Table S5). Among these, the four most abundant OTUs shared 100% identity with the sequences of, respectively, T. eleuthera, T. pacifica, T. laevis and T. mediterranea. Very clear signatures for these four clades are present in the V9 region of the 18S rRNA gene, which would therefore be suitable for the design of fluorescence in situ hybridization or quantitative real-time PCR probes (Supplementary Figure S2). A single other OTU (no. 11) with 647 reads matched an environmental sequence (KJ758075) from the Ross Sea (Antarctica) (Supplementary Table S4). In contrast, all other OTUs did not match any known sequences, not even those from the two environmental clades defined on the basis of the full 18S rRNA gene sequences (Figure 2 and Supplementary Table S4).
We estimated the contribution of the six most abundant OTUs (regrouping the other OTUs into a single ‘other' category) in the 0.8–5 μm fraction in surface and DCM waters. T. pacifica and T. eleuthera were the most abundant and ubiquitous clades, occurring at more than 79% of the stations (Supplementary Table S5). T. eleuthera was more widespread in surface waters than at the DCM. In surface waters (Figures 4b and 5a), T. pacifica seemed to be absent in the Mediterranean Sea (Stns TARA_007 to TARA_030), whereas both species were absent in the Southern Ocean (Stns TARA_082 to TARA_085). In contrast, surface waters of the Indian Ocean and Pacific Ocean were completely dominated by these two OTUs (Figures 4b and 5a). At the DCM (Figures 4c and 5b), the situation was roughly similar. The two OTUs were absent from cold waters below 15 °C, but present up to 30 °C (Supplementary Figure S7B).
Figure 5.
Relative contribution of the major groups of Bolidophyceae OTUs in the 0.8–5 μm plankton size fraction at Tara Oceans stations sampled in surface waters (a) and at the DCM (b). OTUs 7–36 (Supplementary Table S4) have been regrouped in the category ‘B. others'.
T. mediterranea was much less ubiquitous, occurring at about 50% of stations (Supplementary Table S5). It was more characteristic of surface waters but was also found very deep, beyond 150 m (Supplementary Figure S7A). It dominated Mediterranean Sea surface waters (Figures 4b and 5a) and was also present in subtropical South Atlantic and South Pacific waters. It was much less abundant at the DCM (Figures 4c and 5b), although it dominated at some stations, again in the South Atlantic and South Pacific. In contrast to T. eleuthera and T. pacifica, T. mediterranea appears to have a narrower temperature range (Supplementary Figure S7B).
The T. laevis clade was also present in ~50% of stations and was more characteristic of surface waters (Figure 5 and Supplementary Table S5). Its distribution contrasted to that of T. mediterranea. Its highest contributions in surface waters were off South Africa and in the Southern Ocean (Figure 4b and Figure 5a). At the DCM, the T. laevis clade provided significant contributions in the Mediterranean Sea and in the Pacific offshore Costa Rica (Figure 4c and Figure 5b). In agreement with its geographical localization, it extended to very cold water (Supplementary Figure S7B).
The first of the two other abundant OTUs (no. 5 named ‘B. uncult 5') that did not match any cultured Bolidophyceae sequence was absent in surface waters at most stations except one offshore Costa Rica (Stn TARA_102) where it was dominant (Figures 4b and 5a). At the DCM, it was only present at a few stations, but at these stations it was the dominant metabarcode (Figures 4c and 5b). The last abundant OTU (no. 6 or ‘B. uncult 6') was present at a few stations in the Indian Ocean near South Africa and in the Atlantic Ocean off Brazil where it dominated (Figures 4b and 5a). It was much more prevalent at the DCM and was dominant at the same stations where it dominated in surface waters (Figures 4c and 5b). These two OTUs appeared to have a narrower temperature ranges than the four dominant OTUs (Supplementary Figure S7). Finally, OTU no. 11 matching an Antarctica GenBank sequence, represented in Figure 2 by the environmental sequence clone SGYI402 (KJ758075), was most abundant at Stn TARA_085, which was the southern-most station sampled by the Tara Oceans expedition.
Discussion
Multigene phylogenetic analyses including nuclear, plastidial and mitochondrial markers revealed that all silicified Triparma species isolated so far form a monophyletic lineage within the Bolidophyceae, strongly supporting the previous hypothesis that the Parmales and Bolidophyceae belong to the same lineage, sister to the diatoms (Ichinomiya et al., 2011).
Each of the three flagellated Triparma species that were previously classified in the genus Bolidomonas form monophyletic groups according to all gene markers studied. T. pacifica and T. eleuthera were originally thought to belong to the same species based on the absence of clear morphological differences (Guillou et al., 1999b). In the present study, they were never monophyletic for any of the genes analyzed, leading to the description of a new species T. eleuthera. The branching order of the different cultivated Bolidophyceae clades was not consistent across all genes, making it difficult to draw any firm conclusions about their genetic relationships. However, the hypothesis that silicified Triparma species are most closely related to T. eleuthera was supported by all gene phylogenies except for that of the plastid 16S rRNA gene.
We observed two distinct environmental clades (env. I and II; Figure 2). The monophyly of these clades with the Bolidophyceae group is well supported by neighbor-joining bootstrap, ML and Bayesian analyses. As these clades do not contain any cultured representatives, it is difficult to say whether these groups represent other Bolidophyceae species or distinct heterokont forms. These sequences were recovered from similar locations (Figure 4) to where silicified Parmales have been observed by microscopy, in particular polar regions (Silver et al., 1980; Marchant and McEldowney, 1986; Nishida, 1986; Booth and Marchant, 1987; Kosman et al., 1993), supporting the idea that they could correspond to species from this group. In contrast, no environmental clade related to the Bolidophyceae was observed with the plastid 16S rRNA gene, which is probably explained by the lower number of eukaryotic surveys using this gene.
All genes analyzed, with the exception of the 18S rRNA, allowed discrimination of two subclades within silicified Triparma species, one corresponding to the two forms of T. laevis and the other to T. strigata and T. aff. verrucosa. Triparma sp. RCC1657, although being a flagellated organism with no evidence of possessing a silica covering (Figure 1e), was always affiliated to silicified strains in all analyses, grouping with T. strigata and T. aff. verrucosa for the plastid 16S rRNA and rbcL, the nuclear ITS rRNA and the mitochondrial nad1 genes. Interestingly, RCC1657 is the flagellated strain that has been isolated from the highest latitude so far (North Sea), in waters whose temperature range is similar to that where silicified forms are observed (Ichinomiya and Kuwata, 2015). Flagellated cells have occasionally been observed in cultures of silicified T. laevis (MHN, unpublished observations). Triparma could therefore have a life cycle that switches between diploid silicified non-flagellated and haploid naked flagellated stages. This hypothesis is supported by the fact that the flagellated T. pacifica (RCC205) has been hypothesized to be haploid based on the absence of heterozygous alleles in its transcriptome (Kessenich et al., 2014). Life-cycle switching between non-flagellated and flagellated forms is known in some algal classes. The coccolithophore Emiliania huxleyi can exist in a diploid non-motile coccolith-bearing form and in a haploid flagellated non-calcifying form, both stages capable of growing vegetatively (Rokitta et al., 2011), whereas Calyptroshaera sphaeroidea exhibits flagellated holococcolith-bearing and non-flagellated heterococcolith-bearing forms (Noël et al., 2004). The 18S rRNA gene sequence of the small coccoid prasinophyte Pycnococcus provasolii is known to be almost 100% identical to that of the small planktonic flagellate Pseudoscourfieldia marina, leading to speculation that these two forms could be alternate life-cycle stages of the same species (Fawley et al., 1999; Guillou et al., 2004). Extant centric diatoms, which have a diploid vegetative stage, produce naked flagellated haploid male gametes for sexual reproduction (Drebes, 1977). Mann and Marchant (1989) proposed that the ancestral diatom could have been a haploid flagellate that formed a diploid silicified zygote. The mitotic division of the zygote might have taken place preferentially to give rise to the centric diatoms, which are the most ancient diatom group (Kooistra et al., 2007). The conditions that trigger the alternation are often not clear and specific strains may have lost the genetic ability to convert from one form to the other. This is the case of E. huxleyi for which isolates from the oligotrophic ocean have lost the genes necessary to form flagella (von Dassow et al., 2015). Future culture studies of the life cycle of the Bolidophyceae, perhaps modulating important environmental factors such as temperature, light or nutrients to attempt to provoke life-cycle phase changes, will shed light on the relationships between motile and non-motile silicified forms and thus on the early evolutionary history of diatoms.
The Tara Oceans V9 metabarcode data set confirms that the Bolidophyceae are mainly limited to the smallest size fraction below 5 μm, which matches the size range of all silicified and flagellated Bolidophyceae strains isolated in culture to date. Their presence in the larger size fractions, in particular the 180–2000 μm fraction where their contribution increases in comparison with the 5–20 and 20–180 μm fractions (Table 2), could be explained by their consumption by predators and subsequent inclusion into large sinking particles. This hypothesis is reinforced by the fact that parmalean siliceous plates have been observed in the fecal pellets and gut contents of zooplankton (Booth et al., 1980; Marchant and Nash, 1986; Urban et al., 1992; Konno and Jordan, 2012) as well as in marine snow (Marchant et al., 1996). Sequences have also been found in deep-sea sediments and fish microbiota (Supplementary Table S2).
For all size fractions, the contribution of Bolidophyceae to phytoplankton remains in general limited (below 3%), matching previous estimates for Bolidophyceae using 18S rRNA probes (<1% Guillou et al., 1999b; Not et al., 2005) and the low cell concentration (10–100 cell ml−1) usually recorded for silicified Parmales in oceanic waters (Ichinomiya and Kuwata, 2015). However, this group is very widely distributed throughout the ocean, being present at over 90% of the Tara Oceans stations (Figure 4 and Supplementary Table S5). The global distribution of this group is further stressed by a number of recent metabarcoding studies that reported their presence in the English Channel, the Arctic and the Antarctic Oceans (Kilias et al., 2014; Luria et al., 2014; Taylor and Cunliffe, 2014).
The Tara Oceans metabarcoding data set also allows assessment of the distribution of the four phylogenetic clades that can be distinguished based on very clear 18S rRNA signatures. Two OTUs are clearly ubiquitous, T. eleuthera and T. pacifica. The former has been isolated several times from a wide range of oceanic locations (Pacific and Atlantic Oceans, Mediterranean Sea; Table 1), while in contrast T. pacifica has been isolated from a single cruise in the Pacific Ocean (OLIPAC in 1994; Table 1). T. mediterranea is particularly well named as this species dominates in surface waters of the Mediterranean Sea (Figures 4 and 5), the only region from which it has been isolated into culture, during a single cruise and at a single station. The T. laevis clade appears dominant in cold waters similar to those from which it was first described (Booth and Marchant, 1987) and subsequently isolated (Ichinomiya et al., 2011). In the western North Pacific, the silicified forms only appear to grow actively in low temperature waters that occur during winter–spring and in culture T. laevis and T. strigata are unable to grow above 10 °C (Ichinomiya et al., 2013; Ichinomiya and Kuwata, 2015). However, reads from the T. laevis clade were also present and abundant in the tropical Pacific near the Costa Rica dome at 30 m (Figure 4c) where the recorded water temperature was 25 °C (Supplementary Figure S7B) and environmental sequences have been found in the South East Pacific Ocean (Supplementary Table S2). Silicified Parmales have also been reported by microscopy in upwelling areas of the tropical Pacific Ocean. Therefore, sequences found in tropical waters could correspond to Triparma species for which we have no isolates such as T. laevis subsp. mexicana or T. retinervis (Silver et al., 1980; Kosman et al., 1993; Bravo-Sierra and Hernández-Becerril, 2003).
The genetic diversity observed for the metabarcodes with no matching GenBank sequences could correspond to uncultured taxa, either among the silicified forms for which many morphotypes, in particular corresponding to the genera Tetraparma and Pentalamina, have not yet been cultured. These unknown metabarcodes could also correspond to flagellated forms previously classified as Bolidomonas, which are also quite difficult to bring into culture as demonstrated by the fact that T. mediterranea has only be isolated once, almost 20 years ago. Some of these metabarcodes could be more abundant in areas under-represented in this data set such as polar regions, as suggested by OTU no. 11, which could be typical of the Southern Ocean as it matches sequences recovered in the Ross Sea (KJ758075).
The data presented in this paper shed some light on the enigmatic Bolidophyceae group, which is of particular interest given it is phylogenetically close to the diatoms, which constitute the most successful extant phytoplankton group. For the first time, we establish the possibility that some species of Bolidophyceae may have a life cycle with motile flagellated and non-motile silicified stages. However, the Tara Oceans metabarcode data clearly reveal distinct niches between the silicified T. laevis clade and the other flagellated species described so far, the former being mostly a cold-water group while the latter have temperate and tropical distributions. One species, T. mediterranea, seems to be very characteristic of the Mediterranean Sea, a region known to harbor endemic species (Gómez, 2006). This work demonstrates the power of mixing classical culture and phylogenetic studies with novel massive metabarcoding approaches.
Taxonomic Appendix
The multigene approach applied to the new strains of Bolidomonas and Triparma (Figures 2, 3 and Supplementary Figures S4–S6) confirmed that the genus Bolidomonas (Guillou et al., 1999a) is no longer monophyletic and that the class Bolidophyceae must include the order Parmales and the family Triparmaceae (Konno et al., 2007) that were originally classified within the class Chrysophyceae. Therefore, the followed emendations to the original definitions of the class, order, family, genus and description of one new species are required.
Class Bolidophyceae Guillou et Chrétiennot-Dinet 1999
emend. Ichinomiya et Lopes dos Santos.
Diagnosis: This class includes motile naked and non-motile silicified cells. Motile cells with two unequal flagella, ventrally inserted. Long flagellum directed forward, with tubular flagellar hairs. Short flagellum naked and acronemated. Basal apparatus reduced to basal bodies. Transitional helix absent. No eyespot. Non-motile cells with silicified cell wall composed of five or eight plates, all fitting edge to edge. All cell walls composed of shield plates, girdle plates and a ventral plate. In addition, a dorsal plate is usually present. For both motile and non-motile cells, one chloroplast with a girdle lamella, lamellae with two to three appressed thylakoids. Mitochondria with tubular cristae. Pigment composition includes chlorophyll a, c1+c2 and c3 and fucoxanthin as a major carotenoid. 18S rRNA gene sequences place this class as a sister group to the diatoms.
Type genus: Triparma Booth et Marchant
Order Parmales Booth et Marchant 1987 emend. Konno et Jordan 2007.
emend. Ichinomiya et Lopes dos Santos.
Diagnosis as for the class.
Family Triparmaceae Booth et Marchant 1987 emend. Konno et Jordan 2007 emend. Ichinomiya et Lopes dos Santos.
Diagnosis: Motile cells 1–2 μm in diameter, spherical or heart shaped. Two unequal flagella, ventrally inserted. Long flagellum 4–7 μm directed forward, with laterally inserted tubular flagellar hairs. Short flagellum 0.9–2.2 μm naked with a marked acronema. Basal apparatus reduced to basal bodies. Dorsal chloroplast occupies about half the cell. Non-motile cells with silicified cell wall. Cell wall composed of three shield plates, three oblong girdle plates, a triradiate dorsal plate with rounded ends and a large ventral plate. For both motile and non-motile cells, one chloroplast with a girdle lamella, lamellae with two to three appressed thylakoids. Mitochondria with tubular cristae. Pigment composition includes chlorophyll a, c1+c2 and c3 and fucoxanthin as a major carotenoid.
Type genus: Triparma Booth et Marchant
Genus Triparma Booth et Marchant 1987 emend. Konno et Jordan 2007
emend. Ichinomiya et Lopes dos Santos.
Diagnosis as for the family.
Type species: Triparma columacea Booth et Marchant.
Synonym: Bolidomonas Guillou et Chrétiennot-Dinet in Guillou et al., 1999a
Triparma eleuthera Ichinomiya et Lopes dos Santos sp. nov.
Diagnosis: Characters of the motile type of the genus. Cells 1.3–2 μm in size, swimming speed variable, with the long flagellum pulling the cell and changes in direction. Two flagella, inserted at 109°. Short flagellum 2–3 μm. Long flagellum 5–7 μm. The combined nucleotide sequences of the nuclear 18S rRNA (KF422629), rRNA ITS (KU160636), plastid 16S rRNA (LN735356) rbcL (KR998422) and mitochondrial nad1 genes (KR998413) are characteristic of this species.
Etymology: The specific epithet ‘eleuthera' is derived from the feminine form of the Greek adjective eleutheros, meaning freedom and refers to the frequent changes in swimming direction.
Authentic strain: RCC2347 collected by L Garczarek during the BOUM cruise (July 2008), in the Mediterranean Sea at 35°40′N 14°06′E, deposited in the Roscoff Culture Collection, Roscoff, France.
Holotype: Figure 1e in this publication.
Triparma pacifica (Guillou et Chrétiennot-Dinet) Ichinomiya et Lopes dos Santos. comb. nov. Basionym: Bolidomonas pacifica Guillou et Chrétiennot-Dinet (in Guillou et al., 1999a, p 371)
Triparma mediterranea (Guillou et Chrétiennot-Dinet) Ichinomiya et Lopes dos Santos. comb. nov. Basionym: Bolidomonas mediterranea Guillou et Chrétiennot-Dinet (in Guillou et al., 1999a, p 371)
Acknowledgments
We thank Masanobu Kawachi for help with the SEM pictures and Yukiko Taniuchi for help in sequencing the nad1 gene. Two anonymous referees provided very useful comments. Ian Probert provided invaluable help and advice for the taxonomic treatment. Financial support for this work was provided by Grants-in-Aid for Scientific Research 22657027, 23370046, 26291085 and 15K14784 from the Japan Society for the Promotion of Science (JSPS), the Canon Foundation and Core Research for Evolutional Science and Technology from Japan Science and Technology as well as by the European Union programs MicroB3 (UE-contract-287589) and MaCuMBA (FP7-KBBE-2012-6-311975), and the French ‘Investissements d'Avenir' programs OCEANOMICS (ANR-11-BTBR-0008) and EMBRC-France.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Armbrust EV. (2009). The life of diatoms in the world's oceans. Nature 459: 185–192. [DOI] [PubMed] [Google Scholar]
- Booth BC, Lewin J, Norris RE. (1980). Siliceous nanoplankton I. Newly discovered cysts from the Gulf of Alaska. Mar Biol 58: 205–209. [Google Scholar]
- Booth BC, Marchant HJ. (1987). Parmales, a new order of marine chrysophytes, with descriptions of three new genera and seven new species. J Phycol 23: 245–260. [Google Scholar]
- Booth BC, Marchant HJ. (1988). Triparmaceae, a substitute name for a family in the order Parmales (Chrysophyceae). J Phycol 24: 124. [Google Scholar]
- Bravo-Sierra E, Hernández-Becerril DU. (2003). Parmales (Chrysophyceae) from the Gulf of Tehuantepec, Mexico, including the description of a new species, Tetraparma insecta sp Nov., and a proposal to the taxonomy of the group. J Phycol 39: 577–583. [Google Scholar]
- von Dassow P, John U, Ogata H, Probert I, Bendif EM, Kegel JU et al. (2015). Life-cycle modification in open oceans accounts for genome variability in a cosmopolitan phytoplankton. ISME J 9: 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugbjerg N, Guillou L. (2001). Phylogenetic analyses of Bolidophyceae (Heterokontophyta) using rbcL gene sequences support their sister group relationship to diatoms. Phycologia 40: 153–161. [Google Scholar]
- Decelle J, Romac S, Stern R, Bendif EM, Zingone A, Guiry M et al. (2015). PhytoREF: a reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Mol Ecol Resour 15: 1435–1445. [DOI] [PubMed] [Google Scholar]
- Drebes G. (1977). Sexuality. In: Werner D (ed). The Biology of Diatoms. Blackwell Scientific Publications: Oxford, UK, pp 250–283.
- Fawley MW, Qin MB, Yun Y. (1999). The relationship between Pseudoscourfieldia marina and Pycnococcus provasolii (Prasinophyceae, Chlorophyta): evidence from 18S rDNA sequence data. J Phycol 35: 838–843. [Google Scholar]
- Gómez F. (2006). Endemic and Indo-Pacific plankton in the Mediterranean Sea: a study based on dinoflagellate records. J Biogeogr 33: 261–270. [Google Scholar]
- Guillard RRL, Ryther JH. (1962). Studies of marine diatoms. I. Cyclotella nana Husdedt and Detonula confervacea Gran. Can J Microbiol 8: 229–239. [DOI] [PubMed] [Google Scholar]
- Guillou L. (2011). Characterization of the Parmales: much more than the resolution of a taxonomic enigma. J Phycol 47: 2–4. [DOI] [PubMed] [Google Scholar]
- Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L et al. (2013). The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small subunit rRNA sequences with curated taxonomy. Nucleic Acids Res 41: D597–D604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, Chrétiennot-Dinet M-J, Medlin LK, Claustre H, Loiseaux-de Goër S, Vaulot D. (1999. a). Bolidomonas: a new genus with two species belonging to a new algal class, the Bolidophyceae (Heterokonta). J Phycol 35: 368–381. [Google Scholar]
- Guillou L, Eikrem W, Chrétiennot-Dinet M-J, Le Gall F, Massana R, Romari K et al. (2004). Diversity of picoplanktonic prasinophytes assessed by direct nuclear SSU rDNA sequencing of environmental samples and novel isolates retrieved from oceanic and coastal marine ecosystems. Protist 155: 193–214. [DOI] [PubMed] [Google Scholar]
- Guillou L, Moon-van der Staay SY, Claustre H, Partensky F, Vaulot D. (1999. b). Diversity and abundance of Bolidophyceae (Heterokonta) in two oceanic regions. Appl Environ Microbiol 65: 4528–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, Viprey M, Chambouvet A, Welsh RM, Kirkham AR, Massana R et al. (2008). Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata). Environ Microbiol 10: 3349–3365. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M. (2010). New algorithms and methods to estimate maximum-likelihoods phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Ichinomiya M, Kuwata A. (2015). Seasonal variation in abundance and species composition of the Parmales community in the Oyashio region, western North Pacific. Aquat Microb Ecol 75: 207–223. [Google Scholar]
- Ichinomiya M, Nakamachi M, Shimizu Y, Kuwata A. (2013). Growth characteristics and vertical distribution of Triparma laevis (Parmales) during summer in the Oyashio region, western North Pacific. Aquat Microb Ecol 68: 107–116. [Google Scholar]
- Ichinomiya M, Yoshikawa S, Kamiya M, Ohki K, Takaichi S, Kuwata A. (2011). Isolation and characterization of Parmales (Heterokonta/Heterokontophyta/Stramenopiles) from the Oyashio region, Western North Pacific. J Phycol 47: 144–151. [DOI] [PubMed] [Google Scholar]
- Iwai T, Nishida S. (1976). The distribution of modern coccolithophores in the North Pacific. News Osaka Micropaleontol 5: 1–11. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenich CR, Ruck EC, Schurko AM, Wickett NJ, Alverson AJ. (2014). Transcriptomic insights into the life history of bolidophytes, the sister lineage to diatoms. J Phycol 983: 977–983. [DOI] [PubMed] [Google Scholar]
- Kilias ES, Nöthig E-M, Wolf C, Metfies K. (2014). Picoeukaryote plankton composition off West Spitsbergen at the entrance to the Arctic Ocean. J Eukaryot Microbiol 61: 569–579. [DOI] [PubMed] [Google Scholar]
- Komuro C, Narita H, Imai K, Nojiri Y, Jordan RW. (2005). Microplankton assemblages at Station KNOT in the subarctic western Pacific, 1999–2000. Deep-Sea Res Part II 52: 2206–2217. [Google Scholar]
- Konno S, Jordan RW. (2007). An amended terminology for the Parmales (Chrysophyceae). Phycologia 46: 612–616. [Google Scholar]
- Konno S, Jordan RW (2012). Parmales. In: eLS, Oren, A, Pettis, GS (eds), John Wiley & Sons Ltd: Chichester, UK; doi:10.1002/9780470015902.a0023691.
- Konno S, Ohira R, Harada N, Jordan RW. (2007). Six new taxa of subarctic Parmales (Chrysophyceae). J Nannoplankt Res 29: 108–128. [Google Scholar]
- Kooistra WHCF, Gersonde R, Medlin K, Mann DG (2007). The origin and evolution of the diatoms: their adaptation to a planktonic existence. In: Falkowski PG, Knoll AH (eds). Evolution of Primary Producers in the Sea. Academic Press: Burlington, UK, pp 207–249.
- Kopf A, Bicak M, Kottmann R, Schnetzer J, Kostadinov I, Lehmann K et al. (2015). The ocean sampling day consortium. Gigascience 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman CA, Thomsen HA, Østergaard JB. (1993). Parmales (Chrysophyceae) from Mexican, Californian, Baltic, Arctic and Antarctic waters with the description of a new subspecies and several new forms. Phycologia 32: 116–128. [Google Scholar]
- Lepère C, Demura M, Kawachi M, Romac S, Probert I, Vaulot D. (2011). Whole genome amplification (WGA) of marine photosynthetic eukaryote populations. FEMS Microbiol Ecol 76: 516–523. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. (2006). Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- Logares R, Audic S, Bass D, Bittner L, Boutte C, Christen R et al. (2014). Patterns of rare and abundant marine microbial eukaryotes. Curr Biol 24: 813–821. [DOI] [PubMed] [Google Scholar]
- Luria C, Ducklow H, Amaral-Zettler L. (2014). Marine bacterial, archaeal and eukaryotic diversity and community structure on the continental shelf of the western Antarctic Peninsula. Aquat Microb Ecol 73: 107–121. [Google Scholar]
- Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. (2014). Swarm: robust and fast clustering method for amplicon-based studies. PeerJ 2: e593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DG, Marchant HJ (1989). The origins of the diatom and its life cycle. In: Green JC, Leadbeater BSC, Diver WL (eds). The Chromophyte Algae: Problems and Perspectives. Clarendon Press: Oxford, UK, pp 307–323.
- Marchant H, Nash G. (1986). Electron microscopy of gut contents and faeces of Euphausia superba Dana. Mem Natl Inst Polar Res Spec issue 40: 167–177. [Google Scholar]
- Marchant HJ, McEldowney A. (1986). Nanoplanktonic siliceous cysts from Antarctica are algae. Mar Biol 92: 53–57. [Google Scholar]
- Marchant HJ, Watanabe K, Kawachi M. (1996). Marine snow in Antarctic coastal waters. Proc NIPR Symp Polar Biol 9: 75–83. [Google Scholar]
- Nishida S. (1986). Nanoplankton flora in the Southern Ocean, with special reference to siliceous varieties. Mem Natl Inst Polar Res Spec Issu 40: 56–68. [Google Scholar]
- Noël M-H, Kawachi M, Inouye I. (2004). Induced dimorphic life cycle of a coccolithophorid, Calyptrosphaera sphaeroidea (Prymnesiophyceae, Haptophyta). J Phycol 40: 112–129. [Google Scholar]
- Not F, Massana R, Latasa M, Marie D, Colson C, Eikrem W et al. (2005). Late summer community composition and abundance of photosynthetic picoeukaryotes in Norwegian and Barents Seas. Limnol Oceanogr 50: 1677–1686. [Google Scholar]
- Pawlowski J, Audic S, Adl S, Bass D, Belbahri L, Berney C et al. (2012). CBOL Protist working group: barcoding eukaryotic richness beyond the Animal, Plant, and Fungal kingdoms. PLoS Biol 10: e1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesant S, Not F, Picheral M, Kandels-Lewis S, Le Bescot N, Gorsky G et al. (2015). Open science resources for the discovery and analysis of Tara Oceans data. Sci Data 2: 150023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. (2008). jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2015). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria. Available from https://www.R-project.org/.
- Rokitta SD, de Nooijer LJ, Trimborn S, de Vargas C, Rost B, John U. (2011). Transcriptome analyses reveal differential gene expression patterns between the life-cycle stages of Emiliania huxleyi (Haptophyta) and reflect specialization to different ecological niches. J Phycol 47: 829–838. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Silver MW, Mitchell JG, Ringo DL. (1980). Siliceous nanoplankton. II. Newly discovered cysts and abundant choanoflagellates from the Weddell Sea, Antarctica. Mar Biol 58: 211–217. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi A, Suzuki T, Shimada S. (1995). Growth characteristics of Parmales (Chrysophyceae) observed in bag cultures. Mar Biol 123: 631–638. [Google Scholar]
- Taylor JD, Cunliffe M. (2014). High-throughput sequencing reveals neustonic and planktonic microbial eukaryote diversity in coastal waters. J Phycol 50: 960–965. [DOI] [PubMed] [Google Scholar]
- Urban J, McKenzie C, Deibel D. (1992). Nannoplankton found in fecal pellets of macrozooplankton in coastal Newfoundland waters. Bot Mar 36: 267–281. [Google Scholar]
- de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348: 1261605. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR Protocols: A Guide to Methods and Applications. Academic Press: Orlando, FL, USA, pp 315–322.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.