Abstract
The Australian macropodids (kangaroos and wallabies) possess a distinctive foregut microbiota that contributes to their reduced methane emissions. However, methanogenic archaea are present within the macropodid foregut, although there is scant understanding of these microbes. Here, an isolate taxonomically assigned to the Methanosphaera genus (Methanosphaera sp. WGK6) was recovered from the anterior sacciform forestomach contents of a Western grey kangaroo (Macropus fuliginosus). Like the human gut isolate Methanosphaera stadtmanae DSMZ 3091T, strain WGK6 is a methylotroph with no capacity for autotrophic growth. In contrast, though with the human isolate, strain WGK6 was found to utilize ethanol to support growth, but principally as a source of reducing power. Both the WGK6 and DSMZ 3091T genomes are very similar in terms of their size, synteny and G:C content. However, the WGK6 genome was found to encode contiguous genes encoding putative alcohol and aldehyde dehydrogenases, which are absent from the DSMZ 3091T genome. Interestingly, homologs of these genes are present in the genomes for several other members of the Methanobacteriales. In WGK6, these genes are cotranscribed under both growth conditions, and we propose the two genes provide a plausible explanation for the ability of WGK6 to utilize ethanol for methanol reduction to methane. Furthermore, our in vitro studies suggest that ethanol supports a greater cell yield per mol of methane formed compared to hydrogen-dependent growth. Taken together, this expansion in metabolic versatility can explain the persistence of these archaea in the kangaroo foregut, and their abundance in these ‘low-methane-emitting' herbivores.
Introduction
Methane emissions from anthropogenic sources are viewed as a significant contributing factor to climate change and global warming, and ruminant livestock populations are estimated to contribute between 13% and 19% of these emissions (Johnson and Johnson, 1995; Lowe, 2006; IPCC, 2007; Lassey, 2007; Leahy et al., 2010), which is a driving force behind global research efforts to productively alter livestock methane emissions. Recent studies have demonstrated that methane emissions by ruminant livestock is a heritable trait (Lassey et al., 1997; Pinares-Patiño et al., 2011a, 2011b; Pinares-Patino et al., 2013) and that animals selected for a ‘low-methane' phenotype do develop and sustain an altered rumen bacterial population (Shi et al., 2014). Interestingly, although the structure of the methanogenic community changes in these animals, metagenomic and other cultivation-independent methods suggest the abundance of methanogens remains largely unchanged. Instead, there appears to be a reduction in the expression of genes that coordinate the hydrogenotrophic (autotrophic) pathway of methanogenesis in the ‘low emitters', suggesting that the archaeal populations need to adapt to changing environmental conditions via metabolic niche expansion.
Kangaroos and wallabies are the most numerous members of the Macropodidae and produce substantially less methane per unit of digestible organic matter intake than ruminant livestock when fed the same diet (Engelhardt et al., 1978), and this difference was recently confirmed with captive zoo populations in Europe (Madsen and Bertelsen, 2012). Although both are foregut digesters, the digestive anatomy and associated physiology of macropodids and ruminants is quite different (Hume, 1984), and as expected the microbial communities that have evolved to colonize and persist in these digestive chambers are also different.
In that context, recent applications of metagenomic methods have revealed novel insights into the macropodid foregut microbiota that further explain their ‘low-methane' emissions (Morrison, 2013). Evans et al. (2009) were the first to show that the macropodid foregut is colonized by methanogenic archaea, albeit at numbers substantially less than typically reported in ruminants (105 to 106 cells per gram of macropodid digesta as compared to 108 to 109 cells per gram of ruminant digesta). Although the taxonomic assignments of the macropodid archaeal populations are largely similar to those from ruminant livestock virtually nothing is known about their metabolic capabilities and adaptations to the different gut environments. Here, we show that a Methanosphaera sp. isolated from the Western grey kangaroo (Macropus fuliginosus) expands the metabolic versatility of this genus, to include the utilization of short-chain alcohols in the absence of hydrogen gas, to fuel carbon reduction and methanogenesis.
Materials and methods
Animal sampling protocol and digesta preservation
The digesta samples used were collected from 17 Western grey kangaroos (estimated to be between 1 month and 5 years of age) during a culling exercise in 2008 by licensed hunters, as part of the state's control procedures, on a Commonwealth Scientific and Industrial Research Organisation (CSIRO) managed farm in Yallenbee (Western Australia). Digesta samples were collected from the anterior region of the sacciform forestomach, and representative contents were added directly to completely fill a 50 ml sterile Falcon tube and stored at −80 °C. The remainder of the digesta sample was strained through muslin cloth, with ~2 ml of the strained liquid immediately mixed with 2 ml of an anaerobic, pre-sterilized solution of 30% (vol/vol) glycerol. These samples were all stored at −80 °C and used for the subsequent PCR-denaturing gradient gel electrophoresis (DGGE) and cultivation-based studies.
DNA extraction and PCR-DGGE analysis of Western grey kangaroo digesta samples
Samples of whole digesta (~0.5 g) were mixed with a disassociation buffer (Whitehouse et al., 1994) to desorb the microbial cells adherent to plant particles, and the resulting liquid was used for total DNA extraction by the RBB+C method (Yu and Morrison, 2004). The DNA was used as a template for nested PCR, with the initial PCR amplification performed using the archaea-specific primers (Supplementary Table S1) 86F/1340R (Wright and Pimm, 2003). The second, nested PCR was performed using 2 μl aliquots of the primary amplification mixture; combined with touchdown PCR amplification (Hwang et al., 2008) using primers 344F (Raskin et al., 1994) and 519R (Lane et al., 1985). The primer 344F also contained a 38 nt G:C ‘clamp' (Vetriani et al., 1999) for DGGE analyses. Of the original 17 samples only 14 yielded PCR products, which were subjected to DGGE using the Bio-Rad D-code System (Bio-Rad, Hercules, CA, USA; Ouwerkerk et al., 2008) and the gel images were captured using a Typhoon scanner and Typhoon TRIO Variable Mode Imager (GE Healthcare Life Sciences, Parramatta, NSW, Australia).
Enrichment and isolation of Methanosphaera sp. WGK6
The glycerol stock from two animals (#2 and #6) was selected for culture work, based on their distinctive PCR-DGGE patterns (see Results and Supplementary Figure S1). The isolation process was conducted over a period of 2 months, and was initiated by setting up enrichment cultures based on RF30 medium (Joblin et al., 1990) with a mixture of hydrogen and carbon dioxide gases (80:20 vol/vol) added to the headspace within each tube to a final pressure of 150 kPa. In addition, a selection of these starter cultures were also supplied with methanol (1% vol/vol). Penicillin and streptomycin were added to final concentrations of 600 and 200 μg ml−1, respectively, and were incubated at 39 °C. Samples were taken aseptically from the cultures every 2 or 3 days, to check for methane in headspace gases and the presence of autofluorescent cells, as biomarkers of methanogen enrichment. Positive cultures were then further treated with erythromycin (100 μg ml−1 final concentration) and serially diluted to extinction. The highest dilutions still exhibiting autofluorescent growth and methane production were then serially diluted and used to inoculate RF30 agar roll tubes for the generation of single colonies. Several colonies were picked using a sterile glass pipette and then transferred to fresh medium and incubated at 39 °C. Following growth, each culture was confirmed to produce methane and to contain autofluorescent cells. Cell biomass from the candidate axenic cultures was collected by centrifugation and DNA extracted using the RBB+C method (Yu and Morrison, 2004). The DNA samples were then used as templates for PCR amplifications with either archaea- (86F/1340R) or bacteria-specific (27F/1492R) primers (Supplementary Table S1; Wright and Pimm, 2003) and the products were sequenced using an Applied Biosystems 3130 × l Genetic Analyser (Applied Biosytems, Carlsbad, CA, USA).
Substrate utilization tests for strain WGK6 and Methanosphaera stadtmanae DSMZ 3091T
A basal medium of BRN-RF10 ‘medium 1' (Balch et al., 1979) was used for these studies, with the headspace purged with oxygen-free N2 gas to remove any carryover H2, and with the various substrates aseptically added to individual tubes. The individual and substrate combinations tested were ethanol alone, ethanol and H2:CO2 (80:20 vol/vol), methanol alone, methanol and H2:CO2 (80:20 vol/vol), methanol and ethanol, 20 mm each of mono-, di- and trimethylamine, propanol alone and H2:CO2 (80:20 vol/vol). In all cases, the alcohols were added to give a final concentration of 1% (vol/vol), and when required the tubes were pressurized to 202 kPa with H2:CO2 (80%:20%). The two strains were also evaluated for growth and methane formation with acetate or formate as the principal carbon sources. Here, basal BRN-RF10 ‘medium 1' lacking these substrates was prepared and they were subsequently added as sterilized solutions post autoclaving to a final concentration of 20 mm. For all these studies, culture tubes containing the different media were inoculated in triplicate with 0.1 ml aliquots of 6-day-old cultures of either WGK6 or DSMZ 3091T, both of which possessed an optical density at 600 nm (OD600) ~1.0. Growth was monitored over time by measuring OD600 of all the cultures at fixed intervals over a 24-h period (alternating between 9 and 15 h apart) for 7 days. At the end of the incubation period, a sample of the headspace gases were collected from all the cultures and subjected to GC analysis (Gagen et al., 2014).
Comparative growth studies with M. stadtmanae strain DSMZ 3091T
The growth yields, substrate utilization and methane production profiles of WGK6 and DSMZ 3091T were then examined in more detail using large-scale batch fermentations. Here, the medium was prepared in custom-made 1.2-l serum bottles with a H2-free headspace, as described above. The bottles were modified with a Balch tube (Bellco, Vineland, NJ, USA) fused to the wall of the bottle, so that the OD600 measurements could be made. Each bottle also contains a serum bottle closure to support aseptic sampling from the cultures. Each bottle was prepared to contain 500 ml of BRN-RF10 medium and was inoculated with 2-day-old-diluted cultures of either WGK6 or DSMZ 3091T to give a starting optical density at 600 nm (OD600) of ~0.02. The alcohols were added to these media to a final concentration of 1% (vol/vol), and when necessary, pressurized to 202 kPa with the H2:CO2 gas mix. The bottles were placed within a shaking incubator cabinet and agitated at 100 r.p.m. for 6 days. Growth was monitored by OD600 measurements, taken at fixed intervals over each 24 h period (5, 5 and 14 h apart) using SpectraMax Plus 384 spectrophotometer (Molecular Devices, Sunnyvale, CA, USA). Samples (2 ml) were taken daily (at the time of the second OD600 measurement) from which cell biomass was collected by centrifugation. Cell yields were calculated according to dry weight of cells per mol of methane measured at the peak of the exponential phase of growth (Miller and Wolin, 1985). The supernatants from these samples were stored at −80 °C prior to their analyses for methanol, ethanol and short-chain fatty acids, using published GC procedures (Pontes et al., 2009), with the exception that the column temperature gradient was altered slightly to improve peak separation. Samples of the headspace gases were also removed each day, and the hydrogen, CO2 and methane concentrations were also measured by GC analysis (Gagen et al., 2014).
Taxonomic assignment of strain WGK6, genome sequencing and analysis
WGK6 cells were collected after 5 days of growth and the DNA was extracted using the RBB+C method (Yu and Morrison, 2004). The DNA was then quantified using the Quant-iT dsDNA BR assay kit according to the manufacturer's instructions (Invitrogen, Mulgrave, VIC, Australia); and the integrity of the DNA was determined by agarose gels and electrophoresis. The gene encoding 16S rRNA was amplified with primers 86F and 1340R using methods as described above. The individual sequence reads were trimmed and assembled using Vector NTI (InforMax Inc, North Bethesda, MD, USA) to produce a near-complete 16S rRNA gene sequence and used for phylogenetic analysis using MEGA5 (Tamura et al., 2011). The stability of the neighbor-joining tree was evaluated by 1000 bootstrap replications and Kimura 2-parameter modeling (Kimura, 1980).
Genomic DNA from strain WGK6 was subjected to shotgun sequencing at the Australian Centre of Ecogenomics (ACE) using a 454 Roche GS-FLX system (Roche, Branford, CT, USA) with titanium chemistry. The sequence data were quality checked, filtered and then de novo assembled using the Velvet assembler (EMBL-EBI, Saffron Walden, UK; Zerbino and Birney, 2008). The WGK6 contigs were re-ordered using Mauve (Darling et al., 2010) and with the DSMZ 3091T genome sequence as a reference. Mauve is a genome alignment algorithm that not only allows an anchored (and rapid) alignment of two or more genomes, but also supports the rearrangement of these alignment anchors to support the identification of any genome rearrangements. By doing so, it provides information relating to genome synteny, xenologous regions, and genome rearrangements as evolutionary marks. The Mauve-generated assembly was submitted to the xBASE annotation pipeline (Chaudhuri et al., 2008), and the resulting output files examined using the Artemis genome browser (Rutherford et al., 2000). The WGK6 draft genome was further examined using JGI IMG/ER (Berkeley, CA, USA; Markowitz et al., 2012), which facilitated more genes being assigned to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and orthologous groups (Kanehisa and Goto, 2000; Kanehisa et al., 2006; Kanehisa et al., 2010). The draft genome has been deposited at JGI IMG/ER under the accession 2595698213 and the whole genome shotgun project has also been deposited at DDBJ/EMBL/GenBank under the accession JRWK00000000. The version described in this paper with regard to annotations and locus tags is JGI IMG/ER version 2595698213.
RT-PCR analysis of WGK6 alcohol (walc) and aldehyde (wald) dehydrogenase gene expression
Strain WGK6 was cultured for 48 h (OD600~0.6) with RF30 medium supplemented with either methanol and hydrogen, or methanol and ethanol, as previously described. The cells were collected by centrifugation and subjected to phenol:chloroform treatment and bead beating to lyse the cells, followed by RNA purification with the Qiagen RNeasy mini kit (Qiagen, Buranda, QLD, Australia). Residual DNA was removed from the extracts with the Ambion Turbo DNase ‘rigorous' protocol. The quality of the RNA was determined using Agilent RNA 6000 Nano kit (Agilent Technologies, Mulgrave, VIC, Australia) and those samples with an RNA integrity number (RIN) >9 were reverse transcribed using Invitrogen SuperScript III reverse transcriptase (Invitrogen) and random hexamers, according to manufacturer's specifications. The cDNA was then used as a template for qPCR, with primers targeting either the candidate alcohol (walc) or aldehyde (wald) dehydrogenase genes, as well as the intergenic transcribed space (Supplementary Table S1). The primers were all designed using the online software Primer3, and were cross-checked to ensure non-specific binding to other regions of the WGK6 genome by using NCBI, BLAST and pDRAW32. The primers were all designed using the online software Primer3 (Rozen and Skaletsky, 2000) The cDNA was also used with 16S rRNA gene-specific primers. All these reaction mixtures were treated with RNaseH at 37 °C for 20 min, and aliquots (1 μl) were added to 12.5 μl of power SYBR Green PCR master mix (Applied Biosystems), 0.25 μl each of forward and reverse primer pair and then made up to a final volume of 25 μl. The PCR conditions used were one hold at 50 °C for 2 min and then one hold at 95 °C for 2 min, followed by 40 cycles at 95 °C for 15 s, and a 60 °C elongation for 1 min. The real-time PCR efficiency (E) was calculated for all three primer sets, by setting up qPCRs with varying amounts of cDNA added to the reaction mixtures (0.016, 0.08, 0.4 and 2 ng, each done in triplicate). The E-values were calculated as described by Rasmussen (2001) where E =(10(–1/slope)−1) × 100, and for both dehydrogenase primer sets was 90%, and 102% for 16S rRNA gene. Then, the fold change for each of the dehydrogenase gene transcripts was calculated using (Pfaffl, 2001)
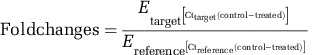 |
Here, ‘target' refers to either the walc- or wald-derived transcript and ‘reference' refers to the 16S rRNA-derived cDNA. ‘Control' and ‘treated' conditions were growth of strain WGK6 with methanol:H2 and methanol:ethanol, respectively.
Taxonomic assignment of WGK6 alcohol and aldehyde dehydrogenase genes
The phylogeny of protein-coding sequences for WGK6 alcohol and aldehyde dehydrogenase genes was constructed with sequences recovered from the NCBI and JGI databases. MEGA5 (Tamura et al., 2011) was used to assess phylogeny for each dehydrogenase gene respectively, ClustalW (Thompson et al., 1994) aligned the sequences and the stability of the Poisson modeled (Rosset, 2007) neighbor-joining tree was evaluated by 1000 bootstrap replications.
Results
Analysis of foregut digesta samples and isolation of Methanosphaera sp. WGK6
The PCR-DGGE analysis of the digesta samples showed there were two dominant profiles (Supplementary Figure S1) and based on these results, digesta samples from two animals (#2 and #6 in Supplementary Figure S1) were selected for methanogen enrichment and isolation. The digesta sample from animal #2 produced cultures with only weak growth (OD600<0.2) and a small amount of methane could only be measured in headspace gases when H2 and CO2 were provided for growth and without antibiotic additions, so were not pursued further. However, the sample from animal #6 produced more actively growing cultures and more methane in headspace gases when H2 and methanol were provided for growth; and autofluorescent cells were also present upon UV-transilluminated microscopy. Interestingly, the addition of erythromycin solubilized in ethanol appeared to further improve growth of the culture. Upon microscopic examination, the culture appeared to be comprised exclusively of coccoid-shaped cells forming clumps, which were autofluorescent during UV transillumination (Supplementary Figure S2). Four single colonies were propagated in broth cultures and were confirmed to share the same morphology, autofluorescence and growth characteristics. Furthermore, each of these four cultures produced amplicons with archaea-domain-specific primers, with no detectable amplicon produced with bacteria-domain-specific primers. The PCR amplicon produced with archaeal primers was sequenced and found to be 100% identical to each other, and share 97% sequence identity with the 16S rRNA gene sequence of M. stadtmanae (DSMZ 3091T). At this stage, the individual cultures were presumed to be axenic and siblings of each other, so only one of these cultures, hereafter referred to WGK6, was selected for further study.
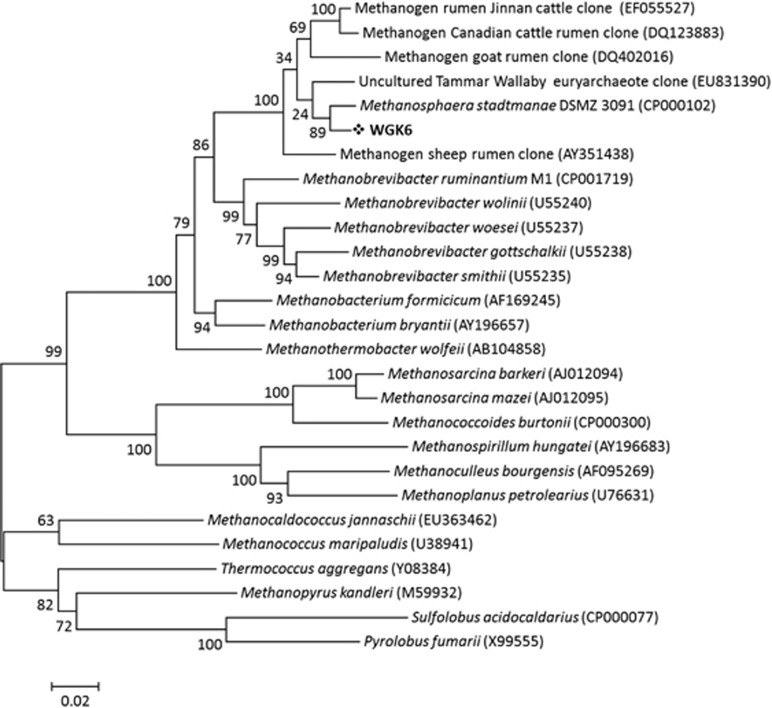
Cell biomass was collected from a WGK6 culture to provide template DNA for 16S rRNA gene-specific PCR analysis. The phylogenetic analysis of the WGK6 and gut methanogen sequences showed that it is assigned to a cluster containing DSMZ 3091T, with other sequences recovered from the rumen microbiomes of cattle in North America (Wright et al., 2007) and China (Pei et al., 2009), as well as other macropodids (Evans et al., 2009; Figure 1). Of these, DSMZ 3091T and Methanosphaera cuniculi (DSMZ 4103T, originally isolated from rabbit rectum) appear to be the only members of this cluster that are available as axenic cultures; and to our knowledge DSMZ 3091T is the only one to have its genome sequenced and publicly available.
Figure 1.
Phylogenetic analysis of the 16S rRNA gene sequence for strain WGK6 shows that it is most similar to the human gut isolate M. stadtmanae DSMZ 3091T and both are clustered with other clone sequences representing uncultured methanogens derived from ruminant and macropodid sources. Only bootstrap values >50% are shown and the scale bar represents 2% sequence divergence, with S. acidocaldarius and P. fumarii used as the outgroups.
Alcohol-fueled methanogenesis by Methanosphaera sp. WGK6
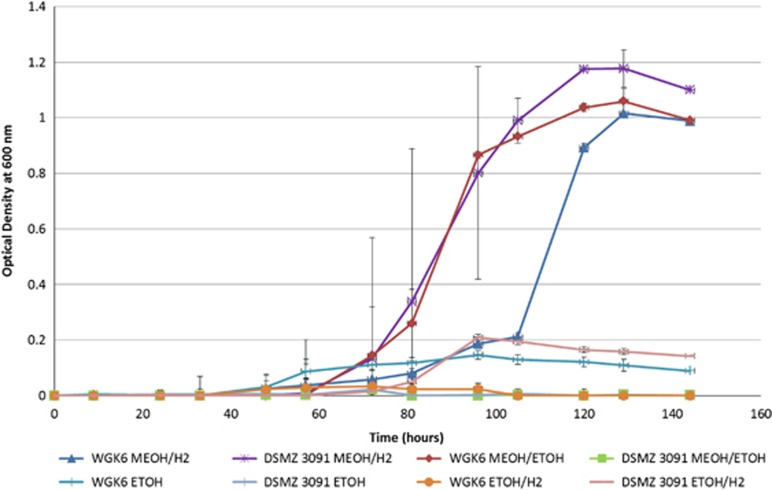
M. stadtmanae DSMZ 3091T has been shown by a combination of culture-based and genomic methods to be a hydrogen-dependent methylotroph restricted to using methanol and H2 (Miller and Wolin, 1983, 1985; Fricke et al., 2006), and incapable of autotrophic growth. Conversely, growth of the WGK6 enrichment culture appeared to be stimulated by the addition of ethanol (and/or erythromycin), suggesting the strain may possess a greater metabolic versatility compared to that of DSMZ 3091T. Substrate utilization tests confirmed that neither strain WGK6 nor DSMZ 3091T was capable of growth when methylated amines, acetate, formate, ethanol, propanol or CO2 were provided as carbon sources in the presence of hydrogen (data not shown). However, differences between the two strains were observed in terms of their ability to utilize combinations of methanol, ethanol and hydrogen to support growth (Figure 2). As expected, both strain DSMZ 3091T and WGK6 grew well when provided with both methanol and H2. Furthermore, neither strain was capable of growth with ethanol alone, nor with ethanol and H2. However, growth of strain WGK6 with the combination of short-chain alcohols occurred sooner than in cultures provided methanol:H2 and the headspace gas pressure was also higher (280 kPa c.f. 145 kPa at 120 h growth). As anticipated, strain DSMZ 3091T was not capable of growth under these conditions. On the basis of these results, we hypothesized that strain WGK6 can use ethanol as a source of reducing power coupled to methanol reduction and methane formation.
Figure 2.
Growth of Methanosphaera sp. WGK6 and M. stadtmanae DSMZ 3091T when provided with different combinations of methanol, ethanol and H2 added to BRN-RF10 basal medium, as described in the Materials and methods. Note that measurable growth is only apparent for strain DSMZ 3091T when provided with a combination of methanol and hydrogen, whereas growth of strain WGK6 can be supported by methanol and ethanol, as well as methanol and hydrogen. Individual values represent the mean (±s.e.m.) produced from triplicate cultures.
Alcohol-fueled methanogenesis is coupled with ethanol utilization and acetate formation by Methanosphaera sp. WGK6
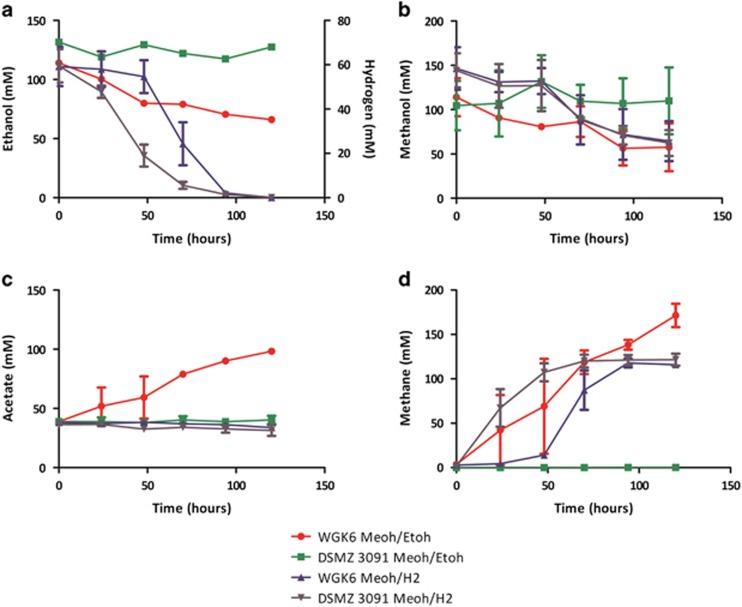
Next, large-volume batch cultures of both WGK6 and DSMZ 3091T provided with either methanol:H2 or methanol:ethanol were sampled longitudinally to determine substrate utilization, product (methane and acetate) formation and growth kinetics, and the results of these experiments are illustrated in Figure 3. Methanol utilization, methane formation and growth was observed for strain WGK6 under both conditions and the cell yields were estimated to be 2.4 and 4.0 g mol−1 methane produced with methanol:H2 and methanol:ethanol, respectively. In addition, ethanol utilization by strain WGK6 (Figure 3a) was coupled with the accumulation of acetate in these cultures only (Figure 3c), suggesting strain WGK6 has the capacity to coordinate a two-stage oxidation of ethanol to provide the reducing power used for methanol reduction and methanogenesis. As expected, methanol utilization, methane formation and growth of strain DSMZ 3091T occurred only with methanol:H2 with a cell yield of 4.0 g mol−1 methane, which is virtually identical to the findings reported by Miller and Wolin (1985). Taken together, these results suggested that strain WGK6 has an expanded metabolic versatility compared to strain DSMZ 3091T via a capacity to catalyse ethanol oxidation via acetaldehyde to acetate.
Figure 3.
Longitudinal monitoring of the utilization of ethanol or H2 (a) and methanol (b), as well as the formation of acetate (c) and methane (d) in batch cultures of Methanosphaera sp. WGK6 and M. stadtmanae DSMZ 3091T when provided with either methanol and hydrogen or methanol and ethanol. As expected methanol and hydrogen utilization, as well as methane formation, was observed for both WGK6 and DSMZ 3091T, when cultured using this substrate combination (▴ and ▾ for WGK6 and DSMZ 3091T, respectively). However, only strain WGK6 (•) and not DSMZ 3091T (▪) demonstrated an ability to utilize both methanol and ethanol to support methane formation (and growth, as depicted in Figure 2). In addition, acetate accumulation over time was only evident when strain WGK6 was cultured with both methanol and ethanol, suggesting a two-step oxidation of ethanol can provide the reducing power required for methanol reduction and methanogenesis (and growth) in the absence of H2. With the exception of the results for ethanol measurements presented in Figure 2a, the results represent the mean values obtained from duplicate experiments.
The WGK6 genome encodes dehydrogenase genes that are absent from the DSMZ 3091T genome
The WGK6 genome was assembled into 37 large contiguous sequences at 68 × coverage, with its genome size estimated to be 1.732 Mbp. The WGK6 draft genome sequence was then further assessed for both genome completeness and contamination using CheckM v1.0.3 (ACE, Brisbane, QLD, Australia), an automated method which examines a draft genome against a broad set of marker genes, specific to an inferred lineage within a reference genome tree (Parks et al., 2014). These analyses suggested that the WGK6 draft genome sequence is 96.8% complete and has a contamination level of 0%. In comparison, CheckM analysis of the DSMZ 3091T closed genome produced values of 97.6% completeness and 0% contamination. In addition, both genomes are calculated to possess G:C content of 28% and are also very similar in size, with the closed DSMZ 3091T genome being 1.767 Mbp (Fricke et al., 2006). The basic annotation tables for both WGK6 and DSMZ 3091T are provided in Supplementary Table S2, and with the exception of the number of 16S rRNA genes found (one copy in strain WGK6 c.f. four copies in DSMZ 3091T) the relative proportions of protein-coding sequences with (and without) function predictions and/or KEGG pathway designations, are virtually identical. However, the Mauve alignment of the two genomes, while showing there is extensive amount of synteny with only a few genome rearrangements between the WGK6 and DSMZ 3091T genomes, did reveal a region of genomic difference and found only in the WGK6 genome (Supplementary Figure S3). A closer examination of the annotation results for this region produced by JGI IMG/ER pipeline and KEGG-based pathway analysis showed this region to encode genes encoding putative alcohol (walc) and aldehyde (wald) dehydrogenases, and are positioned contiguously. No orthologs of these genes could be identified anywhere else within the DSMZ 3091T genome. The walc and wald sequences were then used to query the NCBI non-redundant BLAST databases, to find their closest matches with biochemically validated gene products. The closest for the walc gene product is with the NADPH-dependent butanol dehydrogenase from Clostridium saccharobutylicum (63% amino acid identity and 100% coverage (Youngleson et al., 1989; Keis et al., 2001)); and the wald gene product best matches with an aldehyde dehydrogenase from Alcaligenes eutrophus (47% amino acid identity and 99% coverage (Priefert and Steinbuchel, 1992)).
On the basis of these comparative genome-wide analyses, we conclude that strain WGK6 is capable of utilizing ethanol as a source of reducing power because it possesses genes encoding for an alcohol and aldehyde dehydrogenase, whereas DSMZ 3091T lacks both these genes and thereby is incapable of utilizing ethanol to support methanol reduction, methanogenesis and growth.
The walc and wald genes are constitutively expressed and cotranscribed by Methanosphaera sp. WGK6
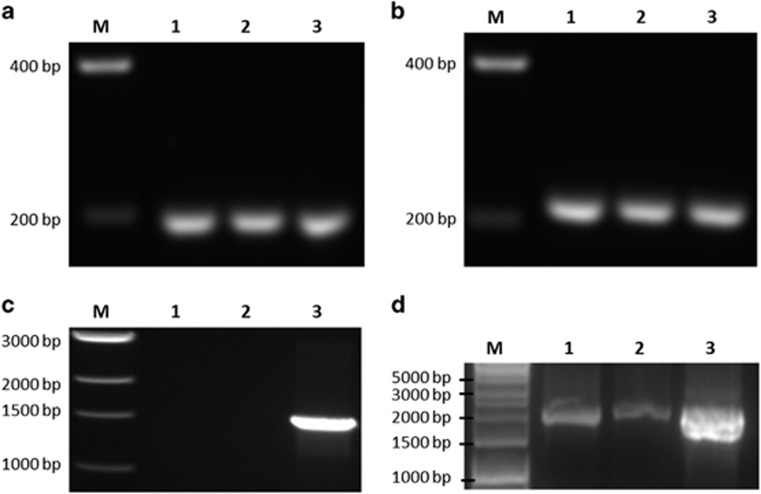
The expression of the walc and wald genes was assessed by qRT-PCR following growth of strain WGK6 with either methanol and hydrogen or methanol and ethanol, and the results are shown in Figure 4. Both genes are expressed constitutively (Figures 4a and b) and are also cotranscribed under both growth conditions (Figure 4c). The qPCR analyses showed there was an ~twofold increase in transcript abundance in response to growth with methanol and ethanol, when using cDNA produced from 16S rRNA as a reference. On the basis of these results, we conclude these genes can support ethanol oxidation and alcohol-fueled methanogenesis by strain WGK6 and underpin its growth in the presence of both methanol and ethanol.
Figure 4.
RT-PCR analysis of the walc and wald transcripts from Methanosphaera sp. WGK6 during growth with either methanol and hydrogen, or methanol and ethanol, as described in the Materials and methods. Both the walc transcript (a) and wald transcript (b) are detectable with both growth conditions. An amplicon was also produced using the walc-F and wald-R primers (c), suggesting the two genes are cotranscribed. In (a–c), lane M shows a DNA standard ladder (sizes annotated), lanes 1 and 2 use cDNA produced from the RNA extracted from cultures provided with ethanol or hydrogen, respectively; and lane 3 shows the results of PCR only, using genomic DNA. In (d), lane M shows a DNA standard ladder, lanes 1 and 2 show PCRs using 16S rRNA gene-specific primers and the two RNA samples, but with no prior RT step; and lane 3 shows the results with the same primers but using WGK6 genomic DNA as a template.
Orthologs of the dehydrogenase genes from strain WGK6 are also present in other members of the Methanobacteriales
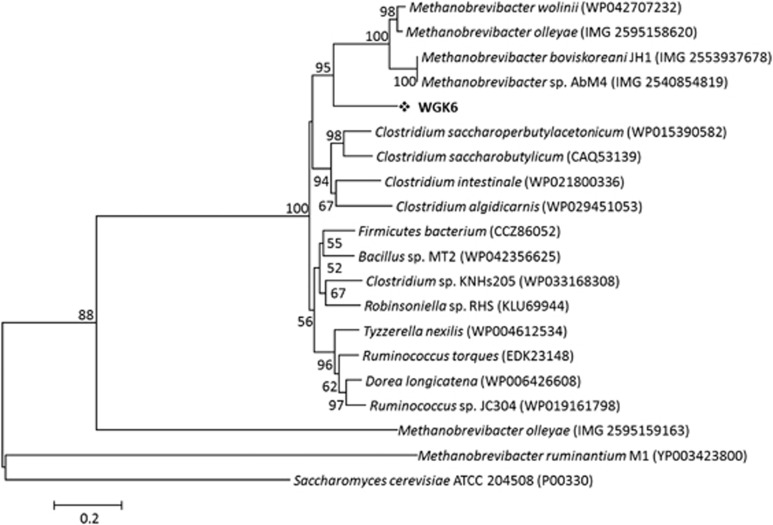
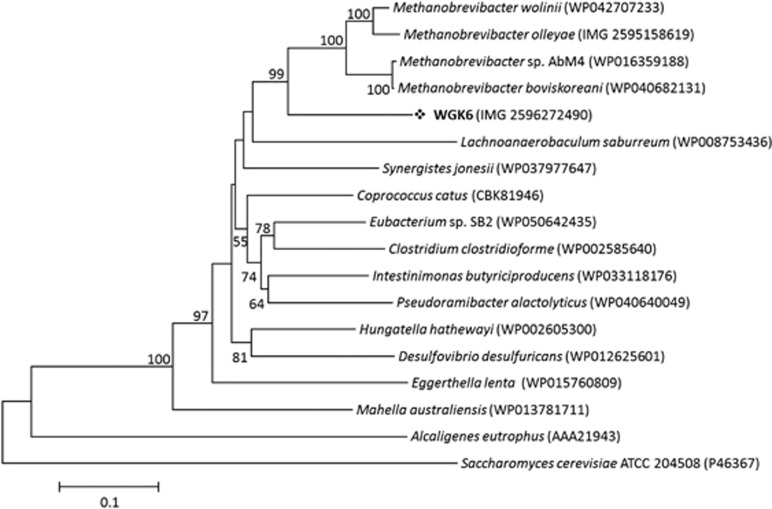
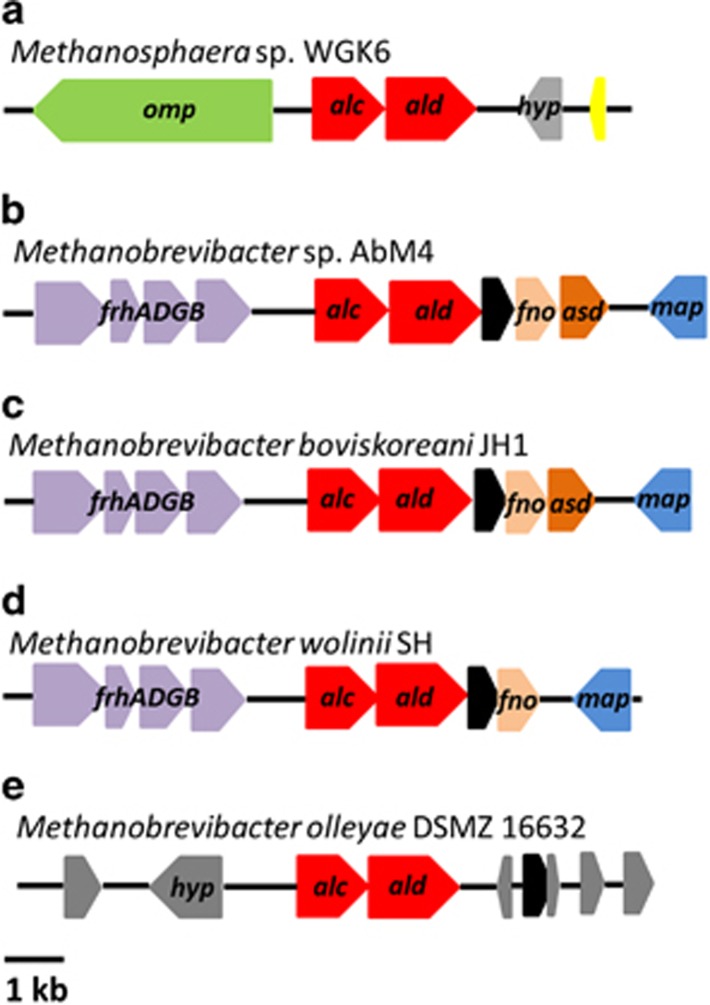
We next chose to examine the phylogeny of the alcohol and aldehyde dehydrogenases from strain WGK6 using protein-coding sequences recovered from the NCBI and JGI databases. In addition to a variety of bacterial sequences used to construct the trees, we also queried the draft genome sequences publicly available for all Methanobrevibacter spp. (that is, all 26 Mbb. smithii draft genomes, Mbb. ruminantium M1, Mbb. wolinii SH, Mbb. olleyae DSMZ 16632, Mbb. boviskoreani JH1, Mbb. millerae DSMZ 16643, Mbb. arboriphilus ANOR1 and Mbb sp. AbM4). First, none of the Mbb. smithii draft genomes were found to possess orthologs or partial hits to either dehydrogenase-coding gene. In addition, no aldehyde dehydrogenase gene has been identified from the Mbb. ruminantium M1 or Mbb. olleyae genomes, and there were no strong hits using the WGK6 gene as the query sequence. Although the M1 genome has been annotated to possess no less than four putative alcohol dehydrogenase genes (Leahy et al., 2010), none of these genes produce strong hits with the WGK6 aldehyde dehydrogenase gene. In that context, the phylogenetic tree constructed for the alcohol dehydrogenase genes (Figure 5) clearly reveals a bifurcation of the alcohol dehydrogenases from these gut-derived methanogenic archaea: one branch represented by Mbb. ruminantium M1 and the second including Methanosphaera WGK6 and the other Methanobrevibacter spp. Similarly, the phylogenetic tree of the aldehyde dehydrogenase gene retains this clustering of Methanobrevibacter spp. (Figure 6). Furthermore, there appears to be a strong degree of gene synteny among Mbb. AbM4, Mbb. boviskoreani JH1 and Mbb. wolinii SH (Figure 7), with the genes upstream of the dehydrogenases encoding for the coenzyme F420-reducing hydrogenase complex (frhADGB) and downstream genes encoding a putative transposase, F420:NADP oxidoreductase, aspartate dehydrogenase (AbM4 and M. boviskoreani only) and a methionine aminopeptidase.
Figure 5.
The phylogenetic tree constructed with the Jones-Taylor-Thornton (JTT) model, using BLASTp alignments of alcohol dehydrogenase genes sampled from NCBI non-redundant BLAST databases. The scale bar represents 20% sequence divergence and the Saccharomyces cerevisiae ATCC 204508 alcohol dehydrogenase gene was used as the outgroup. Numbers represent the relative frequency of branch clustering based on 1000 bootstrap runs, bootstrap values <50% are removed. Note that the WGK6 gene is closely grouped with similar genes recovered from the draft genomes of four different Methanobrevibacter spp. Notably, the putative alcohol dehydrogenase genes from the Mbb. ruminantium M1 genome are only distantly related (only a single representative was chosen for display within the phylogenetic tree).
Figure 6.
The phylogenetic tree constructed with Jones-Taylor-Thornton (JTT) model, using BLASTp alignments of aldehyde dehydrogenase genes sampled from NCBI non-redundant BLAST databases. The scale bar represents 10% sequence divergence and the Saccharomyces cerevisiae ATCC 204508 aldehyde dehydrogenase gene was used as the outgroup. Numbers represent the relative frequency of branch clustering based on 1000 bootstrap runs, bootstrap values <50% are removed. The WGK6 gene is again closely grouped with similar genes recovered from the draft genomes of four different Methanobrevibacter spp., but notably no putative aldehyde dehydrogenase gene(s) have been identified from the Mbb. ruminantium M1 genome.
Figure 7.
Comparison of the genes flanking the alcohol (alc) and aldehyde (ald) dehydrogenase genes (red) in Methanosphaera sp. WGK6, Methanobrevibacter sp. AbM4, Methanobrevibacter boviskoreani JH1, Methanobrevibacter wolinii SH and Methanobrevibacter olleyae DSMZ 16632 (a–e, respectively). A high degree of gene conservation is seen among strains AbM4, JH1 and SH, each possessing a coenzyme F420-reducing hydrogenase complex upstream (frhADGB—purple); and a transposase (black), F420:NADP oxidoreductase (fno—orange) and methionine aminopeptidase (map—blue) located downstream, with strains AbM4 and JH1 also possessing an aspartate dehydrogenase (asd—brown). Methanobrevibacter olleyae DSMZ 16632 is surrounded by hypothetical proteins (hyp—gray); however, there is a transposase (black) downstream of the dehydrogenase genes, while strain WGK6 is flanked by an membrane protein (omp—green), hypothetical proteins (hyp—gray) and a putative histone (yellow).
Discussion
Although Methanosphaera spp. have long been recognized to colonize the gastrointestinal tracts of warm-blooded animals, their metabolic niche has remained largely enigmatic, due in large part to the limited number of strains available for culture-based studies. Here, we demonstrate the ability of a new axenic isolate of Methanosphaera spp. to use ethanol as a source of reducing power, and also show that biomass yield per mol methane produced is greater when ethanol rather than hydrogen is used as reductant. Furthermore, although previous studies have shown that ethanol can stimulate the growth of hydrogenotrophic methanogens such as Methanobrevibacter smithii (Samuel et al., 2007) and Mbb. ruminantium (Leahy et al., 2010), neither are capable of growth in the absence of hydrogen.
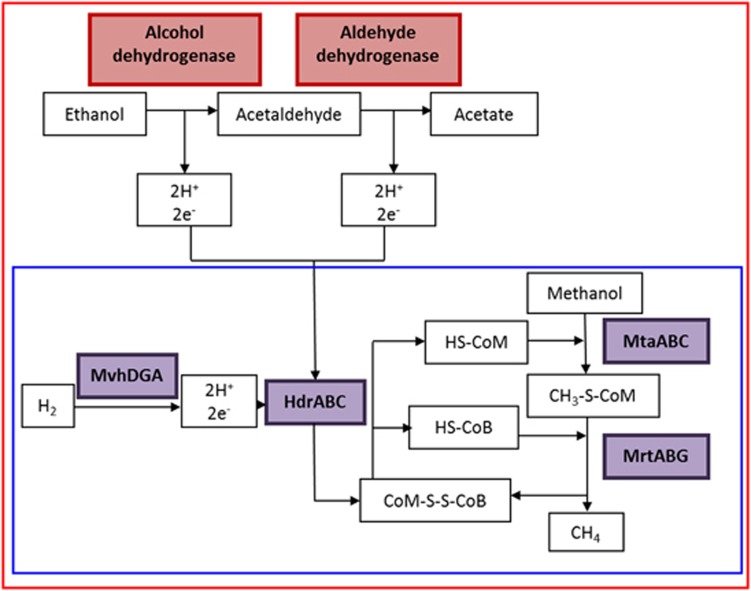
A proposed scheme involving a two-step oxidation of ethanol coupled with methanol reduction and methanogenesis by strain WGK6 is illustrated in Figure 8. Thermodynamically, the four protons and electrons released by the oxidation of ethanol to acetate via acetaldehyde could support the reduction of two mols of methanol, compared to one mol when only hydrogen is present. The calculated free energy available (values and equation provided in Supplementary Materials; Supplementary Table S3) is more favorable with ethanol, calculated to be −199.93 kJ mol−1 (Equation (1)) compared to methanol and hydrogen at −121.16 kJ mol−1 (Equation (2)). In addition, it is possible that the acetate produced from ethanol oxidation can also provide a source of cell carbon, not available via hydrogen-dependent reduction of methanol.
Figure 8.
A model for methanol reduction via hydrogen (blue box) or ethanol (red box) present in Methanosphaera sp. WGK6, based on the results of the genomic and culture-based results presented in this study. The reduction of methanol is initiated by coenzyme M methyltransferase (MtaABC) and coenzyme M (HS-CoM), which produces 2-(methylthio)ethanesulfonic acid (CH3-S-CoM). Methyl-coenzyme M reductase (MrtABG) reduces the product CH3-S-CoM with coenzyme B (HS-CoB) to methane and coenzyme M-HTP heterodisulfide (CoM-S-S-CoB). Heterodisulfide reductase (HdrABC) then acts on CoM-S-S-CoB using 2e− and 2H+ to reduce the disulfide bond thus regenerating HS-CoM and HS-CoB. The necessary electrons and hydrogen protons are generated by non-F420-reducing hydrogenase (MvhADG) from hydrogen. Alternatively, Methanosphaera sp. WGK6 can use a two-step oxidation process with ethanol, performed by the cotranscribed alcohol and aldehyde dehydrogenases, which would result in the provision of four protons and electrons used to reduce two methanol groups to methane and with acetate as an end product.
The demonstration of an alcohol- rather than hydrogen-fueled methanogenesis pathway suggests Methanosphaera spp. have adapted to survive in environments with small amounts of free hydrogen gas, and provides an explanation for their persistence in presumptive ‘low hydrogen' and/or ‘low-methane' environments. For instance, the nutritional ecology of the macropodids would favor the formation of methanol during pectin hydrolysis, as well as a microbiota capable of the production of some short-chain alcohols during fermentation (Kempton et al., 1976; Hume, 1984; Smith, 2009). So even though the relative numbers of methanogenic archaea in these animals are much smaller than found in ruminant livestock, and it has been demonstrated that reductive homoacetogens are effective hydrogen scavengers in the macropodid foregut (Gagen et al., 2010; Godwin et al., 2014), our findings do provide a basis for the persistence of methanogenic archaea in these communities via alcohol-fueled methanogenesis.
In ruminant livestock, a combination of metagenomic and metatranscriptomic methods have now showed that total methanogen numbers are similar between the ‘low' and ‘high' methane-producing sheep, but there are differences in the relative abundances of the methylotrophic Methanosphaera spp. (increased in ‘low-methane' sheep) and the hydrogenotrophic Methanobrevibacter gottschalkii clade (increased in ‘high-methane' sheep; Shi et al., 2014). The abundance of transcripts encoding most functions, coordinating the hydrogenotrophic pathway was also significantly increased in high-methane-producing sheep. The ‘high-methane'-emitting animals have been postulated to possess a longer retention of feed within the rumen as well as alterations in the bacterial ‘ruminotype' increasing the levels of ruminal hydrogen, with coordinate elevated expression of genes encoding the hydrogenotrophic pathway and greater methane yield (Janssen, 2010; Kittelmann et al., 2014). So it seems intuitive then to further suggest that the increased relative abundance of methylotrophic methanogens like Methanosphaera spp. in ‘low-methane' animals may relate to their capacity for alcohol-fueled methanogenesis when the bacterial ruminotype favors less hydrogen production during fermentation (Janssen and Kirs, 2008; Attwood et al., 2011).
It is also notable that recent studies of the human large bowel microbiota in patients suffering from non-alcoholic fatty liver disease and more severe conditions suggest that the microbiota is not only dysbiotic, but also fermentation schemes favouring ethanol formation are more active (Zhu et al., 2013). Coincidentally, other studies have remarked that there is also a shift from hydrogenotrophic to methylotrophic methanogens under such conditions, as well as in patients suffering from inflammatory bowel diseases (Blais Lecours et al., 2014).
In conclusion, the presence of these dehydrogenase genes in multiple members of the Methanobacteriales suggests that the metabolic versatility of these gut-derived methanogens is broader than what has long been accepted, and that alcohol-fueled methanogenesis may be an important evolutionary adaptation to persist in the gut environment.
Acknowledgments
This research has been supported by an Australian Postgraduate Award and a Meat and Livestock Australia Technical Assistance Grant (to ECH) as well as funds available to MM via CSIRO's Science Leader scheme, and The University of Queensland Diamantina Institute. We are especially grateful to the Australian Centre of Ecogenomics (ACE) for the technical expertise and support with the sequencing of the WGK6 genome. We also thank Jagadish Padmanabha and Jane Cheung (CSIRO, Australia) for their valued technical assistance.
Author contributions
ECH planned and performed research and helped write the paper; POC, PNE and SED assisted with research and helped write the paper; WJMS performed research; CSM planned and supervised research; and MM planned and supervised research and wrote the paper.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Attwood GT, Altermann E, Kelly WJ, Leahy SC, Zhang L, Morrison M. (2011). Exploring rumen methanogen genomes to identify targets for methane mitigation strategies. Anim Feed Sci Technol 166–167: 65–75. [Google Scholar]
- Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. (1979). Methanogens: reevaluation of a unique biological group. Microbiol Rev 43: 260–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais Lecours P, Marsolais D, Cormier Y, Berberi M, Haché C, Bourdages R et al. (2014). Increased prevalence of Methanosphaera stadtmanae in inflammatory bowel diseases. PLoS One 9: e87734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Loman NJ, Snyder LA, Bailey CM, Stekel DJ, Pallen MJ. (2008). xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res 36: D543–D546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt WV, Wolter S, Lawrenz H, Hemsley JA. (1978). Production of methane in two non-ruminant herbivores. Comp Biochem Physiol A Mol Integr Physiol 60: 309–311. [Google Scholar]
- Evans PN, Hinds LA, Sly LI, McSweeney CS, Morrison M, Wright AD. (2009). Community composition and density of methanogens in the foregut of the Tammar wallaby (Macropus eugenii. Appl Environ Microbiol 75: 2598–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WF, Seedorf H, Henne A, Kruer M, Liesegang H, Hedderich R et al. (2006). The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol 188: 642–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagen EJ, Denman SE, Padmanabha J, Zadbuke S, Al Jassim R, Morrison M et al. (2010). Functional gene analysis suggests different acetogen populations in the bovine rumen and tammar wallaby forestomach. Appl Environ Microbiol 76: 7785–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagen EJ, Wang J, Padmanabha J, Liu J, de Carvalho IP, Liu J et al. (2014). Investigation of a new acetogen isolated from an enrichment of the tammar wallaby forestomach. BMC Microbiol 14: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin S, Kang A, Gulino L-M, Manefield M, Gutierrez-Zamora M-L, Kienzle M et al. (2014). Investigation of the microbial metabolism of carbon dioxide and hydrogen in the kangaroo foregut by stable isotope probing. ISME J 8: 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume I. (1984). Microbial fermentation in herbivorous marsupials. Bioscience 34: 435–440. [Google Scholar]
- Hwang K, Shin SG, Kim J, Hwang S. (2008). Methanogenic profiles by denaturing gradient gel electrophoresis using order-specific primers in anaerobic sludge digestion. Appl Microbiol Biotechnol 80: 269–276. [DOI] [PubMed] [Google Scholar]
- IPCC. (2007) Climate Change 2007: The Physical Science Basis, Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press: Cambridge. [Google Scholar]
- Janssen PH, Kirs M. (2008). Structure of the Archaeal Community of the Rumen. Appl Environ Microbiol 74: 3619–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen PH. (2010). Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim Feed Sci Technol 160: 1–22. [Google Scholar]
- Joblin KN, Naylor GE, Williams AG. (1990). Effect of Methanobrevibacter smithii on xylanolytic activity of anaerobic ruminal fungi. Appl Environ Microbiol 56: 2287–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Johnson DE. (1995). Methane emissions from cattle. J Anim Sci 73: 2483–2492. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S et al. (2006). From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34: D354–D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. (2010). KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38: D355–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keis S, Sullivan JT, Jones DT. (2001). Physical and genetic map of the Clostridium saccharobutylicum (formerly Clostridium acetobutylicum NCP 262 chromosome. Microbiology 147: 1909–1922. [DOI] [PubMed] [Google Scholar]
- Kempton TJ, Murray RM, Leng RA. (1976). Methane production and digestability measurements in the grey kangaroo and sheep. Aust J Biol Sci 29: 209–214. [DOI] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kittelmann S, Pinares-Patiño CS, Seedorf H, Kirk MR, Ganesh S, McEwan JC et al. (2014). Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS One 9: e103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA 82: 6955–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassey KR, Ulyatt MJ, Martin RJ, Walker CF, David Shelton I. (1997). Methane emissions measured directly from grazing livestock in New Zealand. Atmospheric Environ 31: 2905–2914. [Google Scholar]
- Lassey KR. (2007). Livestock methane emission: From the individual grazing animal through national inventories to the global methane cycle. Agric Forest Meteorol 142: 120–132. [Google Scholar]
- Leahy SC, Kelly WJ, Altermann E, Ronimus RS, Yeoman CJ, Pacheco DM et al. (2010). The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One 5: e8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe DC. (2006). Global change: a green source of surprise. Nature 439: 148–149. [DOI] [PubMed] [Google Scholar]
- Madsen J, Bertelsen MF. (2012). Methane production by red-necked wallabies (Macropus rufogriseus. J Anim Sci 90: 1364–1370. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40: D115–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TL, Wolin MJ. (1983). Oxidation of hydrogen and reduction of methanol to methane is the sole energy source for a methanogen isolated from human feces. J Bacteriol 153: 1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TL, Wolin MJ. (1985). Methanosphaera stadtmaniae gen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen. Arch Microbiol 141: 116–122. [DOI] [PubMed] [Google Scholar]
- Morrison M. (2013). Looking large, to make more, out of gut metagenomics. Curr Opin Microbiol 16: 630–635. [DOI] [PubMed] [Google Scholar]
- Ouwerkerk D, Turner AF, Klieve AV. (2008). Diversity of methanogens in ruminants in Queensland. Aust J Exp Agric 48: 722–725. [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. (2014). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 2: e554v551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei C-X, Mao S-Y, Cheng Y-F, Zhu W-Y. (2009). Diversity, abundance and novel 16S rRNA gene sequences of methanogens in rumen liquid, solid and epithelium fractions of Jinnan cattle. Animal 4: 20–29. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45–e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinares-Patino CS, Hickey SM, Young EA, Dodds KG, MacLean S, Molano G et al. (2013). Heritability estimates of methane emissions from sheep. Animal 7(Suppl 2): 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinares-Patiño CS, Ebrahimi SH, McEwan JC, Dodds KG, Clark H, Luo D. (2011. a). Is rumen retention time implicated in sheep differences in methane emission? Proc NZ Soc Anim Prod 71: 219–222. [Google Scholar]
- Pinares-Patiño CS, Lassey KR, Martin RJ, Molano G, Fernandez M, MacLean S et al. (2011. b). Assessment of the sulphur hexafluoride (SF6) tracer technique using respiration chambers for estimation of methane emissions from sheep. Anim Feed Sci Technol 166: 201–209. [Google Scholar]
- Pontes H, Guedes de Pinho P, Casal S, Carmo H, Santos A, Magalhaes T et al. (2009). GC determination of acetone, acetaldehyde, ethanol, and methanol in biological matrices and cell culture. J Chromatogr Sci 47: 272–278. [DOI] [PubMed] [Google Scholar]
- Priefert H, Steinbuchel A. (1992). Identification and molecular characterization of the acetyl coenzyme A synthetase gene (acoE) of Alcaligenes eutrophus. J Bacteriol 174: 6590–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin L, Stromley JM, Rittmann BE, Stahl DA. (1994). Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol 60: 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen R Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara K-I (eds). Rapid Cycle Real-Time PCR. Springer: Berlin, Heidelberg, 2001, pp 21–34.
- Rosset S. (2007). Efficient inference on known phylogenetic trees using Poisson regression. Bioinformatics 23: e142–e147. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945. [DOI] [PubMed] [Google Scholar]
- Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R et al. (2007). Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA 104: 10643–10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Moon CD, Leahy SC, Kang D, Froula J, Kittelmann S et al. (2014). Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res 24: 1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA. (2009). Macropod nutrition. Vet Clin North Am Exot Anim Pract 12: 197–208. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetriani C, Jannasch HW, MacGregor BJ, Stahl DA, Reysenbach AL. (1999). Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl Environ Microbiol 65: 4375–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse NL, Olson VM, Schwab CG, Chesbro WR, Cunningham KD, Lykos T. (1994). Improved techniques for dissociating particle-associated mixed ruminal microorganisms from ruminal digesta solids. J Anim Sci 72: 1335–1343. [DOI] [PubMed] [Google Scholar]
- Wright AD, Pimm C. (2003). Improved strategy for presumptive identification of methanogens using 16S riboprinting. J Microbiol Methods 55: 337–349. [DOI] [PubMed] [Google Scholar]
- Wright AD, Auckland CH, Lynn DH. (2007). Molecular diversity of methanogens in feedlot cattle from Ontario and Prince Edward Island, Canada. Appl Environ Microbiol 73: 4206–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngleson JS, Jones WA, Jones DT, Woods DR. (1989). Molecular analysis and nucleotide sequence of the adh1 gene encoding an NADPH-dependent butanol dehydrogenase in the Gram-positive anaerobe Clostridium acetobutylicum. Gene 78: 355–364. [DOI] [PubMed] [Google Scholar]
- Yu Z, Morrison M. (2004). Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36: 808–812. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD et al. (2013). Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57: 601–609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.